Abstract
Psoriasis affects the health of myriad populations around the world. The pathogenesis is multifactorial, and the exact driving factor remains unclear. This condition arises from the interaction between hyperproliferative keratinocytes and infiltrating immune cells, with poor prognosis and high recurrence. Better clinical treatments remain to be explored. There is much evidence that alterations in the skin and intestinal microbiome play an important role in the pathogenesis of psoriasis, and restoration of the microbiome is a promising preventive and therapeutic strategy for psoriasis. Herein, we have reviewed recent studies on the psoriasis-related microbiome in an attempt to confidently identify the “core” microbiome of psoriasis patients, understand the role of microbiome in the pathogenesis of psoriasis, and explore new therapeutic strategies for psoriasis through microbial intervention.
Keywords: immunity, microbial interventions, psoriasis, gut microbiota, skin microbiota
Introduction
Over the past two decades, numerous studies have revealed the physiological and metabolic effects of the microbiome on multicellular organisms, with implications for both health and disease. The human microbiome has become an area of utmost interest. Traditional culture-dependent assays have limited the study of the human microbiome and have severely underestimated the diversity of microbial communities in the context of the human body (Tlaskalova-Hogenova et al., 2004). The application of next-generation sequencing, including phylogenetic marker gene-based amplicon studies [e.g., bacterial 16S or fungal internal transcribed spacer (ITS) sequencing] and shotgun metagenomic sequencing, has greatly facilitated research on the human microbiota, allowing us to uncover the mysteries of our second genome, acquired after birth, and to explore the interactions of the microbiota with the host. The internal and external surfaces of many parts in the human body, including the gastrointestinal tract, skin, oral mucosa, vagina and airway, are niches that are colonized by microbes. Among these niches, the colon is the site that most commensal microbes colonize, harboring an estimated 1014 bacterial cells (Savage, 1977), followed by the skin, which harbors approximately two orders of magnitude fewer bacterial cells than the colon (Berg, 1996).
The gut, as the anatomical site with the highest distribution of microorganisms, is rich in nutrients and has a pH close to neutral. Some bacteria are not only prevalent across individuals but also are high in abundance, for example, the genus Bacteroides and anaerobic cocci, whereas some bacteria are abundant in individual fecal samples but not prevalent among individuals, such as the genus Clostridium, members of the genera Bifidobacterium, Eubacterium, Lactobacillus, Streptococcus, and some facultative anaerobes like Escherichia (Lloyd-Price et al., 2016). In the past, the diversity and functions of gut microbes have been severely underestimated due to the reliance on culture-based microbial research. Compared with the nutrient-rich gut, which is well-suited to microbial growth, skin has a low microbial biomass. This unique ecosystem also harbors specific skin-resident microbes, which are predominated by Gram-positive bacteria and facultatively anaerobic microbes (Grice et al., 2009; Bewick et al., 2019), such as Actinobacteria, Firmicutes, Proteobacteria, and Bacteroidetes. The most dominant genera seen on skin include Corynebacterium, Propionibacterium, Streptococcus, and Staphylococcus (Grice and Segre, 2011; Christensen and Bruggemann, 2014; Byrd et al., 2018). The functional and taxonomic compositions of skin-resident microbial communities, characterized by topographical and temporal diversity, are influenced by multiple factors, including lipid content, pH, sweat, sebum secretion and local skin anatomy. Over the last 40 years, much evidence has shown that skin microbes are involved in the development of non-infectious skin diseases, such as acne vulgaris (Holland et al., 1977), rosacea (Thomsen et al., 1980; Till et al., 2000) and psoriasis (Paulino et al., 2006). However, the contribution of the gut microbiome to common skin disorders has not been studied extensively. The recent rapid progression of NGS and bioinformatics technologies has provided access to the composition and genetic functions of the entire gut microbiome and thus enabled systematic investigation of its role in the pathogenesis, flare-up and prognosis of dermatosis.
Psoriasis, a common chronic inflammatory dermatosis, impacts 1–3% of the world’s population. The exact factors that drive psoriasis are not fully understood, but it is considered to be a complicated immune-mediated disease and is affected by both human genetics and environmental factors such as diet, lifestyle and health history. Psoriasis is associated with increased skin inflammation, hyperproliferation of keratinocytes, and overactive IL-17- and IFN-γ-producing T cells and inflammatory dendritic cells (DCs) (Roberson and Bowcock, 2010; Lowes et al., 2014; Afifi et al., 2017). There is no effective cure for this condition. Psoriasis is often commonly accompanied by systematic diseases such as IBDs, psoriatic arthritis, obesity and insulin resistance, which severely influence patients’ quality of life. Strengthening the understanding of metabolic comorbidities associated with psoriasis is of great significance for understanding the pathogenesis of psoriasis and developing novel therapeutic strategies. In particular, the relationship between psoriasis and obesity is bidirectional, with obesity favoring psoriasis and psoriasis predisposing individuals to obesity (Shipman and Millington, 2011; Cooksey et al., 2018; Dauden et al., 2018). Adipokines, synthesized and secreted by adipocytes, which are regulated by the gut microbiota, play an important role in linking the pathophysiology of psoriasis and obesity (Suarez-Zamorano et al., 2015; Kong et al., 2019). Much evidence has shown that the skin and gut microbiota play a role in host homeostasis and immune response, particularly in Th17 cell development (Nemoto et al., 2013; Geem et al., 2014; Cicerone et al., 2015; Bellone et al., 2020). It has been reported that an IBD-like decrease of Faecalibacterium prausnitzii together with an increase of Escherichia coli in psoriasis (Eppinga et al., 2016). However, there has been no comparative study of the gut microbiota of people in psoriasis and obesity. A lot of research has reported the gut microbes of obese people, characterized by lower level of F. prausnitzii than the healthy controls, while the changes in E. coli abundance varied among different research (Million et al., 2013; Fernandes et al., 2014; Yasir et al., 2015).
This review attempts to discuss recent insights into skin and gut microbial communities, including their interaction with host metabolism and the immune system, their composition and functional association with human health and psoriasis, and diagnostic or therapeutic approaches targeting these communities, with a focus on examining the evolution of microbiological intervention and reviewing potential microbial biomarkers associated with the occurrence, transmission, flare-up and severity of the disease, in addition to describing the therapeutic potential of modulating microbial compositions and skin immunology.
Microbiome Alteration: A Potential Pathogenic Factor for Psoriasis
There is a close association between psoriatic attacks and microbiome alterations (dysbiosis), characterized by altered diversity and composition, as well as blooms of opportunistic pathogens. Psoriasis can be provoked or exacerbated by specific pathogens, including bacteria (Staphylococcus aureus and Streptococcus pyogenes), viruses (human papillomavirus and endogenous retroviruses), and fungi (Malassezia and Candida albicans) (Fry and Baker, 2007; Wang and Jin, 2018). Malassezia, the most common fungal species present on normal human skin, show lower abundance in comparison with the healthy controls (Fernandes et al., 2014; Takemoto et al., 2015). Candida species are prevalent on the skin in psoriasis lesions and in the feces of patients with psoriasis (Ovcina-Kurtovic et al., 2016), while interleukin-17, overexpressed in patients with psoriasis, can protect against infections, especially those due to Candida spp. (Lonnberg et al., 2014; Saunte et al., 2017), suggesting that the induced overactive Th17 response is a potential pathogenic factor of psoriasis. We need to identify which comes first: abnormal colonization by C. albicans or psoriasis, that is, the chicken or the egg. If the former occurs first, specific antibiotics can be used to eliminate the abnormal colonization by the pathogen to prevent the occurrence or development of the disease. S. aureus, another common pathogen, may also be a pathogenic factor of psoriasis, given the increase in Th17 polarization and exacerbated cutaneous inflammation during early colonization of newborn mouse skin (Chang et al., 2018).
Skin Microbiome and Psoriasis
Cutaneous Homeostasis: A Result of the Interaction Between Skin-Resident Commensals and Cutaneous Immune Cells
Diverse body sites in healthy humans representing distinct skin niches are generally divided into three microenvironment types: dry sites, moist sites and sebaceous sites. Microbial features and disease susceptibility vary across different skin microenvironments (Grice et al., 2009; Belkaid and Segre, 2014; Byrd et al., 2018). Microbial communities colonizing the skin, cells resident in the epidermis and dermis, such as keratinocytes and fibroblasts, as well as skin immune cells together shape the cutaneous immune system. Commensal microorganisms that colonize the cutaneous surface have an effect on maintaining human skin homeostasis, in addition to educating and priming the cutaneous immune response in the state of pathogen invasion. Commensals can directly inhibit pathogen growth by capturing nutrients and space in the skin and producing antimicrobial peptides (AMPs) or bactericidal compounds; for example, colonization by the commensal Staphylococcus epidermidis inhibits exposure to the pathogen S. aureus during early life, preventing inflammatory diseases (Kennedy et al., 2017; Nakatsuji et al., 2017; Parlet et al., 2019; Gonzalez et al., 2020). Moreover, myriad immune cells are induced by commensals to provide protection against pathogens. Corynebacterium accolens dominates the skin microbial communities, or its membranous derivative mycolic acid protects against the common microbial pathogens S. epidermidis and C. albicans through the expansion of IL-17-producing dermal γδ T cells (Ridaura et al., 2018). Interleukin-17A (IL-17A)-producing T cells [CD8+ (TC17) and CD4+ (TH17)], which are commensal-specific T cells, can be induced upon encountering commensalism under homeostasis and then function as lasting tissue-resident memory cells. Harrison et al. showed that commensal-specific type T cells can mediate antimicrobial defense in the steady state but rapidly adapt to injury and promote tissue repair (Harrison et al., 2019). Colonization by S. epidermidis also shapes the skin’s T cell network. Monoassociation of S. epidermidis has the ability to strengthen cutaneous CD8+ T cells to produce IFNγ and IL-17 effector functions that protect against the skin pathogens Leishmania and C. albicans (Naik et al., 2012). In addition, cutaneous bacteria can be involved in immune tolerance to commensals and their metabolites. Exposure to S. epidermidis in early life contributes to skin homeostasis by triggering an influx of CD4+ T regulatory cells, thus mediating immune tolerance to commensal antigens (Ags) (Scharschmidt et al., 2015). Moreover, skin microbes can also be involved in the development and maturation of cutaneous immune cells. For example, the maturation of mast cells in mice, leaving the bone marrow and eventually settling in the skin, is dependent on skin microbes that promote stem cell factor (SCF) production in KCs (Wang et al., 2017). Another study found that the recruitment of MCs and SCF expression in mouse skin were regulated by the hairless-skin microorganisms (Wu C. C. et al., 2019). Early colonization by microorganisms is critical to the development of the human immune system and has a long-term impact on human health. Mucosal-associated invariant T (MAIT) cells, controlling tissue repair and homeostasis, are imprinted by microbes and induced in response to riboflavin-synthesizing commensals during a specific early life window (Constantinides et al., 2019). In turn, cutaneous immune cells such as DCs, mast cells, natural killer cells, macrophages and various T cells, involved in both innate and adaptive immune responses, shape resident microbial communities in skin. For instance, skin-resident innate lymphoid cells (ILCs) can tune the microbial commensal equilibrium to control the homeostasis of sebaceous glands. The absence of ILCs leads to sebaceous hyperplasia, accompanied by increased production of antibacterial lipids and the restriction of commensals of Gram-positive bacterial communities (Kobayashi et al., 2019). In short, skin commensals maintain cutaneous homeostasis by balancing immune defense against pathogens and immune tolerance to commensal antigens.
Skin, both a physical barrier and an immunologic barrier to external threats, is tightly controlled by its resident microbiota (Chen Y. E. et al., 2018). Psoriasis as an inflammatory skin disease showed decreased barrier function, but the underlying mechanisms remain unclear (Madison, 2003). There is much evidence that psoriatic fares are associated with skin microbiota alterations and the microbiota is a major player in the etiology of this condition (Fahlen et al., 2012; Alekseyenko et al., 2013; Zakostelska et al., 2016). Repeated infection of the skin surface is also closely associated with the development and progression of psoriasis (Alekseyenko et al., 2013; Hurabielle et al., 2020). Disruption of epidermal barrier integrity due to disturbance of the composition and function of the resident microbial community in the skin is significantly related to psoriasis, including some skin fungal flora (Candida albicans) and bacterial flora (Corynebacterium species), which might induce the accumulation of Th17 cells (Zielinski et al., 2012; Tauch et al., 2016). Skin resident microbiota and epidermal immune cells work together to maintain the skin barrier structure and function, and this structure might be damaged under pathological conditions, thus contributing to inflammatory responses. In most cases, however, it is not clear that abnormal immunity to the microorganisms is a bystander to the inflammatory process or a driver and/or magnifier of the disease states, which remains under investigation.
Skin Dysbiosis in Psoriasis
Microbiome perturbations are associated with several immune-mediated dermatoses, such as psoriasis, atopic dermatitis and acne vulgaris. Each disease has distinct skin microbiological characteristics. Many studies have shown that the composition and function of the skin microbiome vary between patients with psoriasis and healthy controls and psoriatic flares were associated with skin microbiome alterations; however, the lack of standardized sampling and profiling protocols also leads to conflicting results. Here, we review recent advances in the study of the close, non-causal association between microbial alteration and psoriasis, attempting to gain insight into the core microbial composition associated with psoriasis and to assess the potential diagnostic value of dysbiosis.
A search performed on October 2020 in the databases Pubmed and Web of Science, using the terms: “skin microbiota,” “skin microbiome,” “psoriasis,” and “humans.” Observational human studies and clinical trials that evaluated the skin microbiota or microbiome composition in psoriasis patients and in those of healthy controls using cultivation-independent methods, written in English, were included. Studies with animal models; studies not compared with healthy controls or unaffected skin regions of the same individual; and studies with a high risk of bias (poor quality), were excluded. Fifty three studies have been found in the databases Pubmed and Web of Science after the removal of duplicates and screening of the title and abstract, and the full texts of only 24 articles were read after the removal of reviews, systematic reviews. Here we describe the whole selected studies in which skin microbes of psoriatic in different disease states were compared and analyzed. Studies compared with healthy controls by rRNA community profiling, are included and described in Table 1.
TABLE 1.
Reported skin dysbiosis in psoriasis.
| Sample size | Amplification area | Skin dysbiosis in psoriasis | Sampling methods | References |
| 28 psoriasis patients and 26 healthy subjects | 16S rRNA V1–V3 variable region | At the phylum level, Actinobacteria↓ Proteobacteria↑ At the genus level, Propionibacterium, Ethanoligenens, and Macrococcus↓ Pseudomonas↑ At the species level, Staphylococcus epidermidis, Propionibacterium acnes, and Propionibacterium granulosum↓ Staphylococcus aureus and Staphylococcus pettenkoferi↑ | Swab | Chang et al., 2018 |
| 54 patients with psoriasis and 37 healthy controls | 16S rRNA V1–V3 variable region | At the phylum level, Proteobacteria↓ Actinobacteria and Firmicutes↑ | Swab | Alekseyenko et al., 2013 |
| 10 patients with psoriasis and 12 healthy controls | 16S rRNA V3–V4 variable region | At the phylum level, Proteobacteria↑ Firmicutes and Actinobacteria↓ At the genus level, Streptococci and Propionibacteria↓ Staphylococci↑ | Biopsy | Fahlen et al., 2012 |
| 3 male first cousins | V2–4–8 and V3–6, 7–9 regions of the bacterial 16S rRNA genes | At family level, Streptococcaceae, Rhodobacteraceae, Campylobacteraceae, and Moraxellaceae At the phylum level, Firmicutes↓ Proteobacteria↑ At the genus level, Staphylococcaceae and Propionibacteriaceae↓ At the species level, Propionibacterium acnes↓ | Swab | Drago et al., 2016 |
| 82 patients with atopic dermatitis, 119 patients with psoriasis and 115 healthy controls | 16S rRNA V1-V4 variable region | At the species level, Corynebacterium simulans, Corynebacterium kroppenstedtii, Finegoldia and Neisseriaceae↑ Lactobacilli, Burkholderia spp. and Propionibacterium acnes↓ | Swab | Fyhrquist et al., 2019 |
| 27 patients with psoriasis and 19 healthy controls | 16S rRNA V3–V4 variable region | At the phylum level, Deinococcus-Thermus↓ At the genus level, Propionibacterium↓ Corynebacterium↑ | Swab | Quan et al., 2020 |
| 50 patients with plaque-type psoriasis and 77 healthy controls | 16S rRNA V3-V4 variable region | At the phylum level, Firmicutes and Protebacteria↑ Fusobacteria and Cyanobacteria↓ | swab | Assarsson et al., 2020 |
Based on sequencing of the 16S rRNA V1-V3 variable region, Chang et al. (2018) revealed that different disease states (healthy, psoriatic lesional, and psoriatic non-lesional) can be discriminated by the taxonomic composition at the phylum and genus levels. Strikingly, the genera Staphylococcus as a whole was not significantly discriminated with any skin condition, while the relative abundance of the Staphylococcus species across all samples is associated with different disease states (see Table 1), suggesting that the dynamic inter-microbe relationship between different Staphylococcus species might lead to the various microbial communities associated with healthy and psoriatic skin. The genus Pseudomonas, enriched with psoriatic skin, includes many opportunistic pathogens and contributes to the treatment response of psoriasis patients treated with narrowband ultraviolent B (UVB) (Assarsson et al., 2018). For example, the most prominent bacteria of Gram-negative toe-web infection, Pseudomonas aeruginosa, is closely associated with the incidence of psoriasis (Goiset et al., 2019). Chang et al. (2018) also found that microbial communities in psoriatic lesions tended to display higher alpha diversity and greater heterogeneity but lower stability than healthy skin microbial communities. In contrast, Alekseyenko et al. (2013) reported that the microbiome of psoriatic lesions displayed decreased taxonomic diversity and was enriched with Firmicutes and Actinobacteria. What’s more, two taxa were found to be correlated best with psoriasis status, Acidobacteria Gp4 and Schlegelella, which were either not found in the HMP subjects (Gp4), or found only rarely. The combination of these two taxa might serve as potential markers for distinguishing skin from different disease states. This study ensures the matching of skin sites among different groups during sampling to avoid the influence of different skin sites and skin types on the skin microbial community. Gao et al. (2008) also showed that the Firmicutes phylum was overrepresented, while the Actinobacteria phylum and Propionibacterium species were underrepresented, in psoriatic lesions by applying RT-PCR. Fyhrquist et al. (2019) applied a larger-scale sample to profile the skin microbiota associated with atopic dermatitis, psoriasis and healthy volunteers and found that reduced abundance of members of Corynabacterium might play a regulatory role in psoriasis. Corynebacterium species of the human microbiome have often been assigned as opportunistic pathogens and Corynebacterium kroppenstedtii, showing a higher level of abundance in skin microbiota of psoriasis patients than healthy subjects, has occasionally been related to human infections, mainly granulomatous mastitis and breast abscesses (Tauch et al., 2016). Quan et al. (2020) also revealed that the higher level of Corynebacterium and the lower level of Cutibacterium were closely associated with psoriatic lesions. Based on sequencing of the 16S rRNA V3-V4 variable region, Fahlen et al. (2012) observed a lower diversity in psoriatic lesions compared to the control group. Both Staphylococci and Propionibacterium were present at lower levels, while Proteobacteria was present at higher levels, in psoriatic subjects than in controls. It is worth mentioning that the region of amplification and the method of sampling used in this study are different from others, which might contribute to the distinct results. Drago et al. (2016) found an increase in Proteobacteria abundance and a decrease in the abundances of Streptococcaceae, Rhodobacteraceae, Campylobacteraceae, and Moraxellaceae as well as Firmicutes in psoriatic subjects compared to healthy controls. The sample size of the study was so small that the reliability of the results remains to be further confirmed, despite strict dietary controls on the included individuals. Another study, applying shotgun metagenomics to profile the microbiome of psoriatic and unaffected skin from 28 patients with plaque psoriasis, found that the microbial communities of psoriatic and unaffected sites showed little difference at the species level while strain heterogeneity colonization and functional variability were revealed, which showed that higher-resolution analyses would be needed to clarify the pathogenesis of psoriasis and identify new therapeutic targets for it (Tett et al., 2017). Paulino et al. (2006) revealed that Malassezia microbiota was host-specific and relatively stable over time and there was no significant difference between samples from healthy skin and psoriatic lesions, using multiplex real-time PCR.
After analyzing most of the studies on the relationship between skin microbes and psoriasis, we found that the variation trends of the Firmicutes, Actinobacteria, and Proteobacteria abundances in various studies were highly controversial. In addition, there was no convincing conclusion regarding whether the diversity of the microbial community on psoriatic lesional skin lower than that on healthy skin. What is certain, however, is that increased S. aureus abundance and decreased S. epidermis abundance were observed in psoriatic lesions (Ng et al., 2017; Liu S. H. et al., 2019). The reports mentioned above are qualitative in nature, and the loss of quantitative definition directly limits insights into what features are reproducible across studies. What makes the results so different is that skin microbes are affected by a variety of host and environmental factors, such as daily hygiene regimens, use of cosmetic products, exposure to antimicrobials, friction, climate, and UV irradiation (Babeluk et al., 2014; Lee H. J. et al., 2018; Burns et al., 2019; McBain et al., 2019). Therefore, these confounders must be strictly controlled during study so that the results across samples and studies can be comparable. In addition, distinct skin sites and skin types, the selection of primer to amplify regions for sequencing and sampling methods including swab, scrape and punch biopsy, contribute to differences in experimental results, particularly on the diversity of the microbial community (Grice et al., 2008; Claesson et al., 2010; Alekseyenko et al., 2013; Castelino et al., 2017; Chang et al., 2018; Stehlikova et al., 2019).
The reports mentioned above are qualitative in nature, and the loss of quantitative definition directly limits insights into what features are reproducible across studies. To gain insight into the characteristic changes in psoriatic lesions, studies with large sample sizes, standardized protocols and sufficient sequencing depth are required. Alternatively, a scientific meta-analysis of publicly available data from studies on the relationship between skin microbes and psoriasis can help address discrepancies in an unbiased manner, and after devising valid statistical methods to exclude experimental and human confounding factors, the microbial alteration associated with psoriasis may be reliably identified.
Gut Microbiome and Psoriasis
The Role of the Gut Microbiota in Maintaining a Healthy Gut Ecosystem
Gut microbes and their metabolites contribute to host health by digesting food and maintaining immune system homeostasis. The gut flora helps digest and break down complex polysaccharides and is crucial for the production of some important nutrients, such as vitamin K and B-group vitamin (Hill, 1997; Karasov et al., 2011; LeBlanc et al., 2013; Heintz-Buschart and Wilmes, 2018). The intestinal microbiota plays a vital role in maintaining epithelial barrier integrity and forming a mucosal immune system to protect against invasion by exogenous pathogens and for balancing host defense and tolerance of dietary and environmental antigens through bacterial metabolites and components. The gut microbiota and its metabolites, intestinal epithelial cells (IECs) and immune cells participate in the maintenance of a healthy gut ecosystem. Indeed, the gut microbiota is a non-self entity and would be eliminated if the immune cells residing in the intestinal mucosa recognized it. The intestinal epithelium serves as a barrier to isolate immune cells residing in the intestinal mucosa from the microbiota present in the intestinal lumen but allows microbial metabolism to gain access to and interact with host cells and thus regulate immune responses.
The interactions between the microbiota and the host are mediated mainly through bacterial components and microbial metabolites. The gut microbiota contributes to innate immune responses via interactions with Toll-like receptors (TLRs) and pattern recognition receptors (PRRs) on the surface of innate immune cells (Rooks and Garrett, 2016; Weaver et al., 2019). In addition, gut microbes participate in the adaptive immune response by inducing the secretion of immunoglobulin A (IgA) and affecting effector T cells (Th1, Th2, Th17), regulatory T cells and Tfh cell member and function (Bunker et al., 2015; Honda and Littman, 2016; Teng et al., 2016; Yissachar et al., 2017). For example, spore-forming bacteria, such as Clostridia, can induce the colonic Tregs, rebalance Th1/Th2/Th17 cells and change to a less proinflammatory immunological milieu in the gut (Jia et al., 2017). Bacteroides fragilis and segmented filamentous bacteria (SFB) have been reported to induce intestinal Tregs and Th17 cell differentiation, respectively, thus affecting the host resistance against infections and promote systemic autoimmunity (Ivanov et al., 2009; Round and Mazmanian, 2010; Wu et al., 2010).
Microbiota-produced metabolites in the intestine, such as short-chain fatty acids (SCFAs), secondary bile acids and tryptophan, balance the activation and suppression of the immune system and affect multiple organs throughout the body. For example, SCFAs, especially butyrate, inhibit immune responses by inhibiting the proliferation, migration, adhesion, and cytokine production of inflammatory cells (Meijer et al., 2010; Schwarz et al., 2017). In addition, SCFAs inhibit histone deacetylase and inactivate NF-κB signaling pathways to tune the activation and apoptosis of immune cells (Meijer et al., 2010; Alekseyenko et al., 2013; Schwarz et al., 2017). There is much evidence of the important role of microbial metabolites of tryptophan in balancing host defense and microbiota homeostasis. Indole-3-aldehyde (IAld), an indole derivative of tryptophan catabolism by the gut microbiota, tunes mucosal reactivity through IL-22 and protects against colonization by C. albicans. The microbial metabolites of tryptophan also affect central nervous system inflammation and astrocyte activity. Moreover, IAld, a tryptophan metabolite derived from the skin microbiota, attenuates inflammation in patients with atopic dermatitis through the aryl hydrocarbon receptor. None of these phenomena would have been observed in germ-free mice (Zelante et al., 2013; Rothhammer et al., 2016). The upregulated tryptophan metabolism pathway is also observed in patients with psoriasis, while the role of microbes in the pathway remains to be explored (Harden et al., 2016). Other microbial metabolites, such as polysaccharide A and retinoic acid produced by commensal Clostridia, Faecalibacterium prausnitzii, and Bacteroides fragilis, induce the accumulation of regulatory T (Treg) cells, which suppress inflammation (Forbes et al., 2016).
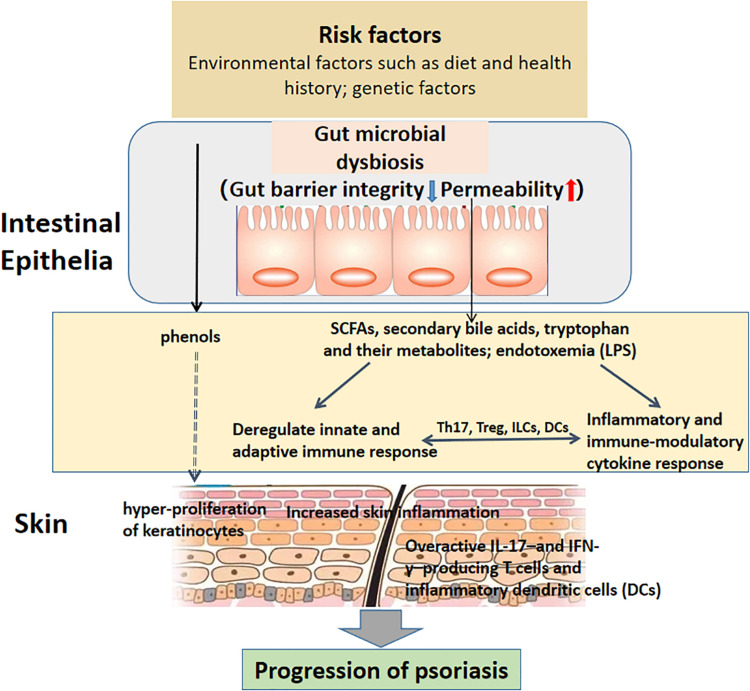
The gut microbiota has a regulatory effect on systemic immunity, causing the functioning and dysfunction of distant organ systems. Dysbiosis, involving alteration in the composition and function of microbial communication, may result in increased gut barrier permeability, which contributes to immune activation by translocation of microbial antigens and their metabolites into the blood circulation (Wells et al., 2017). Destroyed intestinal integrity and increased gut permeability, consequences of the systemic inflammation fueled by gut dysbiosis, has been implicated as a factor contributing to local and systemic immune response, and the pathogenesis of psoriasis (Lin and Zhang, 2017). The skin has a particularly complicated connection with the gut, but the underlying mechanism is still completely understood, and this phenomenon is at least partially attributed to the disruption of the intestinal barrier (Kim et al., 2014; Kosiewicz et al., 2014; Camilleri, 2019). Oral administration of S. aureus and Streptococcus danieliae, which are abundant in the inflammatory skin mouse model, exacerbated skin inflammation associated with imiquimod-induced psoriasis-like dermatitis with elevated levels of TNF-α, IL-17A, IL-17F and IL-22, also suggesting a potential role of gut dysbiosis in the development of psoriasis (Okada et al., 2020). What’s more, Sikora et al. (2018) assessed non-invasive markers of intestinal barrier integrity in psoriasis patients, including concentrations of claudin-3, intestinal fatty acid binding protein (I-FABP) in the blood in psoriasis patients and healthy controls. They found that psoriasis patients had more elevated concentration of plasma claudin-3 and I-FABP, supporting the hypothesis that dysfunction of the intestinal barrier in psoriasis disturbs the homeostatic equilibrium between the microbiota and immune system. In another study conducted by Sikora confirmed that I-FABP is associated with severity of psoriasis (Sikora et al., 2019). The putative relationship between gut dysbiosis and psoriasis development and progression have been shown in Figure 1.
FIGURE 1.
The putative relationship between gut dysbiosis and psoriasis onset and progression. Intestinal barrier function is maintained by group 3 innate lymphoid cells (ILC3s) and IL-17- and IL-22-producing T helper 17 (Th17) cells, which modulate antimicrobial peptide secretion by intestinal epithelial cells (IECs) and IgA production in the gut (Hirota et al., 2013; Kruglov et al., 2013). Moreover, dendritic cells (DCs) participate in microbiota sensing via the Mincle-Syk axis to regulate IL-17 and IL-22 production and promote intestinal barrier integrity (Martinez-Lopez et al., 2019). Host- and microbiota-derived factors induce gut microbe dysbiosis, including disruption of gut barrier integrity and increased permeability, as well as alterations in microbial metabolites such as SCFAs, secondary bile acids, tryptophan, lipopolysaccharides (LPSs) and phenols, thus disturbing immune homeostasis via a low-grade chronic inflammatory process. For example, the intestinal microbiota promotes psoriasis-like skin inflammation by enhancing the Th17 response, and regulatory T cell (Treg) levels decrease in psoriasis patients, leading to an imbalance between effector T cells and suppressor T cells (Zakostelska et al., 2016; Komine, 2020). Th17 cell differentiation requires IL-6 and transforming growth factor-β (TGF-β) from DCs in an antigen-dependent manner (Ivanov et al., 2006; Persson et al., 2013). In contrast, activation of ILC3s requires the release of IL-23 by myeloid cells to produce IL-22 and/or IL-17, which is antigen dependent (Satpathy et al., 2013; Longman et al., 2014). Increased numbers of ILC3s exist in the circulating blood of psoriatic arthritis patients, as well as the lesional and non-lesional skin of psoriasis patients (Soare et al., 2018). Phenols, as metabolites of aromatic amino acids produced by gut bacteria and regarded as bioactive toxins and serum biomarkers of a disturbed gut environment, have the ability to influence keratinocyte differentiation in the skin, but the underlying mechanism remains to be explored (Miyazaki et al., 2014). The involvement of immune cells and their active factors and intestinal microorganisms and their metabolites promotes the progression of psoriasis.
Intestinal Dysbiosis in Psoriasis
The intestinal microbiota contributes to the balance between Th17 effector cells and their counterpart regulatory T cells (van Beelen et al., 2007), and Zakostelska et al. (2016) showed that imiquimod induced milder psoriasis-like skin inflammation in germ-free mice than in conventional mice by enhancing the Th17 response, suggesting that gut dysbiosis acts as a potential pathogenic factor for psoriasis.
Studies on the correlation between intestinal microorganisms and psoriasis are limited and surged in recent years, and we searched in the databases Pubmed and Web of Science on October 2020, using the terms: “gut microbiota,” “gut microbiome,” “psoriasis,” and “humans.” Observational human studies and clinical trials that evaluated the gut microbiota composition in psoriasis patients and in those of healthy controls using cultivation-independent methods, and written in English (see Table 2), were discussed below.
TABLE 2.
Reported intestinal dysbiosis in psoriasis.
| Sample size | Amplification area | Intestinal dysbiosis in psoriasis | Function prediction | References |
| 32 psoriatic patients and 64 healthy controls | V3–V4 region of bacterial 16S rRNA genes | At the family level, Ruminococcaceae and Lachnospiraceae↑ Bacteroidaceae and Prevotellaceae↓ At the phylum level, Firmicutes↑ Bacteroidetes↓ At the genus level, Ruminococcus and Megasphaera↑ | Functional genes and metabolic pathways involving bacterial chemotaxis and carbohydrate transport were predicted to be over-represented; genes related to cobalamin and iron transport were predicted to be underrepresented. | Chen Y. J. et al., 2018 |
| 35 psoriatic patients and 27 healthy controls | V4 and V5 regions of the bacterial 16S rRNA genes | At the phylum level, Firmicutes, Proteobacteria, and Actinobacteria↑ Bacteroidetes↓ At the genus level, Bacillus, Bacteroides, Sutterella, Lactococcus, Lachnospiraceae_UCG004, Lachnospira, Mitochondria_norank, Cyanobacteria_norank, and Parabacteroides↑ Thermus, Streptococcus, Rothia, Granulicatella, Gordonibacter, Allobaculum, and Carnobacterium↓ At the genus level, Bacteroides↓ Faecalibacterium, Akkermansia and Ruminococcus↑ | / | Huang et al., 2019 |
| 52 psoriatic patients and 300 healthy controls extracted from the Human Microbiome Project | Broad-range PCR amplification of the bacterial 16s rRNA gene conserved region | Genus Bacteroides↓ Akkermansia spp.↑ | / | Codoner et al., 2018 |
| 24 psoriatic patients and 22 healthy controls | V4 region of the bacterial 16S rRNA genes | At the phylum level, Firmicutes and Actinobacteria↑ Bacteroidetes and Proteobacteria↓ At the genus level, Blautia↑ Prevotella↓ At the species level, Ruminoccocus gnavus, Dorea formicigenerans, and Collinsella aerofaciens Prevotella copri↓ | Genes encoding energy metabolism and involved in glutathione synthesis were predicted to be underrepresented; genes encoding butyrate kinase and phosphate butyryltransferase were predicted to be underrepresented; pathways related to lipopolysaccharide (LPS) biosynthesis were predicted to be over-represented. | Shapiro et al., 2019 |
| 16 patients with psoriatic arthritis, 15 patients with psoriasis of the skin and 17 healthy, matched control subjects | V1–V2 region of bacterial 16S rRNA genes | At the genus level, Parabacteroides, Coprobacillus, unclassified Ruminococcaceae, unclassified Lachnospiraceae↓ | Decreased quantities of MCFAs | Scher et al., 2015 |
| 19 patients with psoriasis and 20 healthy controls | V2–V3 region of bacterial 16S rRNA genes | At the family level, Bifidobacteriaceae, Coriobacteriaceae, Lachnospiraceae, Clostridiales Family XIII, Eggerthellaceae, Peptostreptococcaceae, Ruminococcaceae and Erysipelotrichaceae↑ Bacteroidaceae, Barnesiellaceae, Prevotellaceae, Tannerellaceae, Burkholderiaceae, Rikenellaceae, Lactobacillaceae, Streptococcaceae, Desulfovibrionaceae, Veillonellaceae, Marinifilaceae, Victivallaceae↓ and Pasteurellaceae At the phylum level, Actinobacteria and Firmicutes↑ Bacteroidetes and Proteobacteria↓ | / | Hidalgo-Cantabrana et al., 2019 |
Many studies have shown that the gut microbiota associated with psoriasis patients significantly differs from that in healthy subjects. Tan et al. (2018) showed that the abundance of Akkermansia muciniphila was markedly reduced in patients with psoriasis. Chen Y. J. et al. (2018) revealed an increased abundance of the phylum Firmicutes, from which Ruminococcus and Megasphaera were the top two genera of discriminant abundance, and a decreased abundance of the phylum Bacteroidetes was observed in psoriasis patients, which might be associated with suppressed cobalamin and iron transport. Obesity and the drug effect were considered as confounders in this study and a significant difference in bacterial composition between psoriasis and controls was found in those with BMI values less than 25 but not among obese subject (BMI ≥ 25). Huang et al. (2019) confirmed that Bacteroidia was a key factor contributing to the dysbiosis of microbiota in psoriasis patients, while the phylum Firmicutes was the key factor contributing to the distribution of the microbiota in healthy subjects. This finding was supported by a previous study reporting that the ratio of Firmicutes and Bacteroidetes was perturbed in psoriasis patients. There were no significant differences in intestinal microbial composition observed in psoriasis with distinct severity. Codoner et al. (2018) found a different psoriatic gut microbiome (with high diversity), an increased presence of Faecalibacterium and a decrease in the abundance of Bacteroides, as well as higher levels of the genera Akkermansia and Ruminococcus, in psoriatic patients. Three hundred healthy controls extracted from the Human Microbiome Project were involved in this study, which made it impossible to ensure a good match of other confounders (age, gender, dietary habit and BMI index) between healthy controls and psoriatic patients, thus perturbing the results. Shapiro et al. (2019) demonstrated a significant increase in the phyla Firmicutes and Actinobacteria phyla in psoriatic patients compared with matched controls, and psoriatic patients showed significant increases in the abundances of species such as Ruminoccocus gnavus, Dorea formicigenerans, and Collinsella aerofaciens, while species such as Prevotella copri and Parabacteroides distasonis were significantly depleted in psoriasis patients compared with controls. Small sample size, insufficient sequencing depth, possible contamination in the experimental process, and various factors affecting the microbial composition of feces, such as diet, in addition to a lack of quantitative definition, make the results unreliable, and better methods remain to be explored to provide access to the relationship between psoriasis and strain-level intestinal bacteria so that psoriasis can be diagnosed and treated with microbial interventions. Moreover, the intestinal microorganisms of psoriatic patients can be compared with those of non-psoriatic patients with obesity, IBD, irritable bowel syndrome (IBS) or other diseases closely related to psoriasis to further understand the role of microorganisms in the interaction of obesity (IBD, IBS) and psoriasis and propose new microbiota-related treatments for the two conditions in the future.
Modulation of the Gut Microbiota for Prevention and Treatment: From Biology to Clinic
Given the gut microbial contribution to inflammatory disease and the immune system, there exists an opportunity to intentionally modulate the microbiome for therapeutic purposes. The shaping of gut microbial composition and function depends largely on environmental factors, particularly diet, rather than human genetics. Restoration of the gut microbiome is a promising preventive and therapeutic strategy in a number of clinical conditions.
Fecal Microorganism Transplantation (FMT)
Myriad studies have reported alterations in the composition and function of the intestinal microbial community and improvements in disease through long-term oral supplementation with food, prescription medicine or traditional herbal products and explored the underlying mechanism (Sung et al., 2017; Lee H. et al., 2018; Sharma et al., 2018; Ejtahed et al., 2019; Liu M. T. et al., 2019; Sheng et al., 2019; Wu T. R. et al., 2019; Xu et al., 2019; Zhang et al., 2019; Wang et al., 2020). Additionally, there is much evidence from both mouse and human studies that the recipient’s microbial community can be gradually rectified to normalize the microbial composition and function by transplanting fecal microorganisms associated with a “healthy” state, which thus have a therapeutic effect, especially in the treatment of recurrent Clostridium difficile infection (CDI) and in the improvement of obesity. A 36-year-old male patient diagnosed with severe psoriasis for 10 years and IBS for 15 years was subjected to FMT twice via both upper endoscopy and colonoscopy with a 5-week interval. After the second FMT treatment for 5 weeks, the two conditions improved greatly (Lai et al., 2018; Mullish et al., 2018; Sun et al., 2018; Krajicek et al., 2019; Yin et al., 2019). Although FMT is potentially effective in many microbiota-related disorders, helping establish trans-kingdom equilibrium between gut fungi, viruses and bacteria to promote the restoration of microbial homeostasis (McSweeney et al., 2020), the effect of FMT depends on several factors, including the bacterial load and microbial composition and function of the recipient and the donor, the physiological and genetic factors related to the recipient and the donor, and FMT preparation and route of administration (Fischer et al., 2016; Vermeire et al., 2016; Duvallet et al., 2017; Ianiro et al., 2017; Allegretti et al., 2019, 2020; Ding et al., 2019; Paramsothy et al., 2019; Ramai et al., 2019; Wilson et al., 2019; de Groot et al., 2020; McSweeney et al., 2020).
Probiotics and Prebiotics With Beneficial Effects in Psoriasis
Fecal microorganism transplantation is not a one-size-fits-all approach, and studies are needed to identify microbial active components that have specific effects in patients with different diseases. This promotes the therapeutic view of regulating systemic immunity by manipulating the intestinal microbiota community either through stimulating bacterial growth via supplementation with prebiotics (non-viable bacterial components and metabolites) or through expansion via administration of probiotics (live beneficial gut bacteria) and synbiotics (combinations of probiotics and prebiotics) (see Table 3). Probiotic supplementation has a promising potential role in the prevention and management of various skin conditions. Chen et al. (2017) revealed that oral administration of Lactobacillus pentosus GMNL-77, a potential probiotic strain, significantly decreased erythaematous scaling lesions in imiquimod-treated mice with epidermal hyperplasia and psoriasis-like skin inflammation, with decreased tumor necrosis factor-alpha (TNF-α), interleukin (IL)-6, and IL-23/IL-17A axis-associated cytokine (IL-23, IL-17A/F, and IL-22) levels in the skin and reduced IL-17- and IL-22-producing CD4+ T cells in the spleen. Ethanol extract (SEL001), isolated from the potent probiotic strain Lactobacillus sakei proBio-65, has a protective effect on imiquimod-treated psoriasis-like skin inflammation in a mouse model, with decreased gene expression levels of IL-19, IL-17A, and IL-23 (Rather et al., 2018). In a documented case of severe pustular psoriasis that did not respond to steroids, dapathon, and methotrexate, clinical improvement was observed within 2 weeks after initiation of Lactobacillus sporogenes supplementation three times a day, and almost complete resolution was observed at 4 weeks (Sikora et al., 2019). In a separate placebo-controlled study of psoriasis patients, Bifidobacterium infantis 35624 supplementation resulted in significantly reduced plasma levels of TNF-α, IL-6 and C-reactive protein (CRP) in the probiotic-treated group (Groeger et al., 2013).
TABLE 3.
Evidence of the beneficial effects of probiotics in psoriasis.
| Patients or mice | Probiotic intervention | Clinical outcome | References |
| Imiquimod-treated mice with psoriasis-like skin inflammation | Oral administration of Lactobacillus pentosus GMNL-77 | (1) decreased erythaematous scaling lesions; (2) decreased TNF-α, IL-6, IL-23, IL-17A/F, and IL-22 levels in the skin; (3) reduced IL-17- and IL-22-producing CD4+ T cells in the spleen | Chen et al., 2017 |
| Same as above | Ethanol extract (SEL001) | Decreased IL-19, IL-17A, and IL-23 | Rather et al., 2018 |
| A 47-year-old female who presented with crops of pustules all over her body for 20 days prior | Oral administration of Lactobacillus sporogenes, one sachet thrice daily | In 15 days, the fever subsided, lesions started involuting and no new lesions appeared | Vijayashankar and Raghunath, 2012 |
| 26 psoriasis patients in three separate randomized, double-blind, placebo-controlled interventions | Bifidobacterium infantis 35624 | Decreased TNF-α, IL-6 and C-reactive protein (CRP) levels | Groeger et al., 2013 |
| Ninety 18–70-year-old adults with plaque psoriasis | Bifidobacterium longum CECT 7347, B. lactis CECT 8145 and Lactobacillus rhamnosus CECT 8361 with a total of 1*10^9 colony-forming units per capsule | After 12 weeks, more patients in the probiotic group compared to the placebo group showed a reduction in the psoriasis area and severity index of up to 75% | Navarro-Lopez et al., 2019 |
Potential Probiotics and Prebiotics for Psoriasis Treatment
Some probiotics have been shown to ameliorate skin inflammation by modulating immune responses in the host, but there is no evidence of their potential role in treating psoriasis (see Table 4). Oral administration of poly-γ-glutamate, a natural product of a few Gram-positive bacteria, including Staphylococcus and Bacillus species, ameliorates AD-like dermatitis in Nc/Nga mice by suppressing the Th2-biased immune response and production of IL-17A, which may indicate it to be a desirable prebiotic for treating overactive Th17 cells involved in psoriasis (Lee et al., 2014). Oral supplementation with milk fermented with Lactobacillus casei or administration of L. casei alone can decrease skin inflammation by modulating the pool size of cytotoxic CD8+ T cells (Chapat et al., 2004). Further research revealed that L. casei DN-114 001 efficiently ameliorated T cell-mediated skin inflammation via the modulation of cytotoxic CD8+ T cells and the participation of CD4+ Treg cells (Hacini-Rachinel et al., 2009). Lactobacillus paracasei CNCM-I 2116 (ST11) can alleviate skin inflammation in vitro by preventing TNF-α release, mast cell degranulation, vasodilation and oedema, thus accelerating the recovery of barrier function (Gueniche et al., 2010).
TABLE 4.
Potential probiotics and prebiotics for psoriasis treatment.
| Patients or mice | Probiotic intervention | Clinical outcome | References |
| Nc/Nga mice | Oral administration of poly-γ-glutamate | Suppressing the Th2-biased immune response and production of IL-17A | Chen et al., 2017 |
| Female C57Bl/6 (2–4-month-old) mice | Oral treatment with 200 μl of live L. casei DN-114 001 | Alleviated T cell-mediated skin inflammation without causing immune suppression, via mechanisms that include control of CD8+ effector T cells and involve regulatory CD4+ T cells | Vijayashankar and Raghunath, 2012 |
| Specific pathogen-free (SPF) male C57BL/6J mice | (i) Probiotic (B. animalis subsp. lactis and L. paracasei subsp. paracasei DSM 46331), (ii) Prebiotic (oat β-glucan) (iii) Synbiotic (a mixture of i and ii) treatments for 12 weeks | Restored caecal levels of acetate, propionate, and butyrate; reduced body weight and alleviated features of metabolic complications | Ke et al., 2019 |
| Male C57BL/6J mice at 7 weeks of age | A combination of L. mali APS1 and dieting | Accelerated body weight loss, reduced caloric intake, and lowered fat mass; ameliorated hepatic steatosis; affected fecal SCFA excretion and expression of satiety hormones | Chen Y. T. et al., 2018 |
Data on probiotic supplementation in psoriasis treatment are limited, but the pathogeneses of psoriasis and obesity have shown certain overlapping genetic and environmental factors as well as immune pathways. Th17 cells and their cytokines play an important role in psoriasis progression and the pathophysiology of obesity, indicating that probiotics, which are effective for obesity treatment, may also be effective for psoriasis treatment. The gut microbiota of obese individuals is less diverse than that of non-obese individuals, with a reduction in Gram-negative bacteria, specifically members of Bacteroidetes, and an increase in Gram-positive Firmicutes (Ley et al., 2006; Turnbaugh et al., 2009), which can also be observed in psoriatic patients (Ley et al., 2006; Turnbaugh et al., 2009). Specific strains belonging to Lactobacillus [L. casei strain Shirota (LAB13), L. gasseri, L. rhamnosus, and L. plantarum, among others] and Bifidobacterium (mainly B. infantis, B. longum, and B. breve B3) species, as well as other microorganisms, including Pediococcus pentosaceus LP28, Bacteroides uniformis CECT 7771, Akk. muciniphila, and Saccharomyces boulardii Biocodex, have shown anti-obesogenic effects in animals (Ukibe et al., 2015; Alard et al., 2016; Li et al., 2016; Bubnov et al., 2017; Shin et al., 2018; Avolio et al., 2019). Among these, the effects of L. casei and B. infantis strains have been confirmed. Notably, there is no evidence of the long-term effects of probiotics either as human food supplements or as adjunctive therapy. Therefore, the safety of these bacteria needs to be rigorously assessed before application in the treatment of various diseases.
Conclusion and Perspectives
This systematic review revealed that microbiome alterations, such as abnormal colonization by C. albicans and S. aureus, may act as potential pathogenic factors for psoriasis. The skin and intestinal microbes of patients with psoriasis were significantly different from those of healthy subjects. However, due to the lack of standardized protocols, the microbial communities of patients with psoriasis were hardly comparable among the various study results, and it was difficult to confidently identify changes in the microbial communities associated with the psoriasis disease status. More rigorous research or robust statistical techniques might provide a credible answer. Microbiological interventions, including fecal transplants and probiotics, have been used in patients and mice with psoriasis, especially in patients who have not responded to multiple other therapies. However, the long-term effects of these interventions have not been reported and should be carefully evaluated in clinical applications, especially for younger patients. Obesity is closely related to psoriasis, and the two have a bidirectional effect. Understanding the role of microbes in the interaction of pathogenesis in two conditions may provide access to novel treatments.
Author Contributions
LC developed the writing of the manuscript, figure, and tables. CP designed the review and contributed to proofreading and revising it with WZ. All the authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by Grant No. 81830096 from the key project of the National Science Foundation, 2020YFA0112904 from National Key Research and Development Project, and Grant Nos. 81773341, 82073458, 81673065, 81974476, and 81773329 from the National Natural Science Foundation of China.
References
- Afifi L., Danesh M. J., Lee K. M., Beroukhim K., Farahnik B., Ahn R. S., et al. (2017). Dietary behaviors in psoriasis: patient-reported outcomes from a U.S. national survey. Dermatol. Ther. (Heidelb.) 7 227–242. 10.1007/s13555-017-0183-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alard J., Lehrter V., Rhimi M., Mangin I., Peucelle V., Abraham A. L., et al. (2016). Beneficial metabolic effects of selected probiotics on diet-induced obesity and insulin resistance in mice are associated with improvement of dysbiotic gut microbiota. Environ. Microbiol. 18 1484–1497. 10.1111/1462-2920.13181 [DOI] [PubMed] [Google Scholar]
- Alekseyenko A. V., Perez-Perez G. I., De Souza A., Strober B., Gao Z., Bihan M., et al. (2013). Community differentiation of the cutaneous microbiota in psoriasis. Microbiome 1:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegretti J. R., Fischer M., Sagi S. V., Bohm M. E., Fadda H. M., Ranmal S. R., et al. (2019). Fecal microbiota transplantation capsules with targeted colonic versus gastric delivery in recurrent clostridium difficile infection: a comparative cohort analysis of high and lose dose. Dig. Dis. Sci. 64 1672–1678. 10.1007/s10620-018-5396-6 [DOI] [PubMed] [Google Scholar]
- Allegretti J. R., Kassam Z., Mullish B. H., Chiang A., Carrellas M., Hurtado J., et al. (2020). Effects of fecal microbiota transplantation with oral capsules in obese patients. Clin. Gastroenterol. Hepatol. 18 855–863.e2. [DOI] [PubMed] [Google Scholar]
- Assarsson M., Duvetorp A., Dienus O., Soderman J., Seifert O. (2018). Significant changes in the skin microbiome in patients with chronic plaque psoriasis after treatment with narrowband ultraviolet B. Acta Derm. Venereol. 98 428–436. 10.2340/00015555-2859 [DOI] [PubMed] [Google Scholar]
- Assarsson M., Soderman J., Dienus O., Seifert O. (2020). Significant differences in the bacterial microbiome of the pharynx and skin in patients with psoriasis compared with healthy controls. Acta Derm. Venereol. 100:adv00273. 10.2340/00015555-3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avolio E., Fazzari G., Zizza M., De Lorenzo A., Di Renzo L., Alo R., et al. (2019). Probiotics modify body weight together with anxiety states via pro-inflammatory factors in HFD-treated Syrian golden hamster. Behav. Brain Res. 356 390–399. 10.1016/j.bbr.2018.09.010 [DOI] [PubMed] [Google Scholar]
- Babeluk R., Jutz S., Mertlitz S., Matiasek J., Klaus C. (2014). Hand hygiene–evaluation of three disinfectant hand sanitizers in a community setting. PLoS One 9:e111969. 10.1371/journal.pone.0111969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y., Segre J. A. (2014). Dialogue between skin microbiota and immunity. Science 346 954–959. 10.1126/science.1260144 [DOI] [PubMed] [Google Scholar]
- Bellone M., Brevi A., Huber S. (2020). Microbiota-propelled T helper 17 cells in inflammatory diseases and cancer. Microbiol. Mol. Biol. Rev. 84: e00064-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg R. D. (1996). The indigenous gastrointestinal microflora. Trends Microbiol. 4 430–435. 10.1016/0966-842x(96)10057-3 [DOI] [PubMed] [Google Scholar]
- Bewick S., Gurarie E., Weissman J. L., Beattie J., Davati C., Flint R., et al. (2019). Trait-based analysis of the human skin microbiome. Microbiome 7:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubnov R. V., Babenko L. P., Lazarenko L. M., Mokrozub V. V., Demchenko O. A., Nechypurenko O. V., et al. (2017). Comparative study of probiotic effects of Lactobacillus and Bifidobacteria strains on cholesterol levels, liver morphology and the gut microbiota in obese mice. EPMA J. 8 357–376. 10.1007/s13167-017-0117-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker J. J., Flynn T. M., Koval J. C., Shaw D. G., Meisel M., McDonald B. D., et al. (2015). Innate and adaptive humoral responses coat distinct commensal bacteria with Immunoglobulin A. Immunity 43 541–553. 10.1016/j.immuni.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns E. M., Ahmed H., Isedeh P. N., Kohli I., Van Der Pol W., Shaheen A., et al. (2019). Ultraviolet radiation, both UVA and UVB, influences the composition of the skin microbiome. Exp. Dermatol. 28 136–141. 10.1111/exd.13854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd A. L., Belkaid Y., Segre J. A. (2018). The human skin microbiome. Nat. Rev. Microbiol. 16 143–155. [DOI] [PubMed] [Google Scholar]
- Camilleri M. (2019). Leaky gut: mechanisms, measurement and clinical implications in humans. Gut 68 1516–1526. 10.1136/gutjnl-2019-318427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelino M., Eyre S., Moat J., Fox G., Martin P., Ho P., et al. (2017). Optimisation of methods for bacterial skin microbiome investigation: primer selection and comparison of the 454 versus MiSeq platform. BMC Microbiol. 17:23. 10.1186/s12866-017-0927-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. W., Yan D., Singh R., Liu J., Lu X., Ucmak D., et al. (2018). Alteration of the cutaneous microbiome in psoriasis and potential role in Th17 polarization. Microbiome 6:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapat L., Chemin K., Dubois B., Bourdet-Sicard R., Kaiserlian D. (2004). Lactobacillus casei reduces CD8+ T cell-mediated skin inflammation. Eur. J. Immunol. 34 2520–2528. 10.1002/eji.200425139 [DOI] [PubMed] [Google Scholar]
- Chen Y. E., Fischbach M. A., Belkaid Y. (2018). Skin microbiota-host interactions. Nature 553 427–436. 10.1038/nature25177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. H., Wu C. S., Chao Y. H., Lin C. C., Tsai H. Y., Li Y. R., et al. (2017). Lactobacillus pentosus GMNL-77 inhibits skin lesions in imiquimod-induced psoriasis-like mice. J. Food Drug Anal. 25 559–566. 10.1016/j.jfda.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. J., Ho H. J., Tseng C. H., Lai Z. L., Shieh J. J., Wu C. Y. (2018). Intestinal microbiota profiling and predicted metabolic dysregulation in psoriasis patients. Exp. Dermatol. 27 1336–1343. 10.1111/exd.13786 [DOI] [PubMed] [Google Scholar]
- Chen Y. T., Yang N. S., Lin Y. C., Ho S. T., Li K. Y., Lin J. S., et al. (2018). A combination of Lactobacillus mali APS1 and dieting improved the efficacy of obesity treatment via manipulating gut microbiome in mice. Sci. Rep. 8:6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen G. J., Bruggemann H. (2014). Bacterial skin commensals and their role as host guardians. Benef. Microbes 5 201–215. 10.3920/bm2012.0062 [DOI] [PubMed] [Google Scholar]
- Cicerone C., Nenna R., Pontone S. (2015). Th17, intestinal microbiota and the abnormal immune response in the pathogenesis of celiac disease. Gastroenterol. Hepatol. Bed Bench 8 117–122. [PMC free article] [PubMed] [Google Scholar]
- Claesson M. J., Wang Q., O’Sullivan O., Greene-Diniz R., Cole J. R., Ross R. P., et al. (2010). Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38:e200. 10.1093/nar/gkq873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codoner F. M., Ramirez-Bosca A., Climent E., Carrion-Gutierrez M., Guerrero M., Perez-Orquin J. M., et al. (2018). Gut microbial composition in patients with psoriasis. Sci. Rep. 8:3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides M. G., Link V. M., Tamoutounour S., Wong A. C., Perez-Chaparro P. J., Han S. J., et al. (2019). MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366:eaax6624. 10.1126/science.aax6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey R., Brophy S., Kennedy J., Gutierrez F. F., Pickles T., Davies R., et al. (2018). Cardiovascular risk factors predicting cardiac events are different in patients with rheumatoid arthritis, psoriatic arthritis, and psoriasis. Semin. Arthritis Rheum. 48 367–373. 10.1016/j.semarthrit.2018.03.005 [DOI] [PubMed] [Google Scholar]
- Dauden E., Blasco A. J., Bonanad C., Botella R., Carrascosa J. M., Gonzalez-Parra E., et al. (2018). Position statement for the management of comorbidities in psoriasis. J. Eur. Acad. Dermatol. Venereol. 32 2058–2073. 10.1111/jdv.15177 [DOI] [PubMed] [Google Scholar]
- de Groot P., Scheithauer T., Bakker G. J., Prodan A., Levin E., Khan M. T., et al. (2020). Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut 69 502–512. 10.1136/gutjnl-2019-318320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Li Q., Li P., Zhang T., Cui B., Ji G., et al. (2019). Long-term safety and efficacy of fecal microbiota transplant in active ulcerative colitis. Drug Saf. 42 869–880. 10.1007/s40264-019-00809-2 [DOI] [PubMed] [Google Scholar]
- Drago L., De Grandi R., Altomare G., Pigatto P., Rossi O., Toscano M. (2016). Skin microbiota of first cousins affected by psoriasis and atopic dermatitis. Clin. Mol. Allergy 14:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvallet C., Gibbons S. M., Gurry T., Irizarry R. A., Alm E. J. (2017). Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 8:1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejtahed H. S., Soroush A. R., Siadat S. D., Hoseini-Tavassol Z., Larijani B., Hasani-Ranjbar S. (2019). Targeting obesity management through gut microbiota modulation by herbal products: a systematic review. Complement. Ther. Med. 42 184–204. 10.1016/j.ctim.2018.11.019 [DOI] [PubMed] [Google Scholar]
- Eppinga H., Sperna Weiland C. J., Thio H. B., van der Woude C. J., Nijsten T. E., Peppelenbosch M. P., et al. (2016). Similar depletion of protective Faecalibacterium prausnitzii in psoriasis and inflammatory bowel disease, but not in Hidradenitis suppurativa. J. Crohns Colitis 10 1067–1075. 10.1093/ecco-jcc/jjw070 [DOI] [PubMed] [Google Scholar]
- Fahlen A., Engstrand L., Baker B. S., Powles A., Fry L. (2012). Comparison of bacterial microbiota in skin biopsies from normal and psoriatic skin. Arch. Dermatol. Res. 304 15–22. 10.1007/s00403-011-1189-x [DOI] [PubMed] [Google Scholar]
- Fernandes J., Su W., Rahat-Rozenbloom S., Wolever T. M., Comelli E. M. (2014). Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr. Diabetes 4:e121. 10.1038/nutd.2014.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M., Kao D., Mehta S. R., Martin T., Dimitry J., Keshteli A. H., et al. (2016). Predictors of early failure after fecal microbiota transplantation for the therapy of clostridium difficile infection: a multicenter study. Am. J. Gastroenterol. 111 1024–1031. 10.1038/ajg.2016.180 [DOI] [PubMed] [Google Scholar]
- Forbes J. D., Van Domselaar G., Bernstein C. N. (2016). The gut microbiota in immune-mediated inflammatory diseases. Front. Microbiol. 7:1081. 10.3389/fmicb.2016.01081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry L., Baker B. S. (2007). Triggering psoriasis: the role of infections and medications. Clin. Dermatol. 25 606–615. 10.1016/j.clindermatol.2007.08.015 [DOI] [PubMed] [Google Scholar]
- Fyhrquist N., Muirhead G., Prast-Nielsen S., Jeanmougin M., Olah P., Skoog T., et al. (2019). Microbe-host interplay in atopic dermatitis and psoriasis. Nat. Commun. 10:4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Tseng C. H., Strober B. E., Pei Z., Blaser M. J. (2008). Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS One 3:e2719. 10.1371/journal.pone.0002719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geem D., Medina-Contreras O., McBride M., Newberry R. D., Koni P. A., Denning T. L. (2014). Specific microbiota-induced intestinal Th17 differentiation requires MHC class II but not GALT and mesenteric lymph nodes. J. Immunol. 193 431–438. 10.4049/jimmunol.1303167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goiset A., Milpied B., Marti A., Marie J., Leroy-Colavolpe V., Pham-Ledard A., et al. (2019). Characteristics, associated diseases, and management of gram-negative toe-web infection: a French experience. Acta Derm. Venereol. 99 1121–1126. [DOI] [PubMed] [Google Scholar]
- Gonzalez T., Stevens M. L., Baatyrbek Kyzy A., Alarcon R., He H., Kroner J. W., et al. (2020). Biofilm propensity of Staphylococcus aureus skin isolates is associated with increased atopic dermatitis severity and barrier dysfunction in the MPAACH pediatric cohort. Allergy 10.1111/all.14489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E. A., Kong H. H., Conlan S., Deming C. B., Davis J., Young A. C., et al. (2009). Topographical and temporal diversity of the human skin microbiome. Science 324 1190–1192. 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E. A., Kong H. H., Renaud G., Young A. C., Program N. C. S., Bouffard G. G., et al. (2008). A diversity profile of the human skin microbiota. Genome Res. 18 1043–1050. 10.1101/gr.075549.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E. A., Segre J. A. (2011). The skin microbiome. Nat. Rev. Microbiol. 9 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeger D., O’Mahony L., Murphy E. F., Bourke J. F., Dinan T. G., Kiely B., et al. (2013). Bifidobacterium infantis 35624 modulates host inflammatory processes beyond the gut. Gut Microbes 4 325–339. 10.4161/gmic.25487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueniche A., Benyacoub J., Philippe D., Bastien P., Kusy N., Breton L., et al. (2010). Lactobacillus paracasei CNCM I-2116 (ST11) inhibits substance P-induced skin inflammation and accelerates skin barrier function recovery in vitro. Eur. J. Dermatol. 20 731–737. [DOI] [PubMed] [Google Scholar]
- Hacini-Rachinel F., Gheit H., Le Luduec J. B., Dif F., Nancey S., Kaiserlian D. (2009). Oral probiotic control skin inflammation by acting on both effector and regulatory T cells. PLoS One 4:e4903. 10.1371/journal.pone.0004903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden J. L., Lewis S. M., Lish S. R., Suarez-Farinas M., Gareau D., Lentini T., et al. (2016). The tryptophan metabolism enzyme L-kynureninase is a novel inflammatory factor in psoriasis and other inflammatory diseases. J. Allergy Clin. Immunol. 137 1830–1840. 10.1016/j.jaci.2015.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison O. J., Linehan J. L., Shih H. Y., Bouladoux N., Han S. J., Smelkinson M., et al. (2019). Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science 363:eaat6280. 10.1126/science.aat6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz-Buschart A., Wilmes P. (2018). Human gut microbiome: function matters. Trends Microbiol. 26 563–574. 10.1016/j.tim.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Hidalgo-Cantabrana C., Gomez J., Delgado S., Requena-Lopez S., Queiro-Silva R., Margolles A., et al. (2019). Gut microbiota dysbiosis in a cohort of patients with psoriasis. Br. J. Dermatol. 181 1287–1295. 10.1111/bjd.17931 [DOI] [PubMed] [Google Scholar]
- Hill M. J. (1997). Intestinal flora and endogenous vitamin synthesis. Eur. J. Cancer Prev. 6 (Suppl. 1) S43–S45. [DOI] [PubMed] [Google Scholar]
- Hirota K., Turner J. E., Villa M., Duarte J. H., Demengeot J., Steinmetz O. M., et al. (2013). Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat. Immunol. 14 372–379. 10.1038/ni.2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland K. T., Cunliffe W. J., Roberts C. D. (1977). Acne vulgaris: an investigation into the number of anaerobic diphtheroids and members of the Micrococcaceae in normal and acne skin. Br. J. Dermatol. 96 623–626. 10.1111/j.1365-2133.1977.tb05206.x [DOI] [PubMed] [Google Scholar]
- Honda K., Littman D. R. (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535 75–84. 10.1038/nature18848 [DOI] [PubMed] [Google Scholar]
- Huang L., Gao R., Yu N., Zhu Y., Ding Y., Qin H. (2019). Dysbiosis of gut microbiota was closely associated with psoriasis. Sci. China Life Sci. 62 807–815. 10.1007/s11427-018-9376-6 [DOI] [PubMed] [Google Scholar]
- Hurabielle C., Link V. M., Bouladoux N., Han S. J., Merrill E. D., Lightfoot Y. L., et al. (2020). Immunity to commensal skin fungi promotes psoriasiform skin inflammation. Proc. Natl. Acad. Sci. U.S.A. 117 16465–16474. 10.1073/pnas.2003022117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianiro G., Valerio L., Masucci L., Pecere S., Bibbo S., Quaranta G., et al. (2017). Predictors of failure after single faecal microbiota transplantation in patients with recurrent Clostridium difficile infection: results from a 3-year, single-centre cohort study. Clin. Microbiol. Infect. 23 337.e1–337.e3. [DOI] [PubMed] [Google Scholar]
- Ivanov I. I., Atarashi K., Manel N., Brodie E. L., Shima T., Karaoz U., et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. I., McKenzie B. S., Zhou L., Tadokoro C. E., Lepelley A., Lafaille J. J., et al. (2006). The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126 1121–1133. [DOI] [PubMed] [Google Scholar]
- Jia L., Shan K., Pan L. L., Feng N., Lv Z., Sun Y., et al. (2017). Clostridium butyricum CGMCC0313.1 protects against autoimmune diabetes by modulating intestinal immune homeostasis and inducing pancreatic regulatory T cells. Front. Immunol. 8:1345. 10.3389/fimmu.2017.01345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov W. H., Martinez del Rio C., Caviedes-Vidal E. (2011). Ecological physiology of diet and digestive systems. Annu. Rev. Physiol. 73 69–93. 10.1146/annurev-physiol-012110-142152 [DOI] [PubMed] [Google Scholar]
- Ke X., Walker A., Haange S. B., Lagkouvardos I., Liu Y., Schmitt-Kopplin P., et al. (2019). Synbiotic-driven improvement of metabolic disturbances is associated with changes in the gut microbiome in diet-induced obese mice. Mol. Metab. 22 96–109. 10.1016/j.molmet.2019.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E. A., Connolly J., Hourihane J. O., Fallon P. G., McLean W. H. I., Murray D., et al. (2017). Skin microbiome before development of atopic dermatitis: early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J. Allergy Clin. Immunol. 139 166–172. 10.1016/j.jaci.2016.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. G., Udayanga K. G., Totsuka N., Weinberg J. B., Nunez G., Shibuya A. (2014). Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE(2). Cell Host Microbe 15 95–102. 10.1016/j.chom.2013.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Voisin B., Kim D. Y., Kennedy E. A., Jo J. H., Shih H. Y., et al. (2019). Homeostatic control of sebaceous glands by innate lymphoid cells regulates commensal bacteria equilibrium. Cell 176 982–997.e916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine M. (2020). Recent advances in psoriasis research; the clue to mysterious relation to gut microbiome. Int. J. Mol. Sci. 21:2582. 10.3390/ijms21072582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Zhang S., Wu R., Su X., Peng D., Zhao M., et al. (2019). New insights into different adipokines in linking the pathophysiology of obesity and psoriasis. Lipids Health Dis. 18:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosiewicz M. M., Dryden G. W., Chhabra A., Alard P. (2014). Relationship between gut microbiota and development of T cell associated disease. FEBS Lett. 588 4195–4206. 10.1016/j.febslet.2014.03.019 [DOI] [PubMed] [Google Scholar]
- Krajicek E., Fischer M., Allegretti J. R., Kelly C. R. (2019). Nuts and bolts of fecal microbiota transplantation. Clin. Gastroenterol. Hepatol. 17 345–352. 10.1016/j.cgh.2018.09.029 [DOI] [PubMed] [Google Scholar]
- Kruglov A. A., Grivennikov S. I., Kuprash D. V., Winsauer C., Prepens S., Seleznik G. M., et al. (2013). Nonredundant function of soluble LTalpha3 produced by innate lymphoid cells in intestinal homeostasis. Science 342 1243–1246. 10.1126/science.1243364 [DOI] [PubMed] [Google Scholar]
- Lai Z. L., Tseng C. H., Ho H. J., Cheung C. K. Y., Lin J. Y., Chen Y. J., et al. (2018). Fecal microbiota transplantation confers beneficial metabolic effects of diet and exercise on diet-induced obese mice. Sci. Rep. 8:15625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc J. G., Milani C., de Giori G. S., Sesma F., van Sinderen D., Ventura M. (2013). Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr. Opin. Biotechnol. 24 160–168. 10.1016/j.copbio.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Lee H., Lee Y., Kim J., An J., Lee S., Kong H., et al. (2018). Modulation of the gut microbiota by metformin improves metabolic profiles in aged obese mice. Gut Microbes 9 155–165. 10.1080/19490976.2017.1405209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Jeong S. E., Lee S., Kim S., Han H., Jeon C. O. (2018). Effects of cosmetics on the skin microbiome of facial cheeks with different hydration levels. Microbiologyopen 7:e00557. 10.1002/mbo3.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. Y., Kim D. J., Won J. N., Lee I. H., Sung M. H., Poo H. (2014). Oral administration of poly-gamma-glutamate ameliorates atopic dermatitis in Nc/Nga mice by suppressing Th2-biased immune response and production of IL-17A. J. Invest. Dermatol. 134 704–711. 10.1038/jid.2013.389 [DOI] [PubMed] [Google Scholar]
- Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444 1022–1023. [DOI] [PubMed] [Google Scholar]
- Li Z., Jin H., Oh S. Y., Ji G. E. (2016). Anti-obese effects of two Lactobacilli and two Bifidobacteria on ICR mice fed on a high fat diet. Biochem. Biophys. Res. Commun. 480 222–227. 10.1016/j.bbrc.2016.10.031 [DOI] [PubMed] [Google Scholar]
- Lin L., Zhang J. (2017). Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 18:2. 10.1186/s12865-016-0187-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. T., Huang Y. J., Zhang T. Y., Tan L. B., Lu X. F., Qin J. (2019). Lingguizhugan decoction attenuates diet-induced obesity and hepatosteatosis via gut microbiota. World J. Gastroenterol. 25 3590–3606. 10.3748/wjg.v25.i27.3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. H., Yu H. Y., Chang Y. C., Chung-Yee Hui R., Huang Y. C., Huang Y. H. (2019). Host characteristics and dynamics of Staphylococcus aureus colonization in patients with moderate-to-severe psoriasis before and after treatment: a prospective cohort study. J. Am. Acad. Dermatol. 81 605–607. 10.1016/j.jaad.2018.05.031 [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J., Abu-Ali G., Huttenhower C. (2016). The healthy human microbiome. Genome Med. 8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman R. S., Diehl G. E., Victorio D. A., Huh J. R., Galan C., Miraldi E. R., et al. (2014). CX(3)CR1(+) mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J. Exp. Med. 211 1571–1583. 10.1084/jem.20140678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonnberg A. S., Zachariae C., Skov L. (2014). Targeting of interleukin-17 in the treatment of psoriasis. Clin. Cosmet. Investig. Dermatol. 7 251–259. 10.2147/ccid.s67534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowes M. A., Suarez-Farinas M., Krueger J. G. (2014). Immunology of psoriasis. Annu. Rev. Immunol. 32 227–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison K. C. (2003). Barrier function of the skin: “la raison d’etre” of the epidermis. J. Invest. Dermatol. 121 231–241. 10.1046/j.1523-1747.2003.12359.x [DOI] [PubMed] [Google Scholar]
- Martinez-Lopez M., Iborra S., Conde-Garrosa R., Mastrangelo A., Danne C., Mann E. R., et al. (2019). Microbiota sensing by Mincle-Syk axis in dendritic cells regulates interleukin-17 and -22 production and promotes intestinal barrier integrity. Immunity 50 446–461.e449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain A. J., O’Neill C. A., Amezquita A., Price L. J., Faust K., Tett A., et al. (2019). Consumer safety considerations of skin and oral microbiome perturbation. Clin. Microbiol. Rev. 32:e00051-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSweeney B., Allegretti J. R., Fischer M., Xu H., Goodman K. J., Monaghan T., et al. (2020). In search of stool donors: a multicenter study of prior knowledge, perceptions, motivators, and deterrents among potential donors for fecal microbiota transplantation. Gut Microbes 11 51–62. 10.1080/19490976.2019.1611153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer K., de Vos P., Priebe M. G. (2010). Butyrate and other short-chain fatty acids as modulators of immunity: what relevance for health? Curr. Opin. Clin. Nutr. Metab. Care 13 715–721. 10.1097/mco.0b013e32833eebe5 [DOI] [PubMed] [Google Scholar]
- Million M., Angelakis E., Maraninchi M., Henry M., Giorgi R., Valero R., et al. (2013). Correlation between body mass index and gut concentrations of Lactobacillus reuteri, Bifidobacterium animalis, Methanobrevibacter smithii and Escherichia coli. Int. J. Obes. (Lond) 37 1460–1466. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Miyazaki K., Masuoka N., Kano M., Iizuka R. (2014). Bifidobacterium fermented milk and galacto-oligosaccharides lead to improved skin health by decreasing phenols production by gut microbiota. Benef. Microbes 5 121–128. 10.3920/bm2012.0066 [DOI] [PubMed] [Google Scholar]
- Mullish B. H., Quraishi M. N., Segal J. P., McCune V. L., Baxter M., Marsden G. L., et al. (2018). The use of faecal microbiota transplant as treatment for recurrent or refractory Clostridium difficile infection and other potential indications: joint British society of gastroenterology (BSG) and healthcare infection society (HIS) guidelines. Gut 67 1920–1941. 10.1136/gutjnl-2018-316818 [DOI] [PubMed] [Google Scholar]
- Naik S., Bouladoux N., Wilhelm C., Molloy M. J., Salcedo R., Kastenmuller W., et al. (2012). Compartmentalized control of skin immunity by resident commensals. Science 337 1115–1119. 10.1126/science.1225152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T., Chen T. H., Narala S., Chun K. A., Two A. M., Yun T., et al. (2017). Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 9:eaah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Lopez V., Martinez-Andres A., Ramirez-Bosca A., Ruzafa-Costas B., Nunez-Delegido E., Carrion-Gutierrez M. A., et al. (2019). Efficacy and safety of oral administration of a mixture of probiotic strains in patients with psoriasis: a randomized controlled clinical trial. Acta Derm. Venereol. 99 1078–1084. [DOI] [PubMed] [Google Scholar]
- Nemoto Y., Kanai T., Takahara M., Oshima S., Okamoto R., Tsuchiya K., et al. (2013). Th1/Th17-mediated interstitial pneumonia in chronic colitis mice independent of intestinal microbiota. J. Immunol. 190 6616–6625. 10.4049/jimmunol.1202930 [DOI] [PubMed] [Google Scholar]
- Ng C. Y., Huang Y. H., Chu C. F., Wu T. C., Liu S. H. (2017). Risks for Staphylococcus aureus colonization in patients with psoriasis: a systematic review and meta-analysis. Br. J. Dermatol. 177 967–977. [DOI] [PubMed] [Google Scholar]
- Okada K., Matsushima Y., Mizutani K., Yamanaka K. (2020). The role of gut microbiome in psoriasis: oral administration of Staphylococcus aureus and Streptococcus danieliae exacerbates skin inflammation of imiquimod-induced psoriasis-like dermatitis. Int. J. Mol. Sci. 21:3303. 10.3390/ijms21093303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcina-Kurtovic N., Kasumagic-Halilovic E., Helppikangans H., Begic J. (2016). Prevalence of Candida species in patients with psoriasis. Acta Dermatovenerol. Croat. 24 209–213. [PubMed] [Google Scholar]
- Paramsothy S., Nielsen S., Kamm M. A., Deshpande N. P., Faith J. J., Clemente J. C., et al. (2019). Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology 156 1440–1454.e1442. [DOI] [PubMed] [Google Scholar]
- Parlet C. P., Brown M. M., Horswill A. R. (2019). Commensal Staphylococci influence Staphylococcus aureus skin colonization and disease. Trends Microbiol. 27 497–507. 10.1016/j.tim.2019.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulino L. C., Tseng C. H., Strober B. E., Blaser M. J. (2006). Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J. Clin. Microbiol. 44 2933–2941. 10.1128/jcm.00785-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson E. K., Uronen-Hansson H., Semmrich M., Rivollier A., Hagerbrand K., Marsal J., et al. (2013). IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity 38 958–969. 10.1016/j.immuni.2013.03.009 [DOI] [PubMed] [Google Scholar]
- Quan C., Chen X. Y., Li X., Xue F., Chen L. H., Liu N., et al. (2020). Psoriatic lesions are characterized by higher bacterial load and imbalance between Cutibacterium and Corynebacterium. J. Am. Acad. Dermatol. 82 955–961. 10.1016/j.jaad.2019.06.024 [DOI] [PubMed] [Google Scholar]
- Ramai D., Zakhia K., Ofosu A., Ofori E., Reddy M. (2019). Fecal microbiota transplantation: donor relation, fresh or frozen, delivery methods, cost-effectiveness. Ann. Gastroenterol. 32 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather I. A., Bajpai V. K., Huh Y. S., Han Y. K., Bhat E. A., Lim J., et al. (2018). Probiotic Lactobacillus sakei proBio-65 extract ameliorates the severity of imiquimod induced psoriasis-like skin inflammation in a mouse model. Front. Microbiol. 9:1021. 10.3389/fmicb.2018.01021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura V. K., Bouladoux N., Claesen J., Chen Y. E., Byrd A. L., Constantinides M. G., et al. (2018). Contextual control of skin immunity and inflammation by Corynebacterium. J. Exp. Med. 215 785–799. 10.1084/jem.20171079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson E. D., Bowcock A. M. (2010). Psoriasis genetics: breaking the barrier. Trends Genet. 26 415–423. 10.1016/j.tig.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks M. G., Garrett W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16 341–352. 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V., Mascanfroni I. D., Bunse L., Takenaka M. C., Kenison J. E., Mayo L., et al. (2016). Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22 586–597. 10.1038/nm.4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J. L., Mazmanian S. K. (2010). Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. U.S.A. 107 12204–12209. 10.1073/pnas.0909122107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy A. T., Briseno C. G., Lee J. S., Ng D., Manieri N. A., Kc W., et al. (2013). Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14 937–948. 10.1038/ni.2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunte D. M., Mrowietz U., Puig L., Zachariae C. (2017). Candida infections in patients with psoriasis and psoriatic arthritis treated with interleukin-17 inhibitors and their practical management. Br. J. Dermatol. 177 47–62. 10.1111/bjd.15015 [DOI] [PubMed] [Google Scholar]
- Savage D. C. (1977). Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31 107–133. [DOI] [PubMed] [Google Scholar]
- Scharschmidt T. C., Vasquez K. S., Truong H. A., Gearty S. V., Pauli M. L., Nosbaum A., et al. (2015). A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43 1011–1021. 10.1016/j.immuni.2015.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher J. U., Ubeda C., Artacho A., Attur M., Isaac S., Reddy S. M., et al. (2015). Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 67 128–139. 10.1002/art.38892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz A., Bruhs A., Schwarz T. (2017). The short-chain fatty acid sodium butyrate functions as a regulator of the skin immune system. J. Invest. Dermatol. 137 855–864. 10.1016/j.jid.2016.11.014 [DOI] [PubMed] [Google Scholar]
- Shapiro J., Cohen N. A., Shalev V., Uzan A., Koren O., Maharshak N. (2019). Psoriatic patients have a distinct structural and functional fecal microbiota compared with controls. J. Dermatol. 46 595–603. 10.1111/1346-8138.14933 [DOI] [PubMed] [Google Scholar]
- Sharma V., Smolin J., Nayak J., Ayala J. E., Scott D. A., Peterson S. N., et al. (2018). Mannose alters gut microbiome, prevents diet-induced obesity, and improves host metabolism. Cell Rep. 24 3087–3098. 10.1016/j.celrep.2018.08.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y., Liu J., Zheng S., Liang F., Luo Y., Huang K., et al. (2019). Mulberry leaves ameliorate obesity through enhancing brown adipose tissue activity and modulating gut microbiota. Food Funct. 10 4771–4781. 10.1039/c9fo00883g [DOI] [PubMed] [Google Scholar]
- Shin J. H., Nam M. H., Lee H., Lee J. S., Kim H., Chung M. J., et al. (2018). Amelioration of obesity-related characteristics by a probiotic formulation in a high-fat diet-induced obese rat model. Eur. J. Nutr. 57 2081–2090. 10.1007/s00394-017-1481-4 [DOI] [PubMed] [Google Scholar]
- Shipman A. R., Millington G. W. (2011). Obesity and the skin. Br. J. Dermatol. 165 743–750. 10.1111/j.1365-2133.2011.10393.x [DOI] [PubMed] [Google Scholar]
- Sikora M., Chrabaszcz M., Maciejewski C., Zaremba M., Waskiel A., Olszewska M., et al. (2018). Intestinal barrier integrity in patients with plaque psoriasis. J. Dermatol. 45 1468–1470. 10.1111/1346-8138.14647 [DOI] [PubMed] [Google Scholar]
- Sikora M., Stec A., Chrabaszcz M., Waskiel-Burnat A., Zaremba M., Olszewska M., et al. (2019). Intestinal fatty acid binding protein, a biomarker of intestinal barrier, is associated with severity of psoriasis. J. Clin. Med. 8:1021. 10.3390/jcm8071021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soare A., Weber S., Maul L., Rauber S., Gheorghiu A. M., Luber M., et al. (2018). Cutting edge: homeostasis of innate lymphoid cells is imbalanced in psoriatic arthritis. J. Immunol. 200 1249–1254. 10.4049/jimmunol.1700596 [DOI] [PubMed] [Google Scholar]
- Stehlikova Z., Kostovcik M., Kostovcikova K., Kverka M., Juzlova K., Rob F., et al. (2019). Dysbiosis of skin microbiota in psoriatic patients: co-occurrence of fungal and bacterial communities. Front. Microbiol. 10:438. 10.3389/fmicb.2019.00438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Zamorano N., Fabbiano S., Chevalier C., Stojanovic O., Colin D. J., Stevanovic A., et al. (2015). Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat. Med. 21 1497–1501. 10.1038/nm.3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Guo Y., Zhang S., Chen Z., Wu K., Liu Q., et al. (2018). Fecal microbiota transplantation can alleviate gastrointestinal transit in rats with high-fat diet-induced obesity via regulation of Serotonin Biosynthesis. Biomed. Res. Int. 2018:8308671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M. M., Kim T. T., Denou E., Soltys C. M., Hamza S. M., Byrne N. J., et al. (2017). Improved glucose homeostasis in obese mice treated with resveratrol is associated with alterations in the gut microbiome. Diabetes 66 418–425. 10.2337/db16-0680 [DOI] [PubMed] [Google Scholar]
- Takemoto A., Cho O., Morohoshi Y., Sugita T., Muto M. (2015). Molecular characterization of the skin fungal microbiome in patients with psoriasis. J. Dermatol. 42 166–170. 10.1111/1346-8138.12739 [DOI] [PubMed] [Google Scholar]
- Tan L., Zhao S., Zhu W., Wu L., Li J., Shen M., et al. (2018). The Akkermansia muciniphila is a gut microbiota signature in psoriasis. Exp. Dermatol. 27 144–149. 10.1111/exd.13463 [DOI] [PubMed] [Google Scholar]
- Tauch A., Fernandez-Natal I., Soriano F. (2016). A microbiological and clinical review on Corynebacterium kroppenstedtii. Int. J. Infect. Dis. 48 33–39. [DOI] [PubMed] [Google Scholar]
- Teng F., Klinger C. N., Felix K. M., Bradley C. P., Wu E., Tran N. L., et al. (2016). Gut microbiota drive autoimmune arthritis by promoting differentiation and migration of Peyer’s patch T follicular helper cells. Immunity 44 875–888. 10.1016/j.immuni.2016.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tett A., Pasolli E., Farina S., Truong D. T., Asnicar F., Zolfo M., et al. (2017). Unexplored diversity and strain-level structure of the skin microbiome associated with psoriasis. NPJ Biofilms Microbiomes 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen R. J., Stranieri A., Knutson D., Strauss J. S. (1980). Topical clindamycin treatment of acne. Clinical, surface lipid composition, and quantitative surface microbiology response. Arch. Dermatol. 116 1031–1034. 10.1001/archderm.116.9.1031 [DOI] [PubMed] [Google Scholar]
- Till A. E., Goulden V., Cunliffe W. J., Holland K. T. (2000). The cutaneous microflora of adolescent, persistent and late-onset acne patients does not differ. Br. J. Dermatol. 142 885–892. 10.1046/j.1365-2133.2000.03467.x [DOI] [PubMed] [Google Scholar]
- Tlaskalova-Hogenova H., Stepankova R., Hudcovic T., Tuckova L., Cukrowska B., Lodinova-Zadnikova R., et al. (2004). Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol. Lett. 93 97–108. 10.1016/j.imlet.2004.02.005 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457 480–484. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukibe K., Miyoshi M., Kadooka Y. (2015). Administration of Lactobacillus gasseri SBT2055 suppresses macrophage infiltration into adipose tissue in diet-induced obese mice. Br. J. Nutr. 114 1180–1187. 10.1017/s0007114515002627 [DOI] [PubMed] [Google Scholar]
- van Beelen A. J., Teunissen M. B., Kapsenberg M. L., de Jong E. C. (2007). Interleukin-17 in inflammatory skin disorders. Curr. Opin. Allergy Clin. Immunol. 7 374–381. [DOI] [PubMed] [Google Scholar]
- Vermeire S., Joossens M., Verbeke K., Wang J., Machiels K., Sabino J., et al. (2016). Donor species richness determines faecal microbiota transplantation success in inflammatory bowel disease. J. Crohns Colitis 10 387–394. 10.1093/ecco-jcc/jjv203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayashankar M., Raghunath N. (2012). Pustular psoriasis responding to Probiotics – a new insight. Our Dermatol. Online 3 326–329. 10.7241/ourd.20124.71 [DOI] [Google Scholar]
- Wang J., Wang P., Li D., Hu X., Chen F. (2020). Beneficial effects of ginger on prevention of obesity through modulation of gut microbiota in mice. Eur. J. Nutr. 59 699–718. 10.1007/s00394-019-01938-1 [DOI] [PubMed] [Google Scholar]
- Wang W. M., Jin H. Z. (2018). Skin microbiome: an actor in the pathogenesis of psoriasis. Chin. Med. J. (Engl.) 131 95–98. 10.4103/0366-6999.221269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Mascarenhas N., Eckmann L., Miyamoto Y., Sun X., Kawakami T., et al. (2017). Skin microbiome promotes mast cell maturation by triggering stem cell factor production in keratinocytes. J. Allergy Clin. Immunol. 139 1205–1216.e1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver L. K., Minichino D., Biswas C., Chu N., Lee J. J., Bittinger K., et al. (2019). Microbiota-dependent signals are required to sustain TLR-mediated immune responses. JCI Insight 4:e124370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. M., Brummer R. J., Derrien M., MacDonald T. T., Troost F., Cani P. D., et al. (2017). Homeostasis of the gut barrier and potential biomarkers. Am. J. Physiol. Gastrointest. Liver Physiol. 312 G171–G193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B. C., Vatanen T., Cutfield W. S., O’Sullivan J. M. (2019). The super-donor phenomenon in fecal microbiota transplantation. Front. Cell. Infect. Microbiol. 9:2. 10.3389/fcimb.2019.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. C., Kim J. N., Wang Z., Chang Y. L., Zengler K., Di Nardo A. (2019). Mast cell recruitment is modulated by the hairless skin microbiome. J. Allergy Clin. Immunol. 144 330–333.e336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. J., Ivanov I. I., Darce J., Hattori K., Shima T., Umesaki Y., et al. (2010). Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32 815–827. 10.1016/j.immuni.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. R., Lin C. S., Chang C. J., Lin T. L., Martel J., Ko Y. F., et al. (2019). Gut commensal Parabacteroides goldsteinii plays a predominant role in the anti-obesity effects of polysaccharides isolated from Hirsutella sinensis. Gut 68 248–262. 10.1136/gutjnl-2017-315458 [DOI] [PubMed] [Google Scholar]
- Xu J., Liu T., Li Y., Liu W., Ding Z., Ma H., et al. (2019). Jamun (Eugenia jambolana Lam.) fruit extract prevents obesity by modulating the gut microbiome in high-fat-diet-fed mice. Mol. Nutr. Food Res. 63:e1801307. [DOI] [PubMed] [Google Scholar]
- Yasir M., Angelakis E., Bibi F., Azhar E. I., Bachar D., Lagier J. C., et al. (2015). Comparison of the gut microbiota of people in France and Saudi Arabia. Nutr. Diabetes 5:e153. 10.1038/nutd.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G., Li J. F., Sun Y. F., Ding X., Zeng J. Q., Zhang T., et al. (2019). [Fecal microbiota transplantation as a novel therapy for severe psoriasis]. Zhonghua Nei Ke Za Zhi 58 782–785. [DOI] [PubMed] [Google Scholar]
- Yissachar N., Zhou Y., Ung L., Lai N. Y., Mohan J. F., Ehrlicher A., et al. (2017). An intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell 168 1135–1148.e1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakostelska Z., Malkova J., Klimesova K., Rossmann P., Hornova M., Novosadova I., et al. (2016). Intestinal microbiota promotes psoriasis-like skin inflammation by enhancing Th17 response. PLoS One 11:e0159539. 10.1371/journal.pone.0159539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T., Iannitti R. G., Cunha C., De Luca A., Giovannini G., Pieraccini G., et al. (2013). Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39 372–385. 10.1016/j.immuni.2013.08.003 [DOI] [PubMed] [Google Scholar]
- Zhang W., Xu J. H., Yu T., Chen Q. K. (2019). Effects of berberine and metformin on intestinal inflammation and gut microbiome composition in db/db mice. Biomed. Pharmacother. 118:109131. 10.1016/j.biopha.2019.109131 [DOI] [PubMed] [Google Scholar]
- Zielinski C. E., Mele F., Aschenbrenner D., Jarrossay D., Ronchi F., Gattorno M., et al. (2012). Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature 484 514–518. 10.1038/nature10957 [DOI] [PubMed] [Google Scholar]