Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases
- PMID: 10357851
- PMCID: PMC98966
- DOI: 10.1128/MMBR.63.2.266-292.1999
Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases
Abstract
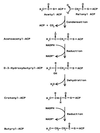
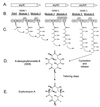
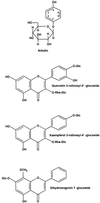
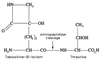
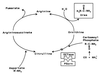
Coronatine, syringomycin, syringopeptin, tabtoxin, and phaseolotoxin are the most intensively studied phytotoxins of Pseudomonas syringae, and each contributes significantly to bacterial virulence in plants. Coronatine functions partly as a mimic of methyl jasmonate, a hormone synthesized by plants undergoing biological stress. Syringomycin and syringopeptin form pores in plasma membranes, a process that leads to electrolyte leakage. Tabtoxin and phaseolotoxin are strongly antimicrobial and function by inhibiting glutamine synthetase and ornithine carbamoyltransferase, respectively. Genetic analysis has revealed the mechanisms responsible for toxin biosynthesis. Coronatine biosynthesis requires the cooperation of polyketide and peptide synthetases for the assembly of the coronafacic and coronamic acid moieties, respectively. Tabtoxin is derived from the lysine biosynthetic pathway, whereas syringomycin, syringopeptin, and phaseolotoxin biosynthesis requires peptide synthetases. Activation of phytotoxin synthesis is controlled by diverse environmental factors including plant signal molecules and temperature. Genes involved in the regulation of phytotoxin synthesis have been located within the coronatine and syringomycin gene clusters; however, additional regulatory genes are required for the synthesis of these and other phytotoxins. Global regulatory genes such as gacS modulate phytotoxin production in certain pathovars, indicating the complexity of the regulatory circuits controlling phytotoxin synthesis. The coronatine and syringomycin gene clusters have been intensively characterized and show potential for constructing modified polyketides and peptides. Genetic reprogramming of peptide and polyketide synthetases has been successful, and portions of the coronatine and syringomycin gene clusters could be valuable resources in developing new antimicrobial agents.
Figures


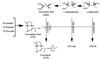



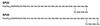
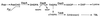

Similar articles
-
Biosynthesis and regulation of coronatine, a non-host-specific phytotoxin produced by Pseudomonas syringae.Subcell Biochem. 1998;29:321-41. doi: 10.1007/978-1-4899-1707-2_10. Subcell Biochem. 1998. PMID: 9594652 Review.
-
Characterization of CorR, a transcriptional activator which is required for biosynthesis of the phytotoxin coronatine.J Bacteriol. 1998 Dec;180(23):6252-9. doi: 10.1128/JB.180.23.6252-6259.1998. J Bacteriol. 1998. PMID: 9829934 Free PMC article.
-
The contribution of syringopeptin and syringomycin to virulence of Pseudomonas syringae pv. syringae strain B301D on the basis of sypA and syrB1 biosynthesis mutant analysis.Mol Plant Microbe Interact. 2001 Mar;14(3):336-48. doi: 10.1094/MPMI.2001.14.3.336. Mol Plant Microbe Interact. 2001. PMID: 11277431
-
A physical map of the syringomycin and syringopeptin gene clusters localized to an approximately 145-kb DNA region of Pseudomonas syringae pv. syringae strain B301D.Mol Plant Microbe Interact. 2001 Dec;14(12):1426-35. doi: 10.1094/MPMI.2001.14.12.1426. Mol Plant Microbe Interact. 2001. PMID: 11768538
-
Control of temperature-responsive synthesis of the phytotoxin coronatine in Pseudomonas syringae by the unconventional two-component system CorRPS.J Mol Microbiol Biotechnol. 2002 May;4(3):191-6. J Mol Microbiol Biotechnol. 2002. PMID: 11931546 Review.
Cited by
-
Distinctiveness of genes contributing to growth of Pseudomonas syringae in diverse host plant species.PLoS One. 2020 Sep 28;15(9):e0239998. doi: 10.1371/journal.pone.0239998. eCollection 2020. PLoS One. 2020. PMID: 32986776 Free PMC article.
-
Pseudomonas syringae pv. actinidiae draft genomes comparison reveal strain-specific features involved in adaptation and virulence to Actinidia species.PLoS One. 2011;6(11):e27297. doi: 10.1371/journal.pone.0027297. Epub 2011 Nov 23. PLoS One. 2011. PMID: 22132095 Free PMC article.
-
Comparative genomics of Pseudomonas syringae pv. syringae strains B301D and HS191 and insights into intrapathovar traits associated with plant pathogenesis.Microbiologyopen. 2015 Aug;4(4):553-73. doi: 10.1002/mbo3.261. Epub 2015 May 4. Microbiologyopen. 2015. PMID: 25940918 Free PMC article.
-
Molecular characterization of an ice nucleation protein variant (inaQ) from Pseudomonas syringae and the analysis of its transmembrane transport activity in Escherichia coli.Int J Biol Sci. 2012;8(8):1097-108. doi: 10.7150/ijbs.4524. Epub 2012 Sep 1. Int J Biol Sci. 2012. PMID: 22991498 Free PMC article.
-
Ornithine Transcarbamylase ArgK Plays a Dual role for the Self-defense of Phaseolotoxin Producing Pseudomonas syringae pv. phaseolicola.Sci Rep. 2015 Aug 10;5:12892. doi: 10.1038/srep12892. Sci Rep. 2015. PMID: 26256666 Free PMC article.
References
-
- Agron P G, Monson E K, Ditta G S, Helinski D R. Oxygen regulation of expression of nitrogen fixation genes in Rhizobium meliloti. Res Microbiol. 1994;145:454–459. - PubMed
-
- Akers C P, Hoff J E. Simultaneous formation of chymopapain inhibitor activity and cubical crystals in tomato leaves. Can J Bot. 1980;58:1000–1003.
-
- Anthamatten D, Hennecke H. The regulatory status of the fixL- and fixJ-like genes in Bradyrhizobium japonicum may be different from that in Rhizobium meliloti. Mol Gen Genet. 1991;225:38–48. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

