Summary
Background
The disease course of amyotrophic lateral sclerosis (ALS) is rapid and, because its pathophysiology is unclear, few effective treatments are available. Genetic research aims to understand the underlying mechanisms of ALS and identify potential therapeutic targets. The first gene associated with ALS was SOD1, identified in 1993 and, by early 2014, more than 20 genes had been identified as causative of, or highly associated with, ALS. These genetic discoveries have identified key disease pathways that are therapeutically testable and could potentially lead to the development of better treatments for people with ALS.
Recent developments
Since 2014, seven additional genes have been associated with ALS (MATR3, CHCHD10, TBK1, TUBA4A, NEK1, C21orf2, and CCNF), all of which were identified by genome-wide association studies, whole genome studies, or exome sequencing technologies. Each of the seven novel genes code for proteins associated with one or more molecular pathways known to be involved in ALS. These pathways include dysfunction in global protein homoeostasis resulting from abnormal protein aggregation or a defect in the protein clearance pathway, mitochondrial dysfunction, altered RNA metabolism, impaired cytoskeletal integrity, altered axonal transport dynamics, and DNA damage accumulation due to defective DNA repair. Because these novel genes share common disease pathways with other genes implicated in ALS, therapeutics targeting these pathways could be useful for a broad group of patients stratified by genotype. However, the effects of these novel genes have not yet been investigated in animal models, which will be a key step to translating these findings into clinical practice.
Where next?
The identification of these seven novel genes has been important in unravelling the molecular mechanisms underlying ALS. However, our understanding of what causes ALS is not complete, and further genetic research will provide additional detail about its causes. Increased genetic knowledge will also identify potential therapeutic targets and could lead to the development of individualised medicine for patients with ALS. These developments will have a direct effect on clinical practice when genome sequencing becomes a routine and integral part of disease diagnosis and management.
Introduction
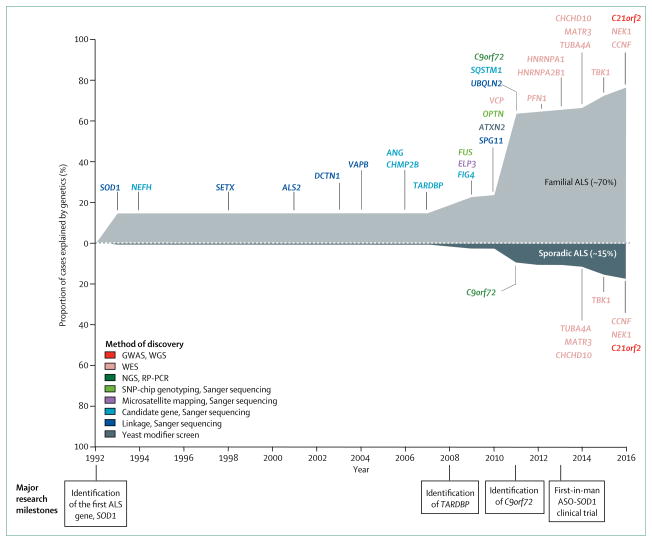
Typically, the disease course of amyotrophic lateral sclerosis (ALS) is rapid, and most patients die within 3–5 years of symptom onset as a result of respiratory failure.1 Although the disease is considered a rare type of motor neuron neurodegeneration, the number of patients with ALS is rapidly increasing because of population ageing. Most patients are aged between 50 and 75 years at diagnosis and, by 2040, an estimated 400 000 patients will be diagnosed with ALS worldwide.2 Approximately 10% of patients with ALS have a family history of disease, whereas the remainder of cases are classified as sporadic.1 The pathophysiology of ALS—familial or sporadic—is unclear, thus few effective treatments are available. Riluzole and edaravone are the current treatments approved for the disease. Riluzole prolongs survival by 2–3 months at best, with little effect on quality of life,3 whereas edaravone mildly improves patient mobility, but the effect on survival is unknown.4 The paucity of effective treatments warrants more genetic and molecular research on the underlying mechanisms of ALS to analyse the disease process at the cellular level. By 2014, 22 genes were implicated in ALS, and mutations in these genes account for about two-thirds of all familial cases and approximately 10% of cases of sporadic ALS.5 Since 2014, seven novel genes associated with ALS—MATR3, CHCHD10, TBK1, TUBA4A, NEK1, C21orf2, and CCNF—have been identified. The rapid identification of multiple novel genes associated with ALS reflects improvements in sequencing technologies and, more importantly, provides an opportunity to better understand the disease (figure 1). Such advances are key to the development of disease-modifying treatments.
Figure 1. Genetic landscape of ALS between 1993 and 2016.
Familial ALS cases constitute about 10% of all cases of ALS. Of this 10%, about 70% can be explained by genetics. Two substantial increases in genetic contribution to ALS were found in 2008 and 2011, corresponding to the identification of TARDBP (contributes to about 4% of familial and 1% of sporadic cases) and C9orf72 (contributing to about 40% of familial cases and 8% of sporadic cases). ALS=amyotrophic lateral sclerosis. GWAS=genome-wide association study. WGS=whole-genome sequencing. WES=whole-exome sequencing. NGS=next-generation sequencing. RP-PCR=repeat-primed polymerase chain reaction. SNP=single nucleotide polymorphism. ASO=antisense oligonucleotide.
In this Rapid Review, we summarise the novel genetic discoveries associated with ALS in chronological order. We focus on the technologies and experimental design used to identify these genes, and have cross-checked genetic variants against the Exome Aggregation Consortium (ExAC) public database, which catalogues more than 7 million variants in the protein coding region of the genome identified in more than 60 000 mostly healthy individuals (ie, those without severe paediatric diseases). Genetic screening is becoming more accessible and common in clinical practice, thus understanding how a variant might cause disease within the context of the larger population could help in making reasonable inference about pathogenicity, especially when family history of disease is unknown. We also discuss the importance of these genes for the development of new therapies.
Novel ALS genes
Frequency data for these seven novel genes identified since 2014 are scarce because few studies have done large-scale screening of independent patient cohorts. The frequency data available for these genes are likely to be inflated, and we hypothesise that the frequency of mutations in these genes in the population will be lower when additional data is obtained. We estimated that for ALS—assuming full penetrance, no founder mutation effect, and disease prevalence of six cases per 100 000 individuals6—a variant observed more than five times per 121 000 alleles in the ExAC database (corresponding to an allele frequency of 0·0033%) is unlikely to cause ALS because it is too common. However, absence of mutations in a gene in the ExAC database does not necessarily infer pathogenicity because rare genetic variants that are unique to one individual or a single family are remarkably common in the human population (about 3–4 million single nucleotide polymorphisms per individual).7
MATR3
In 2014, four mutations (p.S85C, p.F115C, p.P154S, and p.T622A) in MATR3 were identified by exome sequencing in four families of European descent with either ALS alone or with a combination of ALS and dementia.8 Since 2014, 11 additional variants have been described, predominantly occurring in patients with sporadic ALS.9–12 In the ExAC database, three variants (p.E664A, p.N787S, and c.48+1G>T) had a reported allele frequency of 0·03–0·05%, p.F115C was reported once, but none of the other variants were listed. Overall, the contribution of MATR3 to the development of ALS or ALS and frontotemporal dementia is relatively rare, with no significant correlation observed between phenotype and genotype.
In patients with ALS with MATR3 mutations, upper and lower motor neurons are affected and survival duration ranges from 2–12 years.10 The concomitant clinical presentation of ALS and myopathic features in individuals with the p.S85C mutation is important because these patients are initially diagnosed with vocal cord and pharyngeal dysfunction with asymmetric distal myopathy, but the presentation of pyramidal tract signs and progressive respiratory failure at end-stage disease usually warrants re-diagnosis.8 By contrast to TDP43 and FUS, whereby mutations cause relocalisation of the mutant protein from the nucleus to cytoplasm, studies8,13 have shown that the subcellular localisation of mutant MATR3 is generally unaffected. Furthermore, MATR3-positive inclusions were occasionally observed in histopathological sections from patients with MATR3 mutations, and in one individual with C9orf72 expansion.8
MATR3 is a 125 kDa nuclear protein with RNA and DNA binding domains that appears to primarily regulate gene expression.14 Transgenic mice overexpressing human MATR3 protein develop hindlimb paralysis and muscle atrophy, indicating that neuromuscular function is sensitive to MATR3 levels.15 The protein forms a complex with two other ALS-associated RNA-binding proteins, TDP438 and FUS,16,17 in a RNA-dependent manner and the p.S85C mutation enhances this interaction.8 Thus, overlap might occur in the upstream regulatory proteins or downstream effector targets among ALS-RNA binding proteins. Elucidation of this potentially shared set of proteins might identify molecules suitable for therapeutic intervention.
CHCHD10
CHCHD10 was first linked to ALS in a study18 of a large French family who had a complex phenotype of ALS, ataxia, mitochondrial myopathy, parkinsonism, and sensorineural hearing loss. Exome sequencing identified a p.S59L mutation within CHCHD10.18 Subsequently, 20 additional missense variants, clustered in exon 2—which encodes an internal hydrophobic helical segment important for mitochondrial membrane binding19—have been reported in a broad range of neurodegenerative disorders, including ALS and frontotemporal dementia,18,20–28 frontotemporal lobar degeneration,29 parkinsonism,26,27 Alzheimer’s disease,30 autosomal dominant mitochondrial myopathy,31 adult-onset spinal muscular atrophy,32 and Charcot-Marie-Tooth type 2.33 The pathogenicity of p.S59L,18 p.R15L,20–22 and p.G66V20 has been validated in family studies, whereby the mutations were shown to segregate with ALS. Additionally, the mutations were absent in the ExAC database. However, mutations in CHCHD10 appear to be a relatively rare cause of ALS, but might be more frequent among patients diagnosed with frontotemporal dementia.27,34
CHCHD10 is a 14 kDa nuclear-encoded, mitochondrial protein localised to the mitochondrial intermembrane space. The protein is important for the maintenance of mitochondrial dynamics and cellular bioenergetics.35 Patient fibroblasts expressing mutant CHCHD10 protein (p.S59L) have a fragmented mitochondrial network and disrupted mitochondrial cristae.18 These effects are similar to abnormalities in mitochondrial dynamics induced by mutations in TDP43.36 CHCHD10 also interacts with TDP43, which promotes retention of TDP43 in the nucleus,37 but this localisation is disrupted in the presence of CHCHD10 mutations, causing an accumulation of TDP43 in the cytoplasm and synaptic damage.37 Further study is necessary to investigate the mechanistic association between these proteins and their involvement in mitochondrial dysfunction and TDP43 proteinopathy. This insight could identify therapeutic targets susceptible to manipulation by small molecules, to rescue the observed cellular defects involved in ALS.
TUBA4A
TUBA4A was implicated as a novel gene for familial ALS on the basis of exome sequencing data obtained from a large cohort of European and American patients with ALS and controls.38 This finding was replicated in an independent Belgian cohort,26 but not in Asian patients with ALS.39 All variants were absent or had very low frequency in the ExAC database and had adequate segregation data, with the exception of p.K430N. The overall frequency of TUBA4A mutations suggests it is a rare cause of ALS. Little information is available about the clinical presentation, prognosis, or neuropathological evaluation of patients with TUBA4A variants, and although patients often present with typical features of ALS, some also present with features of frontotemporal dementia.26,38
The main cytoskeletal scaffold in cells is comprised of microtubules, composed of polymerised α-tubulin and β-tubulin subunits. In primary motor neurons, expression of missense mutation TUBA4A interferes with tubulin dimerisation, resulting in a weakened microtubule network.38 Mutations have been found to cluster in the protein domain responsible for the interaction with other tubulin subunits and the axonal transport proteins dynein and kinesin.40 This finding highlights the crucial role of cytoskeletal and axonal transport defects in the pathogenesis of ALS. Therapeutic approaches enhancing cytoskeletal integrity might be crucial for halting progression or reversing the disease course.
TBK1
A whole exome sequencing study41 revealed that TBK1 was implicated in ALS. Enrichment of nonsynonymous variants in patients with ALS compared with healthy controls was found across the entire coding region.41 This finding was validated by another whole exome sequencing study,42 which reported segregation of the pathogenic variants within affected families. Mutations in TBK1 are found in about 1% of patients with familial ALS and in approximately 1% of patients with sporadic ALS.42–53 The clinical phenotypes associated with TBK1 mutations are heterogeneous, with variable age of onset, differing progression, and irregular length of survival time.39,43–46 Extrapyramidal symptoms, ataxia, and psychiatric symptoms have also been reported in some patients with TBK1 mutations.46 Neuropathological examination of CNS tissue from patients with a TBK1 mutation showed SQSTM1/p62 and TDP43-positve inclusions,42,45,46 which are indicative of abnormal TDP43 protein aggregation and defective protein clearance pathways.47 Since these inclusions are also observed in other patients with ALS without TBK1 mutations,48 this suggests that a common disease mechanism might exist, and a broad treatment approach to restore defective proteostasis might also benefit patients with TBK1 mutations.49
TBK1 is a homodimeric multidomain protein with a kinase domain, a ubiquitin-like domain, and two coiled-coil domains.50 The protein acts as an interaction platform for multiple proteins and regulates the activities of downstream protein targets involved in key cellular processes that have been implicated in ALS, including neuroinflammation, ubiquitin-proteasome systems, and autophagy pathways involving other genes also associated with ALS—ie, OPTN, SQSTM1/p62, VCP, and UBQLN2.50 Most pathogenic variants identified in TBK1 are concentrated within the kinase and the coiled-coiled domains,42 suggesting that these mutations might operate by altering these downstream regulatory pathways. We identified some variants (p.K291E, p. I305T, p.L306I, p.H322Y, p.T3221I, p.R444Q, and p.A535T) in the ExAC database that had a frequency higher than our estimated threshold of 0·0033%, suggesting that they are unlikely to be pathogenic. Pathogenicity of the other variants will require further investigation in families and in cells or animal models.
NEK1 and C21orf2
Heterozygous loss-of-function mutations in NEK1 have been implicated in sporadic ALS.41 NEK1 interacts with two proteins known to be associated with ALS, ALS2 and VAPB,45 which are involved in endosomal and endoplasmic reticulum lipid trafficking; this interaction provides some functional evidence of NEK1 involvement in ALS pathogenesis. Two independent case-control studies51,52 provided corroborative evidence that NEK1 is associated with ALS, and indicated that it might account for up to 2% of all ALS cases. Clinical descriptions of patients with NEK1 mutations are scarce, but patients appear to present with typical ALS without dementia.51,52
Concomitant with the identification of NEK1, a large case-control study53 using the genome-wide association approach found that C21orf2 was associated with increased ALS risk. NEK1 and C21orf2 interact with each other and are involved in microtubule assembly, DNA damage response and repair, and mitochondrial function.54,55 Additional genetic replication studies in independent cohorts and functional and clinical studies for both NEK1 and C21orf2 are required to fully understand the contribution of these variants in the pathogenesis of ALS.
CCNF
CCNF was identified as a causative gene for ALS on the basis of exome sequence analysis56 of a large family of European descent who had ALS, frontotemporal dementia, or both diseases, with an autosomal dominant pattern of inheritance. The authors reported additional, potentially pathogenic variants in CCNF in familial cases (all absent or less than the 0·0033% threshold in the ExAC database), with an overall mutation frequency that ranged between 0·6 and 3·3% in white populations.57 Clinically, these patients presented with either typical ALS, ALS with frontotemporal dementia, or frontotemporal dementia alone.56
CCNF is the substrate-recognising component of the Skp1-cullin-F-box E3 ubiquitin-ligase complex, which is responsible for tagging proteins with ubiquitin and marking them for degradation via the ubiquitin-proteasome system.57 Neuronal cells overexpressing mutant CCNF show an increase in ubiquitin-tagged proteins, which include TDP43. This increase suggests that these variants affect the proteosomal degradation pathway by either aberrantly tagging all proteins with ubiquitin or failing to transfer ubiquitin-tagged proteins to the proteasome complex for removal.56 This finding indicates that mutations in CCNF might lead to abnormal proteostasis, which might be exacerbated by TDP43 proteinopathy. Therefore, therapies that enhance protein clearance or reduce ubiquitination might be viable approaches to treatment.
Role of genetics in therapy development
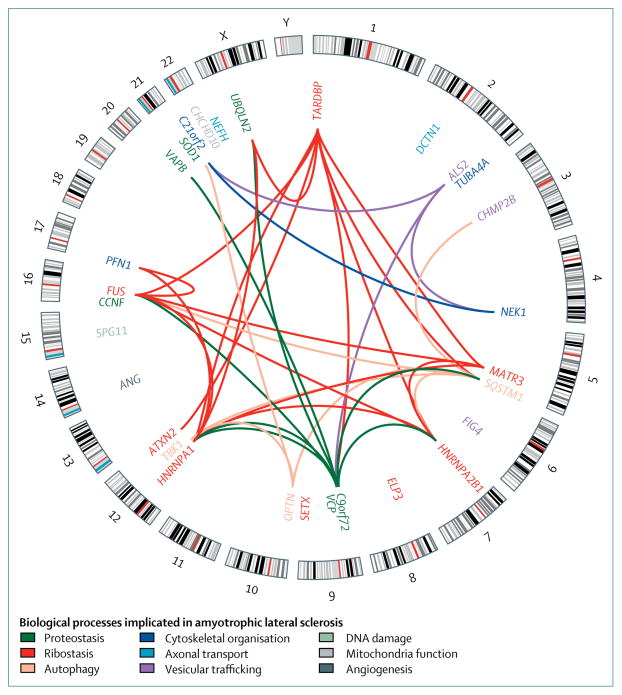
With the exception of riluzole, which was shown to prolong survival for 2–3 months,3 and edavarone, which was shown to decrease the rate of patient immobility,4 currently no treatments are available for ALS that can effectively stop or reverse the disease progression. Diagnosis of ALS is only possible through assessment of clinical symptoms after a substantial number of motor neurons have died. Thus, for a drug to be effective, early or presymptomatic diagnosis would be necessary to prevent further motor neuron degeneration and to preserve the function of remaining motor neurons. However, this presents a challenge because no reliable molecular biomarkers have been identified for presymptomatic diagnosis or for patient stratification in clinical trials. The genetic landscape of ALS is slowly evolving in response to novel genetic discoveries, helping to identify pathogenic cellular pathways (figure 2, table), and to provide both potential biomarkers and targets for drug discovery.
Figure 2. Interactions between genes associated with amyotrophic lateral sclerosis.
The outer circle is a karyotype ideogram showing 24 chromosomes (22 autosomal chromosomes, X chromosome, and Y chromosome); the inner circle shows the location of each gene. Links between genes represent interactions at the protein or gene level. Interaction data was obtained from the Biological General Repository for Interaction Datasets. Black lines indicate cytogenetic band patterns. Biological processes implicated in either the gene or interactions are indicated by colour.
Table.
Genes associated with amyotrophic lateral sclerosis identified between 1993 and 2016
| Gene | Loci | Genetic effect |
Familial amyotrophic lateral sclerosis (%)* |
Sporadic amyotrophic lateral sclerosis (%)* |
Implicated amyotrophic lateral sclerosis pathway |
Disease features† | Other associated allelic disorders |
DNA, RNA, and proteins found to interact with target gene‡ |
|
|---|---|---|---|---|---|---|---|---|---|
| 2016 | C21orf2 | 21q22.3 | Autosomal dominant, risk factor | NA | NA | Cytoskeletal organisation | Typical amyotrophic lateral sclerosis; frontotemporal dementia | Down syndrome; spondylometaphyseal dysplasia; retinal degeneration | ALS2, NEK1 |
| 2016 | CCNF | 16p13.3 | Autosomal dominant | 4 | 2 | Proteostasis | NA | None | None |
| 2016 | NEK1 | 4q33 | Autosomal dominant, autosomal recessive | 2 | 2 | Cytoskeletal organisation; DNA damage and cell cycle | Typical amyotrophic lateral sclerosis | Short-rib thoracic dysplasia 6 with or without polydactylyism; polycystic kidney disease | ALS2, C21orf2 |
| 2015 | TBK1 | 12q14.2 | Autosomal dominant, de novo | <1 | <1 | Autophagy; neuroinflammation | Typical amyotrophic lateral sclerosis; frontotemporal dementia | None | OPTN, SQSTM1 |
| 2014 | CHCHD10 | 22q11.23 | Autosomal dominant | <1 | <1 | Mitochondrial function; cellular bioenergetics | Typical amyotrophic lateral sclerosis; frontotemporal dementia; ataxia; myopathy | Spinal muscular atrophy (Jokela type); autosomal dominant isolated mitochondrial myopathy | None |
| 2014 | MATR3 | 5q31.2 | Autosomal dominant | <1 | <1 | Ribostasis | Slow, progressive typical or atypical amyotrophic lateral sclerosis; frontotemporal dementia; myopathy | Vocal cord and pharyngeal dysfunction with distal myopathy | HNRNPA2B1, HNRNPA1, TARDBP, FUS |
| 2014 | TUBA4A | 2q35 | Autosomal dominant | <1 | <1 | Cytoskeletal organisation; axonal transport | Typical amyotrophic lateral sclerosis; frontotemporal dementia | None | None |
| 2013 | HNRNPA1 | 12q13 | Autosomal dominant, de novo, risk factor | NA | NA | Ribostasis | Typical amyotrophic lateral sclerosis; myopathy; cognitive impairment | Inclusion body myositis with early-onset Paget disease without frontotemporal dementia 3; multisystem proteinopathy | HNRNPA2,B1, TARDBP, UBQLN2, MATR3, FUS, PFN1 |
| 2013 | HNRNPA2B1 | 7p15 | Autosomal dominant, risk factor | NA | NA | Ribostasis | Typical amyotrophic lateral sclerosis; myopathy; cognitive impairment | Inclusion body myositis with early-onset Paget disease with or without frontotemporal dementia 2; multisystem proteinopathy | HNRNPA1, TARDBP, FUS, MATR3, SQSTM1 |
| 2012 | PFN1 | 17p13 | Autosomal dominant | <1 | <1 | Cytoskeletal organisation; axonal growth and transport | Typical amyotrophic lateral sclerosis | None | FUS, HNRNPA |
| 2011 | C9orf72 | 9p21 | Autosomal dominant | 40 | 7 | Intracellular trafficking; autophagy; proteostasis; global RNA alterations; nucleocytoplasmic transport | Typical amyotrophic lateral sclerosis; frontotemporal dementia | None | HNRNPA1, HNRNPA2B1, UBQLN2, SQSTM1 |
| 2011 | SQSTM1 | 5q35 | Autosomal dominant | 1 | <1 | Autophagy; neuroinflammation | Typical amyotrophic lateral sclerosis | Frontotemporal dementia; amyotrophic lateral sclerosis; distal myopathy; childhood-onset neurodegeneration with ataxia, dystonia, and gaze palsy; Paget disease of bone-3 | TBK1, TARDBP, OPTN, CHMP2B, FUS, HNRNPA2B1, NEFM |
| 2011 | UBQLN2 | Xp11 | X-linked autosomal dominant | <1 | <1 | Proteostasis | Typical juvenile or adult onset amyotrophic lateral sclerosis | None | HNRNPA1, TARDBP |
| 2010 | ATXN2 | 12q24 | Autosomal dominant, risk factor | Ribostasis | Typical amyotrophic lateral sclerosis | Spinocerebellar ataxia type 2 | TARDBP | ||
| 2010 | OPTN | 10p13 | Autosomal dominant, autosomal recessive | <1 | <1 | Autophagy; neuroinflammation | Slow, progressive atypical amyotrophic lateral sclerosis | Adult-onset primary open angle glaucoma | TBK1, SQSTM1, SOD1 |
| 2010 | SPG11§ | 15q14 | Autosomal recessive | NA | NA | DNA damage | Slow, progressive juvenile amyotrophic lateral sclerosis, mainly affecting upper motor neurons | Hereditary spastic paraplegia; Charcot-Marie-Tooth disease, axonal, type 2X; spastic paraplegia type 11 | None |
| 2010 | VCP | 9p13 | Autosomal dominant, de novo | 1 | 1 | Proteostasis | Typical amyotrophic lateral sclerosis; frontotemporal dementia | Charcot-Marie-Tooth disease type 2Y; inclusion body myopathy with early-onset Paget disease and frontotemporal dementia 1 | FUS, TARDBP, VAPB, HNRNPA1 |
| 2006 | ANG | 14q11 | Risk factor | NA | NA | Angiogenesis | Typical amyotrophic lateral sclerosis; frontotemporal dementia | None | None |
| 2009 | ELP3 | 8p21 | Undefined | NA | NA | Ribostasis; cytoskeletal integrity | Typical amyotrophic lateral sclerosis | None | None |
| 2009 | FUS | 16p11 | Autosomal dominant, autosomal recessive, de novo | 4 | 1 | Ribostasis | Juvenile and adult onset typical or atypical amyotrophic lateral sclerosis; frontotemporal dementia; dementia | Hereditary essential tremor-4 | MATR3, TARDBP, TAF15, SMN, VCP, SQSTM1, PFN1 |
| 2008 | TARDBP | 1p36 | Autosomal dominant, autosomal recessive, de novo | 4 | 1 | Ribostasis | Typical amyotrophic lateral sclerosis; frontotemporal dementia | None | ATXN2, MATR3, UBQLN2, HNRNPA, 2B1, HNRNPA1, SQSTM1, UBQLN2, VCP, FUS |
| 2006 | CHMP2B | 3p11 | Autosomal dominant | NA | NA | Vesicular trafficking; proteostasis | Typical amyotrophic lateral sclerosis | Frontotemporal dementia | None |
| 2004 | VAPB | 20q13 | Autosomal dominant | NA | NA | Proteostasis | Typical or atypical amyotrophic lateral sclerosis | Finkel type spinal muscular atrophy | VCP |
| 2003 | DCTN1 | 2p13 | Autosomal dominant, risk factor | NA | NA | Axonal transport | Slow, progressive juvenile amyotrophic lateral sclerosis | Distal hereditary motor neuropathy type VIIB; Perry syndrome | None |
| 2001 | ALS2§ | 2q33 | Autosomal recessive | NA | NA | Vesicular trafficking | Slow, progressive, infantile and juvenile amyotrophic lateral sclerosis (mainly affecting upper motor neurons) | Juvenile primary lateral sclerosis; infantile hereditary spastic paraplegia | C21orf2, VCP, NEK1 |
| 1998 | SETX | 9q34 | Autosomal dominant | NA | NA | Ribostasis | Slow, progressive, juvenile amyotrophic lateral sclerosis | Autosomal recessive spinocerebellar ataxia type 1 | None |
| 1994 | NEFH | 22q12 | Autosomal dominant, risk factor | NA | NA | Axonal transport | Typical amyotrophic lateral sclerosis | Axonal Charcot-Marie-Tooth disease type 2CC | None |
| 1993 | SOD1 | 21q22 | Autosomal dominant, autosomal recessive, de novo | 12 | 1–2 | Proteostasis; oxidative stress | Typical amyotrophic lateral sclerosis; cognitive impairment (very rare) | None | OPTN |
NA=not available.
Proportion of cases attributed to mutations in the corresponding genes.
Data extracted from the Online Mendelian Inheritance in Man (OMIM) database.
Data extracted from the Biological General Repository for Interaction Datasets (BioGrid).
ALS2 and SPG11 genes are associated with a phenotype that differs from typical amyotrophic lateral sclerosis, which mainly affects the upper motor neurons, with disease onset at a young age.
Pathogenicity in some cases is likely to be driven by the acquisition of a toxic function through genetic mutation, which forms the basis for antisense oligonucleotide treatment.58 By reducing the production of toxic species, the pathogenic process driven by these species can be modified or stopped to prevent further cellular damage. In animal models of SOD1-associated ALS,59 antisense oligonucleotide treatment significantly delayed disease onset, improved neuromuscular function, and prolonged survival. These effects were accompanied by a corresponding decrease in SOD1 in cerebrospinal fluid, indicating that the concentration of SOD1 in this compartment might be a pharmacodynamic biomarker for future prognostic and efficacy assessments.60 The first clinical trial61 of antisense oligonucleotide treatment in human beings had favourable safety outcomes, and a trial to assess the safety, tolerability, and pharmacokinetics of a second generation SOD1 antisense oligonucleotide is currently in progress (ClinicalTrials.gov, NCT02623699). Phase 2 and phase 3 trials are needed to establish whether the efficacy observed in experimental models can be achieved in human beings.
Similar biomarker development and antisense oligonucleotide studies targeting C9orf72 are in development.62–64 Although it remains unclear which toxic species drive pathogenicity,65 a single dose of antisense oligonucleotide that specifically targets the expanded allele was sufficient to alleviate behavioural symptoms in transgenic C9orf72 mice and reduce the number of RNA foci and dipeptide repeat proteins.66,67 Patients with C9orf72 expansion also showed increased toxic RNA accumulation in tissues and circulating dipeptide repeat proteins in blood and cerebrospinal fluid,67 suggesting that C9orf72 could be a candidate biomarker of disease diagnosis, treatment efficacy evaluation, and prognosis.62
Genetic discoveries have been directly applied in clinical settings to alleviate disease—eg, riboflavin therapy for Brown-Vialetto-Van Laere syndrome,68,69 which is an inherited variant of ALS. The syndrome is a rare progressive neurodegenerative disorder that typically manifests as childhood ALS in combination with sensorineural deafness.70 Brown-Vialetto-Van Laere syndrome is caused by mutations in two riboflavin transporter genes (SLC52A2 and SLC52A3)70 that result in a reduction of plasma flavin and acylcarnitine concentrations.68 Patients treated with high-dose oral riboflavin had marked motor improvements and an overall alleviation of clinical symptoms.68
TBK1 is a key regulatory molecule upstream of OPTN, SQSTM1/p62, and IRF3 in the autophagy and neuroinflammatory pathways that are implicated in ALS.42,50 Manipulation of TBK1 might potentially compensate for defects caused by other ALS-associated proteins in these pathways—eg, VCP and UBQLN2. NEK1 and C21orf2 are known to interact at the protein level and, in addition to TUBA4A, PFN1, NEFH, and PRPH, they represent the building blocks of the cellular scaffold. Administration of small molecules that enhance cytoskeletal integrity could represent a viable therapy for stopping progression or reversing the disease course in patients with these mutations.
Conclusions and future directions
ALS research has been largely driven by advances in our understanding of the genetics underlying the disease. This, in turn, has been fuelled by technological developments in next generation sequencing. Since 2014, seven novel genes—MATR3, CHCHD10, TBK1, TUBA4A, NEK1, C21orf2, and CCNF—associated with ALS have been identified using these techniques. However, the precise disease mechanisms attributed to these genes are unclear, and further elucidation from in-vivo and in-vitro functional studies is required. The collective identification of these novel genes is important within the context of other established genes that are associated with ALS to enable investigation of the disease process at the cellular level (figure 2).
The considerable advances in genetic identification seen in the past decade are likely to continue as whole genome sequencing becomes more accessible. Such progress will facilitate the analysis of larger cohorts leading to a better understanding of the molecular defects that cause motor neuron degeneration. In particular, these techniques will help to identify rare polymorphisms in the non-coding intergenic regions of the genome and structural variants, such as repeat expansions, copy number variants, and indels that might contribute to ALS. The availability of well phenotyped cohorts and efforts in large-scale genomic sequencing are essential to improve our understanding of ALS pathophysiology, and thus, to identify therapeutic targets.
Increased knowledge about the genetic profiles that protect or confer disease risk in patients with ALS will change the way clinical trials are done and how therapy is prescribed to patients. The most important change will be the stratification of patient and control cohorts by genotype, which will increase the success rate of clinical trials. Because ALS is a genetically heterogeneous and complex disease, a personalised medicine approach is emerging, whereby treatment is tailored to the specific mutation that causes disease in an individual patient. Thus, genetic screening for known variants or mutations will be integral to diagnosis, treatment, and prevention of ALS. Many advances have been achieved in the past 5 years, such as the application of gene silencing for SOD1 and C9orf72, the development of viable biomarkers for the diagnosis of patients with ALS who have mutations in those genes, and the evaluation of the efficacy of potential treatments. More breakthroughs are expected in the future when more genes are identified through these large-scale genetic studies.
Search strategy and selection criteria.
We searched PubMed for articles published in English between Dec 1, 2013, and Aug 31, 2017, using the search terms “ALS AND genetics” and “motor neuron disease AND genetics”. We selected articles that reported the identification of the novel amyotrophic lateral sclerosis genes MATR3, CHCHD 10, TBK1, TUBA4A, NEK1, C21orf2, and CCNF. We also searched for articles describing the function and implications of mutations in these selected genes in neurological and non-neurological diseases, and associations with known amyotrophic lateral sclerosis genes identified before 2014. We selected the most relevant articles on the basis of subjective appraisal of their quality and mechanistic insight that could be relevant to amyotrophic lateral sclerosis.
Footnotes
Contributors
All authors contributed equally to the preparation and writing of the manuscript. All authors approved the final version.
Declaration of interests
AC received personal fees from Biogen Idec, Mitsubishi, and Neuraltus. BJT received grants from Merck, Microsoft Research, the Amyotrophic Lateral Sclerosis Association, the Packard Center for Amyotrophic Lateral Sclerosis Research, the Muscular Dystrophy Association, the Center for Disease Prevention and Control, the Myasthenia Gravis Foundation, the Italian Football Association, and the Italian Amyotrophic Lateral Sclerosis Association; and holds the European patent (16204118.0-1404) on the clinical testing and therapeutic intervention for the hexanucleotide repeat expansion of C9orf72 and the USA patent is pending. RC declares no competing interests.
References
- 1.Talbott EO, Malek AM, Lacomis D. The epidemiology of amyotrophic lateral sclerosis. Handb Clin Neurol. 2016;138:225–38. doi: 10.1016/B978-0-12-802973-2.00013-6. [DOI] [PubMed] [Google Scholar]
- 2.Arthur KC, Calvo A, Price TR, Geiger JT, Chiò A, Traynor BJ. Projected increase in amyotrophic lateral sclerosis from 2015 to 2040. Nat Commun. 2016;7:12408. doi: 10.1038/ncomms12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco H, Patin F, Andres CR, Corcia P, Gordon PH. Amyotrophic lateral sclerosis, 2016: existing therapies and the ongoing search for neuroprotection. Expert Opin Pharmacother. 2016;17:1669–82. doi: 10.1080/14656566.2016.1202919. [DOI] [PubMed] [Google Scholar]
- 4.Writing Group, Edaravone (MCI-186) ALS 19 Study Group. Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: a randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2017;16:505–12. doi: 10.1016/S1474-4422(17)30115-1. [DOI] [PubMed] [Google Scholar]
- 5.Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2013;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh R, Thomson KL, Ware JS, et al. Reassessment of Mendelian gene pathogenicity using 7855 cardiomyopathy cases and 60 706 reference samples. Genet Med. 2017;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auton A, Abecasis GR, Altshuler DM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson JO, Pioro EP, Boehringer A, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17:664–66. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leblond CS, Gan-Or Z, Spiegelman D, et al. Replication study of MATR3 in familial and sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2016;37:209.e17–209.e21. doi: 10.1016/j.neurobiolaging.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Marangi G, Lattante S, Doronzio PN, et al. Matrin 3 variants are frequent in Italian ALS patients. Neurobiol Aging. 2017;49:218.e1–218. e7. doi: 10.1016/j.neurobiolaging.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 11.Lin K-P, Tsai P-C, Liao Y-C, et al. Mutational analysis of MATR3 in Taiwanese patients with amyotrophic lateral sclerosis. Neurobiol Aging. 2015;36:2005.e1–2005. e4. doi: 10.1016/j.neurobiolaging.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Xu L, Li J, Tang L, Zhang N, Fan D. MATR3 mutation analysis in a Chinese cohort with sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2016;38:218.e3–218.e4. doi: 10.1016/j.neurobiolaging.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 13.Gallego-Iradi MC, Clare AM, Brown HH, Janus C, Lewis J, Borchelt DR. Subcellular localization of Matrin 3 containing mutations associated with ALS and distal myopathy. PLoS One. 2015;10:e0142144. doi: 10.1371/journal.pone.0142144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coelho MB, Attig J, Bellora N, et al. Nuclear matrix protein Matrin3 regulates alternative splicing and forms overlapping regulatory networks with PTB. EMBO J. 2015;34:653–68. doi: 10.15252/embj.201489852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moloney C, Rayaprolu S, Howard J, et al. Transgenic mice overexpressing the ALS-linked protein Matrin 3 develop a profound muscle phenotype. Acta Neuropathol Commun. 2016;4:122. doi: 10.1186/s40478-016-0393-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Kamelgarn M, Chen J, Kuang L, et al. Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS. Biochim Biophys Acta. 2016;1862:2004–14. doi: 10.1016/j.bbadis.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi A, Takanashi K. FUS interacts with nuclear matrix-associated protein SAFB1 as well as Matrin3 to regulate splicing and ligand-mediated transcription. Sci Rep. 2016;6:35195. doi: 10.1038/srep35195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bannwarth S, Ait-El-Mkadem S, Chaussenot A, et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain. 2014;137:2329–45. doi: 10.1093/brain/awu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Modjtahedi N, Tokatlidis K, Dessen P, Kroemer G. Mitochondrial proteins containing coiled-coil-helix-coiled-coil-helix (CHCH) domains in health and disease. Trends Biochem Sci. 2016;41:245–60. doi: 10.1016/j.tibs.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Müller K, Andersen PM, Hübers A, et al. Two novel mutations in conserved codons indicate that CHCHD10 is a gene associated with motor neuron disease. Brain J Neurol. 2014;137:e309. doi: 10.1093/brain/awu227. [DOI] [PubMed] [Google Scholar]
- 21.Kurzwelly D, Krüger S, Biskup S, Heneka MT. A distinct clinical phenotype in a German kindred with motor neuron disease carrying a CHCHD10 mutation. Brain. 2015;138:e376. doi: 10.1093/brain/awv014. [DOI] [PubMed] [Google Scholar]
- 22.Johnson JO, Glynn SM, Gibbs JR, et al. Mutations in the CHCHD10 gene are a common cause of familial amyotrophic lateral sclerosis. Brain. 2014;137:e311. doi: 10.1093/brain/awu265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaussenot A, Le Ber I, Ait-El-Mkadem S, et al. Screening of CHCHD10 in a French cohort confirms the involvement of this gene in frontotemporal dementia with amyotrophic lateral sclerosis patients. Neurobiol Aging. 2014;35:2884.e1–2884.e4. doi: 10.1016/j.neurobiolaging.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 24.Ronchi D, Riboldi G, Del Bo R, et al. CHCHD10 mutations in Italian patients with sporadic amyotrophic lateral sclerosis. Brain. 2015;138:e372. doi: 10.1093/brain/awu384. [DOI] [PubMed] [Google Scholar]
- 25.Chiò A, Mora G, Sabatelli M, et al. CHCH10 mutations in an Italian cohort of familial and sporadic amyotrophic lateral sclerosis patients. Neurobiol Aging. 2015;36:1767.e3–1767. e6. doi: 10.1016/j.neurobiolaging.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perrone F, Nguyen HP, Van Mossevelde S, et al. Investigating the role of ALS genes CHCHD10 and TUBA4A in Belgian FTD-ALS spectrum patients. Neurobiol Aging. 2017;51:177.e9–177. e16. doi: 10.1016/j.neurobiolaging.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Dols-Icardo O, Nebot I, Gorostidi A, et al. Analysis of the CHCHD10 gene in patients with frontotemporal dementia and amyotrophic lateral sclerosis from Spain. Brain J Neurol. 2015;138:e400. doi: 10.1093/brain/awv175. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Q, Chen Y, Wei Q, et al. Mutation screening of the CHCHD10 gene in Chinese patients with amyotrophic lateral sclerosis. Mol Neurobiol. 2016;54:3189–94. doi: 10.1007/s12035-016-9888-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhang M, Xi Z, Zinman L, et al. Mutation analysis of CHCHD10 in different neurodegenerative diseases. Brain J Neurol. 2015;138:e380. doi: 10.1093/brain/awv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao T, Jiao B, Zhang W, et al. Identification of CHCHD10 mutation in Chinese patients with Alzheimer Disease. Mol Neurobiol. 2017;54:5243–47. doi: 10.1007/s12035-016-0056-3. [DOI] [PubMed] [Google Scholar]
- 31.Ajroud-Driss S, Fecto F, Ajroud K, et al. Mutation in the novel nuclear-encoded mitochondrial protein CHCHD10 in a family with autosomal dominant mitochondrial myopathy. Neurogenetics. 2015;16:1–9. doi: 10.1007/s10048-014-0421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Penttilä S, Jokela M, Bouquin H, Saukkonen AM, Toivanen J, Udd B. Late onset spinal motor neuronopathy is caused by mutation in CHCHD10. Ann Neurol. 2015;77:163–72. doi: 10.1002/ana.24319. [DOI] [PubMed] [Google Scholar]
- 33.Auranen M, Ylikallio E, Shcherbii M, et al. CHCHD10 variant p. (Gly66Val) causes axonal Charcot-Marie-Tooth disease. Neurol Genet. 2015;1:e1. doi: 10.1212/NXG.0000000000000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiao B, Xiao T, Hou L, et al. High prevalence of CHCHD10 mutation in patients with frontotemporal dementia from China. Brain J Neurol. 2016;139:e21. doi: 10.1093/brain/awv367. [DOI] [PubMed] [Google Scholar]
- 35.Genin EC, Plutino M, Bannwarth S, et al. CHCHD10 mutations promote loss of mitochondrial cristae junctions with impaired mitochondrial genome maintenance and inhibition of apoptosis. EMBO Mol Med. 2016;8:58–72. doi: 10.15252/emmm.201505496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Wang L, Lu J, et al. The inhibition of TDP-43 mitochondrial localization blocks its neuronal toxicity. Nat Med. 2016;22:869–78. doi: 10.1038/nm.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woo J-AA, Liu T, Trotter C, et al. Loss of function CHCHD10 mutations in cytoplasmic TDP-43 accumulation and synaptic integrity. Nat Commun. 2017;8:15558. doi: 10.1038/ncomms15558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith BN, Ticozzi N, Fallini C, et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron. 2014;84:324–31. doi: 10.1016/j.neuron.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai P-C, Liu Y-C, Lin K-P, et al. Mutational analysis of TBK1 in Taiwanese patients with amyotrophic lateral sclerosis. Neurobiol Aging. 2016;40:191.e11–16. doi: 10.1016/j.neurobiolaging.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 40.Howes SC, Alushin GM, Shida T, Nachury MV, Nogales E. Effects of tubulin acetylation and tubulin acetyltransferase binding on microtubule structure. Mol Biol Cell. 2014;25:257–66. doi: 10.1091/mbc.E13-07-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cirulli ET, Lasseigne BN, Petrovski S, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–41. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freischmidt A, Wieland T, Richter B, et al. Haploinsufficiency of TBK1 causes familial ALS and frontotemporal dementia. Nat Neurosci. 2015;18:631–36. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 43.Borghero G, Pugliatti M, Marrosu F, et al. TBK1 is associated with ALS and ALS-FTD in Sardinian patients. Neurobiol Aging. 2016;43:180.e1–5. doi: 10.1016/j.neurobiolaging.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caroppo P, Camuzat A, De Septenville A, et al. Semantic and nonfluent aphasic variants, secondarily associated with amyotrophic lateral sclerosis, are predominant frontotemporal lobar degeneration phenotypes in TBK1 carriers. Alzheimers Dement. 2015;1:481–86. doi: 10.1016/j.dadm.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gijselinck I, Van Mossevelde S, van der Zee J, et al. Loss of TBK1 is a frequent cause of frontotemporal dementia in a Belgian cohort. Neurology. 2015;85:2116–25. doi: 10.1212/WNL.0000000000002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Mossevelde S, van der Zee J, Gijselinck I, et al. Clinical features of TBK1 carriers compared with C9orf72, GRN and non-mutation carriers in a Belgian cohort. Brain. 2016;139:452–67. doi: 10.1093/brain/awv358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madill M, McDonagh K, Ma J, et al. Amyotrophic lateral sclerosis patient iPSC-derived astrocytes impair autophagy via non-cell autonomous mechanisms. Mol Brain. 2017;10:22. doi: 10.1186/s13041-017-0300-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brettschneider J, Arai K, Del Tredici K, et al. TDP-43 pathology and neuronal loss in amyotrophic lateral sclerosis spinal cord. Acta Neuropathol. 2014;128:423–37. doi: 10.1007/s00401-014-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalmar B, Greensmith L. Cellular chaperones as therapeutic targets in ALS to restore protein homeostasis and improve cellular function. Front Mol Neurosci. 2017;10:251. doi: 10.3389/fnmol.2017.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oakes JA, Davies MC, Collins MO. TBK1: a new player in ALS linking autophagy and neuroinflammation. Mol Brain. 2017;10:5. doi: 10.1186/s13041-017-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brenner D, Müller K, Wieland T, et al. NEK1 mutations in familial amyotrophic lateral sclerosis. Brain J Neurol. 2016;139:e28. doi: 10.1093/brain/aww033. [DOI] [PubMed] [Google Scholar]
- 52.Kenna KP, van Doormaal PTC, Dekker AM, et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2016;48:1037–42. doi: 10.1038/ng.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Rheenen W, Shatunov A, Dekker AM, et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet. 2016;48:1043–48. doi: 10.1038/ng.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wheway G, Schmidts M, Mans DA, et al. An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat Cell Biol. 2015;17:1074–87. doi: 10.1038/ncb3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang X, Lin H, Wang X, Zuo Q, Qin J, Zhang P. The NEK1 interactor, C21ORF2, is required for efficient DNA damage repair. Acta Biochim Biophys Sin. 2015;47:834–41. doi: 10.1093/abbs/gmv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams KL, Topp S, Yang S, et al. CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nat Commun. 2016;7:11253. doi: 10.1038/ncomms11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D’Angiolella V, Donato V, Vijayakumar S, et al. SCF (Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–42. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schoch KM, Miller TM. Antisense oligonucleotides: translation from mouse models to human neurodegenerative diseases. Neuron. 2017;94:1056–70. doi: 10.1016/j.neuron.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biferi MG, Cohen-Tannoudji M, Cappelletto A, et al. A new AAV10-U7-mediated gene therapy prolongs survival and restores function in an ALS mouse model. Mol Ther. 2017;25:2038–52. doi: 10.1016/j.ymthe.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winer L, Srinivasan D, Chun S, et al. SOD1 in cerebral spinal fluid as a pharmacodynamic marker for antisense oligonucleotide therapy. JAMA Neurol. 2013;70:201–07. doi: 10.1001/jamaneurol.2013.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller TM, Pestronk A, David W, et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12:435–42. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang J, Cleveland DW. Bidirectional transcriptional inhibition as therapy for ALS/FTD caused by repeat expansion in C9orf72. Neuron. 2016;92:1160–63. doi: 10.1016/j.neuron.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 63.Su Z, Zhang Y, Gendron TF, et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83:1043–50. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chiò A, Traynor BJ. Motor neuron disease in 2014: Biomarkers for ALS—in search of the promised land. Nat Rev Neurol. 2014;11:72–74. doi: 10.1038/nrneurol.2014.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gitler AD, Tsuiji H. There has been an awakening: emerging mechanisms of C9orf72 mutations in FTD/ALS. Brain Res. 2016;1647:19–29. doi: 10.1016/j.brainres.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang J, Zhu Q, Gendron TF, et al. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 Is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron. 2016;90:535–50. doi: 10.1016/j.neuron.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gendron TF, Chew J, Stankowski JN, et al. Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72-associated amyotrophic lateral sclerosis. Sci Transl Med. 2017;9:eaai7866. doi: 10.1126/scitranslmed.aai7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Foley AR, Menezes MP, Pandraud A, et al. Treatable childhood neuronopathy caused by mutations in riboflavin transporter RFVT2. Brain J Neurol. 2014;137:44–56. doi: 10.1093/brain/awt315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bashford JA, Chowdhury FA, Shaw CE. Remarkable motor recovery after riboflavin therapy in adult-onset Brown-Vialetto-Van Laere syndrome. Pract Neurol. 2017;17:53–56. doi: 10.1136/practneurol-2016-001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson JO, Gibbs JR, Megarbane A, et al. Exome sequencing reveals riboflavin transporter mutations as a cause of motor neuron disease. Brain J Neurol. 2012;135:2875–82. doi: 10.1093/brain/aws161. [DOI] [PMC free article] [PubMed] [Google Scholar]