Significance
Social genomics research has identified a conserved transcriptional response to adversity (CTRA) that may contribute to social disparities in health. This study identified systematic individual differences in natural language use that track CTRA gene expression more closely than do conventional self-report measures of stress, anxiety, or depression. These language style markers may provide a useful behavioral indicator of the neurobiological processes that mediate social influences on gene expression in immune cells.
Keywords: genomics, psychoneuroimmunology, psycholinguistics
Abstract
Adverse social conditions have been linked to a conserved transcriptional response to adversity (CTRA) in circulating leukocytes that may contribute to social gradients in disease. However, the CNS mechanisms involved remain obscure, in part because CTRA gene-expression profiles often track external social–environmental variables more closely than they do self-reported internal affective states such as stress, depression, or anxiety. This study examined the possibility that variations in patterns of natural language use might provide more sensitive indicators of the automatic threat-detection and -response systems that proximally regulate autonomic induction of the CTRA. In 22,627 audio samples of natural speech sampled from the daily interactions of 143 healthy adults, both total language output and patterns of function-word use covaried with CTRA gene expression. These language features predicted CTRA gene expression substantially better than did conventional self-report measures of stress, depression, and anxiety and did so independently of demographic and behavioral factors (age, sex, race, smoking, body mass index) and leukocyte subset distributions. This predictive relationship held when language and gene expression were sampled more than a week apart, suggesting that associations reflect stable individual differences or chronic life circumstances. Given the observed relationship between personal expression and gene expression, patterns of natural language use may provide a useful behavioral indicator of nonconsciously evaluated well-being (implicit safety vs. threat) that is distinct from conscious affective experience and more closely tracks the neurobiological processes involved in peripheral gene regulation.
Across a diverse array of adverse life circumstances such as low socioeconomic status, social isolation, diagnosis with a life-threatening disease, and posttraumatic stress, circulating immune cells have been found to show a conserved transcriptional response to adversity (CTRA) marked by up-regulated expression of proinflammatory genes and down-regulated expression of genes involved in type I IFN antiviral responses and IgG antibody synthesis (1, 2). These effects are mediated peripherally by sympathetic nervous system (SNS) activation of β-adrenergic signaling pathways that regulate gene transcription in existing cells and stimulate hematopoietic development of new myeloid lineage leukocytes (particularly monocytes) (3, 4). Peripheral SNS activity is controlled by a central network of brain structures including the insular and anterior cingulate cortex, extended amygdala, and lateral hypothalamus, which collectively regulate sympathetic outflow from the medulla oblongata (5, 6). Despite the clear role of CNS processes in regulating SNS activity, CTRA gene expression has shown less consistent association with self-report measures of internal affective experience (e.g., stress, depression, or negative emotions) than it has with measures of external social conditions (e.g., socio-economic status, social rank, bereavement) or subjective perceptions of those external conditions (e.g., perceived isolation) (7–11). Adverse environments are clearly “getting under the skin” somehow, and this presumably involves some form of CNS information processing [as neuropharmacologic and behavioral interventions can inhibit the CTRA (3, 12, 13)], but the specific psychological processes involved remain poorly understood. In the present research we sought to identify an objectively observable behavioral indicator of the psychological processes involved in generating stable individual differences in basal CTRA gene expression.
Findings from affective neuroscience suggest that conscious experiences of negative emotional states such as stress, fear, or anxiety are mediated by a neocortical system which is functionally distinct from the more basic automatic threat-detection and -response system that proximally regulates SNS activity (see ref. 6 for more details on the functional and neuroanatomical distinction between the conscious fear system and the automatic threat defense system). If the nonconscious threat defense system is more sensitive to adverse social conditions than is the conscious affect system, that could explain why CTRA gene expression more reliably tracks measures of adverse environmental conditions than it does self-report measures of experienced affect (Fig. S1). However, this raises a significant methodological challenge for efforts to map the central psychological processes that mediate “social signal transduction” (1, 2): If psychometric self-report instruments cannot accurately track the activity of the relevant neural system, then how instead should we measure it?
Recent psycholinguistic analyses have found that patterns of natural language use change systematically under threatening conditions such as social deception (14–17), terrorist attack (18), low social status (19), and personal crisis (20). These changes include alterations in total language output and a shift in the use of specific function words such as pronouns (17). Function words (e.g., articles, adverbs, pronouns) are generated relatively automatically by CNS language systems and serve to map relationships among the more consciously generated meaning words (e.g., nouns or verbs) that carry the primary semantic information speakers intend to convey (17, 21, 22). Function words by themselves have no semantic purpose, but together they help provide the structure of syntax. Given the relatively automatic production of function words and their empirical sensitivity to threat, systematic variations in language structure may provide an implicit behavioral indicator of the nonconscious threat-response system that regulates SNS activity and, by extension, CTRA gene expression (see Fig. S1 for a graphical depiction of this model) (3, 4). We tested this hypothesis using unobtrusive ecological speech sampling (23, 24) and identified systematic individual differences in broad patterns of natural language use that track CTRA gene expression better than do conventional self-report measures of stress, depression, and anxiety.
Results
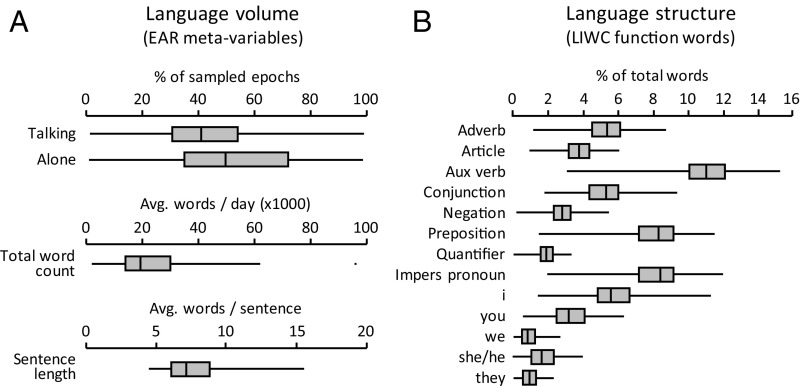
Natural language use was assessed in 22,627 30- to 50-s audio samples collected unobtrusively at 9- to 12.5-min intervals over 2 d from each of 143 healthy community-dwelling adults during waking hours. Speech samples were acquired using the electronically activated recorder (EAR) system (23, 24), which generated an average of 158 (range, 14–260) audio samples per individual. Audio samples were coded for technical validity (i.e., participants being awake and wearing the EAR device) and the presence of others, and all valid samples were transcribed to isolate the study participant’s speech. These analyses yielded individual summary values on four metavariables reflecting total language volume: the fraction of samples in which the individual spoke, the fraction in which the individual was alone, average words spoken, and average sentence length (Fig. 1A). Participant speech transcripts were subsequently processed using the Linguistic Inquiry and Word Count (LIWC) system to quantify individual differences in general language style (25). These analyses yielded 13 dimensions of language structure (Fig. 1B) including the prevalence of eight general categories of function words (adverbs, articles, auxiliary verbs, conjunctions, negations, prepositions, quantifiers, and impersonal pronouns) and five subcategories of personal pronoun [first-person singular, first-person plural, second-person singular, third-person singular, and third-person plural, each represented separately due to their potentially divergent patterns of change under threat (17)].
Fig. 1.
Natural language-use features. Distribution of individual differences in (A) EAR-sampled parameters of language volume and (B) language structure, including eight general categories of function word (capitalized labels) and five subcategories of personal pronoun (lowercase labels). Data represent average values for each parameter computed over a mean of 158 (range 14–260) 30- to 50-s audio samples collected at 9- to 12.5-min intervals over 2 d (resulting in an average of 4,070 ± 272 words per individual). Whiskers indicate the range of individual values across 143 study participants; boxes span 25th–75th percentiles, and internal bars indicate 50th percentile.
Characteristics of the study sample are given in Table S1 and reflect generally healthy young adults (mean age ± SE 34.2 ± 0.7 y; range, 25–56 y) from the Atlanta metropolitan area with a preponderance of females (66%), multiple races (30% African American, 59% white, 7% Asian, 4% other), and low behavioral health risk factors (mean BMI 24.3 ± 0.3, with 8% having a BMI >30 and 8% having a history of regular smoking). Self-report psychometric measures of stress, depression, anxiety, and loneliness were moderately correlated (r’s ranging from 0.44 to 0.64) (Dataset S1) but showed no substantial association with language metrics (all |r| <0.25) (Dataset S1). Several language metrics were also correlated (Dataset S1), and the fraction of audio samples in which the subject spoke (talk frequency) showed particularly strong correlation with total word counts and frequency of being alone (both |r| >0.70, P < 0.0001). Talk frequency was thus analyzed separately to avoid multicollinearity (variance inflation factor = 8.3). No other language metrics showed substantial multicollinearity [variance inflation factors ranged from 1.4 to 2.8, all well below the material threshold of 10 (26)].
CTRA Gene Expression.
Expression of 50 CTRA indicator genes was assessed using microarray-based transcriptome profiling of peripheral blood mononuclear cell samples that were collected under resting conditions following the 2-d language-sampling period. The CTRA profile was quantified as an a priori-defined contrast across the 50 indicator variables [inflammation-related transcripts were weighted +1, and antiviral and antibody-related transcripts were weighted −1 (8, 10, 27, 28)] and was tested for association with five general domains of predictor variables: demographic and health behavior parameters (age, sex, race, BMI, and smoking); language-volume measures (word count, words per sentence, frequency of being alone vs. with others); language-structure measures (relative frequency of 13 function-word classes); self-report psychometric measures (depression, anxiety, perceived stress, and loneliness); and RNA-based measures of leukocyte subset distribution (CD4+ and CD8+ T lymphocytes, B lymphocytes, natural killer cells, and monocytes). Each domain was comprised of multiple subdimensions that were tested simultaneously using a single omnibus partial F test (26) to assess this study’s two a priori substantive hypotheses regarding the potential association of CTRA gene expression with individual differences in language volume and language structure. Following a statistically significant omnibus test, exploratory follow-up analyses assessed the specific subdimension parameters that contributed to the overall domain association with CTRA. Domain associations were tested in an a priori-specified sequence beginning with a null model (predicting similar CTRA expression in all participants), followed by the addition of demographic and behavioral covariates, language-volume measures, language-structure measures, self-report psychometric measures, and leukocyte subset distributions.
Relative to a null model predicting similar CTRA gene expression across all 143 individuals, addition of an a priori-specified set of demographic and health-behavior parameters (age, sex, race, BMI, and smoking) significantly increased predictive power [F(7, 135) = 3.11, P = 0.0045]. Follow-up exploratory analyses of individual subdimension parameter estimates (Dataset S2, model 1) found BMI to be the most significant individual contributor to the overall domain association with gene expression.
The addition of basic language-volume measures to the model significantly enhanced the prediction of CTRA gene expression beyond the level attainable by demographic and behavioral factors alone [F(3, 132) = 12.80, P < 0.0001; alternative likelihood ratio test in Table S2]. Follow-up exploratory analyses of subdimension parameter estimates found low total word count to be the most significant contributor to CTRA prediction by the overall language-volume domain (Dataset S2, model 2).
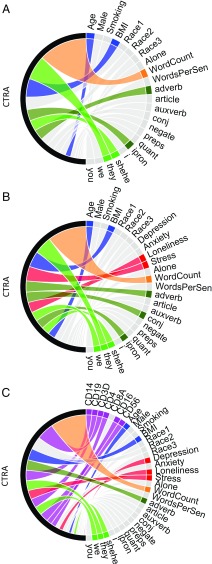
Inclusion of language-structure metrics involving function-word prevalence led to a further increase in CTRA prediction beyond that attainable by demographic/behavioral factors and language volume [F(13, 119) = 6.80, P < 0.0001; see also Fig. 2A]. Follow-up exploratory analyses of individual language-structure parameters (Dataset S2, model 3) found CTRA gene expression to track most strongly with a low prevalence of third-person plural pronouns (e.g., they) and a high prevalence of adverbs (e.g., so, very, really), impersonal pronouns (e.g., it), and third-person singular pronouns (e.g., she, he).
Fig. 2.
Prediction of CTRA gene expression. (A) Prediction of peripheral blood mRNA levels for 50 CTRA indicator transcripts (proinflammatory genes, IFN response genes, and antibody-related transcripts) by demographic and behavioral characteristics (blue chords), language-volume metavariables (orange chords), and language-structure features representing the prevalence of eight categories of function word (dark green chords) and five subcategories of personal pronoun (light green chords). Black arcs indicate total CTRA prediction in each mixed-effect linear model. Chord widths indicate the relative contribution from standardized values of each variable (squared partial regression coefficients, sorted top-to-bottom by descending magnitude in Dataset S2 model 3). Colored chords indicate statistically significant effects (P < 0.05), and gray chords indicate nonsignificant effects. (B) Increment to CTRA prediction from adding self-report measures of depression, anxiety, perceived stress, and perceived social isolation/loneliness (red chords); Dataset S2 model 4. (C) Increment to prediction from adding mRNA markers of leukocyte subsets in the peripheral blood cell pool (purple chords); Dataset S2 model 5.
Ancillary analyses found that language-structure metrics also predicted CTRA gene expression in the absence of language-volume metrics [F(13, 122) = 5.12; P < 0.0001]. Follow-up analyses of individual parameter estimates (Dataset S2, model 3b) again implicated low prevalence of third-person plural pronouns and high prevalence of adverbs, as well as high prevalence of articles and low prevalence of conjunctions (e.g., and, but).
Psychometric Measures.
To determine whether language-use patterns capture the same predictive information as conventional psychometric instruments, additional analyses included self-report measures of depression, anxiety, perceived stress, and perceived social isolation/loneliness. Results (Fig. 2B) continued to show distinct CTRA associations with language volume [(F(3, 112) = 19.24, P < 0.0001] and language structure [F(13, 112) = 7.68, P < 0.0001] above and beyond the significant predictive contribution of psychometric variables [F(4, 112) = 4.70, P = 0.0015]. Follow-up exploratory analysis of individual parameters identified loneliness as the only psychometric variable that consistently predicted gene expression (Dataset S2, model 4). Perceived stress also showed some incremental prediction of gene expression when language metrics were included in the analysis (Dataset S2, model 4) but not in their absence (Dataset S2, model 4b). Neither loneliness nor perceived stress showed any significant association with the specific language features that tracked CTRA gene expression (although both were linked to reduced preposition frequency, and loneliness was also associated with a higher prevalence of audio samples in which the participant was alone) (Dataset S1).
Leukocyte Subsets.
CTRA gene expression is structured in part by hematopoietic influences on the distribution of leukocyte subsets in circulating blood (3, 29). As expected, CTRA gene expression varied with an a priori-specified set of seven variables measuring the prevalence of major leukocyte subset markers [F(7, 105) = 9.51, P < 0.0001] (Fig. 2C). Follow-up analyses of individual parameters (Dataset S2, model 5) found this effect to be carried by mRNA markers of monocytes (CD14), B lymphocytes (CD19), T lymphocytes (CD3D, CD4), and natural killer cells (CD16/FCGR3A, CD56/NCAM1). However, CTRA gene expression continued to show significant additional associations with language volume [F(3, 105) = 30.01, P < 0.0001] and language structure [F(13, 105) = 8.67, P < 0.0001], as well as with psychometric measures [F(4, 105) = 6.02; P < 0.0001], following control for leukocyte subset distributions.
Acute Influences.
This research was motivated by the hypothesis that stable individual differences in threat-related information processing chronically influence both speech production and gene regulation. However, transient environmental events may also acutely modulate speech and gene expression. To discriminate between the relative contributions of acute vs. chronic processes, we compared the strength of association between language patterns and gene expression in all samples (i.e., both acute and chronic influences) with that observed when acute effects were reduced by excluding blood samples collected within 1 wk of language sampling (i.e., chronic influences only). In the absence of acute effects, results continued to show CTRA association with both language volume [F(3, 102) = 23.04, P < 0.0001] and language structure [F(13, 102) = 6.39, P < 0.0001]. In follow-up analyses of the five individual language dimensions that significantly predicted gene expression in the overall sample (Dataset S2, model 3), none showed a significant decrease in predictive strength following the removal of acute influences (change in average association strength: +7.4%) (Table S3).
Discussion
This research identified a relationship between individual differences in natural language use and basal gene-expression profiles in circulating immune cells. Expression of the CTRA transcriptome profile showed verbal correlates in both the volume of speech (total word count and speech frequency) and the structure of speech (pronoun and adverb prevalence). Patterns of speech production predicted CTRA gene expression substantially better than did self-report measures of negative affective states such as stress, depression, and anxiety, and they did so above and beyond the effects of demographic and behavioral factors (age, sex, race, smoking, BMI) and variations in leukocyte subset distributions. Natural language patterns predicted gene expression even when the two variables were sampled more than 1 wk apart, suggesting that their association stems in large part from stable individual differences in processes that jointly influence both speech production and gene regulation. These findings are consistent with the hypothesis that individual differences in automatic CNS threat-detection and -response systems (6, 30) can influence both language processes (17) and leukocyte gene expression (3, 4). The distinct predictive contributions of self-report psychometric instruments and relatively nonconscious speech patterns parallels the distinct neural substrates of consciously experienced negative affect and the more automatic threat-detection and -response systems that proximally regulate SNS activity (Fig. S1) (6). As such, statistical patterns in natural language use may serve as useful indicators of nonconsciously evaluated well-being (i.e., implicit threat vs. safety) that afford greater insight into SNS/β-adrenergic control of peripheral gene expression than do conventional self-report measures of conscious affect.
Exploratory analyses identified several specific language features that predicted CTRA gene expression, but the psychological mechanisms underlying these features remain to be clarified in future research. Low total language output and speech frequency could represent a verbal manifestation of the caution/avoidance and behavioral inhibition responses generated by automatic threat/defense responses (6). CTRA down-regulation with increased language output is also consistent with research linking self-expression to physical health (31, 32) and reciprocal data showing effects of psychological inhibition (reduced expression) on SNS activation (33–36). Consistent with that theory, exploratory analyses of other LIWC metrics besides function words identified inhibition-related language as one of the few significant correlates of CTRA gene expression (SI Results). Language output may also serve as a proxy for social contact rates that could affect gene expression (13, 37). However, direct measures of the presence vs. absence of others failed to predict CTRA gene expression in this sample, whereas language volume continued to associate with gene expression after control for social contact frequency.
Independent of total language volume, CTRA gene expression also tracked stylistic variations in language structure involving patterns of function-word use. We had no a priori hypotheses about the specific types of function word that would associate most strongly with variations in gene expression. Exploratory analyses of that topic found CTRA gene expression to track most strongly with a high prevalence of adverbs, impersonal pronouns, and third-person singular pronouns and with a low prevalence of third-person plural pronouns. Future research will be required to define the psychological mechanisms underlying these specific language features. However, the observed results are consistent with two previous hypotheses regarding the psychological basis for systematic variations in language style. Pronoun use is thought to reflect the deployment of attention (17, 19), and the association of reduced CTRA gene expression with a high prevalence of third-person plural pronouns (they, them) may indicate an outward orientation toward the social world that serves to reduce personal threat or arousal and thereby down-regulate SNS activity (38). In contrast, adverbs often serve as intensifiers (really, very, certainly), and their association with up-regulated CTRA gene expression may indicate greater CNS arousal and consequent increases in SNS activity.
Among the psychometric self-report measures examined, only perceived social isolation (loneliness) consistently predicted CTRA gene expression. However, language volume and language structure continued to predict individual differences in CTRA gene expression above and beyond the effects of loneliness and other psychometric characteristics. Consistent with previous research (9, 10, 29, 37), loneliness was associated with up-regulation of the CTRA. In statistical models that included language metrics, perceived stress also showed a counterintuitive and statistically significant inverse association with the CTRA. Similar inverse stress effects have been observed previously (9, 10) and would be expected in multivariate analyses if gene expression tracks the automatic threat-response system whereas perceived stress tracks the functionally distinct but moderately correlated conscious fear/anxiety system (6). Consistent with that prediction, perceived stress showed no significant association with CTRA gene expression in statistical analyses that omitted language metrics.
The present findings are subject to several limitations. The observed relationships involve spontaneous spoken language use sampled from the everyday life of American adults in the Atlanta metropolitan area, and it is unclear whether similar results would emerge in other language contexts (e.g., in writing or topically directed speech) or in other demographic, socioeconomic, or cultural contexts. This is an observational study, and additional research will be required to define the causal interactions between language and gene expression. This study was motivated by the hypothesis that speech patterns and leukocyte gene-expression profiles come to be associated through their common regulation by stable individual differences in upstream CNS threat-detection and -response systems (Fig. S1) (6). However, language use may also causally affect gene expression [e.g., by influencing perceptual or interpretive processes involved in threat-detection or coping responses (6)], and gene expression may causally influence language use [e.g., via effects of circulating cytokines on CNS affective, cognitive, or social processes (39–41) that subsequently impact speech production]. This study design was not optimal for detecting the effects of acute environmental influence on language and gene expression due to the broad 2-d language-sampling period, the variable lag between language sampling and blood sampling, and the absence of any experimental manipulation of environmental conditions. The CTRA is specific to immune cells, and the language correlates of gene expression in other tissues (e.g., the CNS) remain to be determined in future research, as do the health implications of the preset findings. Future studies will also be required to assess the generalizability of the diagnostic language patterns identified here and to define their underlying neural and psychological mechanisms (e.g., Do they track threat per se or cognitive coping responses that are activated in response to threat? Which specific elements of the nonconscious threat defense system interact with language production? Are language patterns a useful proxy measure of intervention-induced changes in SNS activity?). This study focused on a narrow range of a priori-defined language metrics to avoid capitalizing on chance in statistical association analyses. However, language use can be characterized on many other dimensions (17, 42–44), and some of those may also be found to associate with gene expression in future studies (see SI Results for some initial results). These analyses also tested an a priori genomic hypothesis involving the CTRA gene set as a whole, and no genome-wide discovery analyses were performed to identify specific individual gene transcripts that might relate to language use. Additional genes, gene networks, and epigenetic processes may well be found to relate to language use in future research. These analyses also focused on two a priori hypotheses regarding the general domains of language use that may relate to CTRA gene expression—language volume and language structure—and we had no a priori hypotheses regarding the specific dimensions within each general language domain that might best predict gene expression. Given the exploratory nature of analyses examining specific language features (e.g., word count, adverb prevalence, pronoun frequency), their predictive significance should be regarded as provisional until future studies replicate these findings as a priori hypotheses (45).
The present data indicate a systematic relationship between personal expression and gene expression. As such, statistical pattern analysis of natural language use may provide a useful behavioral indicator of nonconsciously evaluated well-being (implicit safety vs. threat) that is distinct from the information provided by conventional self-report measures and more closely tracks the activity of underlying CNS processes which regulate peripheral physiology, gene expression, and health.
Methods
Study Design and Data Collection.
Language, psychometric, and gene-expression data were collected under basal conditions from 143 healthy (medication-free) adults in the Atlanta metropolitan area. Speech was sampled over 2 d using an iPod Touch implementation of the EAR (worn on a belt) (23), transcribed by trained coders, and processed through LIWC 2007 software (25) to generate language-volume and -structure measures. Audio files were coded by raters to exclude spurious data (e.g., asleep) and to classify participants as being alone vs. with others (intraclass correlation of two independent codings = 0.94) and talking vs. not talking (0.98). Psychometric measures were collected at the time of blood sampling [an average (± SD) of 11 (± 9) d after language sampling] and included the Perceived Stress Scale (46) (Cronbach α = 0.86), Beck Depression Inventory (47) (α = 0.90), Beck Anxiety Inventory (48) (α = 0.84), and UCLA Loneliness Scale (49) (α = 0.91). Total RNA was extracted from resting venous blood samples, tested for mass and integrity, and assayed by Illumina HT-12 v4 BeadArrays in the University of California, Los Angeles (UCLA) Neuroscience Genomics Core Laboratory as previously described (10, 27, 28). Interassay coefficients of variability (CVs) for the 50 analyzed gene transcripts averaged 0.78% (range: 0.34–1.46%). All participants provided written informed consent to participate in the study after the nature and design of the study had been fully described and any related questions had been answered. All procedures were approved by the Institutional Review Board at Emory University.
Analysis.
Quantile-normalized gene-expression data (GSE87656) were log2-transformed and standardized within gene for analysis by mixed-effect linear models (50) testing the association between the average expression of the 50 available CTRA indicator transcripts (10, 27, 28) and a priori-specified sets of demographic and health behavior variables (age, sex, race, BMI, smoking), language-volume metavariables (word count, words per sentence, frequency of being alone vs. with others), language-structure variables (eight general function-word categories and five personal pronoun subcategories listed in Fig. 1B), psychometric variables (stress, depression, anxiety, loneliness), and seven RNA transcripts marking the relative prevalence of major leukocyte subsets (CD14, CD19, CD3D, CD4, CD8A, CD56/NCAM1, CD16/FCGR3A). Predictors were standardized for comparison of association strength, IFN- and antibody-related transcripts were sign-reversed to reflect their inverse contribution to the CTRA (10, 27, 28), and all models included gene-specific intercepts and a fully parameterized (unstructured) covariance matrix to account for correlation among residuals across the 50 CTRA transcripts. Models were estimated using SAS v9.3 PROC MIXED with omnibus partial F tests (26) structured to assess this study’s two primary substantive hypotheses regarding association of the CTRA gene expression contrast with the three-dimensional space of language-volume measures and the 13-dimensional space of language-structure measures (function-word prevalence). Parallel likelihood ratio tests (50) were performed to corroborate omnibus F tests (Table S2). For predictor domains showing a significant omnibus association, exploratory follow-up tests assessed the specific subdimensions contributing to the overall domain association with CTRA. Additional details on measurement and statistical analysis are available in SI Methods.
Supplementary Material
Acknowledgments
We thank Cindy Chung for her insightful discussions and the University of California, Los Angeles Neuroscience Genomics Core Laboratory for assistance. This research was supported by NIH Grants R01-AT004698, P30-AG017265, R01-AG043404, R37-AG033590, and UL1-TR000454.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.L.G. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE87656).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707373114/-/DCSupplemental.
References
- 1.Cole SW. Social regulation of human gene expression: Mechanisms and implications for public health. Am J Public Health. 2013;103(Suppl 1):S84–S92. doi: 10.2105/AJPH.2012.301183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole SW. Human social genomics. PLoS Genet. 2014;10:e1004601. doi: 10.1371/journal.pgen.1004601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powell ND, et al. Social stress up-regulates inflammatory gene expression in the leukocyte transcriptome via β-adrenergic induction of myelopoiesis. Proc Natl Acad Sci USA. 2013;110:16574–16579. doi: 10.1073/pnas.1310655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heidt T, et al. Chronic variable stress activates hematopoietic stem cells. Nat Med. 2014;20:754–758. doi: 10.1038/nm.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beissner F, Meissner K, Bär KJ, Napadow V. The autonomic brain: An activation likelihood estimation meta-analysis for central processing of autonomic function. J Neurosci. 2013;33:10503–10511. doi: 10.1523/JNEUROSCI.1103-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Doux J. Anxious: Using the Brain to Understand and Treat Fear and Anxiety. Viking; New York: 2015. [Google Scholar]
- 7.Knight JM, et al. Low socioeconomic status, adverse gene expression profiles, and clinical outcomes in hematopoietic stem cell transplant recipients. Clin Cancer Res. 2016;22:69–78. doi: 10.1158/1078-0432.CCR-15-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitayama S, Akutsu S, Uchida Y, Cole SW. Work, meaning, and gene regulation: Findings from a Japanese information technology firm. Psychoneuroendocrinology. 2016;72:175–181. doi: 10.1016/j.psyneuen.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Cole SW, et al. Myeloid differentiation architecture of leukocyte transcriptome dynamics in perceived social isolation. Proc Natl Acad Sci USA. 2015;112:15142–15147. doi: 10.1073/pnas.1514249112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole SW, et al. Loneliness, eudaimonia, and the human conserved transcriptional response to adversity. Psychoneuroendocrinology. 2015;62:11–17. doi: 10.1016/j.psyneuen.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller GE, et al. Greater inflammatory activity and blunted glucocorticoid signaling in monocytes of chronically stressed caregivers. Brain Behav Immun. 2014;41:191–199. doi: 10.1016/j.bbi.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antoni MH, et al. Cognitive-behavioral stress management reverses anxiety-related leukocyte transcriptional dynamics. Biol Psychiatry. 2012;71:366–372. doi: 10.1016/j.biopsych.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Creswell JD, et al. Mindfulness-based stress reduction training reduces loneliness and pro-inflammatory gene expression in older adults: A small randomized controlled trial. Brain Behav Immun. 2012;26:1095–1101. doi: 10.1016/j.bbi.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman ML, Pennebaker JW, Berry DS, Richards JM. Lying words: Predicting deception from linguistic styles. Pers Soc Psychol Bull. 2003;29:665–675. doi: 10.1177/0146167203029005010. [DOI] [PubMed] [Google Scholar]
- 15.Hancock JT, Curry LE, Goorha S, Woodworth M. On lying and being lied to: A linguistic analysis of deception in computer-mediated communication. Discourse Process. 2008;41:1–23. [Google Scholar]
- 16.Skillicorn DB, Little A. Patterns of word use for deception in testimony. In: Yang CC, Chau M, Wang JH, Chen H, editors. Security Informatics. Springer; New York: 2010. [Google Scholar]
- 17.Pennebaker JW. The Secret Life of Pronouns: What Our Words Say About Us. Bloomsbury; New York: 2011. [Google Scholar]
- 18.Cohn MA, Mehl MR, Pennebaker JW. Linguistic markers of psychological change surrounding September 11, 2001. Psychol Sci. 2004;15:687–693. doi: 10.1111/j.0956-7976.2004.00741.x. [DOI] [PubMed] [Google Scholar]
- 19.Kacewicz E, Pennebaker JW, Davis M, Jeon M, Graesser AC. Pronoun use reflects standings in social hierarchies. J Lang Soc Psychol. 2013;33:125–143. [Google Scholar]
- 20.Pennebaker JW, Lay TC. Language use and personality during crises: Analyses of Mayor Rudolph Giuliani’s press conferences. J Res Pers. 2002;36:271–282. [Google Scholar]
- 21.Bock JK. Toward a cognitive psychology of syntax: Information processing contributions to sentence formation. Psychol Rev. 1982;89:1–47. [Google Scholar]
- 22.Hickok G. Computational neuroanatomy of speech production. Nat Rev Neurosci. 2012;13:135–145. doi: 10.1038/nrn3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehl MR, Pennebaker JW, Crow DM, Dabbs J, Price JH. The Electronically Activated Recorder (EAR): A device for sampling naturalistic daily activities and conversations. Behav Res Methods Instrum Comput. 2001;33:517–523. doi: 10.3758/bf03195410. [DOI] [PubMed] [Google Scholar]
- 24.Mehl MR, Robbins ML, Deters FG. Naturalistic observation of health-relevant social processes: The electronically activated recorder methodology in psychosomatics. Psychosom Med. 2012;74:410–417. doi: 10.1097/PSY.0b013e3182545470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pennebaker JW, Booth RJ, Francis ME. 2007 Linguistic Inquiry and Word Count: LIWC (2007) (Austin, TX). Available at liwc.wpengine.com/. Accessed October 18, 2017.
- 26.Kutner M, Nachtsheim C, Neter J, Li W. Applied Linear Statistical Models. 5th Ed McGraw-Hill/Irwin; New York: 2005. [Google Scholar]
- 27.Fredrickson BL, et al. A functional genomic perspective on human well-being. Proc Natl Acad Sci USA. 2013;110:13684–13689. doi: 10.1073/pnas.1305419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fredrickson BL, et al. Psychological well-being and the human conserved transcriptional response to adversity. PLoS One. 2015;10:e0121839. doi: 10.1371/journal.pone.0121839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci USA. 2011;108:3080–3085. doi: 10.1073/pnas.1014218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kagan J. Galen’s Prophecy: Temperament in Human Nature. Basic Books; New York: 1994. [Google Scholar]
- 31.Pennebaker JW. Writing about emotional experiences as a therapeutic process. Psychol Sci. 1997;8:162–166. [Google Scholar]
- 32.Gross JJ. Handbook of Emotion Regulation. 2nd Ed Guilford; New York: 2013. [Google Scholar]
- 33.Pennebaker JW, Chew CH. Behavioral inhibition and electrodermal activity during deception. J Pers Soc Psychol. 1985;49:1427–1433. doi: 10.1037//0022-3514.49.5.1427. [DOI] [PubMed] [Google Scholar]
- 34.Gross JJ, Levenson RW. Hiding feelings: The acute effects of inhibiting negative and positive emotion. J Abnorm Psychol. 1997;106:95–103. doi: 10.1037//0021-843x.106.1.95. [DOI] [PubMed] [Google Scholar]
- 35.Gross JJ, Levenson RW. Emotional suppression: Physiology, self-report, and expressive behavior. J Pers Soc Psychol. 1993;64:970–986. doi: 10.1037//0022-3514.64.6.970. [DOI] [PubMed] [Google Scholar]
- 36.Gross JJ. Emotion regulation: Affective, cognitive, and social consequences. Psychophysiology. 2002;39:281–291. doi: 10.1017/s0048577201393198. [DOI] [PubMed] [Google Scholar]
- 37.Cole SW, et al. Social regulation of gene expression in human leukocytes. Genome Biol. 2007;8:R189. doi: 10.1186/gb-2007-8-9-r189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eisenberger NI, Cole SW. Social neuroscience and health: Neurophysiological mechanisms linking social ties with physical health. Nat Neurosci. 2012;15:669–674. doi: 10.1038/nn.3086. [DOI] [PubMed] [Google Scholar]
- 39.Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: When the immune system subjugates the brain. Nat Rev Neurosci. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisenberger NI, Inagaki TK, Mashal NM, Irwin MR. Inflammation and social experience: An inflammatory challenge induces feelings of social disconnection in addition to depressed mood. Brain Behav Immun. 2010;24:558–563. doi: 10.1016/j.bbi.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inagaki TK, Muscatell KA, Irwin MR, Cole SW, Eisenberger NI. Inflammation selectively enhances amygdala activity to socially threatening images. Neuroimage. 2012;59:3222–3226. doi: 10.1016/j.neuroimage.2011.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehl MR, Vazire S, Holleran SE, Clark CS. Eavesdropping on happiness: Well-being is related to having less small talk and more substantive conversations. Psychol Sci. 2010;21:539–541. doi: 10.1177/0956797610362675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz HA, et al. Personality, gender, and age in the language of social media: The open-vocabulary approach. PLoS One. 2013;8:e73791. doi: 10.1371/journal.pone.0073791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iliev R, Dehghani M, Sagi E. Automated text analysis in psychology: Methods, applications, and future developments. Lang Cogn. 2015;7:265–290. [Google Scholar]
- 45.Cao J, Zhang S. Multiple comparison procedures. JAMA. 2014;312:543–544. doi: 10.1001/jama.2014.9440. [DOI] [PubMed] [Google Scholar]
- 46.Cohen S, Kessler RC, Underwood Gordon L. Measuring Stress: A Guide for Health and Social Scientists. Oxford Univ Press; New York: 1995. Perceived stress scale. [Google Scholar]
- 47.Beck A, Steer R, Brown G. Beck Depression Inventory. 2nd Ed Psychological Corporation; San Antonio: 1996. [Google Scholar]
- 48.Beck AT, Epstein N, Brown G, Steer RA. An inventory for measuring clinical anxiety: Psychometric properties. J Consult Clin Psychol. 1988;56:893–897. doi: 10.1037//0022-006x.56.6.893. [DOI] [PubMed] [Google Scholar]
- 49.Russell D, Peplau LA, Cutrona CE. The revised UCLA loneliness scale: Concurrent and discriminant validity evidence. J Pers Soc Psychol. 1980;39:472–480. doi: 10.1037//0022-3514.39.3.472. [DOI] [PubMed] [Google Scholar]
- 50.McCulloch CE, Searle SR, Neuhaus JM. Generalized, Linear, and Mixed Models. John Wiley & Sons; Hoboken, NJ: 2008. [Google Scholar]
- 51.Desbordes G, et al. Effects of mindful-attention and compassion meditation training on amygdala response to emotional stimuli in an ordinary, non-meditative state. Front Hum Neurosci. 2012;6:292. doi: 10.3389/fnhum.2012.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. American Psychiatric Association (2000) Diagnostic and Statistical Manual of Mental Disorders (American Psychiatric Association, Arlington, VA) Ed 4.
- 53.Illumina Inc 2011 Imputing Human HT-12 Expression BeadChip Data. Technical Note: Systems and Software. Available at https://support.illumina.com/content/dam/illumina-marketing/documents/products/technotes/technote_humanht_12_imputation.pdf. Accessed October 17, 2017.
- 54.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.