The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway
- PMID: 10318916
- PMCID: PMC21892
- DOI: 10.1073/pnas.96.10.5522
The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway
Abstract
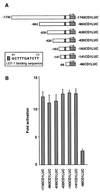
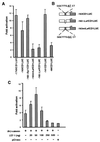
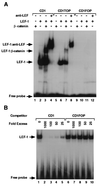
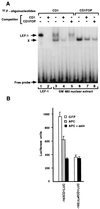
beta-Catenin plays a dual role in the cell: one in linking the cytoplasmic side of cadherin-mediated cell-cell contacts to the actin cytoskeleton and an additional role in signaling that involves transactivation in complex with transcription factors of the lymphoid enhancing factor (LEF-1) family. Elevated beta-catenin levels in colorectal cancer caused by mutations in beta-catenin or by the adenomatous polyposis coli molecule, which regulates beta-catenin degradation, result in the binding of beta-catenin to LEF-1 and increased transcriptional activation of mostly unknown target genes. Here, we show that the cyclin D1 gene is a direct target for transactivation by the beta-catenin/LEF-1 pathway through a LEF-1 binding site in the cyclin D1 promoter. Inhibitors of beta-catenin activation, wild-type adenomatous polyposis coli, axin, and the cytoplasmic tail of cadherin suppressed cyclin D1 promoter activity in colon cancer cells. Cyclin D1 protein levels were induced by beta-catenin overexpression and reduced in cells overexpressing the cadherin cytoplasmic domain. Increased beta-catenin levels may thus promote neoplastic conversion by triggering cyclin D1 gene expression and, consequently, uncontrolled progression into the cell cycle.
Figures

Similar articles
-
Beta-catenin/Tcf-1-mediated transactivation of cyclin D1 promoter is negatively regulated by thyroid hormone.Biochem Biophys Res Commun. 2003 Sep 19;309(2):408-13. doi: 10.1016/j.bbrc.2003.08.019. Biochem Biophys Res Commun. 2003. PMID: 12951064
-
Lymphoid enhancer factor-1 blocks adenomatous polyposis coli-mediated nuclear export and degradation of beta-catenin. Regulation by histone deacetylase 1.J Biol Chem. 2002 Jul 5;277(27):24258-64. doi: 10.1074/jbc.M110602200. Epub 2002 May 1. J Biol Chem. 2002. PMID: 11986304
-
Inhibition of integrin linked kinase (ILK) suppresses beta-catenin-Lef/Tcf-dependent transcription and expression of the E-cadherin repressor, snail, in APC-/- human colon carcinoma cells.Oncogene. 2001 Jan 4;20(1):133-40. doi: 10.1038/sj.onc.1204052. Oncogene. 2001. PMID: 11244511
-
Signaling through beta-catenin and Lef/Tcf.Cell Mol Life Sci. 1999 Oct 30;56(5-6):523-37. doi: 10.1007/s000180050449. Cell Mol Life Sci. 1999. PMID: 11212302 Free PMC article. Review.
-
The Yin-Yang of TCF/beta-catenin signaling.Adv Cancer Res. 2000;77:1-24. doi: 10.1016/s0065-230x(08)60783-6. Adv Cancer Res. 2000. PMID: 10549354 Review.
Cited by
-
Wnt/β-Catenin Signaling Pathway in Pediatric Tumors: Implications for Diagnosis and Treatment.Children (Basel). 2024 Jun 7;11(6):700. doi: 10.3390/children11060700. Children (Basel). 2024. PMID: 38929279 Free PMC article. Review.
-
Suppression of cholangiocarcinoma cell growth by human umbilical cord mesenchymal stem cells: a possible role of Wnt and Akt signaling.PLoS One. 2013 Apr 30;8(4):e62844. doi: 10.1371/journal.pone.0062844. Print 2013. PLoS One. 2013. PMID: 23646150 Free PMC article.
-
A Complex Interplay between Wnt/β-Catenin Signalling and the Cell Cycle in the Adult Liver.Int J Hepatol. 2012;2012:816125. doi: 10.1155/2012/816125. Epub 2012 Sep 3. Int J Hepatol. 2012. PMID: 22973520 Free PMC article.
-
Nuclear IGF1R is a transcriptional co-activator of LEF1/TCF.EMBO Rep. 2012 Mar 1;13(3):244-50. doi: 10.1038/embor.2011.251. EMBO Rep. 2012. PMID: 22261717 Free PMC article.
-
Mitotic and mitogenic Wnt signalling.EMBO J. 2012 Jun 13;31(12):2705-13. doi: 10.1038/emboj.2012.124. Epub 2012 May 22. EMBO J. 2012. PMID: 22617425 Free PMC article. Review.
References
-
- Ben-Ze’ev A, Geiger B. Curr Opin Cell Biol. 1998;10:629–639. - PubMed
-
- Kemler R. Trends Genet. 1993;9:317–321. - PubMed
-
- Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. - PubMed
-
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86:391–399. - PubMed
-
- Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B G, Kemler R. Mech Dev. 1996;59:3–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

