ABSTRACT
The vector-borne flaviviruses cause severe disease in humans on every inhabited continent on earth. Their transmission by arthropods, particularly mosquitoes, facilitates large emergence events such as witnessed with Zika virus (ZIKV) or West Nile virus in the Americas. Every vector-borne flavivirus examined thus far that causes disease in humans, from dengue virus to ZIKV, antagonizes the host type I interferon (IFN-I) response by preventing JAK-STAT signaling, suggesting that suppression of this pathway is an important determinant of infection. The most direct and potent viral inhibitor of this pathway is the nonstructural protein NS5. However, the mechanisms utilized by NS5 from different flaviviruses are often quite different, sometimes despite close evolutionary relationships between viruses. The varied mechanisms of NS5 as an IFN-I antagonist are also surprising given that the evolution of NS5 is restrained by the requirement to maintain function of two enzymatic activities critical for virus replication, the methyltransferase and RNA-dependent RNA polymerase. This review discusses the different strategies used by flavivirus NS5 to evade the antiviral effects of IFN-I and how this information can be used to better model disease and develop antiviral countermeasures.
KEYWORDS: JAK-STAT, NS5, antagonism, dengue virus, flavivirus, interferons, tick-borne encephalitis virus, West Nile virus, yellow fever virus, Zika virus
INTRODUCTION
The cellular antiviral response invoked by type I interferon (IFN-α/β or IFN-I) represents a powerful barrier to virus infection. As shown by mouse models that are deficient in the IFN-I receptor, the presence of an intact IFN-I response can be the difference between a high level of resistance to disease following virus infection and 100% lethality. For example, wild-type mice are resistant to disease caused by Zika virus (ZIKV), but mice deficient for all IFN-I responses due to a lack of the receptor are highly susceptible and can succumb from lethal infection (1–6). The biological effects of IFN-I are mediated through the upregulation of hundreds of genes, termed IFN-stimulated genes or ISGs. Depending on the cell type responding to IFN-I, ISGs function directly through interactions with viral gene products and replication intermediates to disrupt key steps in the virus replication cycle (7–9). Alternatively, in the context of cells of the immune system such as macrophages and dendritic cells (DCs), ISGs are involved in the communication between cells to initiate adaptive responses and promote both viral clearance and the establishment of immune memory (10–12). The power of IFN-I is illustrated by the fact that viruses of vertebrates have evolved multiple diverse strategies to antagonize this response, through the inhibition of IFN-I production, IFN-I signaling, or the action of individual ISGs (13–15). The relative ability of a given virus to antagonize the IFN-I response of specific host species is an important determination of virus tropism and virus virulence (16–19). Therefore, understanding the mechanisms of virus immune evasion associated with IFN-I signaling provides insight into how viruses emerge into new species and how they cause disease. This information can be used in the design of therapeutics or vaccines, particularly if the latter is based on live-attenuated virus vectors. The knowledge of how host species-specific responses are antagonized by a certain virus can also be applied to develop better animal models with increased sensitivity to infection for studies of virus pathogenesis or development of antiviral countermeasures (20). Moreover, understanding the diverse strategies that viruses have evolved in order to suppress IFN-I responses sometimes reveals novel molecules and pathways involved in immunity and can therefore provide insight into human health and disease.
The genus Flavivirus includes 53 currently recognized species (www.ictvonline.org) of which 40 are known to cause disease in humans (21). Some of the more well-known human pathogens from this group are dengue virus (DENV), yellow fever virus (YFV), West Nile virus (WNV), Japanese encephalitis virus (JEV), tick-borne encephalitis virus (TBEV), and ZIKV. The viral single-stranded RNA genome is of positive polarity and comprises one large open reading frame flanked at both the 5′ and 3′ ends by short noncoding sequences termed untranslated regions (UTR) (22). Following virus entry into a cell, the viral RNA is rapidly translated. The resulting single polyprotein has a complex membrane topology and is threaded through the endoplasmic reticulum (ER) directed by multiple transmembrane domains (23). The polyprotein is then cleaved into three structural proteins (capsid [C], envelope [E], and premembrane [M]) that are required for formation of the virion and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) that participate in the formation of specialized membrane rearrangements of the ER and establishment of virus replication complexes (RC) (22). Of particular note, two of the flavivirus nonstructural proteins have enzymatic activity. NS3 comprises the viral RNA helicase and ATPase and together with its cofactor NS2B forms the viral protease. NS5 is the largest (at approximately 900 amino acids) and most conserved of the flavivirus proteins and contains an N-terminal methyltransferase (MTase) that methylates the viral RNA (24) and in so doing disguises the viral RNA as self to prevent host recognition and promote translation (25, 26). The C-terminal two-thirds of NS5 comprises the RNA-dependent RNA polymerase (RdRp) responsible for replication of the viral RNA. NS5 is also the most potent and direct antagonist of IFN-I-dependent JAK-STAT signaling encoded by all of the flaviviruses examined thus far (27). However, despite the evolutionary constraints of NS5 structure and function in maintaining MTase and RdRp enzymatic activities, different flaviviruses have evolved to superimpose IFN antagonist functions on NS5 in different ways (Table 1). The different functions of NS5 in IFN antagonism are discussed below.
TABLE 1.
Functions of flavivirus NS5 in antagonism of IFN-I-dependent JAK-STAT signaling
| Virus | Target in IFN-I JAK-STAT signaling cascade | Host protein coopted for antagonism | Mechanism of antagonism | Additional feature(s) | Reference(s) |
|---|---|---|---|---|---|
| DENV | STAT2 | UBR4 | Proteasomal degradation of STAT2 | Uses N-end rule degradation; species restricted to primates | 53, 54, 57, 58 |
| ZIKV | STAT2 | Unknown | Proteasomal degradation of STAT2 | Species restricted to primates | 63, 64 |
| SPOV | Unknown | Unknown | Suppression of ISG translation | 63 | |
| YFV | STAT2 | TRIM23 | Suppression of ISG translation | Requires IFN-I stimulation for both NS5 function and recognition of ISGF3 | 67 |
| WNV | IFNAR1 | Prolidase | Suppression of IFNAR maturation and cell surface expression | Not highly host species restricted | 27, 83 |
| JEV | Inhibits at or upstream of JAK activation | Unknown | May involve protein tyrosine phosphatases | 81, 85, 86 | |
| TBEV | IFNAR1 | Prolidase | Suppression of IFNAR maturation and cell surface expression | Not highly host species restricted | 27, 83 |
| LGTV | IFNAR1 | Prolidase | Suppression of IFNAR maturation and cell surface expression | Not highly host species restricted | 27, 46, 72, 74, 84 |
INTERFERON RESPONSE TO FLAVIVIRUS INFECTION AND VIRAL ANTAGONISM
Flavivirus infection and replication of viral RNA are initially sensed by the cytosolic host pattern recognition receptor (PRR) known as retinoic acid inducible gene I (RIG-I), resulting in the activation of transcription factors IRF3, AP-1, and NFκB and production of IFN-I (28). The RIG-I-like receptor (RLR) called melanoma differentiation-associated protein 5 (Mda5) also recognizes flavivirus double-stranded RNA (dsRNA), and its activation sustains the IFN-I response (29, 30). Toll-like receptors (TLRs) that recognize virus RNA and may contribute to host IFN-I responses include the endosomal TLR3, TLR7, and TLR8. However, the role of RLRs in IFN-I production following sensing of flaviviruses is greater than that of TLRs and essential for host resistance (31). Once secreted, IFN-I binds to two receptor subunits, IFNAR1 and IFNAR2, in an autocrine and paracrine manner (32). Binding of IFN-I to its receptors activates the Janus kinases (JAK) Tyk2 and JAK1, which are constitutively associated with the receptor intracellular domains. Kinase activation occurs through both auto- and transphosphorylation. Activated JAKs then phosphorylate the IFNAR to create docking sites for signal transducer and activator of transcription 2 (STAT2). STAT2 is then phosphorylated and recruits STAT1 for phosphorylation and the formation of STAT1/STAT2 heterodimers (32, 33). STAT1/STAT2 associate with a third component, IFN-regulatory factor 9 (IRF9), to form the transcriptional activation complex known as IFN-stimulated gene factor 3 (ISGF3). ISGF3 translocates to the nucleus, where it binds to IFN-stimulated response elements (ISRE) in the promoter regions of ISGs to upregulate gene expression. These include a number of ISGs with antiflavivirus activity (e.g., viperin, OAS1a, IFIT1, IFITM proteins, TRIM proteins) as well as both positive and negative regulators of IFN-I signaling (7, 8, 25, 34, 35).
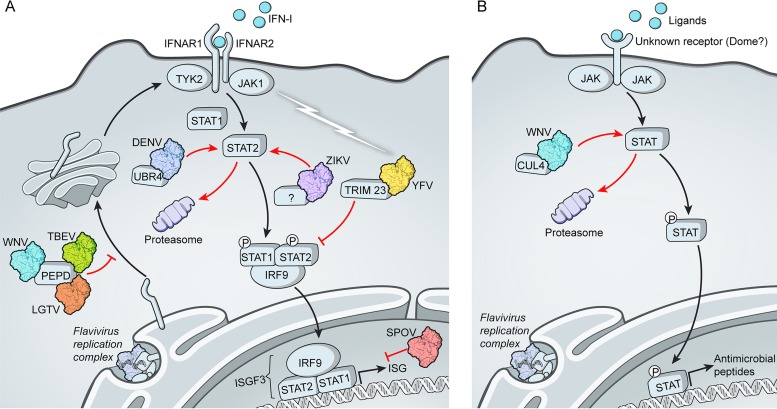
Flaviviruses have evolved a number of mechanisms to delay or impede the initial recognition of viral RNA or prevent RLR signaling. Antagonistic strategies that multiple flaviviruses appear to share include the potential to shield viral dsRNA replicative intermediates by invaginations of the modified ER membranes (28, 36, 37), methylation of the viral RNA at N7 and 2′O positions (38) to prevent recognition by the ISG called IFN-induced protein with tetratricopeptide repeats 1 (IFIT1) and to promote RNA translation (25, 26), and the production of a small subgenomic RNA derived from the 3′UTR termed small flavivirus RNA or sfRNA (39). The sfRNA from multiple flaviviruses counters the IFN-I response (40) and at least in the case of DENV prevents the production of IFN-I by suppressing activation of RIG-I (41). Additional strategies to suppress IFN production that may be more divergent across flaviviruses exist and have been reviewed elsewhere (42–45). However, all flaviviruses that cause disease in humans examined thus far suppress the response of infected cells to secreted IFN-I. This was first suggested by experiments showing that the addition of IFN-I before infection suppressed replication but not when added as little as 4 h after infection (46–48). Early studies examining this phenomenon showed that different flaviviruses suppress signaling at different points in the JAK-STAT cascade. For example, DENV infection has been associated with a loss of STAT2 (49) whereas infection with WNV resulted in a failure of JAK activation (50). The strategies utilized by the different flaviviruses and the specific roles for NS5 are discussed below (Fig. 1).
FIG 1.
Varied mechanisms of NS5-mediated antagonism of IFN-I-dependent signaling in flavivirus-infected cells. (A) The NS5 proteins of DENV, ZIKV, and YFV all bind to STAT2. Binding of STAT2 by DENV or ZIKV NS5 results in STAT2 degradation, whereas YFV NS5 requires IFN-I stimulation to “activate” the antagonist functions of NS5 (depicted by a lightning bolt) resulting in loss of transcriptional activity of ISGF3. Each of these three NS5 proteins utilizes a different E3 ubiquitin ligase in order to antagonize IFN-I signaling (TRIM23, UBR4, or a yet-to-be-identified ligase). The NS5 proteins of TBEV, LGTV, and WNV suppress maturation of the IFN-I receptor subunit IFNAR1 by binding to prolidase (PEPD). (B) Antagonism of JAK-STAT signaling by WNV in mosquitoes involves the host protein cullin 4 (CUL4) to degrade STAT via the proteasome.
DENGUE VIRUS
The four serotypes of DENV (1 to 4) are responsible for an estimated 96 million clinically recognized infections annually, of which 2 million are severe cases resulting in 20,000 deaths (51, 52). In the context of replicating virus, DENV has been shown to suppress activation of IFN-I-stimulated JAK-STAT signaling at multiple levels in human cells, including phosphorylation of Tyk2, STAT1, and STAT2 (48, 49). This can reflect suppression of the upstream signaling molecules, such as the receptor or JAK kinases, which then suppresses all downstream (STAT activation) events, or it may result from the virus individually targeting multiple host proteins in the pathway. The demonstrated role of DENV NS5 is to bind and deplete STAT2 via ubiquitin-dependent proteasomal degradation (53, 54). In infected cells, or in cells expressing a DENV replicon that expresses the nonstructural proteins only, STAT2 is degraded (53). Ectopic expression of NS5 alone is sufficient to suppress IFN-I-stimulated gene expression in gene reporter assays (53) or when examining mRNA upregulation of individual ISGs (54) at a level that is comparable to that of replicating virus. However, while expression of NS5 as a single protein resulted in a block in STAT2 phosphorylation, it was not sufficient to cause STAT2 degradation (53, 54). Loss of STAT2 is observed only if the N terminus of NS5 is produced as the result of a cleavage event such as would occur in the context of the DENV polyprotein (53). During virus replication, cleavage of NS5 to separate it from NS4B is mediated by the viral protease NS2B/3. Expression of an N-terminal cleavable NS5 resulted in STAT2 degradation when that construct was cotransfected together with NS2B/3 (53). However, the cleavage event did not need to be specific to the viral protease. Replacement of the flavivirus protease cleavage site with ubiquitin that is cleaved by endogenous ubiquitin hydrolases after the final two glycine residues resulted in a mature NS5 protein that degraded STAT2. Furthermore, the identity of the N-terminal residue of NS5 was not important, and this residue could be a Gly, as would be present during polyprotein cleavage, or a Met, as is engineered when NS5 is expressed alone from a cDNA plasmid (53). This work showed that it was the act of proteolytic cleavage at the NS5 N terminus that conferred additional functionality on NS5 as an IFN-I antagonist. To date, this is the only example where maturation of the N terminus of NS5 in the context of a polyprotein is required for full function as an IFN-I antagonist, although a more general requirement for polyprotein processing events in flavivirus replication may exist (55).
N-terminal cleavage of proteins followed by proteasomal degradation is a hallmark of the N-end rule pathway of degradation (56). Consistent with this, the first 5 amino acids of DENV NS5 bind to ubiquitin protein ligase E3 component N-recognin 4 (UBR4), an E3 ubiquitin ligase of the N-recognin family (57). N-recognins target substrates that contain a destabilizing N-terminal residue in the context of a structural conformation that exposes the N terminus (56). Interestingly, DENV NS5 bound UBR4 irrespective of whether the NS5 N terminus was generated through a cleavage event, although this interaction was increased by proteolytic processing (57). The binding domain in DENV NS5 for STAT2 is suggested as lying between residues 202 and 306 (53, 54, 57, 58) (Fig. 2). Thus, DENV NS5 utilizes its N terminus together with a central domain to act as a bridge between STAT2 and UBR4. This role for NS5 may result in NS5 degradation along with STAT2 in what could be an act of self-sacrifice to eliminate IFN-I signaling. However, the flavivirus NS5 protein is generated in excess of that required for virus replication, suggesting that different populations of NS5 most likely carry out functions in virus replication and IFN antagonism and that degradation of this population would therefore not adversely impact NS5 replicative functions. Depending on the DENV serotype, NS5 expression is often nuclear, although nuclear localization is not required for STAT2 inhibition, suggesting a cytosolic location for STAT2 binding and degradation (59). Importantly, the relevance of this mechanism to the virus has been demonstrated by the observation that UBR4 enhanced the replication of DENV in an IFN-I-dependent manner, as demonstrated by RNA interference (RNAi)-mediated depletion of UBR4 in IFN-I-treated cells (57). Furthermore, UBR4 was also required for optimal infection of human monocyte-derived DCs and viral suppression of ISG mRNA expression (57).
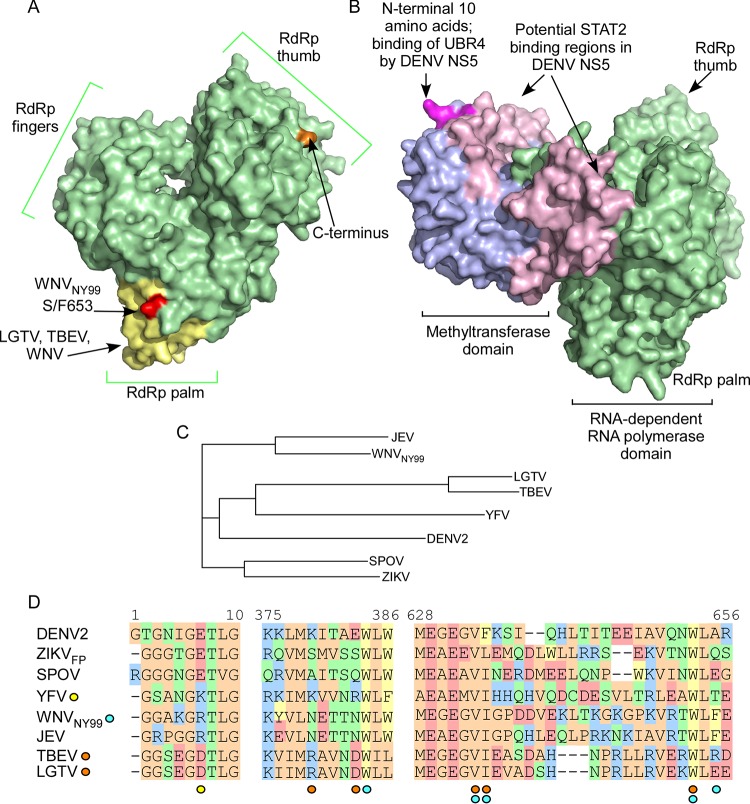
FIG 2.
Structural features of flavivirus NS5 implicated in IFN-I antagonism. (A) Surface-exposed residues of the RdRp domain of WNV (PDB ID 2HCN) are depicted in green, with amino acids important for function of NS5 from WNV, LGTV, and TBEV shown in yellow. The S/F653 residue that determines relative IFN-I antagonism and efficiency of IFNAR1 downregulation in WNV is shown in red. (B) Structure of DENV NS5 that has been rotated 90° in relation to that shown in panel A. The surface-exposed residues of the MTase domain are depicted in blue. The N-terminal 10 amino acids that are important to the function of both DENV and YFV NS5 are shown in magenta. Putative binding regions for human STAT2 on DENV NS5 are shown in light pink. The full-length structure is based on the DENV-4 NS5 crystal structure (PDB ID 5DTO) (24) with images colorized using PyMol. (C) Phylogenetic tree of NS5 sequences discussed in the text. (D) Sequence alignment showing the N-terminal 10 amino acids and the two noncontiguous regions involved in IFN-I antagonism by WNV, LGTV, and TBEV. Residues are color coded according to similar chemistries. Individual amino acids of particular importance are indicated by the colored circles below the alignment for YFV (yellow), WNV (blue), or LGTV and TBEV (orange).
The role of DENV NS5 in IFN-I antagonism is a likely determinant of pathogenesis in a host-specific manner. The historical emergence of DENV in human populations occurred as a result of a number of spillover events from the enzootic transmission cycles between nonhuman primates and mosquito species (51). The evolution of dengue viruses in nonhuman primates may have influenced the ability of DENV to bind and degrade human STAT2 and facilitated the emergence of DENV in humans. However, unlike some of the other flaviviruses, such as WNV and TBEV, DENV cannot antagonize signaling in mice as NS5 cannot bind to mouse STAT2 (58). In contrast, DENV NS5 binds UBR4 equally from human or mouse origin. Hence, the inability of NS5 to bind mouse STAT2 enables IFN-I-dependent responses to severely limit DENV replication in mouse cells (58). This fact underlies one of the major limiting factors in the development of mouse models of DENV for the study of virus pathogenesis and antiviral countermeasures. Advances have been made using the Cre-lox system to generate mice deficient in IFNAR only on DCs, which increases sensitivity to DENV and recapitulates some aspects of human disease (60). However, the development of a completely immunocompetent mouse model of DENV pathogenesis may be further advanced through the replacement of mouse STAT2 with human STAT2 (20).
ZIKA VIRUS
The large-scale emergence of ZIKV in South America, first detected in 2015, is associated with alarming facets of disease not previously attributed to flavivirus infections, including congenital disease associated with mother-to-fetus transmission and virus replication in the fetal central nervous system (CNS), Guillain-Barré syndrome in adults, and sexual transmission (61, 62). Like DENV, ZIKV originally emerged in human populations as a result of spillover transmission from the mosquito-to-nonhuman primate transmission cycle. Also similar to DENV, the replication of ZIKV results in proteasomal degradation of STAT2 mediated by NS5 (63). The function of ZIKV NS5 in IFN-I antagonism is also host species restricted, being functional in the context of human and nonhuman primate STAT2 but not in that of mouse. However, the mechanism of NS5-mediated STAT2 in ZIKV appears to differ from that in DENV. Expression of ZIKV NS5 alone results in STAT2 degradation without the need for the maturation of the NS5 N terminus by a cleavage event, and UBR4 does not participate in the loss of STAT2 (63). Furthermore, the first 10 amino acids of ZIKV NS5 are not critically required for antagonism (64). Thus, although an additional, as yet unidentified, host E3 ubiquitin ligase is most certainly utilized by ZIKV, the N-end rule pathway does not appear to be involved. Additional work is needed to identify the E3 ubiquitin ligase utilized by ZIKV NS5, but the species specificity of NS5 function suggests that development of transgenic mice that express human STAT2 would benefit ZIKV pathogenesis studies in addition to DENV. This is of particular importance, since modeling of congenital ZIKV infection in mice following transmission from the dam currently relies predominantly on the use of immunocompromised mice deficient for IFN-I signaling pathways (5, 6, 65).
The closest known relative of ZIKV is Spondweni virus (SPOV) (66). Similar to the historic distribution of ZIKV, SPOV also circulates in Africa and Southeast Asia, where it causes sporadic but seemingly inconsequential human infections. Interestingly, the NS5 protein of SPOV inhibits JAK-STAT signaling at a point downstream from STAT2 (63). In cells expressing SPOV NS5, both STAT1 and STAT2 become phosphorylated in response to IFN-I stimulation and translocate to the nucleus. SPOV NS5 localizes to the nucleus and prevents ISRE-dependent gene expression but without strong binding to the STATs. Therefore, SPOV must prevent transcriptional activation of ISGs, potentially by binding IRF9, by preventing the DNA binding or transcriptional activity of ISGF3 or through a less direct mechanism like prevention of histone modification. Thus, it is interesting from a biological perspective that ZIKV NS5 function in STAT2 degradation and IFN-I antagonism closely resembles that of the more distantly related DENV and not of SPOV, with which ZIKV shares 65% and 77% amino acid identity in NS5, respectively (63) (Fig. 2C).
YELLOW FEVER VIRUS
Despite the availability of a highly protective live-attenuated vaccine for YFV, approximately 200,000 cases of yellow fever occur annually, resulting in an estimated 30,000 deaths (21). Similar to DENV, YFV evolved in an enzootic transmission cycle between nonhuman primates and mosquitoes, with human infection resulting from spillover events. Today, the urban YFV transmission cycle occurs exclusively in humans and mosquitoes, without the need for a zoonotic amplifying host (21). YFV suppresses gene expression stimulated by type I IFN but not type II (67). Consistent with this specificity by the virus, the target of the YFV NS5 protein is STAT2. However, antagonism of STAT2 by YFV involves some unique features not observed in the context of other flaviviruses. The most striking difference is that binding of NS5 to STAT2 is dependent on cellular stimulation with type I (or type III) IFN (67). The role of IFN-I stimulation is 2-fold: to induce STAT1/STAT2 heterodimers that render STAT2 amenable to NS5 binding and to promote K63-linked ubiquitination of a Lys residue in the NS5 N terminus (at position 6 of the mature protein), which then enables NS5 association with STAT2. The binding of STAT2 does not result in STAT degradation like it does in DENV or ZIKV but instead inhibits binding of ISGF3 to ISRE promoter elements to prevent ISG transcription. The E3 ubiquitin ligase responsible for ubiquitinating NS5 on Lys6 was identified as tripartite motif (TRIM) protein 23 (TRIM23) (67). The TRIM family is a large group of proteins, many of which function in antiviral defense by promoting innate immune signaling (68) or by directly interacting with viral proteins to disrupt their function and restrict virus replication (35, 69, 70). However, depletion of TRIM23 by RNAi resulted in decreased ubiquitination of NS5, decreased binding of NS5 to STAT2, and increased susceptibility of YFV to the antiviral effects of IFN-I (67). A Lys-to-Arg mutation at position 6 of NS5 also increased the susceptibility of recombinant YFV to IFN-I (67). Taken together, these findings revealed a completely new mechanism of viral IFN-I antagonism, where the viral antagonist is activated by the very response it has evolved to suppress. In addition, a member of the TRIM protein family that generally functions in antiviral responses is coopted by YFV to evade the IFN-I response.
VIRUSES IN THE TICK-BORNE ENCEPHALITIS VIRUS COMPLEX
The tick-borne encephalitis complex of viruses includes three subtypes of TBEV (European, Siberian, and Far Eastern), Powassan virus, Kyasanur Forest disease virus, Omsk hemorrhagic fever virus, Louping ill virus, and Langat virus (LGTV), among others (71). An effective killed-virus vaccine against TBEV exists, yet approximately 12,000 cases of encephalitis, meningoencephalitis, or meningitis still occur in Europe, Russia, and parts of Asia annually (21). The Far Eastern TBEV strains are the most virulent, with case fatality rates estimated to be as high as 20%. In contrast, while LGTV shares high amino acid identity with TBEV (>80%), it is considerably less neurovirulent, prompting the application of LGTV as a live-attenuated anti-TBEV vaccine in the 1970s. However, infection with LGTV resulted in meningoencephalitis in approximately 1/12,000 recipients, and its use was discontinued (71).
Cells infected with LGTV are refractory to IFN-I stimulation due to a block in activation of both JAK1 and Tyk2, resulting in failure of all downstream signaling, including STAT1/2 phosphorylation, STAT nuclear localization, and ISRE-dependent gene expression (46). This study was the first to identify NS5 as the dominant IFN-I antagonist encoded by a flavivirus (46). Expression of progressive truncation mutants of LGTV NS5 mapped the minimal linear sequence required for antagonism to residues 355 to 735 within the RdRp domain (72). A random mutagenesis approach identified two short noncontiguous stretches of amino acids within this domain as important for NS5 function, including residues 374 to 380 and 624 to 647 (Fig. 2A and D). Of these, D380 and W647 were of particular importance. Despite the considerable separation of these residues on the linear sequence, the two regions are proximal to each other when modeled on the crystal structure of the flavivirus RdRp (73) (Fig. 2), suggesting that they act cooperatively to antagonize IFN-I signaling (72).
The suppression of JAK activation observed in LGTV-infected cells suggested that antagonism was mediated apically in the JAK-STAT cascade, at the level of the JAKs or receptor subunits. However, LGTV NS5 did not directly suppress JAK1 or Tyk2 activation (74). Instead, cells infected with either LGTV or TBEV lose expression of IFNAR1 but not IFNAR2 (27). IFNAR1 is a common target of many diverse viruses and can be downregulated through increased degradation induced by ER stress pathways or engagement of PRRs (75). Both of these events occur during flavivirus replication (31, 76, 77). However, neither lysosomal nor proteasomal inhibitors rescued IFNAR1 expression in the context of tick-borne flavivirus infection, suggesting a potential novel pathway of receptor antagonism (27). Expression of LGTV NS5 alone downregulated IFNAR1 expression, but not a W647A mutant of NS5, suggesting that loss of IFNAR1 was the major function of NS5 in IFN-I antagonism. Importantly, recombinant TBEV containing a D380A mutation in NS5 exhibited increased sensitivity to IFN-I added after replication was established. This sensitivity was not observed for wild-type TBEV and was a consequence of the inability of the D380A mutant to downregulate IFNAR1, inhibit STAT1 phosphorylation, and suppress ISRE-dependent gene expression. TBEV D380A was also highly attenuated in mice associated with minimal viremia and delayed kinetics of virus reaching the CNS (27). Therefore, this work demonstrated for the first time that the function of flavivirus NS5 as an IFN-I antagonist is an important virulence factor in vivo.
To determine the mechanism of IFNAR1 downregulation, proteomics approaches that identified an interaction between LGTV NS5 and the host protein prolidase (PEPD) were used (27). PEPD is a peptidase that cleaves dipeptides containing Pro to recycle Pro from dietary and endogenous proteins (78) but was not known to have a role in immunity. The interaction between NS5 and PEPD was dependent on residues previously identified as critical for NS5 function as an IFN-I antagonist, raising the possibility that PEPD may function in IFNAR1 expression (27). In support of a role for PEPD in TBEV resistance to IFN-I, TBEV D380A lost sensitivity to IFN-I when either IFNAR1 or PEPD was depleted from cells. Indeed, in the absence of PEPD, IFNAR1 failed to accumulate. However, the half-life of IFNAR1 did not differ in the presence or absence of PEPD, suggesting that PEPD did not affect IFNAR1 stability and consistent with the observation that depletion of IFNAR1 was not through cellular degradation pathways. Instead, PEPD was required for the maturation of IFNAR1 from a partially glycosylated form with high-mannose N-linked carbohydrate to the fully glycosylated complex N-linked oligosaccharide (27). This work also led to the demonstration that IFNAR1 expression and response to IFN-I are low in primary fibroblasts from human patients with prolidase deficiency (PD), identifying PD as a primary immune deficiency in humans (27). Taken together, this work suggests that NS5 from tick-borne flaviviruses has yet another novel way to antagonize type I IFN, by disrupting glycosylation and surface expression of IFNAR1 to prevent downstream signaling and ISG expression.
WEST NILE VIRUS AND JAPANESE ENCEPHALITIS VIRUS
WNV and JEV belong to the same evolutionary clade and share both mosquito vectors (Culex spp.) and vertebrate zoonotic amplifying hosts (birds), and both can cause severe encephalitis or meningitis in humans (21). Furthermore, resistance to the antiviral effects of IFN-I is a determinant of virulence for both WNV and JEV (79, 80). IFN-I signal transduction is blocked in cells infected with either WNV or JEV due to a failure of JAK1 and Tyk2 phosphorylation (50, 81). However, only WNV infection induces a loss of IFNAR1 (27, 81, 82) associated with the function of NS5 (27), making WNV NS5 similar in function to that from LGTV and TBEV (27). Mapping of the residues in NS5 derived from the 1999 New York strain of WNV (NY99) demonstrated that WNV and LGTV share a number of the same amino acids for IFN-I antagonism (83) and that WNV NS5 also binds PEPD (27). In contrast, NS5 from the attenuated WNV strain Kunjin (KUN) was shown to be a relatively poor inhibitor of IFN-I signaling. Recombinant WNVKUN containing a single substitution in NS5 corresponding to the WNVNY99 NS5 residue (S/F653) (Fig. 2A) is more resistant than wild-type WNVKUN to IFN-I (83) associated with an earlier ability to downregulate IFNAR1 and increased NS5/PEPD colocalization by immunofluorescence (27). Thus, there appears to be a surprising conservation of IFN-I antagonism strategies between the tick-borne flaviviruses and WNV. However, it is possible that the NS5 protein from both WNV and TBEV possesses additional functionality that is not present in LGTV. This is because a truncation mutant of LGTV NS5 containing residues 355 to 735 fully antagonizes signaling equivalent to the full-length protein. However, for WNV or TBEV, NS5 can be truncated at the C terminus to residue 735 but NS5 with a truncation at the N terminus does not retain activity (72). In support of this, binding of the TBEV NS5 MTase domain (residues Y222A and S223A) to the host protein Scribble aids in plasma membrane localization of NS5 and IFN-I antagonism (84). While the full role of the NS5 N terminus is currently unknown for WNV and TBEV, it is not required for IFN-I-dependent modification as has been observed for YFV nor for N-end rule processing as occurs in the context of DENV NS5. However, it is possible that this theoretical added functionality in NS5 of TBEV or WNV contributes to the greater virulence of these viruses than of LGTV in humans.
Infection with JEV does not appear to affect IFNAR1 or IFNAR2 expression, despite a block in Tyk2 activation (81). The role for NS5 in this requires the NS5 N-terminal 83 residues but not the C-terminal 143 amino acids (85). The proposed role for JEV NS5 is to activate protein tyrosine phosphatases (PTPs) that have normal roles as negative feedback regulators of JAK activation (85). Additional studies suggest that JEV NS5 expression downregulates calreticulin to inhibit nuclear translocation of STAT1 (86). However, how these activities of NS5 result in inhibition of Tyk2 phosphorylation and suppression of STAT nuclear localization and how they might be coordinated are not known.
FLAVIVIRUS SUPPRESSION OF JAK-STAT IN THE ARTHROPOD VECTOR
JAK-STAT signaling in insects regulates developmental events and cellular immune responses (87, 88). Mosquito STAT proteins most closely resemble human STAT5 and STAT6 (27 to 30% identity) (89). JEV replication in C6/36 mosquito (Aedes albopictus) cells suppressed tyrosine phosphorylation of AeSTAT in response to infection (89). Furthermore, WNV infection or expression of either WNV NS5 or NS1 in Hsu cells (Culex quinquefasciatus) resulted in proteasomal degradation of CxSTAT that is dependent on the Culex E3 ubiquitin ligase cullin 4 (CxCul4) (90) (Fig. 1B). However, it is unknown if the mechanisms of JAK-STAT inhibition observed in mosquito cells are conserved in mammalian cells or if they represent functionally different strategies of antagonism. This is in part because WNV NS5 binding to STAT proteins (63) or STAT degradation in WNV-infected cells (50, 83, 91) has not been observed in the context of mammalian cells.
Ticks have both JAK and STAT molecules that participate in antipathogen responses (92). Nothing is known regarding interactions between tick-borne flaviviruses and the tick vector with respect to NS5 or STAT signaling. However, the tick JAK-STAT pathway can respond to mammalian IFN-γ imbibed with a blood meal when a pathogen is acquired from an infected mammal. This results in upregulation of antimicrobial peptides that reduce the pathogen burden in the tick (shown in the context of Borrelia burgdorferi infection of mice) (93). Vertebrate cytokines can also stimulate the mosquito midgut (94). Thus, it is possible that flavivirus antagonism of JAK-STAT signaling has a role in establishing replication in the arthropod by evading the antiviral effects of IFNs in the blood meal. This may be particularly important for infection of tick species that feed over a number of days. Further work to understand the relationship between IFN antagonism in vertebrate and invertebrate hosts will provide insight into the molecular mechanisms that enable virus maintenance in vectors and whether virus virulence determinants in vertebrate hosts are shaped by evolution in arthropods.
ROLES FOR ADDITIONAL VIRAL PROTEINS IN SUPPRESSION OF JAK-STAT SIGNALING FOLLOWING IFN-I STIMULATION
Although NS5 is clearly the most specific and potent antagonist of IFN-I-dependent JAK-STAT signaling encoded by flaviviruses, additional nonstructural proteins also contribute to suppression of this pathway (91, 95, 96). Indeed, the known functions of NS5 do not always recapitulate the phenotype of suppression observed in virus-infected cells. For example, DENV infection of human DCs prevents Tyk2 phosphorylation (48), which is an event upstream of STAT2 degradation. Some of these additional blocks in signaling may be explained by the induction of more-generalized stress responses that disrupt signaling, such as ER stress, membrane rearrangement, and cholesterol redistribution from the plasma membrane (75-77, 97). The unfolded protein response associated with ER stress and membrane rearrangement is mediated by the small hydrophobic proteins NS4A and NS4B (76, 98), which may explain the ability of these viral proteins to modulate JAK-STAT signaling. Lipid rafts promote cellular signaling events at the plasma membrane, including those by IFNs (97, 99). NS2B and NS3 associate with lipid rafts in the context of DENV replication (100), and WNV recruitment of plasma membrane cholesterol to sites of virus replication in the ER is thought to disrupt IFN-I-dependent JAK-STAT signaling (97). Finally, the engagement of Tyro3/Axl/Mer (TAM) receptors by phosphatidylserine in the lipid membrane of flaviviruses during virus entry triggers the upregulation of suppressor of cytokine signaling (SOCS) proteins (101). SOCS proteins function as negative regulators of JAK activation by targeting their tyrosine kinase activity directly (102). This mechanism of immune suppression is initiated at the time of virus binding to the cell, suggesting that it could contribute to the loss of JAK activity at earlier times in infection before NS5 is present at sufficient amounts to protect the infected cell or in instances where NS5 targets the signaling cascade downstream of JAK activation.
PERSPECTIVES AND OPEN QUESTIONS
The importance of NS5 function in IFN-I antagonism and flavivirus resistance to IFN-I has been demonstrated in the context of replicating virus both in vitro and in vivo (27, 67, 83). The varied strategies utilized by each flavivirus suggests that there is a remarkable plasticity in NS5 for interaction with host proteins, given that it must retain two critical enzymatic activities (MTase and RdRp) for virus replication. These varied strategies also suggest that there is considerable selection pressure driving the evolution of flaviviruses at this critical virus-host interface. However, it is unclear if the different strategies embedded within NS5 to suppress JAK-STAT signaling influences the different pathogenesis of these viruses. This is theoretically possible, given that a critical determinant within the TBEV complex of viruses that dictates the ability of these viruses to cause hemorrhagic fever versus encephalitis was mapped to NS5 (although not in the IFN-I antagonist domain) (103). Regardless of whether the role of NS5 influences disease phenotype, it is most certainly a determinant of the potential of flaviviruses to emerge in humans.
No information exists as to how NS5 is regulated to perform functions in RNA replication and IFN-I antagonism. As NS5 can form dimers with unique conformations (104), it is possible that multiple populations of NS5 exist in an infected cell. Posttranslational modifications, such as phosphorylation (105) or ubiquitination (106), may regulate NS5 self-association, cellular distribution, or function. Furthermore, the orientation of NS5 RdRp and MTase domains from different flaviviruses results in unique conformations that impact replication (107–109) and that may also regulate NS5 function in IFN-I antagonism in a virus species-specific manner. This might be particularly important when the regions of NS5 required for antagonism include both MTase and RdRp domains, as is the case for at least DENV, WNV, and TBEV. Thus, future work is required to determine whether the NS5 function as an IFN-I antagonist is physically separated in the cell from its role in RNA replication and how domain conformation and potential posttranslational modifications might regulate these roles. This information may aid in the development of therapeutics targeting NS5.
The importance of NS5 in IFN-I antagonism provides at least two opportunities for translational research. The first is the incorporation of loss-of-function mutations in NS5 as an attenuating mechanism to increase the safety of live-attenuated anti-flavivirus vaccines. The second is the engineering of improved immunocompetent mouse models in the case of flaviviruses that have restricted host-species tropism. This could be achieved by “humanizing” the viral targets of antagonism in the IFN-I response (20). Using the examples of DENV and ZIKV, it is unlikely that STAT2 alone, or even IFN antagonism in general, is the only species barrier that must be overcome in mice in order to observe all the features of human disease caused by these viruses. Indeed, DENV also antagonizes other pathways of IFN-I production in a species-specific manner (110). However, mice that are transgenic for human homologs of key molecules that are antagonized by multiple viruses, whether they be in pathways of viral nucleic acid sensing or response to IFN-I, would aid in the development of animal models for newly emerging viruses and in the testing of virus countermeasures.
ACKNOWLEDGMENT
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. 2016. A mouse model of Zika virus pathogenesis. Cell Host Microbe 19:720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossi SL, Tesh RB, Azar SR, Muruato AE, Hanley KA, Auguste AJ, Langsjoen RM, Paessler S, Vasilakis N, Weaver SC. 2016. Characterization of a novel murine model to study Zika virus. Am J Trop Med Hyg 94:1362–1369. doi: 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aliota MT, Caine EA, Walker EC, Larkin KE, Camacho E, Osorio JE. 2016. Characterization of lethal Zika virus infection in AG129 mice. PLoS Negl Trop Dis 10:e0004682. doi: 10.1371/journal.pntd.0004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dowall SD, Graham VA, Rayner E, Atkinson B, Hall G, Watson RJ, Bosworth A, Bonney LC, Kitchen S, Hewson R. 2016. A susceptible mouse model for Zika virus infection. PLoS Negl Trop Dis 10:e0004658. doi: 10.1371/journal.pntd.0004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miner JJ, Cao B, Govero J, Smith AM, Fernandez E, Cabrera OH, Garber C, Noll M, Klein RS, Noguchi KK, Mysorekar IU, Diamond MS. 2016. Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell 165:1081–1091. doi: 10.1016/j.cell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yockey LJ, Varela L, Rakib T, Khoury-Hanold W, Fink SL, Stutz B, Szigeti-Buck K, Van den Pol A, Lindenbach BD, Horvath TL, Iwasaki A. 2016. Vaginal exposure to Zika virus during pregnancy leads to fetal brain infection. Cell 166:1247–1256 e1244. doi: 10.1016/j.cell.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacMicking JD. 2012. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat Rev Immunol 12:367–382. doi: 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crouse J, Kalinke U, Oxenius A. 2015. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol 15:231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- 11.González-Navajas JM, Lee J, David M, Raz E. 2012. Immunomodulatory functions of type I interferons. Nat Rev Immunol 12:125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swiecki M, Colonna M. 2015. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol 15:471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee S, Basler CF, Amarasinghe GK, Leung DW. 2016. Molecular mechanisms of innate immune inhibition by non-segmented negative-sense RNA viruses. J Mol Biol 428:3467–3482. doi: 10.1016/j.jmb.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann HH, Schneider WM, Rice CM. 2015. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol 36:124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Versteeg GA, Garcia-Sastre A. 2010. Viral tricks to grid-lock the type I interferon system. Curr Opin Microbiol 13:508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirai-Yuki A, Hensley L, McGivern DR, Gonzalez-Lopez O, Das A, Feng H, Sun L, Wilson JE, Hu F, Feng Z, Lovell W, Misumi I, Ting JP, Montgomery S, Cullen J, Whitmire JK, Lemon SM. 2016. MAVS-dependent host species range and pathogenicity of human hepatitis A virus. Science 353:1541–1545. doi: 10.1126/science.aaf8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maringer K, Fernandez-Sesma A. 2014. Message in a bottle: lessons learned from antagonism of STING signalling during RNA virus infection. Cytokine Growth Factor Rev 25:669–679. doi: 10.1016/j.cytogfr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McFadden G, Mohamed MR, Rahman MM, Bartee E. 2009. Cytokine determinants of viral tropism. Nat Rev Immunol 9:645–655. doi: 10.1038/nri2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Misra A, Thippeshappa R, Kimata JT. 2013. Macaques as model hosts for studies of HIV-1 infection. Front Microbiol 4:176. doi: 10.3389/fmicb.2013.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrison J, Garcia-Sastre A. 2014. STAT2 signaling and dengue virus infection. JAKSTAT 3:e27715. doi: 10.4161/jkst.27715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gubler DJ, Kruno G, Markoff L. 2007. Flaviviruses, p 1153–1252. In Knipe DM, Howley PM (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 22.Lindenbach BD, Thiel HJ, Rice CM. 2007. Flaviviridae: the viruses and their replication, p 1101–1152. In Knipe DM, Howley PM (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- 23.Paul D, Bartenschlager R. 2015. Flaviviridae replication organelles: oh, what a tangled web we weave. Annu Rev Virol 2:289–310. doi: 10.1146/annurev-virology-100114-055007. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y, Soh TS, Lim SP, Chung KY, Swaminathan K, Vasudevan SG, Shi PY, Lescar J, Luo D. 2015. Molecular basis for specific viral RNA recognition and 2′-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5). Proc Natl Acad Sci U S A 112:14834–14839. doi: 10.1073/pnas.1514978112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M Jr, Shi PY, Diamond MS. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyde JL, Diamond MS. 2015. Innate immune restriction and antagonism of viral RNA lacking 2-O methylation. Virology 479-480:66–74. doi: 10.1016/j.virol.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lubick KJ, Robertson SJ, McNally KL, Freedman BA, Rasmussen AL, Taylor RT, Walts AD, Tsuruda S, Sakai M, Ishizuka M, Boer EF, Foster EC, Chiramel AI, Addison CB, Green R, Kastner DL, Katze MG, Holland SM, Forlino A, Freeman AF, Boehm M, Yoshii K, Best SM. 2015. Flavivirus antagonism of type I interferon signaling reveals prolidase as a regulator of IFNAR1 surface expression. Cell Host Microbe 18:61–74. doi: 10.1016/j.chom.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fredericksen BL, Gale M. 2006. West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J Virol 80:2913–2923. doi: 10.1128/JVI.80.6.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M Jr. 2013. The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J Virol 87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M. 2008. Establishment and maintenance of the innate antiviral response to West Nile virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol 82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suthar MS, Diamond MS, Gale M. 2013. West Nile virus infection and immunity. Nat Rev Microbiol 11:115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- 32.Ivashkiv LB, Donlin LT. 2014. Regulation of type I interferon responses. Nat Rev Immunol 14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levy DE, Darnell JE Jr. 2002. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol 3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 34.Mashimo T, Simon-Chazottes D, Guenet JL. 2008. Innate resistance to flavivirus infections and the functions of 2′-5′ oligoadenylate synthetases. Curr Top Microbiol Immunol 321:85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor RT, Lubick KJ, Robertson SJ, Broughton JP, Bloom ME, Bresnahan WA, Best SM. 2011. TRIM79α, an interferon-stimulated gene product, restricts tick-borne encephalitis virus replication by degrading the viral RNA polymerase. Cell Host Microbe 10:185–196. doi: 10.1016/j.chom.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Overby AK, Popov VL, Niedrig M, Weber F. 2010. Tick-borne encephalitis virus delays interferon induction and hides its double-stranded RNA in intracellular membrane vesicles. J Virol 84:8470–8483. doi: 10.1128/JVI.00176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Overby AK, Weber F. 2011. Hiding from intracellular pattern recognition receptors, a passive strategy of flavivirus immune evasion. Virulence 2:238–240. doi: 10.4161/viru.2.3.16162. [DOI] [PubMed] [Google Scholar]
- 38.Decroly E, Ferron F, Lescar J, Canard B. 2011. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol 10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, Khromykh AA. 2008. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 40.Schuessler A, Funk A, Lazear HM, Cooper DA, Torres S, Daffis S, Jha BK, Kumagai Y, Takeuchi O, Hertzog P, Silverman R, Akira S, Barton DJ, Diamond MS, Khromykh AA. 2012. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J Virol 86:5708–5718. doi: 10.1128/JVI.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ, Harris E, Garcia-Blanco MA, Ooi EE. 2015. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diamond MS. 2009. Mechanisms of evasion of the type I interferon antiviral response by flaviviruses. J Interferon Cytokine Res 29:521–530. doi: 10.1089/jir.2009.0069. [DOI] [PubMed] [Google Scholar]
- 43.Green AM, Beatty PR, Hadjilaou A, Harris E. 2014. Innate immunity to dengue virus infection and subversion of antiviral responses. J Mol Biol 426:1148–1160. doi: 10.1016/j.jmb.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morrison J, Aguirre S, Fernandez-Sesma A. 2012. Innate immunity evasion by dengue virus. Viruses 4:397–413. doi: 10.3390/v4030397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye J, Zhu B, Fu ZF, Chen H, Cao S. 2013. Immune evasion strategies of flaviviruses. Vaccine 31:461–471. doi: 10.1016/j.vaccine.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 46.Best SM, Morris KL, Shannon JG, Robertson SJ, Mitzel DN, Park GS, Boer E, Wolfinbarger JB, Bloom ME. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J Virol 79:12828–12839. doi: 10.1128/JVI.79.20.12828-12839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diamond MS, Roberts TG, Edgil D, Lu B, Ernst J, Harris E. 2000. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol 74:4957–4966. doi: 10.1128/JVI.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ho LJ, Hung LF, Weng CY, Wu WL, Chou P, Lin YL, Chang DM, Tai TY, Lai JH. 2005. Dengue virus type 2 antagonizes IFN-alpha but not IFN-gamma antiviral effect via down-regulating Tyk2-STAT signaling in the human dendritic cell. J Immunol 174:8163–8172. doi: 10.4049/jimmunol.174.12.8163. [DOI] [PubMed] [Google Scholar]
- 49.Jones M, Davidson A, Hibbert L, Gruenwald P, Schlaak J, Ball S, Foster GR, Jacobs M. 2005. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J Virol 79:5414–5420. doi: 10.1128/JVI.79.9.5414-5420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo JT, Hayashi J, Seeger C. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J Virol 79:1343–1350. doi: 10.1128/JVI.79.3.1343-1350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vasilakis N, Cardosa J, Hanley KA, Holmes EC, Weaver SC. 2011. Fever from the forest: prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat Rev Microbiol 9:532–541. doi: 10.1038/nrmicro2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ashour J, Laurent-Rolle M, Shi P, García-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol 83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. 2009. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J Infect Dis 200:1261–1270. doi: 10.1086/605847. [DOI] [PubMed] [Google Scholar]
- 55.Khromykh AA, Sedlak PL, Westaway EG. 1999. trans-Complementation analysis of the flavivirus Kunjin ns5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J Virol 73:9247–9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gibbs DJ, Bacardit J, Bachmair A, Holdsworth MJ. 2014. The eukaryotic N-end rule pathway: conserved mechanisms and diverse functions. Trends Cell Biol 24:603–611. doi: 10.1016/j.tcb.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 57.Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, Simon V, Mulder LC, Fernandez-Sesma A, García-Sastre A. 2013. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog 9:e1003265. doi: 10.1371/journal.ppat.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, Bernal-Rubio D, Williams KL, Harris E, Fernandez-Sesma A, Schindler C, García-Sastre A. 2010. Mouse STAT2 restricts early dengue virus replication. Cell Host Microbe 8:410–421. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar A, Buhler S, Selisko B, Davidson A, Mulder K, Canard B, Miller S, Bartenschlager R. 2013. Nuclear localization of dengue virus nonstructural protein 5 does not strictly correlate with efficient viral RNA replication and inhibition of type I interferon signaling. J Virol 87:4545–4557. doi: 10.1128/JVI.03083-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinto AK, Brien JD, Lam CY, Johnson S, Chiang C, Hiscott J, Sarathy VV, Barrett AD, Shresta S, Diamond MS. 2015. Defining new therapeutics using a more immunocompetent mouse model of antibody-enhanced dengue virus infection. mBio 6:e01316-15. doi: 10.1128/mBio.01316-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coyne CB, Lazear HM. 2016. Zika virus—reigniting the TORCH. Nat Rev Microbiol 14:707–715. doi: 10.1038/nrmicro.2016.125. [DOI] [PubMed] [Google Scholar]
- 62.Lazear HM, Diamond MS. 2016. Zika virus: new clinical syndromes and its emergence in the Western Hemisphere. J Virol 90:4864–4875. doi: 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sanchez-Seco MP, Evans MJ, Best SM, Garcia-Sastre A. 2016. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, Power C, Hobman TC. 2016. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, Saucedo-Cuevas L, Regla-Nava JA, Chai G, Sheets N, Tang W, Terskikh AV, Shresta S, Gleeson JG. 2016. Zika virus infects neural progenitors in the adult mouse brain and alters proliferation. Cell Stem Cell 19:593–598. doi: 10.1016/j.stem.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grard G, Moureau G, Charrel RN, Holmes EC, Gould EA, de Lamballerie X. 2010. Genomics and evolution of Aedes-borne flaviviruses. J Gen Virol 91:87–94. doi: 10.1099/vir.0.014506-0. [DOI] [PubMed] [Google Scholar]
- 67.Laurent-Rolle M, Morrison J, Rajsbaum R, Macleod JM, Pisanelli G, Pham A, Ayllon J, Miorin L, Martinez-Romero C, ten Oever BR, Garcia-Sastre A. 2014. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe 16:314–327. doi: 10.1016/j.chom.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, Garcia-Sastre A. 2013. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fletcher AJ, Towers GJ. 2013. Inhibition of retroviral replication by members of the TRIM protein family. Curr Top Microbiol Immunol 371:29–66. doi: 10.1007/978-3-642-37765-5_2. [DOI] [PubMed] [Google Scholar]
- 70.Liu B, Li NL, Wang J, Shi PY, Wang T, Miller MA, Li K. 2014. Overlapping and distinct molecular determinants dictating the antiviral activities of TRIM56 against flaviviruses and coronavirus. J Virol 88:13821–13835. doi: 10.1128/JVI.02505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gritsun TS, Nuttall PA, Gould EA. 2003. Tick-borne flaviviruses. Adv Virus Res 61:317–371. doi: 10.1016/S0065-3527(03)61008-0. [DOI] [PubMed] [Google Scholar]
- 72.Park GS, Morris KL, Hallett RG, Bloom ME, Best SM. 2007. Identification of residues critical for the interferon antagonist function of Langat virus NS5 reveals a role for the RNA-dependent RNA polymerase domain. J Virol 81:6936–6946. doi: 10.1128/JVI.02830-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J. 2007. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol 81:4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valmas C, Grosch MN, Schumann M, Olejnik J, Martinez O, Best SM, Krahling V, Basler CF, Muhlberger E. 2010. Marburg virus evades interferon responses by a mechanism distinct from ebola virus. PLoS Pathog 6:e1000721. doi: 10.1371/journal.ppat.1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fuchs SY. 2013. Hope and fear for interferon: the receptor-centric outlook on the future of interferon therapy. J Interferon Cytokine Res 33:211–225. doi: 10.1089/jir.2012.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ambrose RL, Mackenzie JM. 2011. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J Virol 85:2723–2732. doi: 10.1128/JVI.02050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ambrose RL, Mackenzie JM. 2013. ATF6 signaling is required for efficient West Nile virus replication by promoting cell survival and inhibition of innate immune responses. J Virol 87:2206–2214. doi: 10.1128/JVI.02097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lupi A, Tenni R, Rossi A, Cetta G, Forlino A. 2008. Human prolidase and prolidase deficiency: an overview on the characterization of the enzyme involved in proline recycling and on the effects of its mutations. Amino Acids 35:739–752. doi: 10.1007/s00726-008-0055-4. [DOI] [PubMed] [Google Scholar]
- 79.Keller BC, Fredericksen BL, Samuel MA, Mock RE, Mason PW, Diamond MS, Gale M. 2006. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J Virol 80:9424–9434. doi: 10.1128/JVI.00768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liang J, Liao C, Liao J, Lee Y, Lin Y. 2009. A Japanese encephalitis virus vaccine candidate strain is attenuated by decreasing its interferon antagonistic ability. Vaccine 27:2746–2754. doi: 10.1016/j.vaccine.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 81.Lin RJ, Liao CL, Lin E, Lin YL. 2004. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J Virol 78:9285–9294. doi: 10.1128/JVI.78.17.9285-9294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Evans JD, Crown RA, Sohn JA, Seeger C. 2011. West Nile virus infection induces depletion of IFNAR1 protein levels. Viral Immunol 24:253–263. doi: 10.1089/vim.2010.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, Barrett AD, Mason PW, Bloom ME, García-Sastre A, Khromykh AA, Best SM. 2010. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol 84:3503–3515. doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Werme K, Wigerius M, Johansson M. 2008. Tick-borne encephalitis virus NS5 associates with membrane protein scribble and impairs interferon-stimulated JAK-STAT signalling. Cell Microbiol 10:696–712. doi: 10.1111/j.1462-5822.2007.01076.x. [DOI] [PubMed] [Google Scholar]
- 85.Lin R, Chang B, Yu H, Liao C, Lin Y. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J Vriol 80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang TC, Li SW, Lai CC, Lu KZ, Chiu MT, Hsieh TH, Wan L, Lin CW. 2013. Proteomic analysis for type I interferon antagonism of Japanese encephalitis virus NS5 protein. Proteomics 13:3442–3456. doi: 10.1002/pmic.201300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barillas-Mury C, Han YS, Seeley D, Kafatos FC. 1999. Anopheles gambiae Ag-STAT, a new insect member of the STAT family, is activated in response to bacterial infection. EMBO J 18:959–967. doi: 10.1093/emboj/18.4.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zeidler MP, Bach EA, Perrimon N. 2000. The roles of the Drosophila JAK/STAT pathway. Oncogene 19:2598–2606. doi: 10.1038/sj.onc.1203482. [DOI] [PubMed] [Google Scholar]
- 89.Lin CC, Chou CM, Hsu YL, Lien JC, Wang YM, Chen ST, Tsai SC, Hsiao PW, Huang CJ. 2004. Characterization of two mosquito STATs, AaSTAT and CtSTAT. Differential regulation of tyrosine phosphorylation and DNA binding activity by lipopolysaccharide treatment and by Japanese encephalitis virus infection. J Biol Chem 279:3308–3317. [DOI] [PubMed] [Google Scholar]
- 90.Paradkar PN, Duchemin JB, Rodriguez-Andres J, Trinidad L, Walker PJ. 2015. Cullin4 is pro-viral during West Nile virus infection of Culex mosquitoes. PLoS Pathog 11:e1005143. doi: 10.1371/journal.ppat.1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu WJ, Wang XJ, Mokhonov VV, Shi PY, Randall R, Khromykh AA. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J Virol 79:1934–1942. doi: 10.1128/JVI.79.3.1934-1942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith AA, Pal U. 2014. Immunity-related genes in Ixodes scapularis—perspectives from genome information. Front Cell Infect Microbiol 4:116. doi: 10.3389/fcimb.2014.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smith AA, Navasa N, Yang X, Wilder CN, Buyuktanir O, Marques A, Anguita J, Pal U. 2016. Cross-species interferon signaling boosts microbicidal activity within the tick vector. Cell Host Microbe 20:91–98. doi: 10.1016/j.chom.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Luckhart S, Crampton AL, Zamora R, Lieber MJ, Dos Santos PC, Peterson TM, Emmith N, Lim J, Wink DA, Vodovotz Y. 2003. Mammalian transforming growth factor beta1 activated after ingestion by Anopheles stephensi modulates mosquito immunity. Infect Immun 71:3000–3009. doi: 10.1128/IAI.71.6.3000-3009.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. 2003. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A 100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Evans JD, Seeger C. 2007. Differential effects of mutations in NS4B on West Nile virus replication and inhibition of interferon signaling. J Virol 81:11809–11816. doi: 10.1128/JVI.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mackenzie JM, Khromykh AA, Parton RG. 2007. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2:229–239. doi: 10.1016/j.chom.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 98.Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. 2006. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J Virol 80:4623–4632. doi: 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sehgal PB, Guo GG, Shah M, Kumar V, Patel K. 2002. Cytokine signaling: STATS in plasma membrane rafts. J Biol Chem 277:12067–12074. doi: 10.1074/jbc.M200018200. [DOI] [PubMed] [Google Scholar]
- 100.Garcia Cordero J, Leon Juarez M, Gonzalez YMJA, Cedillo Barron L, Gutierrez Castaneda B. 2014. Caveolin-1 in lipid rafts interacts with dengue virus NS3 during polyprotein processing and replication in HMEC-1 cells. PLoS One 9:e90704. doi: 10.1371/journal.pone.0090704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bhattacharyya S, Zagorska A, Lew ED, Shrestha B, Rothlin CV, Naughton J, Diamond MS, Lemke G, Young JA. 2013. Enveloped viruses disable innate immune responses in dendritic cells by direct activation of TAM receptors. Cell Host Microbe 14:136–147. doi: 10.1016/j.chom.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Porritt RA, Hertzog PJ. 2015. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol 36:150–160. doi: 10.1016/j.it.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 103.Yoshii K, Sunden Y, Yokozawa K, Igarashi M, Kariwa H, Holbrook MR, Takashima I. 2014. A critical determinant of neurological disease associated with highly pathogenic tick-borne flavivirus in mice. J Virol 88:5406–5420. doi: 10.1128/JVI.00421-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Klema VJ, Ye M, Hindupur A, Teramoto T, Gottipati K, Padmanabhan R, Choi KH. 2016. Dengue virus nonstructural protein 5 (NS5) assembles into a dimer with a unique methyltransferase and polymerase interface. PLoS Pathog 12:e1005451. doi: 10.1371/journal.ppat.1005451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bhattacharya D, Mayuri Best SM, Perera R, Kuhn RJ, Striker R. 2009. Protein kinase G phosphorylates mosquito-borne flavivirus NS5. J Virol 83:9195–9205. doi: 10.1128/JVI.00271-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Taylor R, Best SM. 2011. Assessing ubiquitination of viral proteins: lessons from flavivirus NS5. Methods 55:166–171. doi: 10.1016/j.ymeth.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lu G, Gong P. 2013. Crystal structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog 9:e1003549. doi: 10.1371/journal.ppat.1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao Y, Soh TS, Chan KW, Fung SS, Swaminathan K, Lim SP, Shi PY, Huber T, Lescar J, Luo D, Vasudevan SG. 2015. Flexibility of NS5 methyltransferase-polymerase linker region is essential for dengue virus replication. J Virol 89:10717–10721. doi: 10.1128/JVI.01239-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao Y, Soh TS, Zheng J, Chan KW, Phoo WW, Lee CC, Tay MY, Swaminathan K, Cornvik TC, Lim SP, Shi PY, Lescar J, Vasudevan SG, Luo D. 2015. A crystal structure of the Dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog 11:e1004682. doi: 10.1371/journal.ppat.1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, Mulder LC, Barber GN, Fernandez-Sesma A. 2012. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog 8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]