Human monoclonal antibody potency in vitro does not uniformly correlate with its in vivo neutralization of SARS-CoV-2. Antibody Fc-effector function is important for in vivo neutralization, and antibody combinations show superior efficacy compared with single antibodies.
Abstract
SARS-CoV-2, the causative agent of COVID-19, has been responsible for over 42 million infections and 1 million deaths since its emergence in December 2019. There are few therapeutic options and no approved vaccines. Here, we examine the properties of highly potent human monoclonal antibodies (hu-mAbs) in a Syrian hamster model of SARS-CoV-2 and in a mouse-adapted model of SARS-CoV-2 infection (SARS-CoV-2 MA). Antibody combinations were effective for prevention and in therapy when administered early. However, in vitro antibody neutralization potency did not uniformly correlate with in vivo protection, and some hu-mAbs were more protective in combination in vivo. Analysis of antibody Fc regions revealed that binding to activating Fc receptors contributes to optimal protection against SARS-CoV-2 MA. The data indicate that intact effector function can affect hu-mAb protective activity and that in vivo testing is required to establish optimal hu-mAb combinations for COVID-19 prevention.
Introduction
Coronaviruses (CoVs) have a penchant for host range expansion, jumping from reservoir species to different hosts, resulting in newly emerging human infectious diseases. Indeed, in the last 20 yr, three novel human CoVs have emerged, causing epidemic and pandemic diseases most recently exemplified by SARS-CoV-2, the causative agent of COVID-19 (de Wit et al., 2016; Zhou et al., 2020).
Effective therapeutics are desperately needed to address the COVID-19 pandemic, as there is currently only one Food and Drug Administration–approved therapy (remdesivir) and one treatment authorized for emergency use (convalescent plasma; U.S. Food and Drug Administration, 2020). Human mAbs (hu-mAbs) hold great potential for treatment and prevention of COVID-19, and several potent SARS-CoV-2–specific mAbs targeting multiple nonoverlapping epitopes in the receptor-binding domain (RBD) in the spike (S) protein have been reported (Robbiani et al., 2020; Baum et al., 2020; Cao et al., 2020; Hansen et al., 2020; Ju et al., 2020; Liu et al., 2020; Pinto et al., 2020; Wang et al., 2020 Preprint; Zost et al., 2020a; Li et al., 2020). Some of these hu-mAbs have been tested for their ability to prevent or treat SARS–CoV-2 infection in rhesus macaques and hamsters with variable but encouraging results (Rogers et al., 2020; Liu et al., 2020; Shi et al., 2020; Hansen et al., 2020). However, the role of antibody effector function, relative neutralization potency, and combinations in protection have not been examined to date in part because performing experiments in macaques under BSL3 conditions is challenging. In addition to the traditional antibody Fc-effector functions (i.e., antibody-dependent cellular cytotoxicity, phagocytosis, etc.), Fc and cellular Fc-receptor interactions drive aspects of both innate and adaptive immunity including macrophage polarization, antigen presentation, and B cell activation. Thus, the Fc-mediated effector functions of neutralizing antibodies may also play a role in shaping diverse aspects of the adaptive immune response.
Small animal models of SARS-CoV-2 replication and pathogenesis are essential for the preclinical development of vaccines and therapeutics. However, SARS-CoV-2 cannot infect standard laboratory mice due to incompatibility between the RBD and the murine orthologue of the human viral entry receptor, angiotensin-converting enzyme receptor-2 (mACE2; Zhou et al., 2020; Walls et al., 2020; Letko et al., 2020). To obviate this problem, we developed an immune-competent mouse model of COVID-19 by remodeling the SARS-CoV-2 S RBD at the mACE2-binding interface (Dinnon et al., 2020). The recombinant virus, mouse-adapted SARS-CoV-2 (SARS-CoV-2 MA), replicates to high titers in the lungs of laboratory mice and has been used to evaluate COVID-19 vaccines and therapeutics including hu-mAbs (Dinnon et al., 2020; Zost et al., 2020a; Corbett et al., 2020). Here, we examine the role of antibody potency, effector function, and antibody combinations on protection from SARS-CoV-2 MA infection in mice and SARS-CoV-2 infection in Syrian hamsters in vivo.
Results
The effect of mouse adapting spike mutations on antibody neutralization in vitro
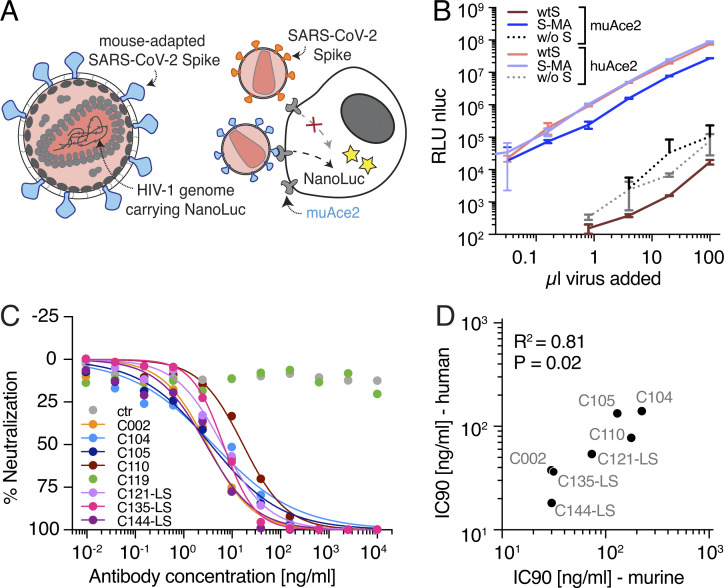
To determine if the two amino acid changes (Q498T/P499Y) in the mouse-adapted SARS-CoV-2 S RBD would affect antibody neutralization, we used HIV-1 virus pseudotyped with the SARS-CoV-2-MA S protein (S-MA). The recombinant pseudovirus was produced by cotransfection of S-MA with a replication-incompetent proviral genome (NL4-3ΔEnv-NanoLuc) that lacks a functional env gene and encodes a nanoluciferase reporter in the place of the nef gene (Fig. 1 A; Robbiani et al., 2020; Schmidt et al., 2020). To maximize S-incorporation, we truncated the C-terminus of S-MA by 19 amino acids. Using nanoluciferase reporter expression as a measure of infection, we compared the ability of pseudotyped virus bearing either WT SARS-CoV-2 S (wtS) or S-MA to infect HT1080 cells stably expressing either human or murine ACE2. Consistent with previous reports (Zhou et al., 2020; Dinnon et al., 2020), wtS did not support pseudovirus infection of murine ACE2–expressing cells (Fig. 1 B), as relative light units (RLUs) remain at background levels observed with viruses lacking spike (Fig. 1 B). In contrast, the SARS-CoV-2 MA supported robust infection of both mouse and human ACE2–expressing cells (Fig. 1 B).
Figure 1.
Antibody potency against the SARS-CoV-2 MA spike. (A) Diagram of the MA SARS-CoV-2 pseudovirus luciferase assay. SARS-CoV-2 S-MA pseudotyped HIV-1 particles carrying the nanoluc gene are used to infect muAce2-expressing HT1080 cells, which will express nanoluc luciferase upon infection, while wtS pseudotyped particles are unable to infect muAce2-expressing cells. Yellow stars indicate visible light released by the nanoluciferase (nLUC) reaction. (B) RLU reads from lysates of muAce2 and human (hu) Ace2–expressing HT1080 cells infected with increasing amounts of SARS-CoV-2 S-MA and wtS pseudovirus, as well as nonpseudotyped control virus (w/o S). Data are mean ± standard deviation of triplicates. One representative experiment is shown. nluc, nanoluc. (C) % neutralization measured for cell lysates of HT1080muAce2 cells 48 h after infection with SARS-CoV-2 S-MA pseudovirus in the presence of increasing concentrations of mAbs. n = 8 samples and 1 isotype control. Data points are shown as circles and curve fits as lines. Data are mean of duplicates, and one representative experiment is shown. ctr, control. (D) IC90 values detected in the SARS-CoV-2 S-MA pseudovirus neutralization assay (IC90-murine) plotted against those detected in the wtS SARS-CoV-2 pseudovirus neutralization assay (IC90-human). R2 = 0.8095; P < 0.02178. Mean values of at least two experiments are shown. ns, not significant.
The S-MA pseudotyped virus was used to measure the neutralizing activity of eight different IgG1 hu-mAbs with variable potencies against SARS-CoV-2 ranging from inhibitory concentration (IC)50/IC90 (half-maximal/90% IC) of 4.4/18 to 26/140 ng/ml (Table 1; Robbiani et al., 2020). C002, C104, C105, C119, C121, and C144 all target the hACE2 interaction surface of the RBD albeit at different angles of approach, and C135 and C110 target a separate nonoverlapping epitope within the RBD (Robbiani et al., 2020; Barnes et al., 2020b). With the exception of C119, the antibody neutralization titers were similar when assayed using S-MA or wtS pseudotyped viruses (R = 0.81, P = 0.02; Table 1 and Fig. 1 C and D). C119 was inactive against S-MA pseudotyped virus because the epitope targeted by this antibody overlaps the mouse-adapting mutations (Fig. 1 C; Dinnon et al., 2020; Barnes et al., 2020a Preprint). Overall, however, mouse-adapting mutations in the S protein do not significantly affect neutralization by most RBD-targeting antibodies tested in vitro.
Table 1. Inhibitory concentrations of mAbs.
| Antibody ID | Murine | Human | ||
|---|---|---|---|---|
| IC50 (ng/ml) | IC90 (ng/ml) | IC50 (ng/ml) | IC90 (ng/ml) | |
| C002 | 3.7 | 29.9 | 8.9a | 37.6a |
| C002-GRLR | 3.8 | 22.7 | ||
| C104 | 6.8 | 223.9 | 23.3a | 140.3a |
| C104-GRLR | 15.4 | 160.5 | ||
| C105 | 8.0 | 129.8 | 26.1a | 133.7a |
| C110 | 22.6 | 176.9 | 18.4a | 77.3a |
| C110-GRLR | 8.1 | 267.5 | ||
| C119 | >1,000 | >1,000 | 9.1a | 97.8a |
| C121-LS | 6.5 | 73.5 | 10.7 | 54.0 |
| C135-LS | 6.9 | 31.5 | 17.3 | 36.3 |
| C144-LS | 3.2 | 30.2 | 4.4 | 18.2 |
In vitro antibody neutralization does not uniformly correlate with its in vivo efficacy
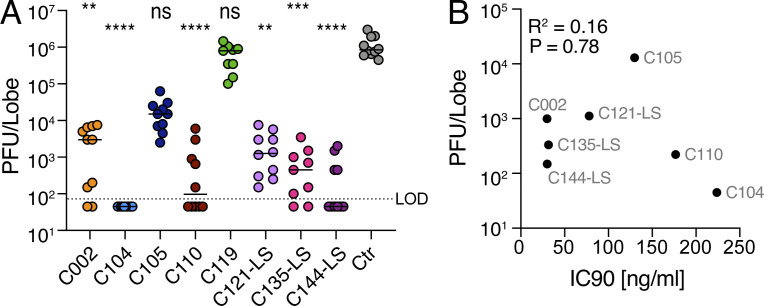
To determine if there is a correlation between in vitro neutralization and in vivo activity, we performed prophylactic efficacy studies in aged BALB/c mice infected with SARS-CoV-2 MA. mAbs (8 mg/kg) were administered by intraperitoneal injection 12 h before intranasal infection with 105 PFU of SARS-CoV-2 MA (Fig. 2 A). Viral lung titers were measured by plaque assay 2 d after infection, which is the kinetic peak of viral replication in this model (Dinnon et al., 2020). Since this is primarily a virus replication model, infected mice did not display overt disease. Mice injected with the isotype control antibody (anti-Zika antibody 3633; Robbiani et al., 2017) had mean viral lung titers of 106 PFU (Fig. 2 A and Table 2). In agreement with the in vitro neutralization data, C119 failed to protect against SARS-CoV-2 MA in vivo (Fig. 1 C, Fig. 2 A, and Table 2). In contrast, the other anti–SARS-CoV-2 antibodies tested protected against infection to varying degrees (Fig. 2 A). C104 (IC90 223 ng/ml) reduced viral loads in the lungs of all mice to below the limit of detection (i.e., 50 PFU). Other antibodies that were more potent than C104 against SARS-CoV-2 MA pseudoviruses in vitro lowered viral loads by 2–4 orders of magnitude (Fig. 2 A; C002, C110, C121-LS, C135-LS, and C144-LS). The LS mutation increases antibody half-life but does not alter potency or effector function (Zalevsky et al., 2010). Interestingly, C105 only reduced the viral loads in vivo by 1–2 orders of magnitude, yet its neutralizing activity against SARS-CoV-2 MA pseudotyped virus in vitro was similar to that of C104 (Fig. 1, Fig. 2, and Table 1). Comparison of the mean viral lung titer and respective IC90 revealed that the in vitro neutralizing activity in pseudovirus assays did not uniformly correlate with in vivo efficacy (Fig. 2 B).
Figure 2.
In vivo potency does not uniformly correlate with in vitro potency. (A) SARS-CoV-2 MA lung titer following antibody prophylaxis. Antibodies (8 mg/kg) were delivered intraperitoneally 12 h before infection with 105 PFU of SARS-CoV-2 MA. Combined data from two independent experiments are shown. All groups are n = 10 mice/group except for C119 and C135-LS. The line is at the geometric mean, and each symbol represents the titer for a single animal. Asterisks indicate statistical differences compared with isotype control by Kruskal-Wallis test with a Dunn’s multiple comparison test (****, P < 0.0001; ***, P = 0.0003; **, P = 0.007–0.004). (B) PFU/lobe values plotted against IC90 values detected in the SARS-CoV-2 S-MA pseudovirus neutralization assay. R2 = 0.1585; P = 0.788. The R and P values in A and B were determined by two-tailed Spearman correlations. ctr, control; LOD, limit of detection; ns, not significant.
Table 2. SARS-CoV-2 MA virus lung titers with single hu-mAb prophylaxis.
| Antibody ID | C002 | C104 | C105 | C110 | C119 | C121-LS | C135-LS | C144-LS | Isotype control |
|---|---|---|---|---|---|---|---|---|---|
| Dose level (mg/kg) | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| Number of animals | 10 | 10 | 10 | 10 | 9 | 10 | 9 | 10 | 10 |
| Minimum (PFU/ml) | 4.5E+01 | 4.5E+01 | 2.5E+03 | 4.5E+01 | 1.0E+05 | 1.5E+02 | 4.5E+01 | 4.5E+01 | 4.5E+05 |
| Maximum (PFU/ml) | 7.5E+03 | 4.5E+01 | 6.3E+04 | 6.0E+03 | 1.5E+06 | 7.5E+03 | 3.5E+03 | 2.0E+03 | 3.0E+06 |
| Range | 7.5E+03 | 0.0E+00 | 6.0E+04 | 6.0E+03 | 1.4E+06 | 7.4E+03 | 3.5E+03 | 2.0E+03 | 2.6E+06 |
| Geometric mean (PFU/ml) | 9.9E+02 | 4.5E+01 | 1.3E+04 | 2.2E+02 | 4.9E+05 | 1.1E+03 | 3.3E+02 | 1.5E+02 | 1.0E+06 |
| Geometric SD factor | 8.6 | 1.0 | 2.6 | 6.9 | 2.5 | 4.0 | 4.8 | 5.0 | 1.8 |
Antibody Fc-effector function contributes to neutralization of SARS-CoV-2 MA in vivo
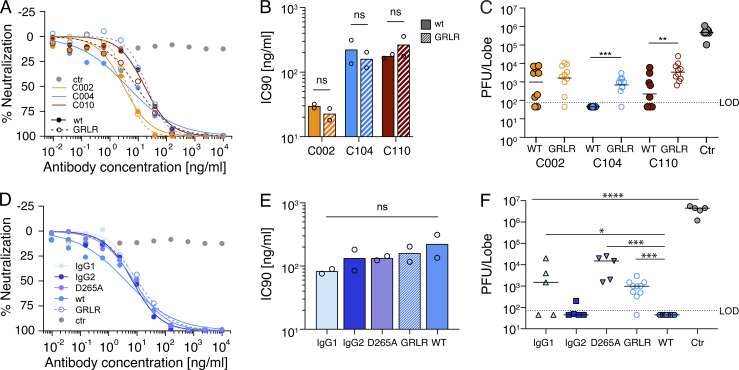
Virus neutralization in vitro is independent of antibody Fc-effector functions that impact in vivo efficacy against other viral infections (Lu et al., 2016; Halper-Stromberg et al., 2014; DiLillo et al., 2014; Bournazos et al., 2019; DiLillo et al., 2016; Bournazos et al., 2014). To examine the role of Fc-effector function on the neutralization of SARS-CoV-2 MA in vivo, we introduced the G236R/L328R (GRLR) mutation that abrogates antibody Fc receptor interaction in C002, C104, and C110 (Fig. 3; Bournazos et al., 2014). C002 and C104 target epitopes on the ACE2-binding interface of RBD, while C110 targets RBD but does not directly overlap the ACE2 interaction surface (Robbiani et al., 2020; Barnes et al., 2020a Preprint). As expected, the three antibody variants (C002GRLR, C104GRLR, and C110GRLR) had IC90 values that were not significantly different from those of WT hu-mAbs in SARS-CoV-2 MA pseudovirus assays in vitro (Fig. 3, A and B). Elimination of Fc-effector function did not significantly affect in vivo protection by C002, the least potent of the three antibodies (Fig. 3 C and Table 3; C002 = 9.9 × 102 PFU vs. C002GRLR = 1.6. × 103 PFU; P = 0.72). In contrast, loss of Fc-effector function significantly decreased the potency of both C104GRLR and C110GRLR (Fig. 3 C). Fc null C104GRLR was 14-fold and C110GRLR sixfold less potent than their Fc effector–sufficient counterparts (P = 0.0001 and P = 0.004, respectively; Fig. 3 C and Table 3). Variable Fc-effector requirements were also observed for anti-influenza antibodies, suggesting that the nature of the antibody–pathogen interaction can influence the ability of the Fc to engage its receptor (DiLillo et al., 2016).
Figure 3.
The variable requirement of Fc-effector function for SARS-CoV-2 neutralization. (A) Antibody potency curves for WT and GRLR mutant antibodies. The % neutralization for cell lysates of HT1080muAce2 cells 48 h after infection with SARS-CoV-2 S-MA pseudovirus in the presence of increasing concentrations of WT (solid lines) or GRLR-modified (dashed lines) mAbs. Data points are shown as closed (WT) or open (GRLR) circles, and corresponding curve fits are shown as continuous (WT) or dashed (GRLR) lines. Data are mean of duplicates, and one representative experiment is shown. ctr, control. (B) IC90 values for antibodies shown in A. Bars represent mean values of two experiments (shown as open circles). (C) SARS-CoV-2 MA lung titer following antibody prophylaxis. WT or GRLR antibodies (8 mg/kg) were delivered intraperitoneally 12 h before infection with 105 PFU of SARS-CoV-2 MA. Combined data from two independent experiments are shown, and all groups are n = 10 mice/group. The line is at the geometric mean, and each symbol represents the titer for a single animal. Asterisks indicate statistical differences compared with isotype control by Mann-Whitney test (**, P < 0.004; ***, P = 0.0001). (D) Antibody potency curves for WT and Fc mutant antibodies performed and displayed similarly to those in A. (E) IC90 values for the Fc mutant antibodies shown in D. Bars represent mean values of two experiments (shown as open circles). (F) SARS-CoV-2 MA lung titer following antibody prophylaxis with chimeric mAb comprised of the variable domains of C104 grafted onto constant regions of mouse IgG1, IgG2b, and IgGD265A. WT C104– and isotype control antibody–treated groups were controls. Antibodies (8 mg/kg) were delivered intraperitoneally 12 h before infection with 105 PFU of SARS-CoV-2 MA. One independent experiment is shown. All groups are n = 5 mice/group. The line is at the geometric mean, and each symbol represents the titer for a single animal. Asterisks indicate statistical differences compared with isotype control by one-way ANOVA with Dunnett’s multiple comparison test or Mann-Whitney test (*, P = 0.022; ***, P = 0.0001–0.0003; ****, P < 0.0001). LOD, limit of detection; ns, not significant.
Table 3. SARS-CoV-2 MA virus lung titers with single hu-mAb prophylaxis using GRLR Fc mutants and murine Fc region grafts.
| Antibody ID | C002 | C002 GRLR | C104 | C104 GRLR | C110 | C110 GRLR | Isotype control |
|---|---|---|---|---|---|---|---|
| Dose level (mg/kg) | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| Number of animals | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| Minimum (PFU/ml) | 4.5E+01 | 4.5E+01 | 4.5E+01 | 4.5E+01 | 4.5E+01 | 6.5E+02 | 1.0E+05 |
| Maximum (PFU/ml) | 7.5E+03 | 1.1E+04 | 4.5E+01 | 3.0E+03 | 6.0E+03 | 2.5E+04 | 1.1E+06 |
| Range | 7.5E+03 | 1.1E+04 | 0.0E+00 | 3.0E+03 | 6.0E+03 | 2.4E+04 | 1.0E+06 |
| Geometric mean (PFU/ml) | 9.9E+02 | 1.6E+03 | 4.5E+01 | 6.9E+02 | 2.2E+02 | 3.4E+03 | 4.8E+05 |
| Geometric SD factor | 8.6 | 5.9 | 1.0 | 3.2 | 6.9 | 3.0 | 1.8 |
| Antibody ID | C104 IgG1 | C104 IgG2 | C104 D265A | C004 GRLR | C004 | Isotype control | |
| Dose level (mg/kg) | 8 | 8 | 8 | 8 | 8 | 8 | |
| Number of animals | 5 | 5 | 5 | 10 | 10 | 5 | |
| Minimum (PFU/ml) | 4.5E+01 | 4.5E+01 | 1.5E+03 | 4.5E+01 | 4.5E+01 | 1.2E+06 | |
| Maximum (PFU/ml) | 2.0E+04 | 2.0E+02 | 2.5E+04 | 3.0E+03 | 4.5E+01 | 5.5E+06 | |
| Range | 2.0E+04 | 1.6E+02 | 2.4E+04 | 3.0E+03 | 0.0E+00 | 4.3E+06 | |
| Geometric mean (PFU/ml) | 7.5E+02 | 6.2E+01 | 7.4E+03 | 6.9E+02 | 4.5E+01 | 3.4E+06 | |
| Geometric SD factor | 15.4 | 1.9 | 3.8 | 3.2 | 1.0 | 1.8 |
To determine which Fc receptors are responsible for enhanced protective activity, we grafted the variable domains of C104 onto the mouse IgG1 or IgG2a Fc or the mouse IgG1 Fc variant IgGD265A, which is a null-binding mutant for all mouse Fc receptors (Clynes et al., 2000). Mouse subclasses display differential affinity for the activating (FcRI, III, and IV) and the inhibitory (FcRIIB) receptors (Nimmerjahn and Ravetch, 2005). Since activating and inhibitory receptors are coexpressed on most effector cells, the in vivo activity of an IgG Fc is the result of the differential affinity of Fc binding to these receptors. Mouse IgG1 binds preferentially to FcRIIB, an inhibitory receptor, while IgG2a binds primarily FcRIV receptor, an activating receptor. FcRIV is expressed on monocytes/macrophages, neutrophils, and dendritic cells but not on murine natural killer cells (Bournazos et al., 2017). As expected, the three mouse antibody variants (i.e., C104-IgG1, IgG2, or D265A) had neutralization potencies that were not significantly different from each other’s or that of their human IgG1 counterpart in SARS-CoV-2 S-MA pseudovirus assays in vitro (Fig. 3, D and E; and Table 3). Whereas mouse C104-IgG2a provided a similar level of protection as WT human C104 (human IgG1, a human subclass that engages mouse FcRIV), mouse C104-IgG1 and the C104-IgGD265A variant were significantly less active (Fig. 3 F and Table 3). We conclude that activating Fc receptors contributes to optimal protection against SARS-CoV-2 MA in vivo.
Antibody combinations potently neutralize SARS-CoV-2 in vivo
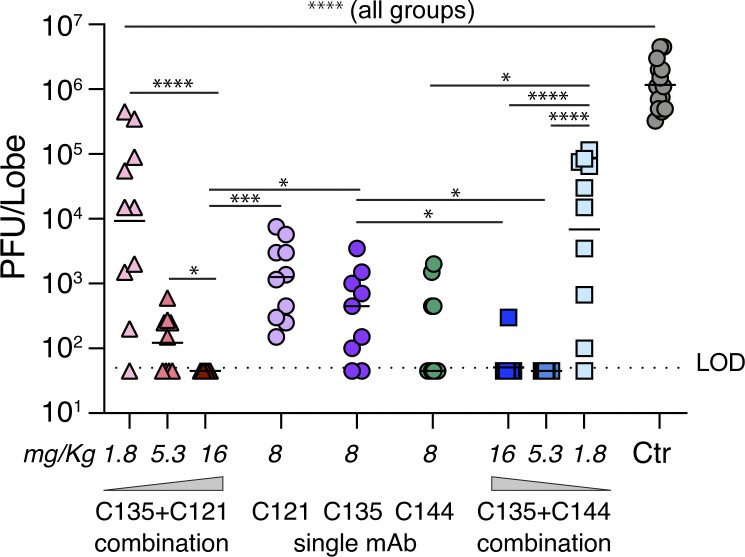
Combinations of antibodies targeting nonoverlapping epitopes have the potential to increase antiviral potency and can prevent the emergence of antibody escape mutations (Hansen et al., 2020; Weisblum et al., 2020 Preprint). To test whether there might be a benefit of antibody combinations for in vivo protection, we tested mixtures of antibodies that target nonoverlapping epitopes on the RBD (C135-LS/C121-LS and C135-LS/C144-LS; Robbiani et al., 2020). The antibody mixtures were tested at combined total doses of 16, or 5.3, or 1.8 mg/kg compared with 8 mg/kg of each antibody alone or 16 mg/kg of isotype control. As expected, the isotype control antibody failed to reduce virus replication (Fig. 4). While 16 mg/kg of C135/C121 provided sterilizing protection (i.e., viral titers below the limit of detection) from SARS-CoV-2 MA replication in all mice tested (n = 15), C135/C144 performed nearly as well with only 1 of 15 mice tested having a low but measurable virus titer (Fig. 4 and Table 4). Whereas C135/C144 was sterilizing at 5.3 mg/kg, half of the C135/C121 mice had low but detectable viral lung titers. Notably, the 1.8-mg/kg dose of either combination significantly reduced titers compared with isotype control antibody but was not as effectivel as 5.3 or 16 mg/kg (Fig. 4 and Table 4). In addition, the antibody combination of C135/C144 at 5.3 mg/kg improved the levels of protection achieved from C135 alone at 8 mg/kg (Fig. 4).
Figure 4.
Antibody combinations increase in vivo potency. SARS-CoV-2 MA lung titer following antibody prophylaxis with isotype control (16 mg/kg) or combinations of C135-LS + C121-LS or C135-LS + C144-LS mixed at a ratio of 1:1 for combined dose levels of 16, 5.3, or 1.8 mg/kg. Antibodies were delivered intraperitoneally 12 h before infection with 105 PFU of SARS-CoV-2 MA. Single antibodies C121-LS, C135-LS, or C144-LS were similarly administered at 8 mg/kg (n = 8/antibody). Combined data from two independent experiments are shown. For 16-mg/kg groups, n = 14 or 15 mice, and all other groups were 9 or 10 mice/group. The line is at the geometric mean, and each symbol represents the titer for a single animal. Asterisks indicate statistical differences compared with isotype control by one-way ANOVA with a Dunnett’s multiple comparison test (****, P < 0.0001) or Kruskal-Wallis test with a Tukey’s multiple comparison test (*, P = 0.02–0.04; ***, P = 0.0002; ****, P < 0.0001). LOD, limit of detection.
Table 4. Comparison of SARS-CoV-2 MA virus lung titers with either single hu-mAb or hu-mAb combination prophylaxis.
| Antibody ID | C121-LS | C135-LS | C144-LS | C135-LS+C121-LS | C135-LS+C144-LS | Isotype control | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dose level (mg/kg) | 8 | 8 | 8 | 16 | 5.3 | 1.8 | 16 | 5.3 | 1.8 | 16 | |
| Number of animals | 10 | 9 | 10 | 15 | 9 | 10 | 15 | 10 | 10 | 14 | |
| Minimum (PFU/ml) | 1.5E+02 | 4.5E+01 | 4.5E+01 | 4.5E+01 | 4.5E+01 | 4.5E+01 | 4.5E+01 | 4.5E+01 | 4.5E+01 | 3.3E+05 | |
| Maximum (PFU/ml) | 7.5E+03 | 3.5E+03 | 2.0E+03 | 4.5E+01 | 6.0E+02 | 4.5E+05 | 3.0E+02 | 4.5E+01 | 1.2E+05 | 4.5E+06 | |
| Range | 7.4E+03 | 3.5E+03 | 2.0E+03 | 0.0E+00 | 5.6E+02 | 4.5E+05 | 2.6E+02 | 0.0E+00 | 1.1E+05 | 4.2E+06 | |
| Geometric mean (PFU/ml) | 1.1E+03 | 3.3E+02 | 1.5E+02 | 4.5E+01 | 1.2E+02 | 9.3E+03 | 5.1E+01 | 4.5E+01 | 6.9E+03 | 1.2E+06 | |
| Geometric SD factor | 4.0 | 4.8 | 5.0 | 1.0 | 2.8 | 21.9 | 1.6 | 1.0 | 18.6 | 2.4 | |
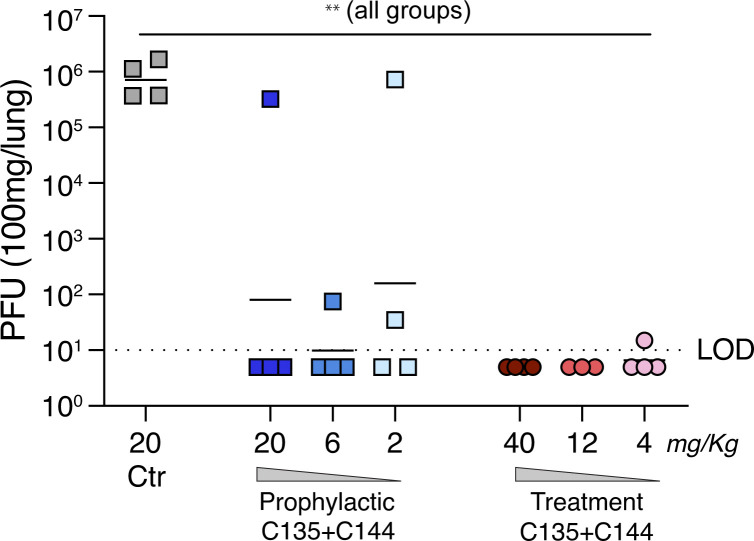
To determine whether the C135/C144 antibody combination is effective against authentic SARS-CoV-2 in vivo, we performed prophylactic and therapeutic efficacy studies in Syrian hamsters. The antibody combination was administered 24 h before infection (i.e., prophylactic) or 12 h after infection (i.e., therapeutic). As a negative control, one group received antibody directed against HIV (10–1074) prophylactically. Viral loads in the lungs were determined 3 d after infection, the peak of replication in this model. In hamsters receiving control antibody, high levels of SARS-CoV-2 replication were observed in the lung tissue (9 × 105 PFU/100 g tissue; Fig. 5). Prophylactic administration of the C135/C144 antibody combination prevented or significantly diminished SARS–CoV-2 replication at all antibody doses tested, including 2 mg/kg (Fig. 5). Similarly, early therapeutic administration of C135/C144 reduced lung viral loads below the limit of detection at the 40- and 12-mg/kg dose levels and decreased viral lung loads >10,000-fold with the lowest dose tested (4 mg/kg; Fig. 5). We conclude that relatively low doses of C135/C144 are effective in neutralizing SARS-CoV-2 both prophylactically and therapeutically.
Figure 5.
Prevention and therapy with C135-LS + C144-LS antibody combination in Syrian hamsters. Hamsters were infected intranasally with 2.6 × 104 PFU SARS-CoV-2, and viral lung titers 3 d after infection were determined by plaque assay on Vero-E6 cells. Antibodies were administered either 24 h before (prophylactic) or 12 h after (treatment) infection. The line is at the geometric mean, and each symbol represents the titer for a single animal. Dotted line indicates the limit of detection. Asterisks indicate statistical differences compared with isotype control by one-way ANOVA with a Dunnett’s multiple comparison test (**, P < 0.01). LOD, limit of detection.
Discussion
To date, there have been >42 million cases and >1 million deaths attributed to COVID-19 globally, 20% of which have occurred in the United States (Johns Hopkins University, 2020). In addition to the many vaccines and drugs now being tested, passive transfer of potent hu-mAbs holds great promise for COVID-19 prevention and therapy. Neutralizing hu-mAbs targeting SARS-CoV-2 promote reduction in viral load and prevent infection in macaques and hamsters (Hansen et al., 2020; Rogers et al., 2020; Ju et al., 2020). Here, we examined the relationship between antibody neutralizing potency in in vitro assays and protection in two small animal models of SARS-CoV-2 infection (Dinnon et al., 2020). The results suggest that engagement of activating Fc receptors can enhance the efficacy of anti–SARS-CoV-2 antibodies in vivo. Moreover, some antibody combinations can be effective for prevention and early therapy for SARS-CoV-2 even at relatively low doses.
The neutralizing activity of antibodies to SARS-CoV-2 has primarily been tested in vitro using pseudotype viruses and microneutralization assays (Schmidt et al., 2020). How these in vitro results translate to in vivo protective activity had not been determined. Our results indicate that the relationship between neutralizing activity against SARS-CoV-2 MA in vitro and antiviral activity in vivo is not linear. Antibody half-life and bioavailability are important variables that can account for this difference. An additional, potentially important factor is the role of leukocyte Fc receptors on viral clearance and infected cell killing in vivo (Bournazos et al., 2020). Indeed, Fc receptors play an important role in viral clearance during HIV-1, Ebola virus, and influenza virus infections (Lu et al., 2016; Halper-Stromberg et al., 2014; DiLillo et al., 2014). Our experiments indicate that Fc receptors also contribute to optimal antibody-mediated protection against SARS-CoV-2. Among this family of receptors, activating Fc receptors on macrophages, neutrophils, and dendritic cells are critical for enhanced antibody protection against SARS-CoV-2 MA (Mercado et al., 2020). Thus far, the evidence from both SARS-CoV-2 vaccine and hu-mAb studies does not support the notion that antibody-dependent enhancement (ADE) of infection driven by Fc-receptor engagement occurs with SARS-CoV-2 as is observed for a feline CoV (feline infectious peritonitis virus; Pedersen, 2014) or flavivirus such as the dengue virus (Halstead and Katzelnick, 2020; Laczkó et al., 2020). In addition, exacerbation of dengue virus infection by ADE is driven by antibody-dependent increases in infection frequency of antigen-presenting cells (monocytes, macrophages, dendritic cells, etc.) normally targeted by dengue virus, while the major cellular targets of SARS-CoV-2 are respiratory epithelial cells (Wang et al., 2017). Thus, there is currently little evidence to suggest that passively transferred immunity via hu-mAb therapy will initiate immune pathologies such as ADE.
An additional nonmutually exclusive explanation for the disparity between the in vivo antiviral activity and the in vitro neutralization results relates to heterogeneity in the way neutralizing antibodies target the SARS-CoV-2 RBD (Barnes et al., 2020a Preprint; Barnes et al., 2020b). Antibodies can neutralize by binding to the ACE2 interaction surface, thereby blocking RBD interaction with its cellular receptor directly or indirectly. Even among the antibodies that directly block the interaction between the RBD and ACE2, there is significant heterogeneity in terms of their angles of approach to binding. Among neutralizing antibodies that target the RBD, C144 belongs to a particularly potent class whose mechanism of action involves blocking RBD–ACE-2 interaction and additionally locking the RBD in a closed configuration, making it inaccessible to ACE2 (Barnes et al., 2020a Preprint; Barnes et al., 2020b). C002, C104, and C105 also bind to the ACE2 interacting surface of the RBD, but they approach the RBD from different angles and bind by different mechanisms (Barnes et al., 2020a Preprint; Barnes et al., 2020b). C104 is the least potent neutralizer of the three antibodies in vitro but the most effective against SARS-CoV-2 MA in vivo. Among other factors, the angle of approach of an antibody to the RBD may alter its in vivo potency by influencing accessibility of the Fc domain to its receptor on effector cells, as suggested for neutralizing antibodies to influenza (DiLillo et al., 2016).
In vitro experiments with chimeric vesicular stomatitis virus (VSV)–SARS-CoV-2 viruses indicate that antibodies can select for escape mutants and that combinations of antibodies targeting nonoverlapping sites can prevent the emergence of resistant variants (Hansen et al., 2020; Weisblum et al., 2020 Preprint). Antibody combinations also have the potential to act synergistically, but there is little evidence for synergy in vitro (Robbiani et al., 2020; Liu et al., 2020; Rogers et al., 2020; Zost et al., 2020b). Nevertheless, we showed that some combinations of antibodies outperform single antibodies in neutralizing SARS-CoV-2 MA in mice. Antibody combinations that target nonoverlapping epitopes may be especially promising for clinical development because they can be dose sparing and also prevent selection of resistant variants.
In summary, the data support the idea that specific combinations of antibodies with intact Fc-effector function should be developed for optimal protection and therapy against SARS-CoV-2.
Material and methods
Cells and viruses
Human Ace2-expressing HT1080 cells (HT1080Ace2cl.14) were described previously (Schmidt et al., 2020). For constitutive expression of murine Ace2 (muAce2) in HT1080 cells, a cDNA-encoding muAce2 was inserted into a lentiviral vector CSIB 3′ to the SFFV promoter. HT1080muAce2 cells were generated by transduction with CSIB-based virus followed by selection with 5 µg/ml blasticidin.
Cells were cultured in DMEM supplemented with 10% FCS at 37°C and 5% CO2. Medium for Ace2-overexpressing cell lines contained 5 µg/ml blasticidin. All cell lines had tested negative for contamination with mycoplasma, and parental cell lines were obtained from the American Type Culture Collection. Recombinant SARS-CoV-2 MA virus was generated as described previously (GenBank accession no. MT844088; Dinnon et al., 2020). For virus titration by plaque assay, the caudal lobe of the right lung was homogenized in PBS, and the resulting homogenate was serial-diluted and inoculated onto confluent monolayers of Vero E6 cells, followed by agarose overlay. Plaques were visualized with overlay of neutral red dye on day 2 after infection (Dinnon et al., 2020).
Antibody production
All antibodies were produced as previously described (Mouquet et al., 2011). Briefly, 293-6E cells that were maintained in Freestyle 293 Expression Medium (Thermo Fisher Scientific) were transiently transfected by using equal amounts of Ig heavy and light chain expression vectors using branched polyethylenimine (Sigma). 7 d after transfection, cells were spun down at 4,200 g for 40 min at 4°C, and supernatants were filtered through a 0.22-µM filter (Millipore). Antibodies were purified from filtered supernatants using Protein G Sepharose 4 Fast Flow (Cytiva). Antibodies were buffer exchanged and concentrated into PBS using an Amicon Ultra centrifugal filter (Millipore).
SARS-CoV-2/SARS-CoV-2 MA pseudotyped reporter virus
SARS-CoV-2 pseudotyped particles were produced by cotransfection of pNL4-3ΔEnv-nanoluc and pSARS-CoV-2-MA-Strunc in 293T cells (Schmidt et al., 2020; Robbiani et al., 2020). For generation of SARS-CoV-2 MA pseudotyped particles, a plasmid expressing the mouse-adapted SARS-CoV-2 S (pSARS-CoV-2-MA-Strunc) was generated by introducing the Q498Y/P499T mutation into pSARS-CoV2-Strunc and was used for cotransfection with pNL4-3ΔEnv-nanoluc.
SARS-CoV-2 MA pseudotyped virus neutralization assay
A 40-µg/ml initial dilution of mAbs was fourfold serially diluted over 11 dilutions. 55 µl of antibody dilutions was incubated with a 55-µl aliquot of SARS-CoV-2 MA pseudotyped virus (containing ∼103 infectious units) for 1 h at 37°C. 100 µl of the mixture was subsequently incubated with HT1080muAce2 cells for 48 h. Consequently, the resulting antibody starting dilution was 10 µg/ml, and each well received 50 µl virus. Following incubation, cells were washed with PBS and lysed with Luciferase Cell Culture 5× reagent (Promega), and nanoluc luciferase activity in cell lysates was measured using the Nano-Glo Luciferase Assay System (Promega). RLUs obtained were normalized to those derived from cells infected with SARS-CoV-2 MA pseudovirus in the absence of mAbs (equivalent to 0% neutralization). The half-maximal and 90% ICs (IC50 and IC90) for mAbs were determined using four-parameter nonlinear regression (GraphPad Prism). In detail, we applied the least squares regression method without weighting, with top and bottom values constrained to 0 (% neutralization) and 100 (% neutralization), respectively, while allowing for a variable HillSlope.
Mouse studies and in vivo infections
All mouse studies were performed at the University of North Carolina (Animal Welfare Assurance #A3410-01) using protocols (19–168, 20–114) approved by the University of North Carolina Institutional Animal Care and Use Committee. All animal work was approved by the Institutional Animal Care and Use Committee at University of North Carolina at Chapel Hill according to guidelines outlined by the Association for the Assessment and Accreditation of Laboratory Animal Care and the US Department of Agriculture. All work was performed with approved standard operating procedures and safety conditions for SARS-CoV-2. Our institutional BSL3 facilities are designed to conform to the safety requirements recommended by Biosafety in Microbiological and Biomedical Laboratories, the US Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, and the National Institutes of Health. Laboratory safety plans have been submitted, and the facility has been approved for use by the University of North Carolina Department of Environmental Health and Safety and the Centers for Disease Control and Prevention. 12-mo-old female BALB/c mice (Envigo; #047) were inoculated with the indicated concentration of antibody intraperitoneally 12 h before infection. For infection, mice were anesthetized with a mixture of ketamine/xylazine and infected with 105 PFU of SARS-CoV-2 MA in 50 µl PBS intranasally. Mice were monitored daily for body weight changes. At 2 d after infection, mice were euthanized, and lung tissue was harvested for viral titer analysis. Samples were stored at −80°C until homogenized and titered by plaque assay as described above.
Hamster studies and in vivo infections
8-wk-old Syrian hamsters obtained from Envigo were housed four per cage under ABSL3 containment. Groups of hamsters were treated with varying doses of a combination of two antibodies to SARS-CoV-2 RBD, an irrelevant (10–1074; anti-HIV) antibody, or PBS. Antibodies were administered by the intraperitoneal route either 24 h before or 12 h after challenge. Virus challenge was conducted under ketamine-xylazine anesthesia by intranasal instillation of 100 μl of SARS-CoV-2 that had been passaged once in Vero and once in Vero E6 cells; the dose determined by back-titration of the inoculum was 2.6e4 PFU. Beginning 2 d before challenge and extending until euthanasia, hamsters were evaluated clinically and weighed and their temperature recorded once daily. An equal number of hamsters from each treatment group was euthanized and necropsied 3 d after challenge. For animals euthanized on day 3, a sample of ∼100 mg of right cranial and right caudal lung lobes was excised, immersed in 0.9 ml of BA1/FBS, and homogenized using a mixer mill with stainless steel balls. Tissue homogenates were frozen to −80°C until assay. Virus titrations were performed using a double-overlay plaque assay on Vero E6 cells in 6-well plates. Briefly, serial 10-fold dilutions of tissue homogenate samples were inoculated onto drained monolayers and incubated 45 min, and 2 ml of a first overlay (0.5% agarose in MEM) without neutral red was added to each well. 1 d later, a second 2-ml overlay containing 0.06 mg/ml neutral red was added to each well, and plaques were counted 1 and 2 d later.
Acknowledgments
We thank Que Dang and Jean Patterson for suggestions on experiments with Syrian hamsters.
These studies were supported by George Mason University Fast Grants to T.P. Sheahan and M.C. Nussenzweig and National Institute of Allergy and Infectious Diseases grants to R.S. Baric (1U19AI142759; Antiviral Drug Discovery and Development Center), M.C. Nussenzweig (P01-AI138398-S1, 2U1 9AI111825), S. Bournazos (R01AI137276), and J.V. Ravetch (U19AI111825). M.C. Nussenzweig and P.D. Bieniasz are Howard Hughes Medical Institute Investigators. The Rockefeller University has utilized the nonclinical and preclinical services program offered by the National Institute of Allergy and Infectious Diseases. This project was supported in part by the North Carolina Policy Collaboratory at University of North Carolina at Chapel Hill with funding from the North Carolina Coronavirus Relief Fund established and appropriated by the North Carolina General Assembly.
Author contributions: A. Schäfer, F. Muecksch, J.C.C. Lorenzi, S.R. Leist, M. Cipolla, S. Bournazos, F. Schmidt, R.M. Maison, A. Gazumyan, D.R. Martinez, R.S. Baric, D.F. Robbiani, T. Hatziioannou, J.V. Ravetch, P.D. Bieniasz, R.A. Bowen, M.C. Nussenzweig, and T.P. Sheahan designed and/or executed the studies described herein. A. Schäfer, F. Muecksch, J.C.C. Lorenzi, S.R. Leist, S. Bournazos, J.V. Ravetch, T. Hatziioannou, P.D. Bieniasz, R.A. Bowen, M.C. Nussenzweig, and T.P. Sheahan wrote the manuscript.
References
- Barnes, C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. 2020a. Structural classification of neutralizing antibodies against the SARS-CoV-2 spike receptor-binding domain suggests vaccine and therapeutic strategies. bioRxiv. 10.1101/2020.08.30.273920 (Preprint posted August 30, 2020) [DOI]
- Barnes, C.O., West A.P. Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. 2020b. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 182:828–842.e16. 10.1016/j.cell.2020.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. 2020. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 369:1014–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos, S., Klein F., Pietzsch J., Seaman M.S., Nussenzweig M.C., and Ravetch J.V.. 2014. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 158:1243–1253. 10.1016/j.cell.2014.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos, S., Wang T.T., Dahan R., Maamary J., and Ravetch J.V.. 2017. Signaling by Antibodies: Recent Progress. Annu. Rev. Immunol. 35:285–311. 10.1146/annurev-immunol-051116-052433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos, S., DiLillo D.J., Goff A.J., Glass P.J., and Ravetch J.V.. 2019. Differential requirements for FcγR engagement by protective antibodies against Ebola virus. Proc. Natl. Acad. Sci. USA. 116:20054–20062. 10.1073/pnas.1911842116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos, S., Gupta A., and Ravetch J.V.. 2020. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 20:633–643. 10.1038/s41577-020-00410-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. 2020. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 182:73–84.e16. 10.1016/j.cell.2020.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynes, R.A., Towers T.L., Presta L.G., and Ravetch J.V.. 2000. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat. Med. 6:443–446. 10.1038/74704 [DOI] [PubMed] [Google Scholar]
- Corbett, K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., et al. 2020. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 586:567–571. 10.1038/s41586-020-2622-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit, E., van Doremalen N., Falzarano D., and Munster V.J.. 2016. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 14:523–534. 10.1038/nrmicro.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo, D.J., Tan G.S., Palese P., and Ravetch J.V.. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 20:143–151. 10.1038/nm.3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo, D.J., Palese P., Wilson P.C., and Ravetch J.V.. 2016. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Invest. 126:605–610. 10.1172/JCI84428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon, K.H. III, Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L. Jr., Hou Y.J., Adams L.E., et al. 2020. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 586:560–566. 10.1038/s41586-020-2708-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halper-Stromberg, A., Lu C.L., Klein F., Horwitz J.A., Bournazos S., Nogueira L., Eisenreich T.R., Liu C., Gazumyan A., Schaefer U., et al. 2014. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell. 158:989–999. 10.1016/j.cell.2014.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead, S.B., and Katzelnick L.. 2020. COVID 19 Vaccines: Should we fear ADE? J. Infect. Dis.:jiaa518. 10.1093/infdis/jiaa518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. 2020. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 369:1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns Hopkins University . 2020. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University. https://coronavirus.jhu.edu/map.html. (Accessed August 28, 2020)
- Ju, B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. 2020. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 584:115–119. 10.1038/s41586-020-2380-z [DOI] [PubMed] [Google Scholar]
- Laczkó, D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaño D., Amanat F., Muramatsu H., Oguin T.H. III, et al. 2020. A Single Immunization with Nucleoside-Modified mRNA Vaccines Elicits Strong Cellular and Humoral Immune Responses against SARS-CoV-2 in Mice. Immunity. 53:724–732.e7. 10.1016/j.immuni.2020.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko, M., Marzi A., and Munster V.. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5:562–569. 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Schäfer A., Kulkarni S.S., Liu X., Martinez D.R., Chen C., Sun Z., Leist S.R., Drelich A., Zhang L., et al. 2020. High potency of a bivalent human VH domain in SARS-CoV-2 animal models. Cell. 183:429–441.e16: E16. 10.1016/j.cell.2020.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. 2020. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 584:450–456. 10.1038/s41586-020-2571-7 [DOI] [PubMed] [Google Scholar]
- Lu, C.L., Murakowski D.K., Bournazos S., Schoofs T., Sarkar D., Halper-Stromberg A., Horwitz J.A., Nogueira L., Golijanin J., Gazumyan A., et al. 2016. Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science. 352:1001–1004. 10.1126/science.aaf1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado, N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., et al. 2020. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 586:583–588. 10.1038/s41586-020-2607-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet, H., Klein F., Scheid J.F., Warncke M., Pietzsch J., Oliveira T.Y., Velinzon K., Seaman M.S., and Nussenzweig M.C.. 2011. Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PLoS One. 6:e24078. 10.1371/journal.pone.0024078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn, F., and Ravetch J.V.. 2005. Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science. 310:1510–1512. 10.1126/science.1118948 [DOI] [PubMed] [Google Scholar]
- Pedersen, N.C. 2014. An update on feline infectious peritonitis: virology and immunopathogenesis. Vet. J. 201:123–132. 10.1016/j.tvjl.2014.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. 2020. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 583:290–295. 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- Robbiani, D.F., Bozzacco L., Keeffe J.R., Khouri R., Olsen P.C., Gazumyan A., Schaefer-Babajew D., Avila-Rios S., Nogueira L., Patel R., et al. 2017. Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico. Cell. 169:597–609.e11. 10.1016/j.cell.2017.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani, D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. 2020. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 584:437–442. 10.1038/s41586-020-2456-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. 2020. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 369:956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, F., Weisblum Y., Muecksch F., Hoffmann H.H., Michailidis E., Lorenzi J.C.C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C., et al. 2020. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 217:e20201181. 10.1084/jem.20201181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. 2020. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 584:120–124. 10.1038/s41586-020-2381-y [DOI] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration . 2020. Coronavirus (COVID-19) Update: FDA Issues Emergency Use Authorization for Potential COVID-19 Treatment. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-potential-covid-19-treatment (accessed August 28, 2020).
- Walls, A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., and Veesler D.. 2020. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 181:281–292.e6. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T.T., Sewatanon J., Memoli M.J., Wrammert J., Bournazos S., Bhaumik S.K., Pinsky B.A., Chokephaibulkit K., Onlamoon N., Pattanapanyasat K., et al. 2017. IgG antibodies to dengue enhanced for FcγRIIIA binding determine disease severity. Science. 355:395–398. 10.1126/science.aai8128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B., Asarnow D., Lee W.H., Huang C.W., Faust B., Ng P.M.L., Ngoh E.Z.X., Bohn M., Bulkley D., Pizzorno A., et al. 2020. Bivalent binding of a fully human IgG to the SARS-CoV-2 spike proteins reveals mechanisms of potent neutralization. bioRxiv. 10.1101/2020.07.14.203414 (Preprint posted July 15, 2020) [DOI]
- Weisblum, Y., Schmidt F., Zhang F., Dasilva J., Poston D., Lorenzi J.C.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., et al. 2020. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. bioRxiv. 10.1101/2020.07.21.214759 (Preprint posted July 22, 2020) [DOI] [PMC free article] [PubMed]
- Zalevsky, J., Chamberlain A.K., Horton H.M., Karki S., Leung I.W., Sproule T.J., Lazar G.A., Roopenian D.C., and Desjarlais J.R.. 2010. Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 28:157–159. 10.1038/nbt.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579:270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost, S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. 2020a. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 584:443–449. 10.1038/s41586-020-2548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost, S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. 2020b. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 26:1422–1427. 10.1038/s41591-020-0998-x [DOI] [PMC free article] [PubMed] [Google Scholar]