Abstract
Human coronaviruses (HCoVs) were first described in the 1960s for patients with the common cold. Since then, more HCoVs have been discovered, including those that cause severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), two pathogens that, upon infection, can cause fatal respiratory disease in humans. It was recently discovered that dromedary camels in Saudi Arabia harbor three different HCoV species, including a dominant MERS HCoV lineage that was responsible for the outbreaks in the Middle East and South Korea during 2015. In this review we aim to compare and contrast the different HCoVs with regard to epidemiology and pathogenesis, in addition to the virus evolution and recombination events which have, on occasion, resulted in outbreaks amongst humans.
Keywords: coronavirus, MERS, SARS, evolution, recombination, pathogenesis
Trends
Six coronaviruses (CoVs) are known to infect humans: 229E, OC43, SARS-CoV, NL63, HKU1, and MERS-CoV.
Many CoVs are simultaneously maintained in nature, allowing for genetic recombination, resulting in novel viruses.
Recombination of CoV in camels has resulted in a dominant MERS lineage that caused human outbreaks in 2015.
A Brief Introduction to Human and Animal Coronaviruses
Coronaviruses (CoVs), of the family Coronaviridae, are enveloped viruses with a single-strand, positive-sense RNA genome approximately 26–32 kilobases in size, which is the largest known genome for an RNA virus [1]. The term ‘coronavirus’ refers to the appearance of CoV virions when observed under electron microscopy, in which spike projections from the virus membrane give the semblance of a crown, or corona in Latin 2, 3. All coronaviruses share similarities in the organization and expression of their genome, in which 16 nonstructural proteins (nsp1 through nsp16), encoded by open reading frame (ORF) 1a/b at the 5′ end, are followed by the structural proteins spike (S), envelope (E), membrane (M), and nucleocapsid (N), which are encoded by other ORFs at the 3′ end. CoVs are separated into four genera based on phylogeny: alpha-CoV (group 1), beta-CoV (group 2), gamma-CoV (group 3) and delta-CoV (group 4) (http://ictvonline.org/proposals/2008.085-122 V.v4.Coronaviridae.pdf). Within the beta-CoV genus, four lineages (A, B, C, and D) are recognized. Distinct from other beta-CoV lineages, lineage A viruses also encode a smaller protein called hemagglutinin esterase (HE), which is functionally similar to the S protein [4].
Since the early 1970s, a variety of pathological conditions in domestic animals have been attributed to CoV infections [5]. With the exception of infectious bronchitis virus (IBV), which causes avian infectious bronchitis in chickens [6], canine respiratory coronavirus (CRCoV), which causes respiratory disease in dogs [7], and mouse hepatitis virus (MHV), which can cause a progressive demyelinating encephalitis in mice 3, 8, other CoV infections typically result in gastrointestinal symptoms. For instance, transmissible gastroenteritis coronavirus (TGEV) 9, 10, bovine coronavirus (BCV) [11], feline coronavirus (FCoV) [12], canine coronavirus (CCoV) [13], and turkey coronavirus (TCV) [14] are known to cause enteritis in their respective hosts [1].
In humans, CoV infections primarily involve the upper respiratory tract and the gastrointestinal tract, and vary from mild, self-limiting disease, such as the common cold, to more severe manifestations, such as bronchitis and pneumonia with renal involvement [15]. The first human coronavirus (HCoV) was isolated during 1965 from the nasal discharge of patients with the common cold and termed B814 [16]. Currently, six different CoV strains are known to infect humans. These include: HCoV-229E (229E), HCoV-OC43 (OC43), severe acute respiratory syndrome coronavirus (SARS-CoV), HCoV-NL63 (NL63), HCoV-HKU1 (HKU1), and Middle East respiratory syndrome coronavirus (MERS-CoV) [17]. 229E and OC43 are the prototype viruses from the two main HCoV lineages (Alpha and Beta, respectively) that cause 15–29% of all common colds, and are the best characterized [18]. SARS-CoV is the aetiological agent that was behind an outbreak of severe respiratory disease through China during 2002–2003 [19], and MERS-CoV is the pathogen responsible for an ongoing outbreak of severe respiratory disease centered in the Middle East since 2012 [20]. In this review, we first compare and contrast the epidemiology and pathogenesis of the known CoVs infecting humans, in which CoVs that cause mild to severe disease in humans are newly emerged from a zoonotic source. Second, we describe the ecology of CoVs, and highlight evidence for viral recombination between the different CoVs within animal populations, which may result in the generation of novel CoVs that are transmissible and lethal to humans. Finally, we summarize the phylogeny, evolution, and genetic recombination of CoVs in detail.
Epidemiology and Pathogenesis of HCoV
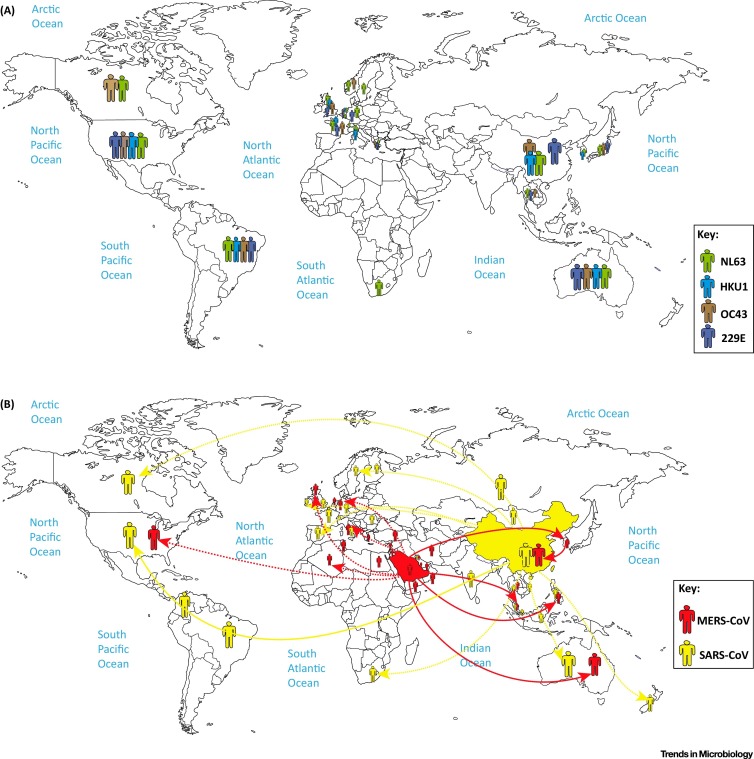
The epidemiology and pathogenesis of each HCoV are discussed within the following sections and summarized in Table 1 and Figure 1 , respectively. In general, 229E, OC43, and NL63 are distributed globally (Figure 1A) and tend to be transmitted predominantly during the winter season in temperate-climate countries [21], while NL63 showed a spring–summer peak of activity from a study in Hong Kong [22].
Table 1.
Comparison of Clinical Symptoms, Incubation Time, Median Time to Death, and Case Fatality Rates of Human Coronaviruses
| HCoV | Clinical Symptoms | Case Fatality Rate | Incubation Period | Median Time to Death | Refs |
|---|---|---|---|---|---|
| 229E | General malaise Headache Nasal discharge Sneezing Sore throat Fever and cough (10–20% of patients) |
N/Aa | 2–5 days | — | 18, 81, 82 |
| OC43 | General malaise Headache Nasal discharge Sneezing Sore throat Fever and cough (10–20% of patients) |
N/A | 2–5 days | — | 18, 82, 83, 84 |
| SARS-CoV | Fever Myalgia Headache Malaise Chills Nonproductive cough Dyspnea Respiratory distress Diarrhea (30–40% of patients) |
9% | 2–11 days | 23 days | 19, 42, 85, 86, 87, 88 |
| NL63 | Cough, Rhinorrhea Tachypnea Fever Hypoxia Obstructive laryngitis (croup) |
N/A | 2–4 days | — | 89, 90, 91, 92 |
| HKU1 | Fever Running nose Cough Dyspnea |
N/A | 2–4 days | — | 2, 32, 33, 34, 93, 94 |
| MERS-CoV | Fever Cough Chills Sore throat Myalgia Arthralgia Dyspnea Pneumonia Diarrhea and vomiting (one-third of patients) Acute renal impairment |
36% | 2–13 days | 14 days | 41, 42, 43, 95, 96 |
N/A, not applicable.
Figure 1.
Global Distribution of Human Coronaviruses. (A) Green, blue, brown, and purple represent the global distribution of the NL63, HKU1, OC43, and 229E human coronaviruses, respectively. (B) Red and yellow represent the global distribution of MERS-CoV and SARS-CoV, respectively.
Human Coronavirus 229E (alphacoronavirus)
Strain 229E was discovered during 1966, when researchers were characterizing five novel agents that were isolated from the respiratory tract of humans who had contracted the common cold [23]. 229E was adapted to grow in WI-38 lung fibroblast cell lines, and was later shown to be morphologically identical to IBV and MHV [24]. Symptoms of 229E infection include general malaise, headache, nasal discharge, sneezing, and a sore throat [25]. A small portion of patients (10–20%) will also exhibit fever and cough. The incubation time is approximately 2–5 days, followed by illness lasting between 2 and 18 days, and clinically indistinguishable from respiratory tract infections caused by other pathogens, such as rhinovirus and influenza A [18]. 229E is distributed globally (Figure 1A).
Human Coronavirus OC43 (betacoronavirus, lineage A)
Strain OC43 was discovered, during 1967, in the nasopharyngeal wash of a patient with the common cold, and is adapted to grow in organ cultures containing suckling mouse brains. Similar to 229E, OC43 is morphologically indistinguishable from IBV and MHV [24], and patients infected with OC43 present the same clinical symptoms as that of 229E [24]. However, there is no serological cross-reactivity between 229E and OC43 [24]. OC43 is also distributed globally (Figure 1A).
SARS-CoV (betacoronavirus, lineage B)
Patients infected with SARS-CoV initially present with fever, myalgia, headache, malaise, and chills, followed by a nonproductive cough, dyspnea, and respiratory distress generally 5 to 7 days later, which may result in death (http://www.who.int/csr/sars/en/WHOconsensus.pdf?ua=1). Other notable features in some cases include infection of the gastrointestinal tract, liver, kidney, and brain. Diffuse alveolar damage, epithelial cell proliferation, and an increase in macrophages is seen in SARS-CoV infection of the lung. Lymphopenia, hemophagocytosis in the lung, in addition to white-pulp atrophy of the spleen observed in SARS patients, are similar to fatal H5N1 influenza virus infections [1]. Diarrhea is observed in approximately 30–40% of SARS infections (http://www.cdc.gov/sars/about/faq.html).
An outbreak of disease caused by SARS-CoV, originating from Guangdong Province in southern China during November 2002, eventually spread to other countries in Asia, in addition to North America and Europe (37 countries/regions in total) over 9 months (http://www.who.int/ith/diseases/sars/en/) (Figure 1B). An eventual 8273 cases were reported, with 775 deaths for a case fatality rate (CFR) of 9%, and the majority of cases and deaths occurred in mainland China and in Hong Kong [19]. The elderly were more susceptible to SARS disease, with a mortality rate of over 50% (http://www.cdc.gov/sars/surveillance/absence.html).
Human Coronavirus NL63 (alphacoronavirus)
NL63 is primarily associated with young children, the elderly, and immunocompromised patients with respiratory illnesses [26]. NL63 was a novel HCoV isolated from a 7-month-old child with coryza, conjunctivitis, fever, and bronchiolitis in the Netherlands during late 2004 [26], and an independent investigation described the isolation of virtually the same virus from a nasal sample collected from an 8-month-old boy suffering from pneumonia in the Netherlands [27]. NL63 was also detected in New Haven, USA, during 2005 amongst 79 of 895 children, and was initially called HCoV-NH (NH), but genetic and phylogenetic analyses showed that NH and HKU1 likely are the same species 26, 28. Infections with NL63 typically result in mild respiratory disease similar to the common cold, characterized by cough, rhinorrhea, tachypnea, fever, and hypoxia, and resolve on their own [29]. Obstructive laryngitis, also known as croup, is frequently observed with NL63 infections. A study estimated that NL63 accounts for an estimated 4.7% of common respiratory diseases [26]. NL63 is distributed globally (Figure 1A).
Human Coronavirus HKU1 (betacoronavirus, lineage A)
HKU1 was first discovered during January 2005 in a 71-year-old patient from Hong Kong who had been hospitalized with pneumonia and bronchiolitis [30]. The symptoms of HKU1 respiratory tract infections are not able to be separated from those caused by other respiratory viruses. Most patients present with fever, running nose, and cough for infections in the upper respiratory tract, whereas a fever, productive cough, and dyspnea are the common symptoms presenting for infections in the lower respiratory tract [31]. Most HKU1 infections are self-limiting, with only two deaths reported in patients with pneumonia due to HKU1 [32]. Although HKU1 is clinically relatively mild in children, HKU1 infection is associated with a high incidence of seizures, and was found in a patient with meningitis 33, 34. HKU1 cases outside Asia were detected in New Haven, USA, in 2 out of 851 children [35], and also in Australia [36], France [37], and Brazil [38], indicating a global distribution for the virus (Figure 1A).
MERS-CoV (betacoronavirus, lineage C)
MERS-CoV was first isolated from the lungs of a 60-year-old patient who had died from a severe respiratory illness in Jeddah, Saudi Arabia, 2012 [39]. Clinical manifestations of MERS-CoV infection range from asymptomatic to severe pneumonia with acute respiratory distress, septic shock, and renal failure resulting in death [40]. A typical disease course begins with fever, cough, chills, sore throat, myalgia, and arthralgia, followed by dyspnea and rapid progression to pneumonia 40, 41, 42, 43. Approximately one-third of patients present with gastrointestinal symptoms, such as diarrhea and vomiting. Acute renal impairment was the most striking feature of disease caused by MERS-CoV, which is thus far unique for human CoV infections 40, 44. Seventy-five percent of patients with MERS disease also had at least one other comorbidity, and patients who died were more likely to have a pre-existing/underlying condition [40]. Countries around the Arabian Peninsula are known to be endemic for MERS-CoV, and Saudi Arabia has reported the most cases, but since its discovery in 2012, cases have been occasionally exported to other countries through travel, sometimes causing clusters of secondary outbreaks (Figure 1B) [45]. As of December 31, 2015, a total of 1621 laboratory-confirmed infections have been reported, with 584 deaths (CFR = 36.0%) over 26 countries (http://www.who.int/csr/disease/coronavirus_infections/en), making MERS-CoV one of the most dangerous viruses known to humans.
Ecology of Human Coronaviruses
Based on currently available evidence, 229E, OC43, NL63, and HKU1 are well adapted to humans, and the viruses widely circulate in the human population (Figure 2 ), with most cases causing mild disease in immunocompetent adults, and none of these viruses have been found to be maintained within an animal reservoir. However, in the case of SARS-CoV and MERS-CoV, the viruses are not as well adapted to being maintained in humans, and thus are likely spread mainly in zoonotic reservoir(s), with occasional spillover into the susceptible human population, possibly via an intermediate host species.
Figure 2.
Intra- and Inter-Species Transmission of Human Coronaviruses. Red, yellow, green, blue, brown, and purple arrows represent transmission of MERS-CoV, SARS-CoV, NL63, HKU1, OC43, and 229E, respectively, between bats, camels, cows, humans, and masked palm civets (shown in a legend on the side of the figure). Unbroken arrows represent confirmed transmission between the two species in question, and broken arrows represent suspected transmission.
Regarding SARS-CoV, epidemiology data implicated masked palm civets (Paguma larvata) from live animal markets (LAM) in Guangdong Province, China, as a route of exposure to SARS-CoV [46]. However, masked palm civets from the wild or from farms without LAM exposure were largely negative for SARS-CoV [47]. This suggests that palm civets were an intermediate host, but not a reservoir for SARS-CoV [48]. Subsequent studies have shown that wild horseshoe bats (Rhinolophidae family), which can also be found in LAM in China and served in some Chinese restaurants in Guangdong, China, have detectable levels of antibodies against SARS-CoV and a SARS-CoV-like virus (SARSr-Rh-Bat CoV) 49, 50, suggesting a bat origin for SARS-CoV. An evolutionary relationship between coronaviruses and bats was proposed, in which the ancestor for SARS-CoV first spread to bats of the Hipposideridae family, then Rhinolophidae, then masked palm civets and eventually humans [51]. Recently, two new SARS-like CoVs were isolated in horseshoe bats, and these viruses showed the highest relationship to SARS-CoV from all known bat coronaviruses [52]. ORF8 analysis of the SARS-like CoVs in bats suggests that Chinese horseshoe bats are the natural reservoirs of SARS-CoV 52, 53, and that intermediate hosts might not be needed for direct human infection, particularly for some bat SARS-CoV-like viruses [52].
In the case of MERS-CoV, studies in Oman, Saudi Arabia, Qatar, United Arab Emirates, and Jordan have shown that dromedary camels are seropositive for neutralizing antibodies against MERS-CoV [54], as well as camels of Middle East origin in Africa, including Egypt, Kenya, Nigeria, Ethiopia, Tunisia, Somalia, and Sudan [55]. Subsequent studies on dromedary camels in Saudi Arabia [56], Qatar [57], and Egypt [58] show that live MERS-CoV can be isolated primarily from the nasal swabs of camels, showing that camels are potential source of MERS-CoV infection. However, many confirmed cases lack contact history with camels [59], suggesting direct human-to-human transmission, or through contact with a yet-to-be-identified animal species that is maintaining MERS-CoV. Studies on HKU4, a coronavirus of bat origin and the most phylogenically closely related to MERS-CoV, had shown that HKU4 is able to utilize the CD26 receptor for virus entry [60]. As CD26 is a known receptor for MERS-CoV [61], the similarity in receptor specificity of these two CoVs supports the hypothesis that MERS-CoVs is also a bat-originated CoV. However, live MERS-CoV has not been isolated from wild bats, nor have viral sequences been detected.
A recent investigation discovered that multiple HCoV species, including MERS-CoV, beta-CoV group A, and a 229E-like virus, circulate amongst dromedary camels in Saudi Arabia [62]. Furthermore, multiple lineages of circulating MERS-CoV in these camels had resulted in a dominant, recombinant MERS-CoV strain that was responsible for the MERS-CoV outbreaks during 2015 [62]. These results show that various CoVs of human and animal origin are currently circulating in the wild, providing ample opportunity for CoVs to undergo evolution and genetic recombination, thereby resulting in recombinant CoVs that may potentially be more deadly to humans. In the following sections, we summarize the evolution and genetic recombination of the current HCoVs.
Phylogeny, Evolution and Genetic Recombination in Human Coronaviruses
Phylogenetic Analysis
The phylogenetic tree for the CoVs of zoonotic and human origin was constructed based on the full genome. It can be observed that 229E and NL63 belong to the alpha-CoV genus and are grouped together with porcine epidemic diarrhea virus (PEDV), TGEV, porcine respiratory coronavirus (PRCV), feline infectious peritonitis virus (FIPV), and some bat-derived coronaviruses (Figure 3 ). The topology of the phylogeny is similar to the tree constructed based on the RNA-dependent RNA polymerase gene [55].
Figure 3.
Phylogenetic Tree of Known Animal and Human Coronaviruses. Phylogenetic analysis of the full-length genome sequences of coronaviruses publicly available in GenBank was performed using RAxML, with 1000 bootstrap replicates. Lines with different colors represent different coronavirus species, Alpha-coronaviruses (blue lines), Beta-coronaviruses (red lines for Beta-coronavirus C, and black lines for Beta-coronavirus A, B, and D), Gamma-coronaviruses (green lines), and Delta-coronaviruses (purple lines). Human isolates are highlighted with different colors, whereas strains from other hosts are shown in black. Numbers at the branches represent bootstrap values obtained in the phylogenetic analysis.
OC43, HKU1, SARS-CoV, and MERS-CoV belong to the beta-CoV genus. Lineage A includes OC43 and HKU1, MHV, porcine hemagglutinating encephalomyelitis virus (PHEV), in addition to equine, rabbit, camel, bovine, antelope-derived animal coronaviruses. Lineage B includes SARS-CoV, SARS-like viruses of bat and palm civet origin, and some bat-derived CoVs. Lineage C includes MERS-CoV and some bat-derived viruses. Lineage D contains only bat-derived coronavirus (Figure 3). In contrast to alpha- and beta-CoVs, gamma-CoVs consist mainly of avian coronaviruses (such as IBV), as well as CoVs isolated from aquatic animals, whales, and dolphins. CoVs of wild-bird origin are clustered into the delta-CoV, which also contains some swine-derived CoVs (Figure 3).
In general, alpha- and beta-CoVs primarily include CoVs from mammals, whereas gamma- and delta-CoVs mostly include CoVs of avian origin. Notably, all currently known HCoVs belong to either the alpha- or betacoronavirus genera, which also contain CoVs of mainly bat origin.
Mutation and Genetic Recombination in Coronaviruses
The estimated mutation rates in CoV are moderate to high compared to other single-stranded RNA (ssRNA) viruses, and the average substitution rate for CoVs was ∼10−4 substitutions per year per site [63]. The nucleotide mutation rate of IBV for the hypervariable region in the S gene was estimated to be 0.3–0.6 × 10–2 per site per year [64]. The S gene of OC43 possessed an average rate of 6.410.58 × 10–4 substitution rates per site per year [65]. Additionally, the S gene of 229E possessed a rate of ∼3 × 10− 4 substitutions per site per year [63]. For SARS-CoV, the mutation rate in the whole genome was estimated to be 0.80–2.38 × 1–3 nucleotide substitutions per site per year, and the nonsynonymous and synonymous substitution rates were estimated to be 1.16–3.30 × 10–3 and 1.67–4.67 × 10–3 per site per year, respectively, which is similar to other RNA viruses [66]. The evolutionary rate for MERS-CoV genomes was estimated as 1.12 × 10−3 substitutions per site per year (95% credible interval [95% CI], 8.76 × 10−4; 1.37 × 10−3) [67]. Furthermore, the large RNA genome in CoV allows for extra plasticity in genome modification by mutations and recombinations, thereby increasing the probability for intraspecies variability, interspecies ‘host jump’, and novel CoVs to emerge under the right conditions 68, 69, 70.
The major reason for recombination may be at the replication step in the virus life cycle. During replication, a set of subgenomic RNAs is generated, increasing the homologous recombination rate among closely related genes from different lineages of CoVs or other viruses by template switching [33]. Concurrently, circulating CoVs in multiple host species likely contribute to increases in the rate of recombination events. However, the exact mechanism of genetic recombination in CoVs remains unclear: recombination site ‘breakpoints’ in the viral genome – the crossing point of the recombinant genes between two distinct viral strains or genotypes – appear to be random, as different recombinant strains have different breakpoints.
Genetic recombination has been previously documented for animal CoVs, for instance: MHV [71], TGEV [72], as well as feline and canine coronaviruses 73, 74. Genetic recombination for other HCoVs including OC43, NL63, HKU1, SARS-CoV [51], and MERS-CoV have also been documented and are discussed in detail below.
OC43 has diverged into five distinct genotypes (A to E). Genotype A is typified by the prototype VR759 strain that was isolated in 1967 [75]. Genotypes B and C are two naturally circulating viruses. Genotype D emerged via recombination between genotypes B and C viruses at the breakpoint of 2500–5000 bp in the NSP2–NSP3 gene and 15500 bp-3′end in NSP12-N genes [75] (Figure 4 , Key Figure). Genotype E, identified in 2014, emerged by recombination amongst the genotype B, C, and D viruses with different breakpoints in the genes compared to the generation of genotype D [76] (Figure 4). A study in France, between 2001 and 2013, demonstrated that OC43 strains have more recombination patterns in Lower Normandy, and circulating OC43 viruses have a high genetic diversity [17].
Figure 4.
Key Figure: Genetic Recombination in Human Coronaviruses
The colored regions of each human coronavirus indicate genetic recombination at the genomic sites with coronaviruses of zoonotic origin. Specific nucleotide locations where recombination occurred are also shown in broken lines.
NL63 also has the potential for genetic recombination, and one such example is between the Amsterdam 1 and 496 strains. Recombination site breakpoints were identified in two parts of the S gene [63] (Figure 4).
Recombination events have been observed for different genotypes of HKU1, in which recombination breakpoints were found on genotypes B and C in the nsp6-nsp7 and nsp16-HE genes, respectively [70] (Figure 4). HKU1 was also shown to be able to recombine with other animal beta-CoVs, such as MHV, at the nsp3, nsp4, nsp6, nsp7, nps9, and nsp10 genes [77] (Figure 4).
SARS-CoV has a possible recombinant history with lineages of alpha- and gamma-CoV [78]. Many specific breakpoints and a large number of smaller recombinant regions had been identified in the RNA-dependent RNA polymerase (RDRP, nsp12 gene) at the following locations: nucleotides 13 392–13 610, 15 259–15 342, and 15 974–16 108 based on the sequence of the SARS-CoV TOR2 strain [78]. Recombination locations were also identified in the nsp9 and most of nsp10 gene located at 12 613–13 344, and parts of nsp14 (18 117–18 980) [78]. Another study analyzing the genome of the SARS-CoV CV7 strain had shown that seven putative recombination locations were found between SARS-CoV and six other coronaviruses: PEDV, TGEV, BCV, 229E, MHV, and IBV (Figure 4). This further suggests that SARS-CoV emerged via serial horizontal transmission and by genetic recombination, enhancing the adaptation process to its new host [79].
Recombination was also detected between the SARS-related bat-associated CoV (SARSr-Bat CoVs), in which recombination between the Rp3 strain from Guangxi Province and Rf1 strain from Hubei Province, China, generated a new strain (SARSr-Civet CoV SZ3) in civets, with a breakpoint at the nsp16/spike and S2 region [51] (Figure 4). Furthermore, the SARS-CoV Rf1/2004 strain is itself a recombinant from the bat CoV 279/2004, bat CoV Rm1/2004 and civet CoV SZ3/2003 [51] (Figure 4).
A recent study showed that the epidemic MERS-CoV experienced recombination events between the different lineages, which occurred in dromedary camels in Saudi Arabia [62]. Lineages 3 and 5 viruses were predominant during July and December 2014; however, lineage 5 became dominant within the camel population by 2015. Lineage 5, which is closely associated with the MERS-CoV causing the outbreak in South Korea in 2015, as well as recent human infections in Riyadh, Saudi Arabia, is a recombinant virus between lineages 3 and 4, or groups 3 and 5 of clade B (Figure 4) 62, 80.
Concluding Remarks
The result from a high frequency of recombination events in CoVs is the generation of novel viruses with a high genetic diversity, with unpredictable changes in virulence during human infections. With multiple species of CoVs circulating in the wild amongst different animal species that may constantly interact with one another, it is likely not a matter of if, but when, the next recombinant CoV will emerge and cause another outbreak in the human population. As such, some crucial future areas of investigation include: (i) the prevalence of HCoVs already circulating within the animal population, (ii) the commonality of coronavirus recombination in animals, (iii) animals which may potentially serve as mixing vessels for the generation of novel recombinant CoVs, and (iv) a surveillance network to monitor and predict the potential emergence of a highly virulent, recombinant CoV from animals (see Outstanding Questions). Furthermore, lessons from the SARS-CoV and MERS-CoV outbreaks must be urgently learned in advance to effectively prepare for the next CoV outbreak.
Outstanding Questions.
How common is coronavirus recombination in animals?
Can coronavirus undergo recombination in animal hosts other than camels?
Is there a way to predict the potential emergence of a novel recombinant coronavirus in the human population?
Author Contributions
Shuo Su and Gary Wong contributed equally to this manuscript.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81461168030), the China National Grand S&T Special Project (2014ZX10004-001-006), and the National Key Program for Infectious Disease of China (2016ZX10004222). G.F.G. is a leading principal investigator of the NSFC Innovative Research Group (grant number 81321063). J.Z and S.S are supported by the Priority Academic Program Development of Jiangsu Higher Education Institution. G.W. is the recipient of a Banting Postdoctoral Fellowship from the Canadian Institutes of Health Research (CIHR), and a President's International Fellowship Initiative from the Chinese Academy of Sciences (CAS). All authors declare no competing interests.
Contributor Information
Shuo Su, Email: shuosu@njau.edu.cn.
Yuhai Bi, Email: beeyh@im.ac.cn.
George F. Gao, Email: gaof@im.ac.cn.
References
- 1.Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai M. Coronaviridae. In: Knipe D.M., Howley P.M., editors. Fields Virology. 5th edn. Lippincott Williams & Wilkins; 2007. pp. 1305–1335. [Google Scholar]
- 3.Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Vir. Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langereis M.A. Attachment of mouse hepatitis virus to O-acetylated sialic acid is mediated by hemagglutinin-esterase and not by the spike protein. J. Virol. 2010;84:8970–8974. doi: 10.1128/JVI.00566-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durham P.J. Rotavirus and coronavirus associated diarrhoea in domestic animals. N. Zealand Vet. J. 1979;27:30–32. doi: 10.1080/00480169.1979.34595. [DOI] [PubMed] [Google Scholar]
- 6.Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 7.Erles K. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology. 2003;310:216–223. doi: 10.1016/S0042-6822(03)00160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiner L.P. Pathogenesis of demyelination induced by a mouse hepatitis. Arch. Neurol. 1973;28:298–303. doi: 10.1001/archneur.1973.00490230034003. [DOI] [PubMed] [Google Scholar]
- 9.Pensaert M. Transmissible gastroenteritis of swine: virus–intestinal cell interactions. II. Electron microscopy of the epithelium in isolated jejunal loops. Arch. Gesamte Virusforsch. 1970;31:335–351. doi: 10.1007/BF01253768. [DOI] [PubMed] [Google Scholar]
- 10.Pensaert M. Transmissible gastroenteritis of swine: virus-intestinal cell interactions. I. Immunofluorescence, histopathology and virus production in the small intestine through the course of infection. Arch. Gesamte Virusforsch. 1970;31:321–334. doi: 10.1007/BF01253767. [DOI] [PubMed] [Google Scholar]
- 11.Bridger J.C. Replication of an enteric bovine coronavirus in intestinal organ cultures. Arch. Virol. 1978;57:43–51. doi: 10.1007/BF01315636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedersen N. Pathogenicity studies of feline coronavirus isolates 79-1146 and 79-1683. Am. J. Vet. Res. 1984;45:2580–2585. [PubMed] [Google Scholar]
- 13.Binn L.N. Recovery and Characterization of a Coronavirus from Military Dogs with Diarrhea. Proc. Annu. Meet. U.S. Anim Health Assoc. 1974:359–366. [PubMed] [Google Scholar]
- 14.Ismail M.M. Pathogenicity of turkey coronavirus in turkeys and chickens. Avian Dis. 2003;47:515–522. doi: 10.1637/5917. [DOI] [PubMed] [Google Scholar]
- 15.Wevers B.A., van der Hoek L. Recently discovered human coronaviruses. Clin. Lab. Med. 2009;29:715–724. doi: 10.1016/j.cll.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyrrell D.A., Bynoe M.L. Cultivation of a novel type of common-cold virus in organ cultures. Br. Med. J. 1965;1:1467–1470. doi: 10.1136/bmj.1.5448.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kin N. Genomic analysis of 15 human coronaviruses OC43 (HCoV-OC43s) circulating in France from 2001 to 2013 reveals a high intra-specific diversity with new recombinant genotypes. Viruses. 2015;7:2358–2377. doi: 10.3390/v7052358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monto A.S. Medical reviews. Coronaviruses. Yale J. Biol. Med. 1974;47:234–251. [PMC free article] [PubMed] [Google Scholar]
- 19.Peiris J.S.M. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raj V.S. MERS: emergence of a novel human coronavirus. Curr. Opin. Virol. 2014;5:58–62. doi: 10.1016/j.coviro.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendley J.O. Coronavirus infections in working adults. Eight-year study with 229 E and OC 43. Am. Rev. Respir. Dis. 1972;105:805–811. doi: 10.1164/arrd.1972.105.5.805. [DOI] [PubMed] [Google Scholar]
- 22.Chiu S.S. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong. China. Clin. Infect. Dis. 2005;40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 24.McIntosh K. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. U.S.A. 1967;57:933–940. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyrrell D.A. Signs and symptoms in common colds. Epidem. Infect. 1993;111:143–156. doi: 10.1017/s0950268800056764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Hoek L. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol. Rev. 2006;30:760–773. doi: 10.1111/j.1574-6976.2006.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fouchier R.A. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esper F. Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J. Infect. Dis. 2005;191:492–498. doi: 10.1086/428138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdul-Rasool S., Fielding B.C. Understanding human coronavirus HCoV-NL63. Open Virol. J. 2010;4:76–84. doi: 10.2174/1874357901004010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woo P.C. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woo P.C. More and more coronaviruses: human coronavirus HKU1. Viruses. 2009;1:57–71. doi: 10.3390/v1010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woo P.C. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J. Infect Dis. 2005;192:1898–1907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau S.K.P. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J. Clin. Microbiol. 2006;44:2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaunt E.R. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esper F. Coronavirus HKU1 infection in the United States. Emerg. Infect. Dis. 2006;12:775–779. doi: 10.3201/eid1205.051316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sloots T.P. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J. Clin. Virol. 2006;35:99–102. doi: 10.1016/j.jcv.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vabret A. Detection of the new human coronavirus HKU1: a report of 6 cases. Clin. Infect. Dis. 2006;42:634–639. doi: 10.1086/500136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goes L.G. Coronavirus HKU1 in children, Brazil, 1995. Emerg. Infect. Dis. 2011;17:1147–1148. doi: 10.3201/eid1706.101381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaki A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 40.Zumla A. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Tawfiq J.A. Middle East respiratory syndrome coronavirus: a case–control study of hospitalized patients. Clin. Infect. Dis. 2014;59:160–165. doi: 10.1093/cid/ciu226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Assiri A. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arabi Y.M. Clinical course and outcomes of critically ill patients with Middle East respiratory syndrome coronavirus infection. Ann. Intern. Med. 2014;160:389–397. doi: 10.7326/M13-2486. [DOI] [PubMed] [Google Scholar]
- 44.Drosten C. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet. Infect. Dis. 2013;13:745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su S. MERS in South Korea and China: a potential outbreak threat? Lancet. 2015;385:2349–2350. doi: 10.1016/S0140-6736(15)60859-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan Y. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 47.Poon L.L. Identification of a novel coronavirus in bats. J. Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tu C. Antibodies to SARS coronavirus in civets. Emerg. Infect. Dis. 2004;10:2244–2248. doi: 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kan B. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005;79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lau S.K. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 2010;84:2808–2819. doi: 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ge X.Y. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Z. ORF8-related genetic evidence for Chinese horseshoe bats as the source of human severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2016;213:579–583. doi: 10.1093/infdis/jiv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reusken C.B. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet. Infect. Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chan J.F.W. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hemida M.G. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg. Infect. Dis. 2014;20:1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raj V.S. Isolation of MERS coronavirus from a dromedary camel, Qatar, 2014. Emerg. Infect. Dis. 2014;20:1339–1342. doi: 10.3201/eid2008.140663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chu D.K. MERS coronaviruses in dromedary camels, Egypt. Emerg. Infect. Dis. 2014;20:1049–1053. doi: 10.3201/eid2006.140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samara E.M., Abdoun K.A. Concerns about misinterpretation of recent scientific data implicating dromedary camels in epidemiology of Middle East respiratory syndrome (MERS) Mbio. 2014;5:e01430–e1514. doi: 10.1128/mBio.01430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Q. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu G. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sabir J.S. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 63.Pyrc K. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J. Mol. Biol. 2006;364:964–973. doi: 10.1016/j.jmb.2006.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cavanagh D. Does IBV change slowly despite the capacity of the spike protein to vary greatly? Adv. Exp. Med. Biol. 1998;440:729–734. doi: 10.1007/978-1-4615-5331-1_94. [DOI] [PubMed] [Google Scholar]
- 65.Ren L. Genetic drift of human coronavirus OC43 spike gene during adaptive evolution. Sci. Rep. 2015;5:11451. doi: 10.1038/srep11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Z. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol. Biol. 2004;4:21. doi: 10.1186/1471-2148-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cotten M. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. Mbio. 2014;5:e01062–e1113. doi: 10.1128/mBio.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woo P.C. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood) 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 69.Woo P.C. Infectious diseases emerging from Chinese wet-markets: zoonotic origins of severe respiratory viral infections. Curr. Opin. Infect. Dis. 2006;19:401–407. doi: 10.1097/01.qco.0000244043.08264.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Woo P.C. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lai M.M. Recombination between nonsegmented RNA genomes of murine coronaviruses. J. Virol. 1985;56:449–456. doi: 10.1128/jvi.56.2.449-456.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tian P.F. Evidence of recombinant strains of porcine epidemic diarrhea virus, United States, 2013. Emerg. Infect. Dis. 2014;20:1735–1738. doi: 10.3201/eid2010.140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Terada Y. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS ONE. 2014;9:e106534. doi: 10.1371/journal.pone.0106534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Decaro N. Recombinant canine coronaviruses related to transmissible gastroenteritis virus of Swine are circulating in dogs. J. Virol. 2009;83:1532–1537. doi: 10.1128/JVI.01937-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lau S.K.P. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J. Virol. 2011;85:11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y. Genotype shift in human coronavirus OC43 and emergence of a novel genotype by natural recombination. J. Infect. 2015;70:641–650. doi: 10.1016/j.jinf.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woo P.C. Phylogenetic and recombination analysis of coronavirus HKU1, a novel coronavirus from patients with pneumonia. Arch. Virol. 2005;150:2299–2311. doi: 10.1007/s00705-005-0573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stanhope M.J. Evidence from the evolutionary analysis of nucleotide sequences for a recombinant history of SARS-CoV. Infect. Genet. Evol. 2004;4:15–19. doi: 10.1016/j.meegid.2003.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang X.W. Testing the hypothesis of a recombinant origin of the SARS-associated coronavirus. Arch. Virol. 2005;150:1–20. doi: 10.1007/s00705-004-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Y. Origin and possible genetic recombination of the Middle East respiratory syndrome coronavirus from the first imported case in China: phylogenetics and coalescence analysis. Mbio. 2015;6:e01280–01215. doi: 10.1128/mBio.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pene F. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin. Infect. Dis. 2003;37:929–932. doi: 10.1086/377612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Papa A. Coronaviruses in children, Greece. Emerg. Infect. Dis. 2007;13:947–949. doi: 10.3201/eid1306.061353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cabeca T.K. Epidemiological and clinical features of human coronavirus infections among different subsets of patients. Influenza Other Respir. Virus. 2013;7:1040–1047. doi: 10.1111/irv.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dijkman R. Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J. Virol. 2013;87:6081–6090. doi: 10.1128/JVI.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cleri D.J. Severe Acute Respiratory Syndrome (SARS) Infect. Dis. Clin. N. Am. 2010;24:175–202. doi: 10.1016/j.idc.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gu J. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsang K.W. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 88.Peiris J.S.M. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moes E. A novel pancoronavirus RT-PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect. Dis. 2005;5:6. doi: 10.1186/1471-2334-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bastien N. Human coronavirus NL63 infection in Canada. J. Infect. Dis. 2005;191:503–506. doi: 10.1086/426869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van der Hoek L. Croup is associated with the novel coronavirus NL63. PLoS Med. 2005;2:764–770. doi: 10.1371/journal.pmed.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chiu S.S. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin. Infect. Dis. 2005;40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wat D. The common cold: a review of the literature. Eur. J. Intern. Med. 2004;15:79–88. doi: 10.1016/j.ejim.2004.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chung J.Y. Detection of viruses identified recently in children with acute wheezing. J. Med. Virol. 2007;79:1238–1243. doi: 10.1002/jmv.20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.WHO Mers-Cov Research Group State of knowledge and data gaps of Middle East respiratory syndrome coronavirus (MERS-CoV) in humans. PLoS Curr. Outbreaks. 2013 doi: 10.1371/currents.outbreaks.0bf719e352e7478f8ad85fa30127ddb8. Published online November 12, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Corman V.M. Viral Shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin. Infect. Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]