Activated Notch1 modulates gene expression in B cells similarly to Epstein-Barr viral nuclear antigen 2
- PMID: 10644343
- PMCID: PMC111648
- DOI: 10.1128/jvi.74.4.1727-1735.2000
Activated Notch1 modulates gene expression in B cells similarly to Epstein-Barr viral nuclear antigen 2
Abstract
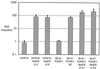
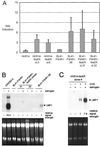
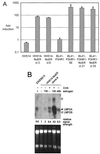
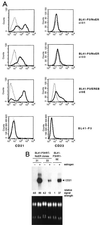
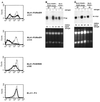
Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch transactivate genes by interacting with the transcription factor RBP-Jkappa. The viral protein EBNA2 may hence be regarded as a functional equivalent of an activated Notch receptor. Until now, nothing has been known about the physiological role of Notch signaling in B cells. Here we investigated whether activated Notch can induce the same phenotypic changes as EBNA2 in Burkitt's lymphoma cells. An estrogen receptor fusion protein of the intracellular part of mouse Notch 1 (mNotch1-IC), mimicking in the presence of estrogen a constitutively active Notch receptor, was stably transfected into the Burkitt's lymphoma cell lines BL41-P3HR1 and HH514. Northern blot analysis revealed that the LMP2A gene is induced by Notch-IC in the presence of estrogen, whereas increased expression of LMP1 could be detected only if cycloheximide was simultaneously added. Concerning the cellular genes regulated by EBNA2, Notch-IC was able to upregulate CD21 but not CD23 expression. Immunoglobulin mu (Igmu) expression, which is downregulated by EBNA2, was also negatively regulated by Notch-IC. Similarly to EBNA2, Notch-IC was able to repress c-myc expression, which is under the control of the immunoglobulin heavy-chain locus in Burkitt's lymphoma cells with a t(8;14) translocation. The data show that Notch-IC is able to participate in gene regulation in B cells.
Figures
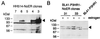
Similar articles
-
Activated mouse Notch1 transactivates Epstein-Barr virus nuclear antigen 2-regulated viral promoters.J Virol. 1999 Apr;73(4):2770-80. doi: 10.1128/JVI.73.4.2770-2780.1999. J Virol. 1999. PMID: 10074124 Free PMC article.
-
Activated Notch1 can transiently substitute for EBNA2 in the maintenance of proliferation of LMP1-expressing immortalized B cells.J Virol. 2001 Mar;75(5):2033-40. doi: 10.1128/JVI.75.5.2033-2040.2001. J Virol. 2001. PMID: 11160707 Free PMC article.
-
Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J kappa.Immunobiology. 1997 Dec;198(1-3):299-306. doi: 10.1016/s0171-2985(97)80050-2. Immunobiology. 1997. PMID: 9442401 Review.
-
The NP9 protein encoded by the human endogenous retrovirus HERV-K(HML-2) negatively regulates gene activation of the Epstein-Barr virus nuclear antigen 2 (EBNA2).Int J Cancer. 2011 Sep 1;129(5):1105-15. doi: 10.1002/ijc.25760. Epub 2011 Jan 12. Int J Cancer. 2011. PMID: 21710493
-
EBNA2 and Notch signalling in Epstein-Barr virus mediated immortalization of B lymphocytes.Semin Cancer Biol. 2001 Dec;11(6):423-34. doi: 10.1006/scbi.2001.0409. Semin Cancer Biol. 2001. PMID: 11669604 Review.
Cited by
-
EBV-associated diseases: Current therapeutics and emerging technologies.Front Immunol. 2022 Oct 27;13:1059133. doi: 10.3389/fimmu.2022.1059133. eCollection 2022. Front Immunol. 2022. PMID: 36389670 Free PMC article. Review.
-
Activation of CD21 and CD23 gene expression by Kaposi's sarcoma-associated herpesvirus RTA.J Virol. 2005 Apr;79(8):4651-63. doi: 10.1128/JVI.79.8.4651-4663.2005. J Virol. 2005. PMID: 15795251 Free PMC article.
-
When Viruses Cross Developmental Pathways.Front Cell Dev Biol. 2021 Aug 5;9:691644. doi: 10.3389/fcell.2021.691644. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34422814 Free PMC article. Review.
-
Cellular target genes of Epstein-Barr virus nuclear antigen 2.J Virol. 2006 Oct;80(19):9761-71. doi: 10.1128/JVI.00665-06. J Virol. 2006. PMID: 16973580 Free PMC article.
-
EBNA2 and activated Notch induce expression of BATF.J Virol. 2003 May;77(10):6029-40. doi: 10.1128/jvi.77.10.6029-6040.2003. J Virol. 2003. PMID: 12719594 Free PMC article.
References
-
- Albert T, Urlbauer B, Kohlhuber F, Hammersen B, Eick D. Ongoing mutations in the N-terminal domain of c-Myc affect transactivation in Burkitt's lymphoma cell lines. Oncogene. 1994;9:759–763. - PubMed
-
- Artavanis Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. - PubMed
-
- Babcock G J, Decker L L, Volk M, Thorley-Lawson D A. EBV persistence in memory B cells in vivo. Immunity. 1998;9:395–404. - PubMed
-
- Braeuninger A, Kuppers R, Strickler J G, Wacker H H, Rajewsky K, Hansmann M L. Hodgkin and Reed-Sternberg cells in lymphocyte predominant Hodgkin disease represent clonal populations of germinal center-derived tumor B cells. Proc Natl Acad Sci USA. 1997;94:9337–9342. . (Erratum, 94:14211.) - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

