Abstract
We monitored the concentration of indicator viruses crAssphage and pepper mild mottle virus (PMMoV) and human pathogen adenovirus (HAdV) in influent from a wastewater treatment plant in Brisbane, Australia in 1-h and 24-h composite samples. Over three days of sampling, the mean concentration of crAssphage gene copies (GC)/mL in 24-h composite samples did not differ significantly (p = 0.72-0.92), while for PMMoV GC/mL (p value range: 0.0002-0.0321) and HAdV GC/mL (p value range: 0.0028-0.0068) significant differences in concentrations were observed on one day of sampling compared to the other two. For all three viruses, the variation observed in 1-h composite samples was greater than the variation observed in 24-h composite samples. For crAssphage, in 54.1% of 1-h composite samples, the concentration was less than that observed in 24-h composite samples; whereas for PMMoV and HAdV the concentration was less in 79.2 and 70.9% of 1-h composite samples, respectively, compared to the relevant 24-h composite samples. Similarly, the concentration of crAssphage in 1-h compared to 24-h composite samples did not differ (p = 0.1082) while the concentrations of PMMoV (p < 0.0001) and HAdV (p < 0.0001) in 1-h composite samples were significantly different from 24-h composite samples. These results suggest that 24-h composite samples offer increased analytical sensitivity and decreased variability compared to 1-h composite samples when monitoring wastewater, especially for pathogenic viruses with low infection rates within a community. Thus, for wastewater-based epidemiology applications, 24-h composite samples are less likely to produce false negative results and erroneous public health information.
Keywords: Enteric viruses, Enveloped viruses, Epidemiology, Human health risks, Surveillance, WBE, Wastewater
Graphical Abstract
1. Introduction
Several groups of non-enveloped enteric viruses, such as human adenovirus (HAdV), astrovirus (AtVs), enterovirus (EV), hepatitis A virus (HAV), hepatitis E virus (HEV), norovirus (NoV), rotavirus (RoV), Aichi virus (AiV), sapovirus (SaV) and torqueteno virus (TTV) are commonly found in wastewater (Xagoraraki and O'Brien, 2020). These viruses are waterborne and known to cause gastroenteritis, respiratory disease, common cold, meningitis, liver disaese, nausea, vomiting and fever in infected individuals (Frankhauser et al., 2002; Clark and McKendrick, 2004; Ganesh and Lin, 2013). Enteric virus transmission occurs mainly through fecal-oral route via human feces (Kotwal et al., 2014). Enveloped viruses, such as influenza A virus (H1N1) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have also been reported to be present in wastewater (Lago et al., 2003; Heijnen and Medema, 2009; Ahmed et al., 2020a; Haramoto et al., 2020; La Rosa et al., 2020a; Medema et al., 2020; Randazzo et al., 2020a; Sherchan et al., 2020).
Viruses present in municipal wastewater originate from human feces with rare examples (e.g., polyomaviruses) that may also be excreted in urine (Xagoraraki and O'Brien, 2020). Therefore, the concentration of human viruses in wastewater is expected to depend on patterns of human defecation and urination. Intraday fluctuations in virus concentration in municipal wastewater are also expected to differ depending on a number of factors, such as the size of both the service population and catchment, the virus type, the type of sewer system, stormwater intrusion, and climatic conditions. Monitoring of human pathogenic and indicator viruses in municipal wastewater has been used to assess the microbiological quality of wastewater treatment plant (WWTP) influent and to monitor viral diseases in a community, known as wastewater-based epidemiology (WBE) (Bivins et al., 2020). WBE is a powerful tool which can be used to detect not only pathogens significant to public health, but also chemicals or illicit drugs at the community level by monitoring wastewater (Choi et al., 2018; Sims and Kasprzyk-Hordern et al., 2020; Devault and Karolak, 2020).
WBE has been attracting much attention as a complementary tool to monitor the degree of viral disease prevalence in a community, including coronavirus disease 2019 (COVID-19). WBE can provide information on the prevalence of the viruses in the community, spatial and temporal trends on the circulation of viruses, and screening of asymptomatic individuals. WBE could be a valuable tool for resource-limited regions where clinical testings are limited or not available. Complementing clinical testing, WBE can be used as an early warning tool or to detect hot spots for better management of on-going and upcoming epidemics (La Rosa et al., 2020b; Medema et al., 2020b; Thompson et al., 2020).
The application of WBE can be greatly impacted by temporal fluctuations of viral loads in influent wastewater at a wastewater treatment plant (WWTP), however, the temporal dynamics of viruses in wastewater are underreported. A recent review paper has identified several sources of uncertainty in WBE, such as sampling approaches (i.e., grab vs. composite), lack of knowledge on the persistence of viral nucleic acid in wastewater, low recovery efficiency of virus concentration methods, and molecular assays that provide the most sensitive detection and enable accurate quantification at low levels (Kitajima et al., 2020).
Currently, inadequate comparison and reporting of the efficacy of wastewater sampling and virus concentration methods for the detection of enteric viruses and enveloped viruses from wastewater impedes WBE applications. Sampling strategies are particularly important, since poorly designed strategies can introduce biases (i.e., false negatives) in data interpretation. Several studies have used grab sampling approaches for monitoring of enteric and enveloped viruses in wastewater (Ahmed et al., 2020a; Haramoto et al., 2020; Tandukar et al., 2020; McCall et al., 2020; Gyawali and Hewitt, 2018). These methods collect an untreated wastewater sample from wastewater influent at a single point in time. Grab sampling methods may be inadequate for detection of low levels of enteric or enveloped viruses in untreated wastewater due to diurnal variations. Composite samples collected using volume, flow or time-based sampling modes pooling the wastewater into a single sample provide the basis for representative sample collection over extended periods of time (Ort et al., 2010). For enteric virus concentrations or other enveloped viruses such as SARS-CoV-2 in wastewater, little information is available on which of these sampling approaches provide the most appropriate means to collect a representative sample.
The aim of this study was to investigate the intraday variability of two indicator viruses, namely crAssphage and pepper mild mottle virus (PMMoV) and a pathogenic virus HAdV in an urban wastewater treatment plant (WWTP) in Brisbane, Australia. The two indicator viruses selected were chosen because, unlike some other viral indicators (e.g., bacteriophages), both crAssphage and PMMoV are human-associated (Ahmed et al., 2019), owing to the consumption of vegetables infected with PMMoV and crAssphage as a commensal member of the human gut microbiota. CrAssphage and PMMoV are highly abundant in untreated wastewater across wide geographic regions, and their concentrations exhibit temporal stability. Both have been recently used to normalize SARS-CoV-2 RNA in wastewater (Medema et al., 2020b). Conversely, HAdV is a human pathogen capable of infecting both the respiratory and gastrointestinal tracts (Lynch et al., 2011). The concentration of HAdV in wastewater is expected to be much lower than crAssphage and PMMoV due to the lower prevalence of shedding populations as well as lower shedding amount from an individual. By considering viruses that are both more and less prevalent among the population and subsequently in wastewater, the observations of the current study can inform the design of appropriate wastewater sampling strategies for sensitive and reliable monitoring of pathogenic viruses in wastewater for WBE or other applications such as recycled water safety monitoring.
2. Materials and methods
2.1. Wastewater sampling
Untreated wastewater samples were collected from the influent of an urban WWTP in Brisbane, Australia, using a conventional autosampler ISCO 3700 (Teledyne ISCO, Inc., Lincoln, NE, USA). The autosampler base contained ice to keep the samples cool during sample collection. The autosampler was programmed to collect a sample every 15 min over a 24-h period that were composited to provide 24 1-h composite over three separate days (June 22, 2020, June 25, 2020, and July 01, 2020), yielding a total of 72 1-h composite samples. In addition, equal volume aliquots from each of the 24 1-h composite samples were pooled to create a one 24-h composite sample for each of the three days yielding a total of three 24-h composite samples for three days. The sewer network is comprised of reticulated, branched and trunk main sewers. No precipitation occurred in the WWTP catchment during wastewater sampling. The hydraulic treatment capacity of the WWTP is approximately 170 mega liter (ML)/day and average dry weather flow is 140 ML/day. Approximately 17% of wastewater was estimated to be from commercial and industrial sources including waste from landfills, hospitals and an airport. The maximum hydraulic residence time in the sewer network is estimated to be 11–12 h, and there are 83 pump stations in the catchment. Samples were transported on ice to the laboratory, stored at 4 °C and processed within 5 days. While five days is a longer holding time compared to customary ~24 h, a recent study reported that SARS-CoV-2 and murine hepatitis virus (MHV) RNA in untreated wastewater at 4 °C can persist for > 30 days, with minimal to no decay observed within the first 5 days (Ahmed et al., 2020b), suggesting no adverse effects from the extended holding times.
2.2. Viral nucleic acid extraction
RNeasy Mini and DNeasy Blood & Tissue Kits (Qiagen, Hilden, Germany) were used to directly extract nucleic acid from 140 μL and 200 μL of untreated wastewater, respectively without any pre-treatment or concentration. A QIAcube Connect platform (Qiagen) was used to eluate 60 μL of RNA and 100 μL of DNA from lysates. All nucleic acid samples were stored at −80 °C and subjected to qPCR and RT-qPCR analyses within 1–3 days after nucleic acid extraction to avoid potential losses associated with storing, as well as freezing and thawing of nucleic acid samples.
2.3. Determination of PCR inhibition
An experiment was conducted to determine the presence of PCR inhibition in nucleic acid extracted from wastewater samples using Sketa22 qPCR and MHV RT-qPCR assays (Besselsen et al., 2002; Haugland et al., 2005). Known gene copy (GC) numbers of Oncorhynchus keta DNA (2 × 102 GC/reaction) and MHV RNA (2 × 104 GC/reaction) were added in the qPCR and RT-qPCR reactions, respectively (without sample) and the quantification cycle (Cq) values obtained acted as reference points. The same amounts of O. keta and MHV were also added into qPCR and RT-qPCR reactions respectively in the presence of wastewater nucleic acid samples. If the Cq value of a wastewater nucleic acid sample substantially increases (i.e., two Cq values), the sample was considered to have PCR inhibitors (Staley et al., 2012).
2.4. qPCR and RT-qPCR assay
Previously published qPCR and RT-qPCR assays were used for the analysis of crAssphage (CPQ_056 assay) (Stachler et al., 2017), HAdV (Heim et al., 2003) and PMMoV (Rosario et al., 2009; Haramoto et al., 2013). The primers and probes for these qPCR assays are shown in Supplementary Table ST1 along with qPCR cycling conditions. For all qPCR assays, positve control synthetic DNA (4 μg) in plasmid cloning vectors or gBlocks gene fragments were purchased from the Integrated DNA Technologies (Coralville, IA, USA). qPCR standards were prepared from the positive control, ranging from 106 to 1 GC/μL of DNA. CrAssphage and HAdV qPCR amplifications were performed in 20 μL reaction mixtures using SsoAdvanced Universal Probes Supermix (Bio-Rad Laboratories, Richmond, CA, USA). CrAssphage qPCR mixtures contained 10 μL of Supermix, 1000 nM of each forward and reverse primer, 100 nM probe and 3 μL of template DNA. HAdV qPCR mixture contained 10 μL of Supermix, 200 nM of each forward, reverse primer, and probe and 3 μL of template DNA. PMMoV RT-qPCR mixtures contained 10 μL of qScript XLT 1-Step RT-qPCR ToughMix (QuantaBio, Beverly, MA, USA), 200 nM of forward and reverse primer, 80 nM probe and 5 μL of template RNA. For each qPCR run, a series of standard (3 × 106 to 3 GC/reaction) and no template controls (n = 3) were included. The qPCR assays were performed using a Bio-Rad CFX96 thermal cycler (Bio-Rad Laboratories). All qPCR reactions were performed in triplicate. The qPCR assay limit of detection (ALOD) values were determined from Cq values of the three separate standard curves run for each assay. qPCR ALOD values were defined as the number of copies that could be detected and quantified in 2/3 qPCR assays (Senkbeil et al., 2019). The sample limit of detection (SLOD) was calculated by dividing the ALOD by the DNA or RNA template volume (i.e., 3 μL for crAssphage and HAdV and 5 μL for PMMoV) added to the qPCR/RT-qPCR well and then multiplying this number by the total volume of DNA or RNA extracted from each sample (100 μL for crAssphage and HAdV and 60 μL for PMMoV) to yield SLOD.
2.5. Quality control
A reagent blank and an extraction blank were included for each batch of nucleic acid extraction to ensure no carryover contamination occurred during extraction. No carryover contamination was observed in reagent blank samples. To minimize potential qPCR and RT-qPCR contamination, nucleic acid extraction and PCR setup were performed in separate laboratories.
2.6. Data analysis
GraphPad Prism 8.3.1 (GraphPad Software, San Diego, CA, USA) was used to conduct one-way analysis of variance (ANOVA) with Tukey's multiple comparison test to evaluate differences in concentrations of each individual virus across different sampling days, and to compare concentrations of three viruses tested in 1-h and 24-h composite samples. The same software was also used to conduct unpaired t-test to test the difference in concentration of each virus in the 1-h composite samples versus 24-h composite samples (α = 0.05 for both tests).
3. Results
3.1. PCR inhibition, performance characteristics and limit of detection
Mean Cq values for O. keta in sterile water ranged from 31.6 to 31.9, while these values ranged from 31.6 to 32.1 for the 1-h composite samples and 31.2 to 32.1 for 24-h composite samples (Supplementary Table ST2). The difference in mean Cq value for O. keta in the 1-h and 24-h composite samples collected in different days compared to sterile water samples were −0.10 to 0.70 and −0.40 to 0.40, respectively. Similarly, mean Cq values for MHV in sterile water ranged from 24.5 to 24.9, while these values ranged from 24.4 to 24.6 for the 1-h composite samples and 24.5 to 24.8 for the 24-h composite samples (Supplementary Table ST2). The differences in mean Cq values for MHV in 1-h and 24-h composite samples collected on different days compared to sterile water samples ranged from −0.50 to 0.10 and −0.40 to 0.30, respectively. Based on the Cq values, all DNA and RNA samples were considered to be free from PCR inhibition and were used for downstream qPCR and RT-qPCR analysis. The amplification efficiencies of crAssphage, HAdV and PMMoV assays were within the prescribed range (96–104%) of MIQE guidelines (Bustin et al., 2009). The coefficient of determination (R2) values for all assays were between 0.96 and 0.99. The slope of the standard curves and Y-intercept values are shown in Supplementary Table ST3. The ALOD values were 2, 1, and 3 GC/μL of nucleic acid for crAssphage, HAdV and PMMoV, respectively. The SLOD values for crAssphage and HAdV were 66.6 and 33.3 GC/200 μL of untreated wastewater sample, respectively. Similarly, the SLOD value for PMMoV was 36 GC/200 μL of untreated wastewater sample.
3.2. Concentration of viruses in untreated wastewater
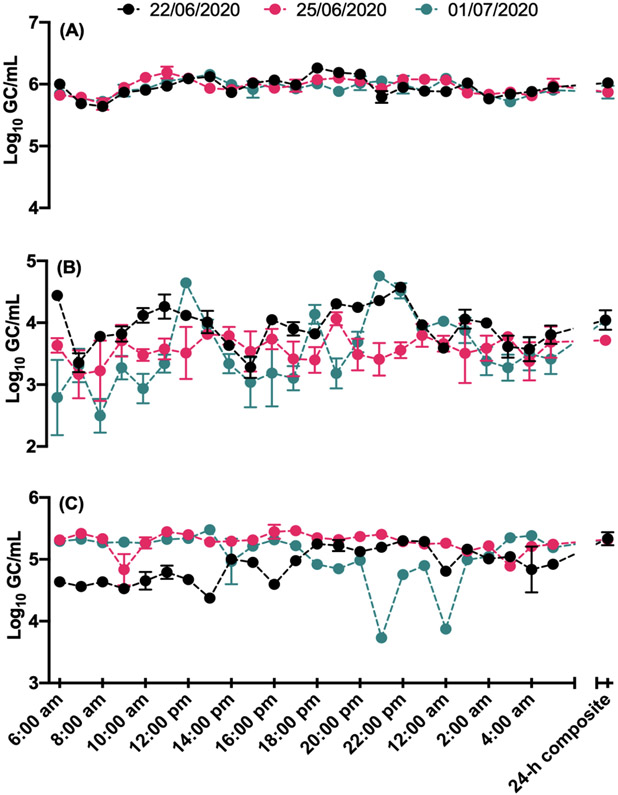
Concentrations of crAssphage, HAdV and PMMoV in the 1-h and 24-h composite samples are shown in Fig. 1. Among the three viruses analysed, the mean concentrations of crAssphage, HAdV and PMMoV in the 1-h composite samples ranged from 5.94 to 5.96, 3.54 to 3.95 and 4.90 to 5.28 log10 GC/mL, respectively, over three sampling dates (Table 1). One-way ANOVA indicated that the pooled mean concentration (5.95 ± 0.13 log10 GC/mL) of crAssphage in the 1-h composite samples over the entire sampling campaign was significantly (p < 0.0001) greater than HAdV (3.69 ± 0.43 log10 GC/mL) and PMMoV (5.08 ± 0.34 log10 GC/mL). The mean concentration of PMMoV was also significantly greater (p < 0.0001) than HAdV.
Fig. 1:
Concentrations (log10 GC/mL) of (A) crAssphage, (B) HAdV, and (C) PMMoV in the 1-h and 24-h time-based composite samples.
Table 1.
Mean (log10 GC/mL), standard deviation (SD), range and 95% confidence interval (CI), coeeficient of variation (CV%) of crAssphage, HAdV and PMMoV in the 1-h composite untreated wastewater samples collected on three separate dates.
| Concentrations (log10 GC/mL) |
Sampling dates | |||
|---|---|---|---|---|
| 22/06/2020a | 25/06/2020a | 01/07/2020a | 22/06-2020 – 01/07/2020b |
|
| CrAssphage | ||||
| Mean | 5.95 | 5.96 | 5.94 | 5.95 |
| SD | 0.15 | 0.12 | 0.11 | 0.13 |
| Min | 5.64 | 5.68 | 5.72 | 5.64 |
| Max | 6.26 | 6.19 | 6.16 | 6.26 |
| Lower 95% CI | 5.89 | 5.91 | 5.89 | 5.92 |
| Upper 95% CI | 6.01 | 6.02 | 5.98 | 5.98 |
| CV (%) | 2.57 | 2.12 | 1.97 | 2.22 |
| HAdV | ||||
| Mean | 3.95 | 3.58 | 3.54 | 3.69 |
| SD | 0.33 | 0.20 | 0.57 | 0.43 |
| Min | 3.28 | 3.17 | 2.50 | 2.50 |
| Max | 4.57 | 4.06 | 4.76 | 4.76 |
| Lower 95% CI | 3.80 | 3.49 | 3.30 | 3.59 |
| Upper 95% CI | 4.09 | 3.66 | 3.79 | 3.79 |
| CV (%) | 8.46 | 5.65 | 16.3 | 11.9 |
| PMMoV | ||||
| Mean | 4.90 | 5.28 | 5.05 | 5.08 |
| SD | 0.27 | 0.15 | 0.43 | 0.34 |
| Min | 4.37 | 4.83 | 3.73 | 3.73 |
| Max | 5.30 | 5.46 | 5.48 | 5.48 |
| Lower 95% CI | 4.78 | 5.22 | 4.87 | 5.00 |
| Upper 95% CI | 5.01 | 5.35 | 5.23 | 5.16 |
| CV (%) | 5.55 | 2.90 | 8.53 | 6.73 |
1-h composite for each day.
1-h composite for three days.
Similarly, the mean concentration (5.92 ± 0.08 log10 GC/mL) of crAssphage in the 24-h composite samples collected over three sampling dates was significantly (p value range: 0.0021 - <0.0001) greater than HAdV (3.93 ± 0.18 log10 GC/mL) and PMMoV (5.33 ± 0.006 log10 GC/mL). The mean concentration of PMMoV was also significantly (p < 0.001) greater than HAdV. The concentrations of all three viruses among three sampling dates were also compared. The mean concentration of crAssphage in the 24-h composite sample collected on June 22, 2020 did not differ significantly from June 25, 2020 and July 01, 2020 (p value range: 0.72–0.92). The mean concentration of HAdV in the 24-h composite sample on June 22, 2020 differed significantly (p value range: 0.002–0.006) from June 25, 2020 and July 01, 2020. However, the mean concentration of HAdV on June 25, 2020 did not differ significantly (p = 0.9536) from July 01, 2020. The mean concentration of PMMoV in the 24-h composite sample on June 22, 2020 differed significantly (p value range: 0.0002–0.03) from June 25, 2020 and July 01, 2020, but there was no statistically significant difference between June 25, 2020 and July 01, 2020 (p = 0.1980).
The variation in concentration of all three viruses in the 24-h composite samples with the concentration in the 1-h individual composite samples was also compared. The standard deviations (SDs) of crAssphage in the 1-h composite samples ranged from 0.11 to 0.15 log10 GC/mL over the three sampling dates, whereas the SD for 24-h time-based composite samples was 0.08 log10 GC/mL over three sampling dates. A greater variation was observed for HAdV in the 1-h individual composite samples, with SDs ranging from 0.20 to 0.57 log10 GC/mL over three sampling dates compared to 24-h time-based composite samples (0.18 log10 GC/mL) over three sampling dates. A greater variation was also observed for PMMoV in the 1-h composite samples, with SDs ranging from 0.15 to 0.43 log10 GC/mL over three sampling dates compared to 24-h composite samples (0.006 log10 GC/mL) over three sampling dates. The coefficient of variation (CV) values for crAssphage over three days were small ranged from 1.97 to 2.57%. However, CV values for HAdV and PMMoV were much greater than crAssphage, ranging from 5.65 to 16.3% and 2.90 to 8.63%, respectively.
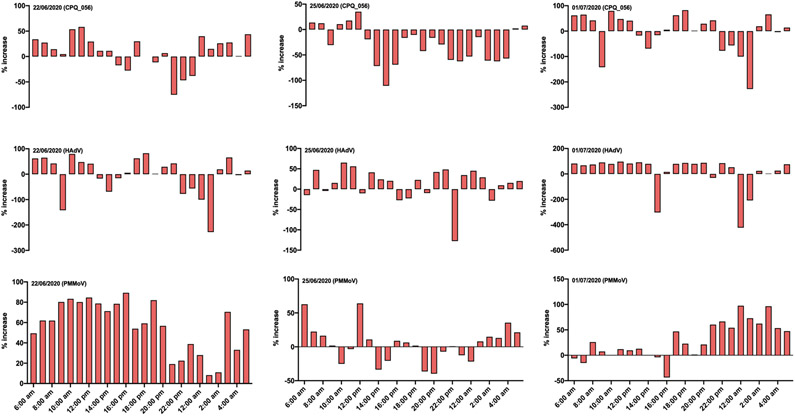
The percentage increase or decrease in concentration was determined by calculating the difference in the concentration of viruses between 1-h and 24-h composite samples and then divideded the increase by the original number and multiplied the value by 100. Overall, the concentration of crAssphage was greater in 24-h composite samples compared to 39/72 (54.1%) of the 1-h composite wastewater samples (Fig. 2). Similarly, concentrations of HAdV and PMMoV were greater in 24-h composite samples compared to 51/72 (70.8%) and 57/72 (79.2%) 1-h composite wastewater samples (Fig. 2), but not all comparisons were statistically significant. Specifically, direct comparison of crAssphage concentrations between the 1-h composite samples and 24-h time-based composite samples indicated no statistically significant difference (p = 0.1082), while concentrations in the latter sample type were significantly higher for both HAdV and PMMoV (p < 0.0001 for both).
Fig. 2:
Percentage increase of crAssphage, HAdV and PMMoV in 24-h composite samples compared to the 1-h composite samples.
4. Discussion
Achieving a representative sample is vital for the successful application of WBE as a sensitive early warning system or to monitor trends of virus circulation in the community. Poorly designed sampling procedures may introduce biases and contribute to false-negative results (WRF, 2020). False negative results (i.e., virus infections present in the community but the wastewater analytical method fails to yield a signal) could have economic and public health consequences in wastewater surveillance. The concentration of viruses, especially pathogenic viruses, in untreated wastewater is expected to vary throughout the day depending on a number of factors, including the infection prevalence in the community, the frequency and timing of shedding into the sewer, concentrations in the body fluids entering the wastewater stream, variations in flow due to water introduced from non-sewer sources (e.g., washing systems or stormwater), proportion of industrial wastewater, and the travel time of wastewater from households to the sampling point (O'Reilly et al., 2020; Polo et al., 2020). The concentration of pathogenic viruses is also typically much lower compared to indicator viruses such as PMMoV and crAssphage (Symonds et al., 2018) necessitating collection of larger sample volumes or creation of a composite sample. The sampling strategies for virus detection in wastewater range from composite samples collected over a 24-h period (La Rosa et al., 2020b; Westhaus et al., 2020) to single grab samples collected during the peak flow (Polo et al., 2020), while other researchers collect grab wastewater samples in the morning (Randazzo et al., 2020b), and all are based on relatively small volumes (typically 50–250 mL). From the available data on the concentrations of enteric viruses and SARS-CoV-2 RNA in wastewater, it is not clear how widely the signal may vary throughout the day or whether different sampling strategies affect the outcome.
In this study, we determined the concentrations of crAssphage, HAdV and PMMoV in samples collected as 1-h composites throughout the day and 24-h composite samples prepared from mixing 24 1-h samples collected from a large urban WWTP over three sampling days in Southeast Queensland, Australia. Even though our study was conducted at a large WWTP (serving > 500,000 people), recent studies indicated no difference in concentrations of various indicators (including crAssphage and other less abundant viral indicators such as human polyomavirus) between urban and rural WWTPs suggest that our findings could be extended to a wide range of sewerage systems (Korajkic et al., 2020; Mayer et al., 2018). While it would have been ideal to determine the concentrations of SARS-CoV-2 RNA in wastewater samples so that their loadings and variability could be compared directly with enteric viruses; the concentrations of SARS-CoV-2 RNA in the study catchment have been low resulting in many non-detects or below the limit of quantification during this pandemic (Ahmed et al., 2020a). As a result, we selected two indicator viruses and a pathogenic virus as potentially similar or greater than SARS-CoV-2 loadings in wastewater samples, had the prevalence of SARS-CoV-2 been higher in the community.
In this study, we did not perform virus concentration; instead viral nucleic acids were directly extracted from a small volume of wastewater. This is because the selected viruses are highly abundant in untreated wastewater throughout the world. The concentration of crAssphage, HAdV and PMMoV in wastewater would have been lower (due to loss during concentration process and variability of recovery) if a virus concentration method was used in this study. However, this does not affect the overall results and data interpretation of this study.
Over the course of three sampling days, concentration of HAdV and PMMoV in 1-h composite samples varied, albeit not in the same manner (e.g., HAdV concentrations were the highest during the first sampling day, while PMMoV concentrations were the highest during the second day). On the other hand, crAssphage concentrations were similar across the sampling days. A recent report suggested using PMMoV concentrations in the wastewater as an internal reference to calibrate SARS-CoV-2 concentrations across the samples (Wu et al., 2020). While this is a promising approach that can help normalize viral targets typically present in low concentrations, such as SARS-CoV-2, caution should be exercised as concentrations of potential normalization viruses (e.g., PMMoV) may not co-vary with the desired viral pathogen target on a daily basis. Future research efforts need to examine intraday variability of SARS-CoV-2 and prospective viral calibrators to determine feasibility of this approach.
Among the three viruses analysed, the concentrations of crAssphage in the 1-h and 24-h composite samples were up to three orders of magnitude greater than HAdV and PMMoV. This is consistent with previous findings where we showed crAssphage concentration was much greater than other indicator and pathogenic viruses in wastewater in Southeast Queensland, Australia and USA (Ahmed et al., 2019). The high concentration of crAssphage in wastewater makes it a more sensitive marker gene for tracking faecal pollution in environmental waters (Ahmed et al., 2019; Korajkic et al., 2020). The intraday variability of crAssphage and PMMoV concentrations were much lower than HAdV. This is because these indicator viruses are typically shed by both healthy and infected people, whereas, pathogenic viruses are only shed by those who are infected. However, the CV of PMMoV concetrations were considerably higher than the CV of crAssphage concentrations. While both indicator viruses are human-associated, crAssphage is a DNA bacteriophage likely infecting Bacteroides intestinalis, a commensal inhabitant of human gastrointestinal tract (Shkoporov et al., 2018) while PMMoV is a plant virus infecting peppers belonging to Capsicum spp. (Fauquet et al., 2005). Therefore, the concentrations of PMMoV RNA in any sewerage system are directly dependent on dietary intake of infected pepper plants by the contributing populations which is likely to be variable throughout the day resulting in the elevated CVs. Therefore, measurements of pathogenic viruses such as HAdV or other enteric viruses such as norovirus or enterovirus, typically present in lower concentrations, are expected to be more variable throughout the day. Of note, HAdV concentrations measured in this study are comparable to those reported by others (Ahmed et al., 2019; Hamza et al., 2019; Elmahdy et al., 2020) suggesting that the greater intraday variability of HAdV compared to indicator viruses present in higher concentrations is likely not limited to this study. Assuming these viruses are indicative of the variability in SARS-CoV-2, the results indicate that collecting a grab sample at a pre-determined time may be inferior to a 24-h composite sample when seeking to achieve sensitive detection of low levels of enteric viruses and SARS-CoV-2 RNA in wastewater and avoid false negatives.
A recent study recommended the collection of grab sample during the peak hour of flow may be a good approach for surveillance of SARS-CoV-2 in wastewater (Polo et al., 2020). However, the travel time of wastewater from households to the WWTP in some catchments can typically range from between 2 and 12 h or greater in some cases, and therefore, it may be difficult to determine when is the best time to collect a grab sample (Castigliono et al., 2013). Furthermore, it is unclear if collecting a grab sample during the predicted peak hour of toilet usage will capture the strongest signal or result in a more diluted signal (Michael-Kordatou et al., 2020; WRF, 2020). Another consideration is the combined sewerage systems, where wastewater is considerably diluted during the heavy rainfalls or periods of snow fall and snow melting. For example, during combined sewer overflow (CSO) events, the contribution of rainwater to wastewater can range from 5 to 40% (Passerat et al., 2011), whereas the seasonal dilution factor during the snow melting period can be as high ~6900 (Madoux-Humery et al., 2013). These circumstances present unusual case scenarios that merit further consideration when selecting the best sampling approach for a given wastewater system.
In this study, the concentrations of all three viruses were greater (54.1-79.2%) in 24-h composite samples when compared with 1-h composite samples, suggesting that composite samples may be preferable over a grab sample taken at a pre-determined time when the concentration of the target virus is expected to be low due to low shedding in the community. The longer the composite sampling duration (i.e., 24-h or 72-h), the more representative the sample will be, however, shorter duration such as 2-h, 4-h and 6-h sampling over 24-h and 72-h period may also be equally representative when the travel time of wastewater from households to the WWTP is shorter. It is important to note that we had observed significant differences in concentrations between the 1-h and 24-h composite samples for PMMoV and HAdV, suggesting that the two sample types are not interchangeable within the study or across the studies.
One drawback of composite sampling is that setting up autosamplers can be expensive and the collection system needs to provide cool storage conditions (refrigeration or filled with ice) to maintain the integrity of the sample. However, recent studies suggested longer persistence of SARS-CoV-2 RNA in water and wastewater even at 15–20 °C. (Ahmed et al., 2020b; Bivins et al., 2020). It should be noted that, results obtained in this study were based on wastewater samples from a large urban WWTP with specific catchment characteristics, and different results may be observed for wastewater systems with significantly different catchment characteristics. The results presented in this study also reflect a scenario over a short duration (i.e., three days) and do not include seasonal and weather variations. Therefore, it would be useful to undertake similar studies for a range of different catchments under variable weather (dry vs. wet) and climatic conditions (sub-tropical vs. temperate). Further studies should focus on comparing time-based vs. flow-based composite sampling and how that influences the results for wastewater surveillance of the SARS-CoV-2 signal. Also, the appropriate volume of composite samples that should be analysed to avoid false-negative detection should be considered carefully especially when the prevalence of the target virus is low.
Representative wastewater sampling along with effective concentration and sensitive detection methods are needed for successful application of WBE at the community level. Our results based on two indicator viruses (PMMoV and crAssphage) and a pathogenic virus (HAdV) indicate that there is intra and interday variability of concentrations, which is especially pronounced with targets present in lower concentrations (e.g., HAdV). Therefore, in this study, time-based 24-h composite sample typically yielded greater concentrations of viruses especially HAdV and PMMoV compared to the 1-h composite samples. Extrapolating our findings to SARS-CoV-2 RNA in wastewater, the 24-h composite samples are likely to be superior to single grab samples or the 1-h composite samples, and would facilitate more sensitive detection of the virus. However, this assertion shouldbe further examined.
Supplementary Material
Acknowledgements
We thank Paul Sherman and Jason Dwyer from Urban Utilities for facilitating wastewater sample collection. S.P. Sherchan was supported by the NIH grant R21AI157434. K. Bibby was supported by NSF grant 2027752.
Footnotes
Disclaimers
The views expressed in this article are those of the author(s) and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. The U.S. Environmental Protection Agency through the Office of Research and Development provided technical direction but did not collect, generate, evaluate, or use the environmental data described herein.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Ahmed W, Gyawali P, Feng S, Mclellan SL, 2019. Host-specificity and sensitivity of established and novel sewage-associated marker genes in human and nonhuman fecal samples. Appl. Environ. Microbiol 85 (14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O’Brien JW, Choi PM, Kitajima M, Simpson SL, Li J, Tscharke B, Verhagen R, Smith WJM, Zaugg J, Dierens L, Hugenholtz P, Thomas KV, Mueller JF, 2020a. First confirmed detection of SARS-CoV-2 un untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ 728, 138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Bertsch PM, Bibby K, Haramoto E, Hewitt J, Huygens F, Gyawali P, Korajkic A, Riddel S, Sherchan SP, Simpson SL, Sirikanchana K, Symonds EM, Verhagen R, Vasan SS, Kitajima M, Bivins A, 2020b. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res 191, 110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besselsen DG, Wagner AM, Loganbill JK, 2002. Detection of rodent coronaviruses by use of fluorogenic reverse transcriptase-polymerase chain reaction analysis. Comp. Med 52 (2), 111–116. [PubMed] [Google Scholar]
- Bivins A, Greaves J, Fischer R, Yinda KC, Ahmed W, Kitajima M, Munster VJ, Bibby K, 2020. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT, 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem 55 (4), 611–622. [DOI] [PubMed] [Google Scholar]
- astiglioni S, Bijslma L, Covaci A, Emke E, Hernandez F, Reid M, Ort C, Thomas KV, van Nuijs ALN, de Voogt P, Zuccato E, 2013. Evaluation of uncertainties associated with the determination of community drug use through the measurement of sewage drug biomarkers. Environ. Sci. Technol 47 (3), 1452–1460. [DOI] [PubMed] [Google Scholar]
- Choi PM, Tscharke BJ, Donner E, O’Brien JW, Grant SC, Kaserzon SL, Mackie R, O’Malley E, Crosbie ND, Thomas KV, Mueller JF, 2018. Wastewater-based epidemiology biomarkers: past, present and future. TrAC Trends Anal. Chem. (Reference Ed.) 105, 453–469. [Google Scholar]
- Clark B, McKendrick M, 2004. A review of viral gastroenteritis. Curr. Opin. Infect. Dis 17 (5), 461–469. [DOI] [PubMed] [Google Scholar]
- Devault DA, Karolak S, 2020. Wastewater-based epidemiology approach to assess population exposure to pesticides: a review of a pesticide pharmacokinetic dataset. Environ. Sci. Pollut. Res 27, 4695–4707. [DOI] [PubMed] [Google Scholar]
- Elmahdy EM, Shaheen MNF, Rizk NM, Saad-Hussein A, 2020. Quantitative detection of human adenovirus and human rotavirus group A in wastewater and El- Rahawy Drainage Canal influencing River Nile in the North of Giza, Egypt. Food Environ. Virol 12 (3), 218–225. [DOI] [PubMed] [Google Scholar]
- Fankhauser RL, Monroe SS, Noel JS, Humphrey CD, Bresee JS, Parashar UD, Ando T, Glass RI, 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis 186, 1–7. [DOI] [PubMed] [Google Scholar]
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, 2005. Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, Amsterdam, The Netherlands. [Google Scholar]
- Ganesh A, Lin J, 2013. Waterborne human pathogenic viruses of publiac health concern. Int. J. Environ. Health Res 23 (6), 544–564. [DOI] [PubMed] [Google Scholar]
- Gyawali P, Hewitt J, 2018. Detection of infectious noroviruses from wastewater and seawater using PEMAXTM treatment combined with RT-qPCR. Water 10 (7), e84. [Google Scholar]
- Hamza H, Rizk NM, Gad MA, Hamza IA, 2019. Pepper mild mottle virus in wastewater in Egypt: a potential indicator of wastewater pollution and the efficiency of the treatment process. Arch. Virol 164 (11), 2707–2713. [DOI] [PubMed] [Google Scholar]
- Haramoto E, Kitajima M, Kishida N, Konno Y, Katayama H, Asami M, Akiba M, 2013. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Environ. Microbiol 79 (23), 7413–7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E, Malla B, Thakali O, Kitajima M, 2020. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ 737, 140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP, 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39 (4), 559–568. [DOI] [PubMed] [Google Scholar]
- Heijnen L, Medema G, 2009. Surveillance of influenza A and the pandemic influenza A (H1N1) 2009 in sewage and surface water in The Netherlands. J. Water Health 9 (3), 434–442. [DOI] [PubMed] [Google Scholar]
- Heim A, Ebnet C, Harste G, Pring-Akerblom P, 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol 70 (2), 228–239. [DOI] [PubMed] [Google Scholar]
- Kitajima M, Ahmed W, Bibby K, Carducci A, Gerba CP, Hamilton KA, Haramoto E, Rose JB, 2020. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ 739, 139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korajkic A, McMinn B, Herrmann MP, Sivaganesan M, Kelty CA, Clinton P, Nash MS, Shanks OC, 2020. Viral and bacterial fecal indicators in untreated wastewater across the contiguous United States exhibit geospatial trends. Appl. Environ. Microbiol 86 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwal G, Cannon JL, 2014. Environmental persistence and transfer of enteric viruses. Curr. Opin. Virol 4, 37–43. [DOI] [PubMed] [Google Scholar]
- La Rosa G, Iaconelli M, Mancini P, Ferraro GB, Veneri C, Bonadona L, Lucentini L, Suffredini E, 2020a. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ 736, 139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G, Mancini P, Bonanno G, Veneri C, Iaconelli M, Bonadonna L, Lucentini L, Suffredini E, 2020b. SARS-CoV-2 has been circulating in northen Italy since December 2019: evidence from environmental monitoring. Sci. Total Environ 750, 141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lago PM, Gary HE, Perez LS, caceras V, Olivera JB, Puentes RP, Corredor MB, Jimenez P, Pallansch MA, Cruz RG, Poliovirus detection in wastewater and stools following an immunization campaign in Havana, Cuba. Int. J. Epidemiol 32 (5), 772–777. [DOI] [PubMed] [Google Scholar]
- Lynch JP 3rd, Fishbein M, Echavarria M, 2011. Adenovirus. Semin. Respir. Crit. Care Med 32 (4), 494–511. [DOI] [PubMed] [Google Scholar]
- Madoux-Humery AS, Dorner S, Sauve S, Aboulfadl K, Galarneau M, Servais P, Prevost M, 2013. Temporal variability of combined sewer overflow contaminants: evaluation of wastewater micropolutants as tracers of fecal contamination. Water Res. 47, 4370–4382. [DOI] [PubMed] [Google Scholar]
- Mayer RE, Reischer GH, Ixenmaier SK, Derx J, Blaschke AP, Ebdon JE, Linke R, Egle L, Ahmed W, Blanch AR, Byamukama D, Savill M, Mushi D, Cristobal HA, Edge TA, Schade MA, Aslan A, Brooks YM, Sommer R, Masago Y, Sato MI, Taylor HD, Rose JB, Wuertz S, Shanks OC, Piringer H, Mach RL, Savio D, Zessner M, Farnleitner AH, 2018. Global distribution of human-associated fecal genetic markers in reference reference samples from six continents. Environ. Sci. Technol 52 (9), 5076–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall C, Wu H, Miyani B, Xagoraraki I, 2020. Identification of multiple potential viral diseases in a large urban center using wastewater surveillance. Water Res. 184 (1), e116160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G, Been F, Heijnen L, Petterson S, 2020b. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions. Curr. Opin. Environ. Sci. Health 17, 49–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A, 2020a. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol acs.estlett., 0c00357 [DOI] [PubMed] [Google Scholar]
- Michael-Kordatou I, karaolia P, Fatta-Kassinos D, 2020. Sewage analysis as a tool for the COVID-19 pandemic response andmanagement: the urgent need for optimised protocols for SARS-CoV-2detection and quantification. J. Environ. Chem, Engineer 8, 104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort C, Lawrence MG, Reungoat J, Mueller JF, 2010. Sampling for PPCPs in wastewater systems: comparison of different sampling modes and optimization strategies. Environ. Sci. Technol 44 (16), 6289–6296. [DOI] [PubMed] [Google Scholar]
- Passerat J, Ouattara NK, Mouchel JM, Rocher V, Servais P, 2011. Impact of an intense combined sewer overflow event on the microbiological water quality of the Seine River. Water Res. 45, 893–903. [DOI] [PubMed] [Google Scholar]
- Polo D, Quintela-Baluja M, Cosbishley A, Jones DL, Singer AC, Graham DW, Romalde J, 2020. Making waves: wastewater-based epidemiology for COVID-19 – approaches and challenges for surveillance and prediction. Water Res. 186, 116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W, Cuevas-Ferrando E, Sanjuan R, Domingo-Calap P, S´anchez G, 2020b. Metropolitan wastewater analysis for COVID-19 epidemiological sureveillance. Int. J. Hyg Environ. Health 230, 113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W, Truchado P, Cuevas-Ferrando E, Simon P, Allende A, Sanchez G, 2020a. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 181, 115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K, Symonds EM, Sinigalliano C, Stewart J, Breitbart M, 2009. Pepper mild mottle virus as an indicator of fecal pollution. Appl. Environ. Microbiol 75 (22), 7261–7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkbeil JS, Ahmed W, Conrad J, Harwood VJ, 2019. Use of Escherichia coli genes associated with human sewage to track fecal contamination source in subtropical waters. Sci. Total Environ 686, 1069–1075. [DOI] [PubMed] [Google Scholar]
- Sherchan SP, Shahin S, Ward LM, Tandukar S, Aw TG, Schmitz B, Ahmed W, Kitajima M, 2020. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ 140261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkoporov AN, Khokhlova EV, Fitzgerald CB, Stockdale SR, Draper LA, Ross RP, Hill C, 2018. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun 9 (1), 4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims N, Kasprzyk-hordern B, 2020. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int 139, 105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachler E, Kelty C, Sivaganesan M, Li X, Bibby K, Shanks OC, 2017. Quantitative crAssphage PCR assays for human fecal pollution measurements. Environ. Sci. Technol 51 (16), 9146–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley C, Gordon KV, Schoen ME, Harwood VJ, 2012. Performance of two quantitative PCR methods for microbial source tracking of human sewage and implications for microbial risk assessment in recreational waters. Appl. Environ. Microbiol 78 (20), 7317–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds EM, Nguyen KH, Harwood VJ, Breitbart M, 2018. Pepper mild mottle virus: a plant pathogen with a greater purpose in (waste)water treatment development and public health management. Water Res. 144, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandukar S, Sherchan SP, Haramoto E, 2020. Applicability of CrAssphage, pepper mild mottle virus and tobacco mosaic virus as indicators of reduction of enteric viruses during wastewater treatment. Sci. Rep 10, e3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Nancharaiah V, Gu X, Lee WL, Rajal VB, Haines MB, Girones R, Ng LC, Alm EJ, Wuertz S, 2020. Making waves: wastewater surveillance of SARS-CoV-2 for population-based health management. Water Res. 184, 116181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S, Weber A-F, Schiwy S, Linnemann V, Brinkmann M, Widera M, Greve C, Janke A, Hollert H, Wintgens T, ciesek S, 2020. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ 751, 141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WRF, 2020. Wastewater Surveillance of the COVID-19 Genetic Signal in Sewersheds Recommendations from Global Experts.
- Wu F, Zhang J, Xiao A, Gu X, Lee WL, Armas F, Kauffman K, Hanage W, Matus M, Ghaeli N, Endo N, Duvallet C, Poyet M, Moniz K, Washburne AD, Ericson TB, Chai PR, Thomson J, Alm EJ, 2020. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems 5 (4), 300614–300620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xagoraraki I, O’Brien E, 2020. Wastewater-based Epidemiology for Early Detection of Viral Outbreaks. Women Water Qual. Women in Engineering and Science Springer, Cham. 10.1007/978-3-030-17819-2_5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.