Abstract
Background. Human coronavirus NL63 (HCoV-NL63) is a recently discovered human coronavirus found to cause respiratory illness in children and adults that is distinct from the severe acute respiratory syndrome (SARS) coronavirus and human coronaviruses 229E (HCoV-229E) and OC43 (HCoV-OC43).
Methods. We investigated the role that HCoV-NL63, HCoV-OC43, and HCoV-229E played in children hospitalized with fever and acute respiratory symptoms in Hong Kong during the period from August 2001 through August 2002.
Results. Coronavirus infections were detected in 26 (4.4%) of 587 children studied; 15 (2.6%) were positive for HCoV-NL63, 9 (1.5%) were positive for HCoV-OC43, and 2 (0.3%) were positive for HCoV-229E. In addition to causing upper respiratory disease, we found that HCoV-NL63 can present as croup, asthma exacerbation, febrile seizures, and high fever. The mean age (± standard deviation [SD]) of the infected children was 30.7 ± 19.8 months (range, 6–57 months). The mean maximum temperature (±SD) for the 12 children who were febrile was 39.3°C ± 0.9°C, and the mean total duration of fever (±SD) for all children was 2.6 ± 1.2 days (range, 1–5 days). HCoV-NL63 infections were noted in the spring and summer months of 2002, whereas HCoV-OC43 infection mainly occurred in the fall and winter months of 2001. HCoV-NL63 viruses appeared to cluster into 2 evolutionary lineages, and viruses from both lineages cocirculated in the same season.
Conclusions. HCoV-NL63 is a significant pathogen that contributes to the hospitalization of children, and it was estimated to have caused 224 hospital admissions per 100,000 population aged ⩽6 years each year in Hong Kong.
Coronaviruses are enveloped RNA viruses and are subdivided into 3 groups. Human coronavirus 229E (HCoV-229E) and human coronavirus OC43 (HCoV-OC43) have been recognized since the mid 1960s as causes of upper respiratory disease, including the common cold, and are group 1 and group 2 coronaviruses, respectively. In 2003, a novel coronavirus (SARS-CoV) was identified as the causative agent of the severe acute respiratory syndrome (SARS). Its formal taxonomic status within the coronaviridae is not yet established [1]. Recently, another group 1 human coronavirus, HCoV-NL63, was reported in The Netherlands [2, 3]. In one report, 8 adults and children infected with HCoV-NL63 virus were documented; 3 patients had lower respiratory tract infection, 2 had an unknown clinical presentation, and the others had upper respiratory tract infection. A second report documented the same coronavirus in an 8-month-old boy with pneumonia [3]. One hundred thirty-nine other children and adults with respiratory tract infections whom tested negative for known viral pathogens underwent screening, which revealed the virus in 4 more children. However, a systematic study of the prevalence of HCoV-NL63 infection or its clinical presentation is still lacking. Furthermore, there is little information about the seasonality and clinical role of coronaviruses (other than SARS-CoV) in tropical or subtropical climates.
In 2001 and 2002, we performed prospective clinical and virological studies of children (age, ⩽18 years) with acute respiratory tract infection who were admitted to Queen Mary Hospital (Hong Kong, China). Data on human metapneumovirus (hMPV) infection, influenza, respiratory syncytial virus (RSV) infection, parainfluenza virus infection, and adenovirus infection were reported elsewhere [4]. In the present study, we use the clinical specimens to investigate the role of HCoV-229E, HCoV-OC43, and the newly discovered HCoV-NL63 on childhood respiratory infections.
Patients and Methods
Hong Kong Island has a population of 288,371 persons aged ⩽18 years, 84,018 of whom are ⩽6 years old (based on 2001 census data [5]). Two public hospitals of the Hospital Authority of Hong Kong, Queen Mary Hospital and Pamela Youde Nethersole Eastern Hospital, provide 90% of all acute pediatric hospital care for Hong Kong Island, with an admission ratio of 1 : 1.65.
We studied a systematic sample of children (age, ⩽18 years) with acute respiratory infection admitted to Queen Mary Hospital during the period from August 2001 to March 2002. All children admitted to Queen Mary Hospital with signs and symptoms of respiratory infection on one fixed day each week (Tuesday) were included in the study. From April through August 2002, study enrollment times were increased to Tuesday and Thursday. All children with respiratory infection were included, regardless of other complaints. Because we had previously documented an association between influenza virus A infection and febrile seizures, children with febrile seizures were also included [6]. Nasopharyngeal aspirate (NPA) specimens were collected for virological testing from all subjects. Two aliquots of the NPA specimen were frozen at -70°C for subsequent molecular testing. At the end of the study, all medical records were reviewed by a pediatrician (S.S.C.).
Viral diagnosis. Rapid diagnostic testing for RSV, influenza viruses A and B, adenovirus, and parainfluenza virus types 1–3 was performed with use of immunofluorescent antigen detection and culture, as previously described [4, 7, 8]. hMPV was detected by RT-PCR [4].
Coronavirus RT-PCR. Total nucleic acid was extracted from 220 µL of the NPA clinical specimen using the QIAamp Virus BioRobot 9604 kit (Qiagen), in accordance with the manufacturer's instructions. The total nucleic acid was eluted in 86 µL of the kit elution buffer and stored at -70°C. cDNA was generated using random hexamers, as described elsewhere [9]. PCR was performed using a set of consensus coronavirus-reactive primers, as well as primers specifically designed to amplify HCoV-229E, HCoV-OC43, and HCoV-NL63 [3, 10, 11].
The consensus primers Coro IN6–7 (forward, 5′-GGTTGGGACTATCCTAAGTGTGA-3′; reverse, 5′-CCATCATCAGATAGAATCATCATA-3′) amplified the conserved region of the coronavirus polymerase gene [9]. Two µL of cDNA was added to 50 uL of reaction mixture containing 1× PCR buffer (Roche Diagnostic Manufacturer, Germany), 2.5 µmol/L each primer, 100 µmol/L dNTP (Roche), 3 mmol/L MgCl2, and 2.5 U of AmpliTaq Gold polymerase (Roche). Samples were then heated to 94°C for 10 min, followed by 40 cycles of amplification as follows: 1 min each of denaturation, annealing, and extension at 94°C, 48°C, and 72°C, respectively, followed by final 10 min at 72°C.
RT-PCR was performed in a multiplex format, as described elsewhere [11] with modification [12]. The PCR conditions were the same as those for the Coro IN6 and 7 RT-PCR described above, except that denaturation was for 2 min at 94°C, and annealing was for 1 min at 60°C. Ten µL of PCR products were analyzed in a 2% (w/v) agarose gel by electrophoresis and stained with ethidium bromide, 0.5 µg/mL. The sizes of the PCR-amplified product for the coronavirus consensus (Coro IN6–7), 229E, and OC43 RT-PCR were 440 bp, 294 bp, and 469 bp, respectively.
A real-time quantitative PCR specific to the N region of HCoV-NL63 was used for specific detection and quantitation in NPA specimens [3]. In brief, 2 µL of cDNA was amplified in a 25-µL reaction containing 0.625 U of AmpliTaq Gold polymerase (Roche), TaqMan buffer A, 0.2 mmol/L dNTP, 5.5 mmol/L MgCl2, 0.25 U of AmpErase UNG, 0.8 µmol/L each primer, and 0.4 µmol/L probe. The primer sequences were forward, 5′-AGGACCTTAAATTCAGACAACGTTCT-3′; and reverse, 5′-GATTACGTTTGCGATTACCAAGACT-3′. The probe was 5′-6FAM-TGTTGTTTGGGTTGCTAAG-MGBNFQ-3′.
Reactions were first incubated at 50° for 2 min, followed by 95° C for 10 min, and they were then thermal-cycled for 40 cycles (95° C for 15 s and 60° C for 1 min). Plasmid containing the target was used as a positive control. Quantitative results are expressed as genome copies per reaction. One copy per reaction approximates to 326 copies per 1 mL of NPA.
Standard precautions were taken to prevent PCR cross-contamination during specimen extraction and PCR analysis. Positive and negative controls were included in each RT-PCR experiment.
Phylogenetic analysis of HCoV-NL63. PCR-amplified products of the 1a gene [2] were genetically sequenced, and deduced viral sequences were analyzed and aligned with use of BioEdit, version 5.0.9 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
Statistical analysis. HCoV-NL63 loads per reaction were converted to log10 values and correlated with the days from symptom onset using Pearson correlation. Unpairedt test was used for comparisons between children with high and those with low HCoV-NL63 loads. The χ2 test was used to detect the association between HCoV-NL63 load and the number of pathogens identified. A 2-sidedP value of <.05 was considered to be statistically significant. Data on clinical manifestations in patients with HCoV-NL63, influenza A, and RSV infection were analyzed with 1-way analysis of variance, and significant results were further tested with a post-hoc Tukey-Kramer test.
Results
We detected coronavirus infections in 26 (4.4%) of 587 children studied during the 13-month period. Fifteen children (2.6%) were positive for HCoV-NL63 infection, 9 (1.5%) were positive for HCoV-OC43 infection, and 2 (0.3%) were positive for HCoV-229E infection (table 1). For comparison, influenza A virus was detected in 7.7% of patients, RSV was detected in 8.2%, and adenovirus was detected in 7.2%, although some of these children had multiple viruses detected (table 1). Children aged ⩽6 years accounted for 510 of the 587 specimens studied. From the identification rate of HCoV-NL63 in our representative sample at Queen Mary Hospital, we estimate that 71 children aged ⩽6 years were admitted to this hospital over a 12-month period. With use of the known admission ratio between the 2 hospitals that provide care for 90% of patients on Hong Kong Island, we estimated that 188 children aged ⩽6 years were admitted because of HCoV-NL63 infection, leading to an annual hospitalization rate of 224 hospitalizations per 100,000 population aged ⩽6 years.
Table 1.
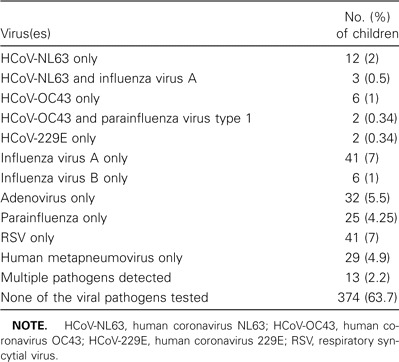
Viruses identified in nasopharyngeal aspirate specimens obtained from 587 hospitalized children.
Table 2 shows the performance of the coronavirus consensus and type-specific RT-PCR. Fourteen patients had cases detected using the consensus coronavirus primers. Genetic sequencing of the PCR-amplified product revealed the infecting virus to be HCoV-NL63 in 7 children, HCoV-OC43 in 5, and 229E in 2. These results were confirmed by the virus type—specific primers in all patients, with the exception of 1 patient for whom the amplified sequence indicated infection with HCoV-NL63 (table 2). This additional patient's case was confirmed using third primer set targeted at the polymerase 1a region used for phylogenetic analysis. The human coronavirus type-specific primers detected an additional 8 and 4 patients infected with HCoV-NL63 and HCoV-OC43, respectively. Therefore, in total, 15 patients had evidence of HCoV-NL63 infection (table 2).
Table 2.
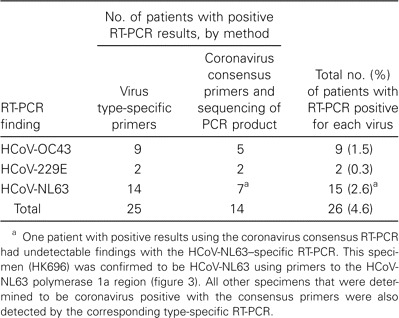
Comparative results of RT-PCR using consensus and specific primers for human coronaviruses NL63 (HCoV-NL63), OC43 (HCoV-OC43), and 229E (HCoV-229E) in nasopharyngeal aspirate specimens of 587 hospitalized children.
HCoV-OC43 infection mainly occurred in the fall and winter months of 2001, whereas HCoV-NL63 infections mainly occurred in the spring and summer months of 2002 (figure 1). The mean age (±SD) of the 15 children infected with HCoV-NL63 was 30.7 ± 19.8 months (range, 6–57 months). Three children had no documented fever in the hospital. Two of them had a history of fever at home. One totally afebrile child was hospitalized for stridor. The mean maximum temperature (±SD) for the 12 children who were febrile in the hospital was 39.3°C ± 0.9°C, and the mean total duration of fever (±SD) for all children who were ever febrile was 2.6 ± 1.2 days (range, 1–5 days). Most children presented with respiratory symptoms, although 1 child presented with febrile seizure without obvious respiratory infection (table 3). Chest radiography was performed for 8 patients, but all findings were normal. Five children (35%) had an alternative microbiologic diagnosis (influenza A H3N2 for 3 children, group AStreptococcus species isolated from a throat culture for 1 child with scarlet fever, andCampylobacter jejuni in the stool of a child with upper respiratory infection and diarrhea). One child with influenza A and HCoV-NL63 coinfection also hadEscherichia coli urinary tract infection. The 11 children who had HCoV-NL63 as the sole cause of infection and the patient with diarrhea who had respiratory symptoms not explained byC. jejuni infection were compared with age-matched children with influenza A or RSV infection. Children admitted for influenza A had a longer mean duration of fever than did those infected with HCoV-NL63 (table 4). Children with HCoV-NL63 infection were less likely than those with RSV infection to have lower respiratory involvement (9% vs. 63.6%;P = .02).
Figure 1.
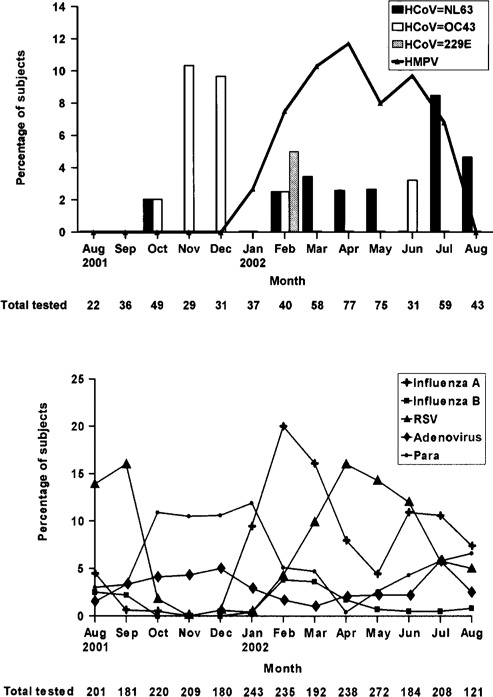
A, Seasonality of human coronavirus infections in Hong Kong, China. The percentage of subjects with RT-PCR positive for human coronavirus NL63 (HCoV-NL63), OC-43 (HCoV-OC43), and 229E (HCoV-229E), as well as human metapneumovirus (hMPV), in the study population of 587 patients is shown.B, The percentage of patients positive for influenza virus A or B, parainfluenza virus (Para), adenovirus, and respiratory syncytial virus (RSV) among all 2684 pediatric admissions during this same period. Adapted from [4].
Table 3.
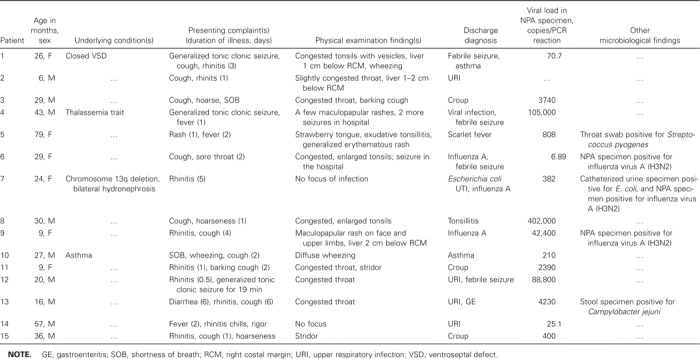
Clinical data on children with human coronavirus NL63 RNA detected in nasopharyngeal aspirate (NPA) specimens.
Table 4.
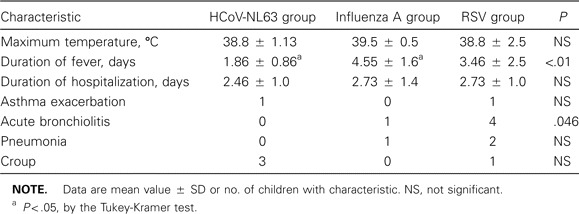
Comparison of clinical manifestations for 11 children infected solely with human coronavirus NL63 (HCoV-NL63), influenza virus A, or respiratory syncytial virus (RSV).
If we excluded patients for whom the presence of other respiratory pathogens was documented, the HCoV-NL63 load in NPA specimens appears to correlate inversely with the time after onset of symptoms (r = -0.8) (figure 2). If patients were categorized into those with high viral load (⩾1000 copies/reaction) and low viral load (<1000 copies/reaction), the mean times (±SD) between onset of symptoms (defined as onset of fever, if the child was febrile, or as onset of other respiratory symptoms, if the child was afebrile) and specimen collection were 1 ± 0.6 and 2.25 ± 0.96 days, respectively (P = .02). Neither peak temperature nor duration of fever was found to be significantly different between the 2 groups. Six of 10 patients with HCoV-NL63 as the sole identified pathogen had a high viral load, compared with 1 of 4 children infected with multiple viruses (P not significant). One child with HCoV-NL63 upper respiratory tract infection had positive results of only the consensus primer PCR, so no viral load could be measured.
Figure 2.
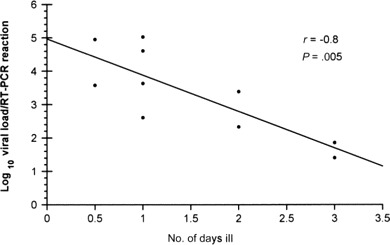
Correlation of human coronavirus NL63 (HCoV-NL63) load and duration of illness. Patients with multiple respiratory pathogens are excluded from this analysis. Viral load is expressed as genome copies per reaction, and 1 genome copy per reaction approximates to 326 copies per 1 mL of nasopharyngeal aspirate (NPA) material. The viral load appears to correlate inversely with time after onset of clinical symptoms, withr = -0.8 (by Pearson Correlation coefficient). The equation log10 (viral load) = -1.09 (day) + 4.96 was found to predict HCoV-NL63 load in the NPA specimen in relation to symptom onset.
Phylogenetic analysis of HCoV-NL63 1a gene recovered from 12 patients indicated that these viruses cluster into 2 evolutionary lineages. Both lineages were represented in Hong Kong and appear to cocirculate in the same season (figure 3).
Figure 3.

Phylogenetic analysis of the human coronavirus NL93 (HCoV-NL63) polymerase 1a gene region. Phylogenetic trees were constructed by the neighbor-joining method, and bootstrap values were determined by 1000 replicates in MEGA 2.1 (available at http://www.megasoftware.net). Viral sequences generated from this study are in bold. Two complete viral genome sequences, HCoV-NL63 and human group 1 coronavirus associated with pneumonia (human group 1 coronavirus), and other reference sequences were obtained in GenBank (accession numbers AY518894, AY567488-AY567494, NC 005831, and AY675541–675553). Genetic sequences from 4 other patients could not be obtained, because the primers for the polymerase 1a region did not amplify a PCR product of sufficient quantity to allow reliable genetic sequencing analysis.
The 9 patients with HCoV-OC43 infection had a mean age (±SD) of 33.6 ± 14.5 months (range, 14–57 months) (table 5). Two children remained afebrile while in the hospital: one was a 28-month-old boy with stridor who never had fever, even while at home, and the other was a 23-month-old girl with known early asthma who presented with exacerbation of asthma and a 6-day history of fever (temperature, ⩾39.4°C) before hospital admission. The mean maximum temperature (±SD) of febrile children in the hospital was 39.3°C ± 0.4°C, with a mean total duration of fever (±SD) of 5 ± 2.4 days (range, 1.5–8 days). The mean duration of hospitalization (±SD) for this group of children was 2.6 ± 1.5 days. Chest radiography was performed for 7 patients, and 3 had evidence of infiltrates. One of these children also had evidence of parainfluenza type 1 infection. The discharge diagnoses of the other 2 were viral pneumonia and atypical pneumonia (table 4).
Table 5.
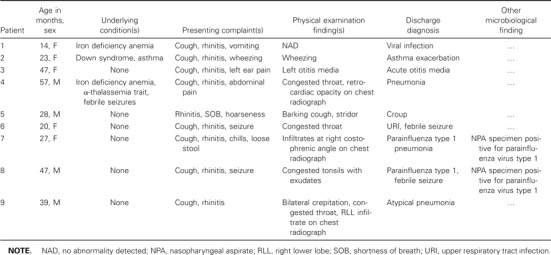
Characteristics of children with human coronavirus OC43 infection.
Two children had HCoV-229E identified in their NPA specimens. One was a 23-month-old girl who presented with rhinitis, cough, and fever and who was found to have acute otitis media and right lower lobe pneumonia during examination. Blood culture subsequently yieldedStreptococcus pneumoniae. The other patient was a 13-year-old boy who had a brain tumor and who had undergone renal transplantation, with prolonged fever. Tests of his NPA were positive for HCoV-229E on 3 occasions over the subsequent 3 months. He had minimal respiratory symptoms throughout and had no abnormalities detected by chest radiography.
Discussion
We documented that human coronavirus infection was a significant cause of hospitalization for children aged ⩽18 years, accounting for 4.4% of all admissions for acute respiratory infections. The newly discovered HCoV-NL63 was the most common coronavirus identified and was estimated to be associated with a hospitalization rate of 210.6 hospitalizations per 100,000 population aged ⩽6 years.
A previous study showed that 8.2% of children aged <18 months who were admitted to a Chicago, Illinois, hospital with lower respiratory tract diseases had serological evidence of HCoV-229E or HCoV-OC43 infection [13]. Of these children infected with coronavirus, 59% had pneumonia, 29% had bronchiolitis, and 5.8% had croup. In another study that used an ELISA to detect HCoV-229E and HCoV-OC43 antigen in respiratory tract secretions obtained from 108 children with respiratory infection and 51 siblings, human coronavirus was found in 30% and 29% of the samples from the index children and their siblings, respectively [14]. This high level of detection could be biased by testing during the winter months and by performing tests mostly on specimens that tested negative for other organisms. Of the 45 children infected, only 2 children had pneumonia.
HCoV-OC43 and HCoV-229E are reported to have a winter seasonality in temperate regions, and preliminary data on the new HCoV-NL63 from The Netherlands also suggests a winter predominance [2, 10, 15]. In Hong Kong, we found that HCoV-OC43 was predominant in the winter, whereas HCoV-NL63 had a spring-summer peak of activity. There was little cocirculation of these 2 viruses. Our data suggest that the seasonality of coronaviruses in tropical and subtropical regions is not restricted to the winter.
We have described the clinical spectrum of this newly discovered HCoV-NL63 in a cohort of hospitalized children. In addition to causing upper respiratory disease, we found that HCoV-NL63 infection can present as croup, asthma exacerbation, and febrile seizures. The peak age for hospitalization with HCoV-NL63 infection was 2–3 years, which is similar to that for HCoV-OC43 infection. In comparison, the mean ages (±SD) of children admitted to the hospital with influenza, hMPV infection, and RSV infection in the same cohort were 30.2 ± 22.6, 31.7 ± 18.7, and 19.5 ± 17.2 months, respectively [4]. In contrast to an earlier report, the majority of children with HCoV-NL63 infection in our study had no major underlying disorders [3]. A coinfecting pathogen was identified in 5 of the 15 children. The child withC. jejuni diarrhea also had symptoms of an upper respiratory tract infection that were not explained by the bacterial infection but were probably attributable to HCoV-NL63. In the other 4 children, the coinfecting pathogen could fully explain the clinical presentation, and it is difficult to comment on the contribution of HCoV-NL63. When age-matched children with influenza A or RSV infection and HCoV-NL63 infection were compared, children with HCoV-NL63 infection had a shorter duration of fever than did those with influenza A, and they were less likely to have lower respiratory tract involvement than were those with RSV infection. The duration of hospitalization seemed to be equivalent for all 3 infections. Rapid virological diagnosis of RSV infection and influenza were available to clinicians, but diagnosis of HCoV-NL63 infection was retrospective. Availability of rapid diagnosis has been shown to result in shorter hospital stay [7].
The HCoV-NL63 load in the nasopharynx is high during the first 1–2 days of symptoms and decreases in the next few days (figure 2). No correlation with disease severity was observed. These viral load data derived from cross-sectional data for a small cohort of patients and need to be further confirmed with sequential specimens. This pattern, with peak viral load around the onset of disease, is similar to that seen for influenza but different from that observed for SARS [16, 17]. The quantitative RT-PCR missed 1 of the patients with HCoV-NL63, suggesting that further optimization of the primer and probe sequence is required.
HCoV-OC43 was associated with exacerbation of asthma and pneumonia, as well as upper respiratory tract disease, in this study. In 1 study of 11 hospitalized US military recruits who only had evidence of coronavirus seroconversion, 36% had pneumonia [18]. Asthma exacerbation was found in 14% of children with HCoV-NL63 infection and in 11% of children with HCoV-OC43 infections in this study [19].
HCoV-229E was repeatedly detected in 1 immunocompromised child over 3 months. Genetic analysis of a 305-bp region of the spike protein region of 229E revealed that these 3 specimens had identical genetic sequence (unpublished data). However, more contemporary HCoV-229E sequences from Hong Kong would be needed to conclude whether the sequence identity in these 3 specimens provided evidence of virus persistence rather than reinfection. Both reinfection with human coronavirus and prolonged shedding have been previously reported [14].
A number of children with human coronavirus infection had other pathogens detected in the same clinical specimen. Multiple pathogens in the respiratory tract of patients with respiratory disease are being increasingly recognized [13]. It is difficult to establish whether these are true coinfections or whether one of the pathogens represents continued shedding from a previous infection. It was previously shown that human coronavirus was detectable by PCR in 0.4% of patients without symptoms of respiratory tract infection and that HCoV RNA could be detected by real-time RT-PCR up to 14 days after infection [12]. In patients with coinfection, the apparent association between influenza virus A with HCoV-NL63 and parainfluenza virus type 1 with HCoV-OC43 is likely due to the coincidence of the seasonality of these viruses.
In conclusion, we demonstrate that human coronaviruses contribute to hospitalization of children with respiratory disease and that the newly recognized HCoV-NL63 is at least as important as—if not more important than—HCoV-229E and HCoV-OC43 in this regard. We confirm earlier findings from The Netherlands [2] that there are at least 2 lineages of HCoV-NL63 and demonstrate that both variants may cocirculate in the tropics. A better understanding of human coronavirus epidemiology is of particular importance in view of potential interactions in pathogenesis and serodiagnosis of SARS.
Acknowledgments
We thank Bonnie Wong and Chiu Mei Pang, for technical support, and Wilfred H. S. Wong, for statistical analysis.
Financial support. Research Grants Council of Hong Kong (HKU 7396/03M) and The Ellison Medical Foundation (grant ID-IA-0036-02).
Potential conflicts of interest. All authors: no conflict.
References
- 1.Gorbalenya AE, Snijder EJ, Spaan WJ. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J Virol. 2004;78:7863–6. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Hoek L, Pyrc K, Jebbink MF, et al. Identification of a new human coronavirus. Nat Med. 2004;10:368–73. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fouchier RA, Hartwig NG, Bestebroer TM, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci USA. 2004;101:6212–6. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peiris JS, Tang WH, Chan KH, et al. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis. 2003;9:628–33. doi: 10.3201/eid0906.030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Census & Statistics Department . 2001 Population census. 26 October. 2001. Available at: http://www.info.gov.hk/censtatd/eng/hkstat/fas/01c/cd0272001_index.html. [Google Scholar]
- 6.Chiu SS, Tse CY, Lau YL, Peiris M. Influenza A infection is an important cause of febrile seizures. Pediatrics. 2001;108:63. doi: 10.1542/peds.108.4.e63. [DOI] [PubMed] [Google Scholar]
- 7.Woo PC, Chiu SS, Seto WH, Peiris M. Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J Clin Microbiol. 1997;35:1579–81. doi: 10.1128/jcm.35.6.1579-1581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan KH, Maldeis N, Pope W, et al. Evaluation of the Directigen FluA+B test for rapid diagnosis of influenza virus type A and B infections. J Clin Microbiol. 2002;40:1675–80. doi: 10.1128/JCM.40.5.1675-1680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poon LL, Wong OK, Chan KH, et al. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS) Clin Chem. 2003;49:953–5. doi: 10.1373/49.6.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–76. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 11.Pitkaranta A, Arruda E, Malmberg H, Hayden FG. Detection of rhinovirus in sinus brushings of patients with acute community-acquired sinusitis by reverse transcription-PCR. J Clin Microbiol. 1997;35:1791–3. doi: 10.1128/jcm.35.7.1791-1793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Elden LJ, van Loon AM, van Alphen F, et al. Frequent detection of human coronaviruses in clinical specimens from patients with respiratory tract infection by use of a novel real-time reverse-transcriptase polymerase chain reaction. J Infect Dis. 2004;189:652–7. doi: 10.1086/381207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McIntosh K, Chao RK, Krause HE, Wasil R, Mocega HE, Mufson MA. Coronavirus infection in acute lower respiratory tract disease of infants. J Infect Dis. 1974;130:502–7. doi: 10.1093/infdis/130.5.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isaacs D, Flowers D, Clarke JR, Valman HB, MacNaughton MR. Epidemiology of coronavirus respiratory infections. Arch Dis Child. 1983;58:500–3. doi: 10.1136/adc.58.7.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vabret A, Mourez T, Gouarin S, Petitjean J, Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin Infect Dis. 2003;36:985–9. doi: 10.1086/374222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser L, Briones MS, Hayden FG. Performance of virus isolation and Directigen Flu A to detect influenza A virus in experimental human infection. J Clin Virol. 1999;14:191–7. doi: 10.1016/s1386-6532(99)00058-x. [DOI] [PubMed] [Google Scholar]
- 17.Peiris JS, Chu CM, Cheng VC, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–72. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wenzel RP, Hendley JO, Davies JA, Gwaltney JM., Jr Coronavirus infections in military recruits: three-year study with coronavirus strains OC43 and 229E. Am Rev Respir Dis. 1974;109:621–4. doi: 10.1164/arrd.1974.109.6.621. [DOI] [PubMed] [Google Scholar]
- 19.McIntosh K, Ellis EF, Hoffman LS, Lybass TG, Eller JJ, Fulginiti VA. The association of viral and bacterial respiratory infections with exacerbations of wheezing in young asthmatic children. J Pediatr. 1973;82:578–90. doi: 10.1016/S0022-3476(73)80582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]


