Abstract
Human coronaviruses (HCoVs), including SARS-CoV and MERS-CoV, are zoonotic pathogens that originated in wild animals. HCoVs have large genomes that encode a fixed array of structural and nonstructural components, as well as a variety of accessory proteins that differ in number and sequence even among closely related CoVs. Thus, in addition to recombination and mutation, HCoV genomes evolve through gene gains and losses. In this review we summarize recent findings on the molecular evolution of HCoV genomes, with special attention to recombination and adaptive events that generated new viral species and contributed to host shifts and to HCoV emergence.
Video Abstract
Keywords: human coronavirus, molecular evolution, gene gain/loss, positive selection, recombination, host shift
Trends
Human coronaviruses (HCoVs) are zoonotic pathogens with large and complex genomes. Some HCoV accessory proteins were acquired from host genes, and some were lost or split during HCoV evolution. Most likely SARS-CoV ORF8 became dispensable during the shift to the human/civet host.
HCoV spike proteins adapted to use diverse cellular receptors. This occurred by divergence followed, in some cases, by convergent evolution to bind the same receptor.
Recombination and positive selection shaped the diversity of CoV genomes, especially the S gene. Positive selection in the S gene of MERS-CoV and related CoVs mainly acted on the heptad repeats.
In MERS-CoV and other lineage C beta-CoVs, positive selection targeted the nonstructural components, particularly ORF1a. Most adaptive events occurred in nsp3, which acts as a viral protease and contributes to suppression of interferon responses.
Human Coronaviruses Are Zoonotic Pathogens
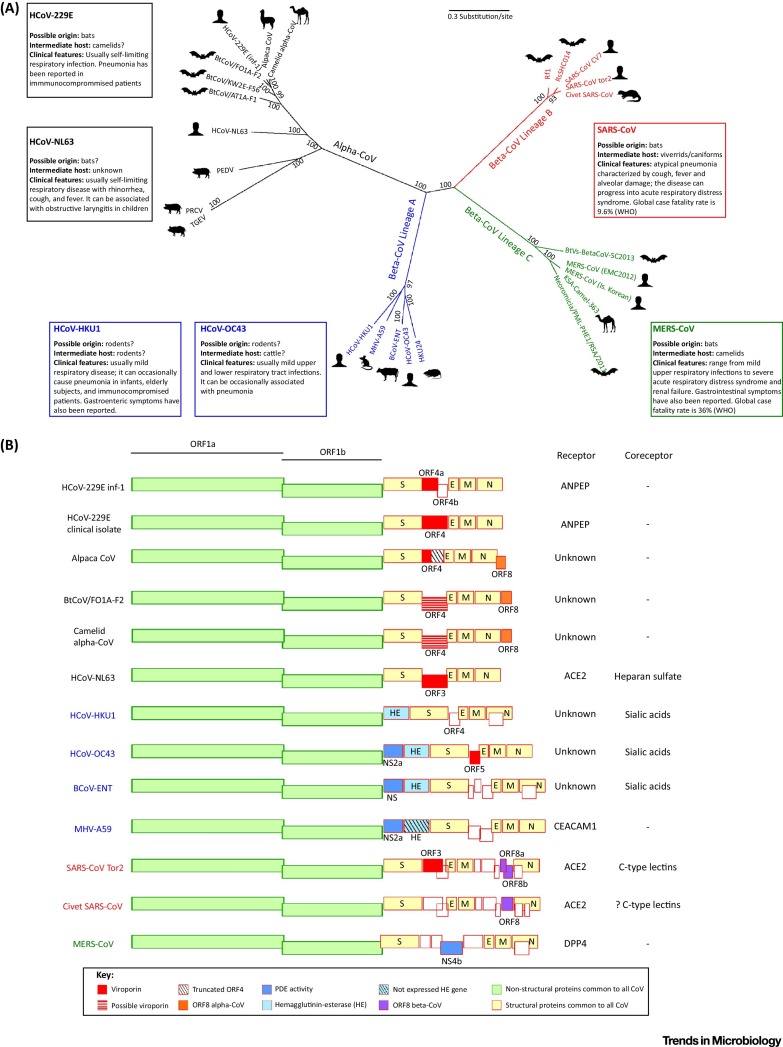
The recent emergence of severe acute respiratory syndrome-related coronavirus (SARS-CoV) and of Middle East respiratory syndrome-related Coronavirus (MERS-CoV) (order Nidovirales, family Coronaviridae, subfamily Coronavirinae) as dangerous zoonoses stirred great interest in the ecology and evolution of coronaviruses. Before the SARS-CoV epidemic only two HCoVs were known: HCoV-229E and HCoV-OC43. Two additional HCoVs, HCoV-NL63 and HCoV-HKU1, were discovered in 2004–2005 from clinical specimens [1]. These viruses originated in animals and are mainly responsible for respiratory diseases in humans (Figure 1A, Key Figure). Specifically, all HCoVs are thought to have a bat origin, with the exception of lineage A beta-CoVs, which may have reservoirs in rodents [2]. The phylogenetic relationships of HCoVs and other animal CoVs mentioned in this review are summarized in Figure 1A.
Figure 1.
Key Figure: Phylogenetic Relationships and Genome Organization of Human and Animal Coronaviruses (CoVs)
CoVs that infect nonhuman mammals are included only if they are mentioned in the text for comparative purposes. (A) The phylogenetic tree of complete genome sequences of HCoVs and selected mammalian CoVs was obtained with RAxML 8.2.4 [68]. Numbers indicate bootstrap support. CoVs are colored according to genus and lineage. Information about origin, intermediate host, and clinical presentation is reported for the six HCoVs 1, 2, 3, 4, 5, 89. Data about case fatality rate were derived from the World Health Organization website (http://www.who.int/mediacentre/factsheets/mers-cov/;http://www.who.int/csr/sars/country/table2004_04_21/en/). (B) CoV genome organization is schematically reported together with information on receptor/coreceptor usage. Virus names are colored according to their genus or lineage, as in (A). Only ORFs mentioned in the text are colored or shaded. Empty boxes represent accessory ORFs that are not described in the text.
A number of field studies identified and sequenced viruses related to HCoVs in wildlife reservoirs, and phylogenetic reconstruction provided important clues on the most likely events that led to the introduction of HCoVs in human populations. Several recent excellent reviews delve into the knowns and unknowns of HCoV origin in terms of reservoir species, amplification host, and, more generally, of CoV ecology 1, 3, 4, 5. In this review we instead focus on the molecular evolution of HCoV genomes. The general concepts of evolutionary analyses in viruses are outlined in Box 1 , whereas the most common approaches that were applied to the analysis of CoV sequence evolution in terms of phylogenetic reconstruction, detection of recombination, and identification of selection signatures are summarized in Boxes 1 and 2 .
Box 1. Molecular Evolution in Viruses: General Concepts.
RNA viruses are rapidly evolving pathogens that can accumulate considerable genetic diversity in relatively short time periods. This is mostly due to their high nucleotide mutation rates (but see text for CoVs). The diversity of extant viral sequences can be analyzed to construct phylogenetic relationships among species/strains and to infer the underlying evolutionary patterns. In the presence of recombination a single phylogenetic tree is unable to describe the evolution of homologous sequences. Because recombination is common in many viruses, including HCoVs, the evolution of viral genomes is best modeled by several phylogenies, one for each nonrecombinant fragment (Box 2). By reassorting mutations, recombination has the potential to generate novel viral phenotypes. Thus, not only recombination is of interest per se, but failure to account for its presence can distort phylogeny-based analyses, including estimates of natural selection [66]. Natural selection acts pervasively on viral sequences. When coding regions are concerned, natural selection is commonly estimated in terms of ω (also referred to as dN/dS) – that is, the observed number of nonsynonymous differences per nonsynonymous site (dN) over the observed number of synonymous differences per synonymous site (dS). Under neutral evolution, ω is expected to be equal to 1, as the rate at which amino acid substitutions accumulate is similar to the rate for synonymous changes. Due to the fact that essential protein domains can often tolerate only minor sequence changes, most amino acid replacements are eliminated by selection; this generates ω values <1, a situation referred to as negative (or purifying) selection. Nevertheless, amino acid replacements can be advantageous for a virus in terms, for example, of host adaptation or immune evasion: in this case ω values can be higher than 1 (positive selection). Thus, evaluation of how ω varies from site to site or from branch to branch in a phylogeny is commonly used to describe selective events. Some possible caveats should nevertheless be kept in mind. (i) The saturation of substitution rates (especially dS) may occur and affect evolutionary inference when fast-evolving sequences are analyzed (see Box 3 for an example, and Box 2 for methods to overcome this problem). (ii) In viral genomes synonymous substitutions are not always neutral; this may be due to the presence of overlapping reading frames, conserved RNA secondary structures, packaging signals, and other functional elements (Box 3). (iii) A relaxation in the intensity of both negative and positive selection may occasionally occur (Box 2 and Figure 2A).
Box 2. Molecular Evolution in Viruses: Methods and Caveats.
Phylogenetic tree construction
Distance-based and character-based approaches can be used in phylogenetic tree reconstruction. Distance-based methods measure pairwise differences among sequences and generate the tree from the resultant distance matrix (e.g., UPGMA, Neighbour Joining). Character-based methods evaluate all possible trees and estimate the one that best fits the data. This approach includes maximum likelihood (ML) and Bayesian methods (e.g., phyML [67], RaxML [68], MrBayes [69]).
Recombination detection
The location of recombination breakpoints can be detected using phylogenetic incongruence among segments in a sequence alignment (e.g., GARD [70]) or by evaluating the distribution of nucleotide substitution along genomic regions (e.g., Recco [71]). A common approach is to use different methodologies to identify breakpoint locations and contributing sequences (e.g., RDP [72]).
Positive selection analyses
Positive selection is usually estimated based on ω variation across sites and/or lineages (Box 1). A common approach is to compare ML models that allow or not a class of codons in the alignment to evolve with ω > 1 (e.g., the ‘site models’ in PAML [73]) Likelihood ratio tests are then applied to determine whether the neutral model can be rejected in favor of the positive selection model. Alternatively, branch-site models can be applied to detect episodic positive selection on specific branches of the phylogeny. Different methods allow to test a priori whether a branch is under selection 74, 75 or to model different evolutionary scenarios for each branch [76]. When evolutionary analysis is focused on recent timescales, selection may not have yet fixed the advantageous mutations or removed the deleterious ones. One possibility is to compare the distribution of nonsynonymous and synonymous polymorphisms in a specific lineage with the ratio of nonsynonymous to synonymous fixed differences between lineages/species. The McDonald-Kreitman test [77] has been widely applied for this purpose.
Relaxed selection
When selection is relaxed, smaller ω values tend toward 1, whereas ω values higher than 1 decrease. This phenomenon can be confused with episodic selection. Specific methods allow one to infer whether a branch in the phylogeny is under positive or relaxed selection [18].
Subsitution rate saturation
Saturation of substitution rates can be a serious issue for deep tree branches. Nonetheless, branch-site methods are relatively insensitive to biases introduced by dS saturation, and can be applied to the analysis of distantly related species [78]. Alternatively, specific indexes have been developed to detect substitution saturation [79]; in the presence of dS saturation, third-codon positions can be removed to obtain reliable phylogenies.
Synonymous constraint
Regions with an excess of synonymous constraint can be identified using a recently developed sliding-window ML-based method [80].
HCoV Genome Organization
CoVs are positive-sense, single-strand RNA viruses with a likely ancient origin, and HCoVs repeatedly emerged during the past 1000 years (Box 3 ). All CoVs have nonsegmented genomes that share a similar organization. About two thirds of the genome consists of two large overlapping open reading frames (ORF1a and ORF1b; see Glossary), that are translated into the pp1a and pp1ab polyproteins. These are processed to generate 16 nonstructural proteins (nsp1 to 16). The remaining portion of the genome includes ORFs for the structural proteins: spike (S), envelope (E), membrane (M) and nucleoprotein (N). A variable number of accessory proteins are also encoded by distinct viruses (Figure 1B).
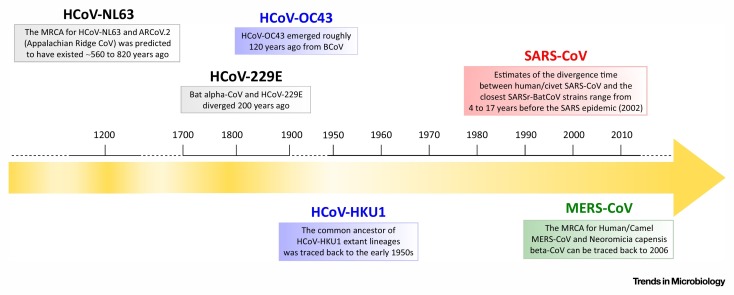
Box 3. Time Origin of CoV Genera and HCoV Emergence.
Coronaviruses are classified into four distinct genera (alpha, beta, gamma, and delta) [81]; alpha-CoVs and beta-CoVs circulate in mammalian hosts, whereas gamma-CoVs and delta-CoVs mainly infect birds [82]. An analysis of the RNA-dependent RNA polymerase (RdRp) gene in 43 CoVs provided a first estimate of around 10 000 years ago for the time of the most recent common ancestor (tMRCA) of the four genera [83]. This result was questioned on the basis of observations suggesting a longstanding interaction between CoVs and their hosts [82]. Thus, Wertheim and coworkers hypothesized that natural selection, in particular negative selection, resulted in a bias of the tMRCA estimate [82]. Indeed, strong negative selection can result in the saturation of substitutions at synonymous sites and consequently in underestimation of branch lengths in a phylogeny. To overcome this issue, the authors applied a branch-site test (see Box 1) [76] to estimate branch lengths while taking into account the effect of different selective pressures among lineages in the CoV phylogeny [82]. Their findings placed the separation of the four CoV genera around 300 million years ago, highlighting the importance of evolutionary models in molecular clock dating. Interestingly, this tMRCA is consistent with the separation time between mammals and aves [84], suggesting a coevolutionary relationship between coronavirus and their hosts. However, the dating estimates obtained for closely related viruses by Wertheim and coworkers were in agreement with previous studies, suggesting that the action of natural selection is not biasing the estimation of more recent divergence times. Although most HCoVs were identified only recently, molecular clock analyses indicate that some of these viruses diverged from closely related CoVs hundreds of years ago. In particular, the emergence of HCoV-NL63 and HCoV-229E has been roughly estimated around 500–800 and 200 years ago, respectively 85, 86. HCoV-OC43 is thought to have shared a common ancestor with BCoV around 120 years ago [87]. As for SARS-CoV and MERS-CoV, molecular dating studies estimated that they diverged from bat CoVs in the last three decades 14, 88. Finally, the MRCA of HCoV-HKU1 extant lineages was estimated to have existed in the 1950s [60]. Clearly, these dates should be regarded as estimates and confidence intervals are often wide. A timeline for the emergence of HCoVs is depicted in Figure I .
Figure I.
Timeline for the Emergence of HCoVs.
Among RNA viruses, CoVs have exceptionally long genomes (up to 32 kb). Genome expansion in CoVs is believed to be at least partially mediated by increased replication fidelity. Although estimates of the mutation rate for CoVs differ, possibly depending on the phase of CoV adaptation to novel hosts, several studies have shown that these viruses may possess an unusually high replication fidelity 6, 7, 8. Indeed, a major step that allowed genome expansion in CoVs and, more generally, in Nidovirales, was the acquisition of a set of RNA-processing enzymes that improved the low fidelity of RNA replication [9]. These enzymes include an RNA 3′-to-5′ exoribonuclease (ExoN) and possibly an endoribonuclease (NendoU) [9]. Additional evidence, though, suggests that features distinct from replication fidelity underlie genome expansion in Nidovirales. These include a peculiar genome organization [9] and a processive replication complex [10].
Importantly, CoV genome expansion allowed the acquisition and maintenance of genes encoding diverse accessory proteins that may promote virus adaptation to specific hosts and often contribute to the suppression of immune responses, as well as to virulence. Accessory proteins differ in number and sequence even among CoVs belonging to the same lineage (Figure 1B), raising interesting questions about their origin and evolution.
Gene Gains and Gene Losses
The acquisition (or loss) of novel protein-coding genes has the potential to drastically modify viral phenotypes. Thus, tracing these gain/loss events may identify important turning points in viral evolution.
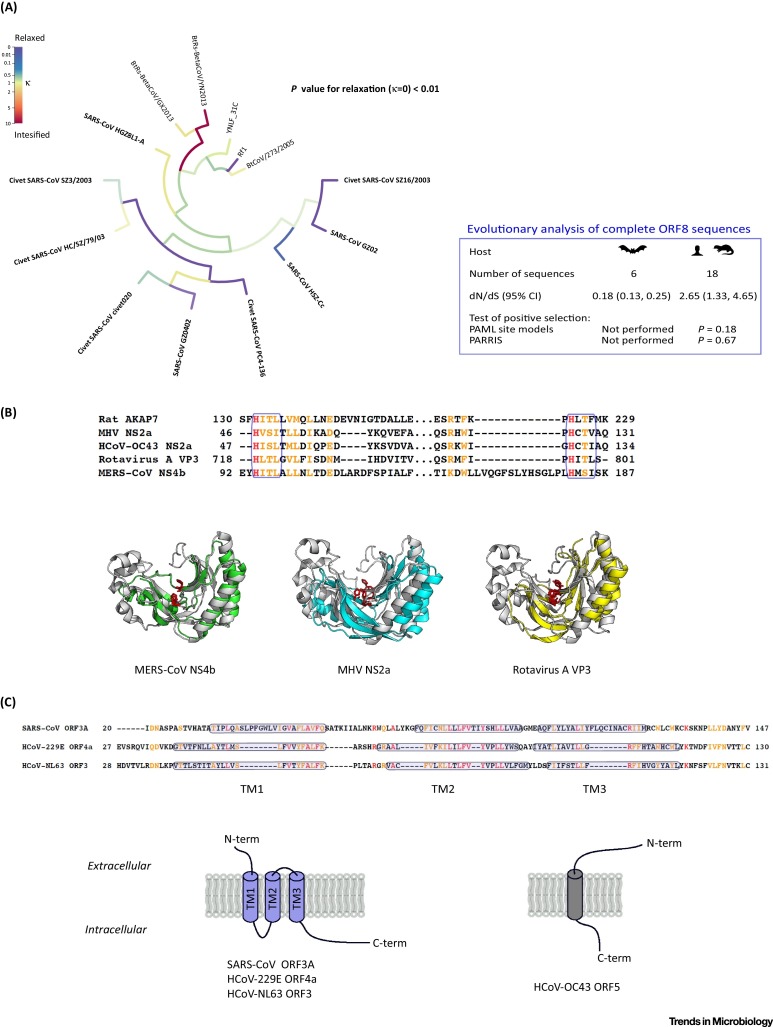
Among SARS-CoV accessory proteins, the origin of ORF8 has remained mysterious for a while, as SARS-CoV-related (SARSr) bat viruses were isolated but found to encode divergent ORF8 proteins (amino acid identity with SARS-CoV ORF8 around 33%) 11, 12, 13. Very recently, SARSr-BatCoVs from Rhinolophus sinicus (Rs) and Rhinolophus ferrumequinum (Rf) were isolated 14, 15. Analysis of the ORF8 region revealed high sequence identity with civet/human SARS-CoV. Two groups came to the conclusion that recombination within SARSr-Rs-CoVs or between SARSr-Rs-CoVs and SARSr-Rf-CoVs led to the acquisition of an ORF8 closely related to that of civet/human SARS-CoV and ultimately originated the virus responsible for the human epidemic 14, 15. Interestingly, Lau and coworkers also reported that the ORF8 region has a dN/dS = 3.5 in SARS-CoVs isolated from humans (but not in SARSr-BatCoVs), indicating the action of positive selection (Box 1) [14]. This finding is interesting per se and becomes even more important considering that, early in the human epidemic, SARS-CoVs acquired a signature 29-nucleotide deletion which split ORF8 into two functional ORFs (ORF8a and b) [16]. These findings suggest that rapid evolution of ORF8 might facilitate host shifts [14]. This possibility is, however, questioned by the presence of additional SARS-CoV human isolates that carry independent and larger deletions in the ORF8 region [16]. Thus, an alternative explanation for these findings is that the activity of ORF8 became dispensable in the human host. If this were the case, relaxed purifying selection rather than positive selection might be responsible for the high dN/dS. To disentangle these alternative possibilities we analyzed ORF8 in human and civet viruses that carry an intact gene, as well as in bat viruses. Although we confirmed that dN/dS is well above 1 for human/civet SARS-CoV ORF8, we detected no evidence of positive selection using the M7/M8 ‘site models’ from PAML (Box 2) or with PARRIS (PARtitioning approach for Robust Inference of Selection) [17] (Figure 2A). Instead, we obtained evidence that relaxation of natural selection [18] in ORF8 accompanied the shift from bats to civets/humans (Figure 2A). These results suggest no major adaptive role for ORF8 during the human SARS-CoV epidemic and support the view that ORF8 is dispensable for virulence and transmission at least in the human/civet host.
Figure 2.
Evolution of Human Coronavirus (HCoV) Accessory Proteins.(A) Test for relaxation of selective strength for SARS-CoV and SARSr-BatCoVs ORF8. Branches are colored according to the selection intensity parameter k. RELAX evaluates if selection on the test branches (bold) is relaxed (k < 1) or intensified (k > 1) compared to background branches. In the evolutionary analysis table the number of sequences differs from that in the tree because RELAX removes identical sequences. Evidence of positive selection was searched for using the M7/M8 ‘site models’ from PAML (see Box 2) and with PARRIS. M7 and M8 represent the null and the positive selection models, respectively. A likelihood ratio test (with 2 degrees of freedom) was applied. (B) An amino acid alignment of rodent AKAP7 and four viral phosphodiesterases (PDEs) is shown. Amino acids are colored red if they are identical, orange if they have very similar properties. PDEs belonging to the 2H family are characterized by two H-Φ-[S/T]-Φ motifs (blue boxes), where Φ is a hydrophobic residue. The structure of rat AKAP7 (gray, PDB ID: 2VFK) is superimposed on MERS-CoV NS4b (green, model generated from 2VFK), MHV NS2a (cyan, PDB ID: 4Z5V), and Rotavirus A VP3 (yellow, PDB ID: 5AF2). Catalytic histidines are shown in red. (C) Sequence and membrane topology comparison of HCoV viroporins. Transmembrane regions (TM1-3) predicted by the TMHMM algorithm [90] are boxed in blue. The corresponding topology model for SARS-CoV ORF3A, HCoV-229E ORF4a (from the Inf-1 strain), and HCoV-NL63 ORF3 is shown. The topology model of HCoV-OC43 OFR5 was derived from recent data [34].
A similar gene loss from the genome of a bat-derived ancestor occurred during the evolution of HCoV-229E. CoVs closely related to HCoV-229E were recently isolated from African hipposiderid bats [19], and a CoV belonging to the same species as HCoV-229E had been described in captive alpacas suffering from an acute respiratory syndrome 20, 21 (Figure 1A). Analysis of these viral genomes indicated that, compared to HCoV-229E, they carry an additional ORF at the genomic 3’ end [20] (Figure 1B). This ORF, which is designated ORF8 but shares no homology with the homonymous SARS-CoV gene, has unknown function and shows limited similarities to any other CoV gene [20]. We analyzed the sequences of recently identified alpha-CoVs from camels [22] and found that ORF8 is encoded by these viruses, as well (Figure 1B). Thus, it is presently unknown whether the loss of ORF8 conferred some advantage during the host shift to humans or, as in the case of ORF8 in SARS-CoV, it became dispensable in the human host.
Another interesting feature of some CoVs is that they encode phosphodiesterases (PDEs) (Figure 1B). These viral enzymes cleave 2’,5’-oligoadenylate, the product of OAS proteins, to prevent activation of the cellular endoribonuclease RNase L and consequently block interferon (IFN)-induced antiviral responses [23]. The PDE activity in the mouse hepatitis virus (MHV) NS2a protein is critical for hepatovirulence [23]. HCoV-OC43, as well as other lineage A nonhuman beta-CoVs, encode NS2a proteins that are characterized by a high degree of sequence similarity to the MHV PDE (Figure 1B). A protein with structure and sequence homology to NS2a is also encoded by an unrelated virus, Group A rotavirus. In this case the PDE activity resides in the C-terminal portion of VP3, a virulence factor [24]. Interestingly, both VP3 and NS2a show two motifs that are characteristic of the 2H-PDE family and share very little sequence similarity to the PDE domain of a cellular protein, AKAP7 [24] (Figure 2B). AKAP7 and the viral PDEs display structural homology (Figure 2B), and murine AKAP7 can complement an inactive MHV NS2a gene [25]. From an evolutionary standpoint, these observations suggest that: (i) beta-CoVs and rotaviruses have independently acquired PDE activities; and (ii) AKAP7 served as the source gene in both viral genera (Figure 2B). More recently, a PDE activity was also discovered in the NS4b protein of MERS-CoV (Figure 1B) and other lineage C beta-CoVs [26]. Similar to those in lineage A beta-CoVs and rotavirus, NS4b belongs to the 2H-phosphoesterase family and displays a predicted structure homologous to AKAP7 [26] (Figure 2B). It remains to be determined whether NS4b was acquired by capturing a vertebrate AKAP7, but the observation that distinct viruses acquired, most likely independently, a PDE activity underscores the importance of these enzymes for viral fitness.
It was recently proposed that CoVs (and other viruses) stole additional genes from their hosts [27]. Hemagglutinin-esterases (HEs) are encoded by lineage A beta-CoVs (e.g., HCoV-HKU1 and HCoV-OC43) (Figure 1B), as well as influenza C virus and toroviruses. Structural analysis suggested that these viral enzymes derive from an ancestral host lectin, although it is unclear whether acquisition occurred in an ancestral virus followed by speciation or multiple times [27]. Incidentally, the N-terminal domain of the CoV spike protein is also believed to derive from a cellular lectin [28]. Unlike the influenza virus C enzyme, CoV HEs lack membrane-fusion activity and are accessory to the spike protein by serving primarily as receptor-destroying enzymes (RDE) – that is, they aid viral detachment from carbohydrates present on infected cells 29, 30. In fact, HEs are present only in the genome of lineage A beta-CoVs, most of which use sialic acids as coreceptors [1] (Figure 1B). These observations suggest that sialic acid-binding spike proteins coevolved with HE genes serving as RDEs. This hypothesis is supported by the observation that the MHV spike protein evolved from an ancestral sugar-binding preference to a protein-binding mode and that several MHV strains lost expression of HE 27, 28 (Figure 1B).
Finally, it is important to notice that artificial selection can lead to unintended changes in viral genomes. Such changes most likely result from passages in culture that, on one hand, relieve the virus from pressures exerted in vivo (e.g., by the host immune system) and, on the other hand, derive from viral adaptation to the in vitro system. An example of these effects is the loss of a full-length ORF4 in the HCoV-229E prototype strain which, due to a two-nucleotide deletion, has a split gene, encoding two proteins (ORF4a and ORF4b) 31, 32 (Figure 1B). Conversely, clinical isolates display a full-length ORF4 sequence [32]. An intact ORF4 is also observed in bat and camel viruses related to HCoV-229E 19, 22, whereas the alpaca alpha-CoV displays a one-nucleotide insertion, resulting in a frameshift [20] (Figure 1B). The availability of only a single alpaca CoV genome makes it impossible to determine whether the inserted sequence is representative of the alpaca CoV population or, else, if it represents a sequencing error.
Overall, these observations suggest that loss of full-length ORF4 is a result of passaging in cell culture, a process that often generates attenuated viruses. An interesting finding on the role of ORF4a came from the observation that its protein product regulates virus production in vitro by functioning as a viroporin [33]. Most likely, the same function is performed by the full-length ORF4 as well.
Viroporins were also detected in SARS-CoV, HCoV-OC43, and HCoV-NL63 34, 35 (Figure 1B). As expected, given the relatedness of the two viruses, the proteins from HCoV-NL63 and HCoV-229E share substantial sequence similarity. Limited similarity is also observed with the SARS-CoV protein, especially in the first and second transmembrane regions, suggesting either a common origin or independent acquisition followed by convergent optimization of residues in the transmembrane helices (Figure 2C). Conversely, the HCoV-OC43 protein (encoded by ORF5, originally denoted NS12.9) is unrelated to the other CoV viroporins, both in terms of sequence and of domain topology [34] (Figure 2C). A protein homologous to the HCoV-OC43 viroporin is instead encoded by MHV (accessory protein NS5a) and functions as an antagonist of IFN-induced antiviral responses 34, 36. Whether the HCoV-OC43 viroporin has the same IFN-antagonizing activity remains to be investigated; however, mutant viruses lacking ORF5 display growth defects in vitro and in vivo, as well as reduced virulence in mice [34]. Interestingly, the viroporins from SARS-CoV, HCoV-NL63, and HCoV-229E can complement the viroporin-defective mutant HCoV-OC43 in vitro [34]. Thus, the conserved function of CoV viroporins was most likely attained by convergent evolution for acquisition of unrelated genes.
Evolution of Structural and Nonstructural Proteins
Clearly, CoV genomes do not only evolve by gene gains and losses, but also via subtler changes that modify protein sequences, and recombination has an important role in reassorting variants.
Several excellent reviews have focused on the evolutionary history of SARS-CoV genomes in terms of recombination and natural selection 37, 38, 39; hereafter, SARS-CoV will be mentioned only to draw comparisons with other CoVs.
From an evolutionary standpoint, nonstructural proteins have attracted less attention than the structural components. This is likely due to the fact that proteins exposed on the virus surface represent the preferential targets of the host immune response. Thus, analyzing and describing their variability and evolutionary dynamics has a clear relevance for the development of preventive strategies (e.g., vaccines) and of treatment options (e.g., administration of neutralizing antibodies). Moreover, structural proteins, and the S protein in particular, determine the first and essential steps in infection and most likely represent the major determinants of host and tissue tropism.
In CoVs, the S protein includes two functionally distinct units: the S1 region contains an N-terminal domain (NTD) and the receptor-binding domain (RBD, also referred to as C-terminal domain or CTD), whereas the S2 region includes the fusion peptide, two heptad repeats (HR1 and HR2), and the transmembrane region (Figure 3A) [38]. A striking feature of HCoV spike proteins is that they have adapted to use diverse cellular receptors and there is no congruence in the phylogeny of HCoV and their receptor usage. In fact, closely related viruses may use different receptors (Figure 1B). For instance, HCoV-229E uses aminopeptidase N (ANPEP), whereas HCoV-NL63 exploits ACE2, this latter representing the receptor for the relatively divergent SARS-CoV (Figure 1B). It is presently unclear how these binding specificities evolved. The latest developments on this topic and, more generally, on the evolution of structural and nonstructural proteins are detailed below for the five known HCoVs.
Figure 3.
Evolution at the Coronavirus (CoV)–Host Interaction Surface.(A) Schematic representation of MERS-CoV spike protein domains. Positively selected sites in MERS-CoV and other lineage C beta-CoVs are shown in red, RBD mutations emerged in the South Korean outbreak are in magenta (see text). A detail of the interaction surface between the MERS-CoV RBD and human DPP4 (PDB ID: 4F5C) is also reported. (B) Ribbon diagram of the interaction surface of human ACE2 with the spike protein of SARS-CoV (PDB ID: 2AJF) and HCoV-NL63 (PDB ID: 3KBH). The binding surface of porcine ANPEP with the TGEV spike protein (PDB ID: 4F5C) is also shown. The location of the HCoV-299E binding site on ANPEP is circled. Red denotes protein regions involved in binding.
MERS-CoV
The evolutionary analysis of MERS-CoV is a rapidly moving field, as sequences from the latest phases of the epidemic have just become available. Analysis of an ever increasing number of viral sequences of both MERS-CoV and of related beta-CoVs revealed that genetic variability in the S gene was shaped by recombination and positive selection. In fact, both ancient and recent intra-spike recombination events were described 22, 40, 41. Interestingly, recombination events with breakpoints within the S gene occurred in camels in Saudi Arabia and originated the MERS-CoV lineage that spread to South Korea.
Analysis of positive selection of MERS-CoV spike genes indicated that several adaptive variants arose in MERS-CoV and in phylogenetically related CoVs [42]. Contrary to common expectation and to what happened during the SARS-CoV host shift to humans, positive selection did not target the RBD. In fact, most adaptive substitutions were detected in the region encompassing the heptad repeats, regions of central importance for virus cell entry (Figure 3A) 42, 43. In other CoVs, variants in the heptad repeats were previously shown to affect host or tissue tropism 44, 45, 46. Interestingly, during the South Korean outbreak, MERS-CoVs that carry point mutations in the spike protein RBD emerged and rapidly spread [47]. These viruses showed decreased binding to the cellular receptor [47] (Figure 3A). Because several immune epitopes are located in the RBD, these findings point to the possibility that MERS-CoV is evolving to avoid the binding of neutralizing antibodies, resulting in a trade-off with receptor-binding affinity [47]. If this were the case, the phases of MERS-CoV adaptation to humans may have consisted of initial events that modulated host tropism through changes in the heptad repeats followed by the emergence of virus variants that escape immune responses. In MERS-CoV and other lineage C beta-CoVs, positive selection also targeted the nonstructural components, particularly ORF1a [48]. Most adaptive events occurred in nsp3, a multifunctional protein which acts as a viral protease and contributes to the suppression of interferon responses through its deubiquitinating and deISGylating activities [49]. Selection in nsp3 is ongoing among MERS-CoV isolated from humans and camels [48]. In analogy to the S protein, though, no major selective event was found to be associated with camel-to-human transmission, although a positively selected change (R911C) in nsp3 was observed among human-derived viruses alone, suggesting that viral adaptation to our species represented the underlying pressure [48].
HCoV-229E
A recent analysis indicated that HCoV-229E may have recombined with the alpaca alpha-CoV virus within the S gene, as also demonstrated by the distinct phylogenetic trees for the S1 and S2 regions [19]. Also, HCoV-229E acquired a deletion in the S gene compared to bat viruses [19]. Recent sequencing of several of such viruses showed that this deletion is also present in the alpaca CoV S gene and in camel-derived alpha-CoVs [22]. This finding is particularly interesting because deletions in the NTD are associated with changes in tissue tropism in TGEV (transmissible gastroenteritis virus): in this porcine virus the spike has dual tropism for the respiratory and intestinal tracts, but the N-terminally deleted variants from PRCV (porcine respiratory coronavirus) only infect the respiratory tract 50, 51. In chiroptera, CoVs are mainly restricted to the gastrointestinal tract, whereas in humans and camelids, the upper and lower respiratory airways are infected. It will be important to determine whether the S gene deletion in HCoV-229E and camelid alphaCoVs is indeed responsible for a change in tissue tropism.
HCoV-NL63
Recombination contributed to shaping the diversity of the S gene among HCoV-NL63 viruses. Recombination between an ancestral HCoV-NL63 virus and the related PEDV was also detected in the M gene that, in its 3′ portion, is more similar to PEDV than to HCoV-229E [52]. Like SARS-CoV, HCoV-NL63 uses its RBD to bind ACE2. The binding site on the cellular receptor is the same for the two viruses but the RDBs show no sequence similarity. Interestingly, the RBDs of SARS-CoV and HCoV-NL63 do not display any structural similarity either: HCoV-NL63 contacts ACE2 with three discontinuous beta-loops, whereas SARS-CoV binds the receptor through a continuous subdomain [53] (Figure 3B). These observations suggest that the two viruses independently acquired the ability to bind the same ACE2 region via convergent evolution or that they shared an ACE2-binding ancestor long ago. Strikingly, TGEV, which is phylogenetically related to HCoV-NL63, uses two regions corresponding to the HCoV-NL63 beta-loops to bind a distinct cellular receptor, ANPEP (Figure 1B, Figure 3B) [54]. Finally, HCoV-229E, sharing sequence homology with HCoV-NL63 and TGEV (Figure 1A), binds ANPEP, but engages a region distinct from that bound by TGEV [55]. Overall, these data highlight the extraordinary plasticity of CoV RBDs, and their complex evolutionary dynamics whereby divergent evolution can be followed by convergent adaptation to the same receptor. This complexity is further expanded by the ability of some CoVs to use other cellular attachment molecules to complement the function of the RBD. Indeed, the S protein of HCoV-NL63 exploits heparan sulfate proteoglycans to adhere to host cells [56]. Interestingly, a similar ability to bind heparan sulfate can be gained by MHV with relatively few in vitro-acquired mutations in the S protein [57]. In line with the view that heparan sulfate is an aspecific receptor, the mutant MHV viruses display expanded host tropism [57], highlighting the potential relevance of combinatorial receptor usage or receptor shifts for interspecies transmission.
HCoV-OC43
Recombination seems to be rampant in HCoV-OC43 viruses and contributed to originate the A to E viral genotypes, as well as viruses that do not belong to these major genotypes 58, 59, 60. To our knowledge, no study has analyzed the fitness of recombinant viruses or, more generally, of viruses belonging to distinct genotypes. Nonetheless, two reports indicated that genotype D has become predominant in the East Asian population 58, 59. Whether this is due to population acquired immunity against the older A and B genotypes or to viral features unrelated to antigenicity remains to be determined.
The active recombination in HCoV-OC43 suggests that inference of natural selection is best performed by analysis of sequences belonging to the same genotype. In one such analysis, positive selection was found to act on the S gene of genotype D viruses [61]. Interestingly, several positively selected sites with high posterior probability of positive selection are located in the NTD, which is involved in the binding of sialic acids.
A positively selected site was located in the CTD, a region that has unclear function in the HCoV-OC43 S protein, as no known protein receptor has been identified to date [61]. However, recent data from HKU1 suggest that, by analogy, a protein receptor for HCoV-OC43 may exist [62] (see below).
HCoV-HKU1
The structure of the S protein of HKU1 was recently solved; the glycan-binding site is located in the NTD and is conserved with bovine coronavirus (BCoV) S1 [63]. Nonetheless, antibodies against the CTD, but not those against the NTD, block HKU1 infection of human tracheal–bronchial epithelial cells, suggesting that the CTD is the major RBD, and that a protein receptor for HKU1 exists [62]. In analogy to HCoV-NL63, glycans may mediate only the initial attachment to the host cells.
A recent survey of HKU1 clinical isolates from different geographic origins indicated that most viruses from Colorado form a subclade in the HKU1 phylogeny and carry three distinctive substitutions in the S protein within the NTD, CTD, and close to the S1/S2 cleavage site (W197F, F613Y, and H716D, respectively) [64]. It will be interesting to assess whether these differences are functional and derive from a selective process.
Concluding Remarks
Thanks to high-throughput techniques, a large number of complete CoV genomes have become available to the scientific community, and many more will be coming in the near future. Field studies have contributed enormously to widen our knowledge on the diversity of CoVs hosted by different vertebrates, and epidemiological surveys have provided CoV sequences from distinct geographic areas and associated with different disease phenotypes. In parallel, resources have been created to store and mine these data (e.g., The Virus Pathogen Database and Analysis Resource, ViPR [65]). These advances have allowed tracing the evolutionary history of the large and complex CoV genomes to an unprecedented detail. The emerging picture indicates that CoV genomes display high plasticity in terms of gene content and recombination. The long CoV genome expands the sequence space available for adaptive mutation, and the spike protein can adapt with relative ease to exploit different cellular receptors. These features are likely to underlie the alarming propensity of CoVs for host jumps. Despite these advances, major challenges remain (see Outstanding Questions). Efforts to underscore the viral genetic determinants that favor interspecies transmission should be pursued as an effective strategy to prevent or prepare for future HCoV emergence.
Outstanding Questions.
Do different genotypes (arising from recombination or mutation) of the same CoV species result in distinct viral phenotypes?
What is the relevance of adaptive changes in nonstructural proteins for host adaptation? And what is the role played by accessory proteins in interspecies transmission and virulence?
Why do similar viruses determine very different disease phenotypes in distinct mammalian hosts? Do the viral and host genome interplay in determining disease? And to what extent?
What is the distribution of CoVs in different mammalian orders? And how genetically diverse are CoVs hosted in wild or domestic mammals?
Can evolutionary information be applied to develop tools to predict which viruses have greater zoonotic potential and may result in the next epidemic?
Glossary
- dN
the observed number of nonsynonymous substitutions per nonsynonymous site.
- dS
the observed number of synonymous substitutions per synonymous site.
- Hemagglutinin-esterases (HEs)
a family of viral proteins that mediate binding to O-acetylated sialic acids.
- Homology
the relationship between elements (e.g., genes, proteins) deriving from a common ancestor.
- Lectins
a group of proteins with carbohydrate recognition activity. Lectins are categorized in many distinct families depending on structural and functional properties.
- Maximum likelihood (ML)
is a statistical method for estimating population parameters from a data sample. Given one or more unknown parameters and a sample data, the ML estimates of the parameters are the values maximizing the probability of obtaining the observed data.
- Open reading frame (ORF)
the part of a reading frame that contains no stop codons. An ORF is a continuous stretch of nucleotide triplets that have the potential to code for a protein or a peptide.
- Phosphodiesterases (PDEs)
are enzymes that break a phosphodiester bond. PDEs belonging to the 2H family are characterized by two H-Φ-[S/T]-Φ motifs (where Φ is a hydrophobic residue) separated by an average of 80 residues.
- Positive selection
the accumulation of favorable amino acid-replacing substitutions, which results in more nonsynonymous changes than expected under neutrality (dN/dS>1).
- Purifying selection
the elimination of deleterious amino acid-replacing substitutions, which results in fewer nonsynonymous changes than expected under neutrality (dN/dS < 1) (it is also referred to as negative selection).
- Viroporins
hydrophobic viral proteins that can promote the formation of channels following insertion into the host cell membrane and oligomerization.
References
- 1.Graham R.L. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lau S.K. Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A. J. Virol. 2015;89:3076–3092. doi: 10.1128/JVI.02420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan J.F. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drexler J.F. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su S. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckerle L.D. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckerle L.D. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vega V.B. Mutational dynamics of the SARS coronavirus in cell culture and human populations isolated in 2003. BMC Infect. Dis. 2004;4:32. doi: 10.1186/1471-2334-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lauber C. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLoS Pathog. 2013;9:e1003500. doi: 10.1371/journal.ppat.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subissi L. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menachery V.D. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. U. S. A. . 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge X.Y. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X.L. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J. Virol. 2015;90:3253–3256. doi: 10.1128/JVI.02582-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau S.K. Severe acute respiratory syndrome (SARS) coronavirus ORF8 protein is acquired from sars-related coronavirus from greater horseshoe bats through recombination. J. Virol. 2015;89:10532–10547. doi: 10.1128/JVI.01048-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Z. ORF8-related genetic evidence for Chinese horseshoe bats as the source of human severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2016;213:579–583. doi: 10.1093/infdis/jiv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 17.Scheffler K. Robust inference of positive selection from recombining coding sequences. Bioinformatics. 2006;22:2493–2499. doi: 10.1093/bioinformatics/btl427. [DOI] [PubMed] [Google Scholar]
- 18.Wertheim J.O. RELAX: detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol. 2015;32:820–832. doi: 10.1093/molbev/msu400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corman V.M. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015;89:11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crossley B.M. Identification and characterization of a novel alpaca respiratory coronavirus most closely related to the human coronavirus 229E. Viruses. 2012;4:3689–3700. doi: 10.3390/v4123689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crossley B.M. Identification of a novel coronavirus possibly associated with acute respiratory syndrome in alpacas (Vicugna pacos) in California, 2007. J. Vet. Diagn. Invest. 2010;22:94–97. doi: 10.1177/104063871002200118. [DOI] [PubMed] [Google Scholar]
- 22.Sabir J.S. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 23.Zhao L. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell. Host Microbe. 2012;11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang R. Homologous 2′,5′-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proc. Natl. Acad. Sci. U. S. A. 2013;110:13114–13119. doi: 10.1073/pnas.1306917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gusho E. Murine AKAP7 has a 2′,5′-phosphodiesterase domain that can complement an inactive murine coronavirus ns2 gene. mBio. 2014;5:e01312–e1314. doi: 10.1128/mBio.01312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thornbrough J.M. Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio. 2016 doi: 10.1128/mBio.00258-16. Published online March 29, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L., Li F. Structural analysis of the evolutionary origins of influenza virus hemagglutinin and other viral lectins. J. Virol. 2013;87:4118–4120. doi: 10.1128/JVI.03476-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng G. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang X. Human coronavirus HKU1 spike protein uses o-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desforges M. The acetyl-esterase activity of the hemagglutinin-esterase protein of human coronavirus OC43 strongly enhances the production of infectious virus. J. Virol. 2013;87:3097–3107. doi: 10.1128/JVI.02699-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dijkman R. Human coronavirus 229E encodes a single ORF4 protein between the spike and the envelope genes. Virol. J. 2006;3:106. doi: 10.1186/1743-422X-3-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farsani S.M. The first complete genome sequences of clinical isolates of human coronavirus 229E. Virus Genes. 2012;45:433–439. doi: 10.1007/s11262-012-0807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang R. The ORF4a protein of human coronavirus 229E functions as a viroporin that regulates viral production. Biochim. Biophys. Acta. 2014;1838:1088–1095. doi: 10.1016/j.bbamem.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang R. The ns12.9 accessory protein of human coronavirus OC43 is a viroporin involved in virion morphogenesis and pathogenesis. J. Virol. 2015;89:11383–11395. doi: 10.1128/JVI.01986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu W. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koetzner C.A. Accessory protein 5a is a major antagonist of the antiviral action of interferon against murine coronavirus. J. Virol. 2010;84:8262–8274. doi: 10.1128/JVI.00385-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao G.P. SARS molecular epidemiology: a Chinese fairy tale of controlling an emerging zoonotic disease in the genomics era. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2007;362:1063–1081. doi: 10.1098/rstb.2007.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu G. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corman V.M. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014;88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J.I. The recent ancestry of Middle East respiratory syndrome coronavirus in Korea has been shaped by recombination. Sci. Rep. 2016;6:18825. doi: 10.1038/srep18825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forni D. The heptad repeat region is a major selection target in MERS-CoV and related coronaviruses. Sci. Rep. 2015;5:14480. doi: 10.1038/srep14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cotten M. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio. 2014 doi: 10.1128/mBio.01062-13. Published online February 18, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada Y. Acquisition of cell-cell fusion activity by amino acid substitutions in spike protein determines the infectivity of a coronavirus in cultured cells. PLoS One. 2009;4:e6130. doi: 10.1371/journal.pone.0006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Navas-Martin S. Murine coronavirus evolution in vivo: functional compensation of a detrimental amino acid substitution in the receptor binding domain of the spike glycoprotein. J. Virol. 2005;79:7629–7640. doi: 10.1128/JVI.79.12.7629-7640.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McRoy W.C., Baric R.S. Amino acid substitutions in the S2 subunit of mouse hepatitis virus variant V51 encode determinants of host range expansion. J. Virol. 2008;82:1414–1424. doi: 10.1128/JVI.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim Y. Spread of mutant Middle East respiratory syndrome coronavirus with reduced affinity to human CD26 during the South Korean outbreak. mBio. 2016 doi: 10.1128/mBio.00019-16. Published online March 1, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forni D. Extensive positive selection drives the evolution of nonstructural proteins in lineage C betacoronaviruses. J. Virol. 2016;90:3627–3639. doi: 10.1128/JVI.02988-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baez-Santos Y.M. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasschaert D. Porcine respiratory coronavirus differs from transmissible gastroenteritis virus by a few genomic deletions. J. Gen. Virol. 1990;71:2599–2607. doi: 10.1099/0022-1317-71-11-2599. [DOI] [PubMed] [Google Scholar]
- 51.Sanchez C.M. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pyrc K. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J. Mol. Biol. 2006;364:964–973. doi: 10.1016/j.jmb.2006.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu K. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reguera J. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. 2012;8:e1002859. doi: 10.1371/journal.ppat.1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen L. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc. Natl. Acad. Sci. U. S. A. 2012;109:17966–17971. doi: 10.1073/pnas.1210123109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milewska A. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 2014;88:13221–13230. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Haan C.A. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J. Virol. 2005;79:14451–14456. doi: 10.1128/JVI.79.22.14451-14456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lau S.K. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J. Virol. 2011;85:11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y. Genotype shift in human coronavirus OC43 and emergence of a novel genotype by natural recombination. J. Infect. 2015;70:641–650. doi: 10.1016/j.jinf.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Al-Khannaq M.N. Molecular epidemiology and evolutionary histories of human coronavirus OC43 and HKU1 among patients with upper respiratory tract infections in Kuala Lumpur, Malaysia. Virol. J. 2016;13 doi: 10.1186/s12985-016-0488-4. 33-016-0488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ren L. Genetic drift of human coronavirus OC43 spike gene during adaptive evolution. Sci. Rep. 2015;5:11451. doi: 10.1038/srep11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qian Z. Identification of the receptor-binding domain of the spike glycoprotein of human betacoronavirus HKU1. J. Virol. 2015;89:8816–8827. doi: 10.1128/JVI.03737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kirchdoerfer R.N. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dominguez S.R. Isolation, propagation, genome analysis and epidemiology of HKU1 betacoronaviruses. J. Gen. Virol. 2014;95:836–848. doi: 10.1099/vir.0.059832-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pickett B.E. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anisimova M. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics. 2003;164:1229–1236. doi: 10.1093/genetics/164.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guindon S. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 68.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 70.Kosakovsky Pond S.L. Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 71.Maydt J., Lengauer T. Recco: recombination analysis using cost optimization. Bioinformatics. 2006;22:1064–1071. doi: 10.1093/bioinformatics/btl057. [DOI] [PubMed] [Google Scholar]
- 72.Martin D., Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 73.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 74.Zhang J. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 75.Murrell B. Gene-wide identification of episodic selection. Mol. Biol. Evol. 2015;32:1365–1371. doi: 10.1093/molbev/msv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kosakovsky Pond S.L. A random effects branch-site model for detecting episodic diversifying selection. Mol. Biol. Evol. 2011;28:3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McDonald J.H., Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 78.Gharib W.H., Robinson-Rechavi M. The branch-site test of positive selection is surprisingly robust but lacks power under synonymous substitution saturation and variation in GC. Mol. Biol. Evol. 2013;30:1675–1686. doi: 10.1093/molbev/mst062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xia X. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 2003;26:1–7. doi: 10.1016/s1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- 80.Sealfon R.S. FRESCo: finding regions of excess synonymous constraint in diverse viruses. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0603-7. 38-015-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Groot R.J. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wertheim J.O. A case for the ancient origin of coronaviruses. J. Virol. 2013;87:7039–7045. doi: 10.1128/JVI.03273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Woo P.C. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blair J.E., Hedges S.B. Molecular phylogeny and divergence times of deuterostome animals. Mol. Biol. Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- 85.Huynh J. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012;86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pfefferle S. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg. Infect. Dis. 2009;15:1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vijgen L. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Z. Evolutionary dynamics of MERS-CoV: potential recombination, positive selection and transmission. Sci. Rep. 2016;6:25049. doi: 10.1038/srep25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gralinski L.E., Baric R.S. Molecular pathology of emerging coronavirus infections. J. Pathol. 2015;235:185–195. doi: 10.1002/path.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krogh A. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.