Abstract
Studies have demonstrated that ~60%–80% of emerging infectious diseases (EIDs) in humans originated from wild life. Bats are natural reservoirs of a large variety of viruses, including many important zoonotic viruses that cause severe diseases in humans and domestic animals. However, the understanding of the viral population and the ecological diversity residing in bat populations is unclear, which complicates the determination of the origins of certain EIDs. Here, using bats as a typical wildlife reservoir model, virome analysis was conducted based on pharyngeal and anal swab samples of 4440 bat individuals of 40 major bat species throughout China. The purpose of this study was to survey the ecological and biological diversities of viruses residing in these bat species, to investigate the presence of potential bat-borne zoonotic viruses and to evaluate the impacts of these viruses on public health. The data obtained in this study revealed an overview of the viral community present in these bat samples. Many novel bat viruses were reported for the first time and some bat viruses closely related to known human or animal pathogens were identified. This genetic evidence provides new clues in the search for the origin or evolution pattern of certain viruses, such as coronaviruses and noroviruses. These data offer meaningful ecological information for predicting and tracing wildlife-originated EIDs.
Introduction
Emerging infectious diseases (EIDs) pose a great threat to global public health. Approximately 60%–80% of human EIDs originate from wildlife, as shown by the typical examples of hemorrhagic fever, avian influenza and henipavirus-related lethal neurologic and respiratory diseases that originated from rodents, wild birds and bats (Jones et al., 2008; Wolfe et al., 2007; Lloyd-Smith et al., 2009; Marsh and Wang, 2012; Smith and Wang, 2013). The primary issues associated with the prevention and control of EIDs are how to quickly identify the pathogen, determine where it originated and control the chain of transmission. These issues, along with the limited knowledge of the viral population and ecological diversity of wildlife, complicate the study of EIDs. Therefore, meaningful information afforded by the understanding of the viral community present in wildlife, as well as the prevalence, genetic diversity and geographical distribution of these viruses, is very important for the prevention and control of wildlife-borne EIDs (Anthony et al., 2013; Daszak et al., 2000).
Bats are mammals with a wide geographical distribution, extensive species diversity, unique behaviors (characteristic flight patterns, long life spans and gregarious roosting and mobility behaviors) and intimate interactions with humans and livestock (Calisher et al., 2006). They are natural reservoirs of a large variety of viruses, including many important zoonotic viruses that cause severe diseases in humans and domestic animals, including henipaviruses, Marburg virus and Ebola virus (Luis et al., 2013; Quan et al., 2013; O'Shea et al., 2014). The severe acute respiratory syndrome (SARS) outbreak in 2003, which resulted in nearly 8000 cases and 800 deaths worldwide, was suspected to have originated in bats and then spread to humans (Li et al., 2005). All of these examples reveal that zoonotic viruses carried by bats can be transmitted directly or via certain intermediate hosts from bats to humans or domestic animals with high virulence.
As most bat-borne pathogens are transmitted by four routes (airborne, droplet, oral–fecal and contact transmission) from the respiratory tracts, oral cavities or enteric canals of bats to other species, it is particularly important to determine the viral communities present at these locations. In this study, bat individuals of 40 representative species across China were sampled by pharyngeal and anal swabbing, to assess the variety of viruses residing in bat species. Metagenomic analysis was then conducted to screen the viromes of these samples. Here we outline the viral spectrum within these bat samples and the basic ecological and genetic characteristics of these novel bat viruses. The identification of novel bat viruses in this study also provides genetic evidence for cross-species transmission between bats or between bats and other mammals. These data offer new clues for tracing the sources of important viral pathogens such as SARS coronavirus (SARS-CoV) and Middle East respiratory syndrome CoV (MERS-CoV).
Materials and methods
Bat samples
Pharyngeal and anal swab samples were collected separately and then immersed in virus sampling tubes (Yocon, Beijing, China) containing maintenance medium and were temporarily stored at −20 °C. The samples were then transported to the laboratory and stored at −80 °C.
Viral nucleic acid library construction, next-generation sequencing and taxonomic assignments
A tube with either a pharyngeal or anal swab sample in maintenance medium was vigorously vortexed to re-suspend the sample in solution. Samples from the same bat species and from the same site were then pooled. The pooled samples were processed with a viral particle-protected nucleic acid purification method and then amplified by sequence-independent reverse transcriptase-PCR as described previously (Wu et al., 2012; Wu et al., 2014). Briefly, the samples were centrifuged at 10 000 g for 10 min at 4 °C. Supernatant from each sample was filtered through a 0.45-μm polyvinylidene difluoride filter (Millipore, Darmstadt, Germany), to remove eukaryotic and bacterial-sized particles. The filtered samples were then centrifuged at 100 000 g for 3 h at 4 °C. The pellets were re-suspended in Hank's balanced salt solution. To remove naked DNA and RNA, the re-suspended pellet was digested in a cocktail of DNase and RNase enzymes, including 14 U of Turbo DNase (Ambion, Austin, TX, USA), 20 U of benzonase (Novagen, Darmstadt, Germany), and 20 U of RNase One (Promega, Madison, WI, USA) at 37 °C for 2 h in 1 × DNase buffer (Ambion). The viral nucleic acids were then isolated using a QIAmp MinElute Virus Spin Kit (Qiagen, Valencia, CA, USA). Viral first-strand cDNA was synthesized using the primer K-8N (5′-GACCATCTAGCGACCTCCAC–NNNNNNNN-3′) and a Superscript III system (Invitrogen, Carlsbad, CA, USA). To convert first-strand cDNA into double-stranded DNA, the cDNA was incubated at 37 °C for 1 h in the presence of 5 U of Klenow fragment (NEB, Ipswich, MA, USA) in 1 × NEB buffer 2 (final volume of 25 μl). Sequence-independent PCR amplification was conducted using 1 μM primer K (5′-GACCATCTAGCGACCTCCAC-3′) and 0.5 U Phusion DNA polymerase (NEB). The PCR products were analyzed by agarose gel electrophoresis. A DNA smear of larger than 500 bp was excised and extracted with a MinElute Gel Extraction Kit (Qiagen).
The amplified viral nucleic acid libraries were then analyzed using an Illumina GA II sequencer (Illumina, Sandiego, CA, USA) for a single read of 81 bp in length. The raw sequence reads were filtered using previously described criteria (Wu et al., 2012; Yang et al., 2011), to obtain valid sequences. Each read was evaluated for viral origin by conducting alignments with the NCBI non-redundant nucleotide (nt) database (NT) and protein database (NR) using BLASTn and BLASTx (with parameters -e 1e-5 –F T). Reads with no hits in NT or NR were further assembled using the Velvet software (v1.2.10, Pittsburgh Supercomputing Center, Pittsburgh, PA, USA) and the contigs were again aligned with NT and NR to identify any viruses that were present. The taxonomies of the aligned reads with the best BLAST scores (E-value <10−5) from all lanes were parsed and exported with MEGAN 4–MetaGenome Analyzer (Wu et al., 2012). Besides the alignment-first analytical strategy, we also tested assembly-first strategy to analyze the sequence data. The reads were firstly assembled by metagenomics assemblers (for example, MetaVelvet and IDBA-UD) (Namiki et al., 2012; Peng et al., 2012) and the output contigs were then aligned to NT and NR. As the percentage of viral reads was small in the whole data sets and most of the assembled contigs were thus of bacterial and host origins, the assembly-first strategy generally had lower sensitivity than the alignment-first strategy in this study.
Identification of the prevalence and positive rate of each virus
According to the molecular clues provided by metagenomic analyses, the sequence reads classified into the same virus family or genus by MEGAN 4 were extracted and then assembled with SeqMan program (Lasergene, DNAstar, Madison, WI, USA). A draft genome with several or a large number of single-nt polymorphisms of each virus was obtained. Based on the partial genomic sequences of the viruses obtained by the assembly, we designed specific nested primers for PCR or reverse trancriptase-PCR to screen for each virus in individual samples from each bat species (the primer sequences for each virus are available in Supplementary Table S2).
Genome sequencing of each virus in positive samples by PCR
The accurate locations of the reads and the relative distances between reads of the same virus were determined based on the alignment results exported using MEGAN 4. Representative positive samples for each virus were selected for genome sequencing as viral quasi-species. The reads with accurate genomic locations were then used for reads-based PCR to identify partial genomes. Based on the partial genomic sequences obtained by specific nested PCR, the remaining genomic sequences were determined using inverse PCR, genome walking, and 5′- and 3′ rapid amplification of cDNA ends.
Genome sequences and phylogenetic and evolutionary analyses
All genome sequences have been submitted to GenBank. The accession numbers for all sequences are listed in Supplementary Table S2. The GA II sequence data have been deposited into the NCBI sequence reads archive under the accession number SRA051252. Routine sequence alignments were performed using MegAlign program (Lasergene) or T-coffee with manual curation. MEGA5.0 (Phoenix, AZ, USA) was used to align the nt and amino acid (aa) sequences using MUSCLE package with the default parameters. The best substitution model was then evaluated using Model Selection package. Finally, a maximum-likelihood method with an appropriate model was used to conduct phylogenetic analyses with 1000 bootstrap replicates.
Results
Sampling, metagenomic analysis and experimental verification of bat viruses
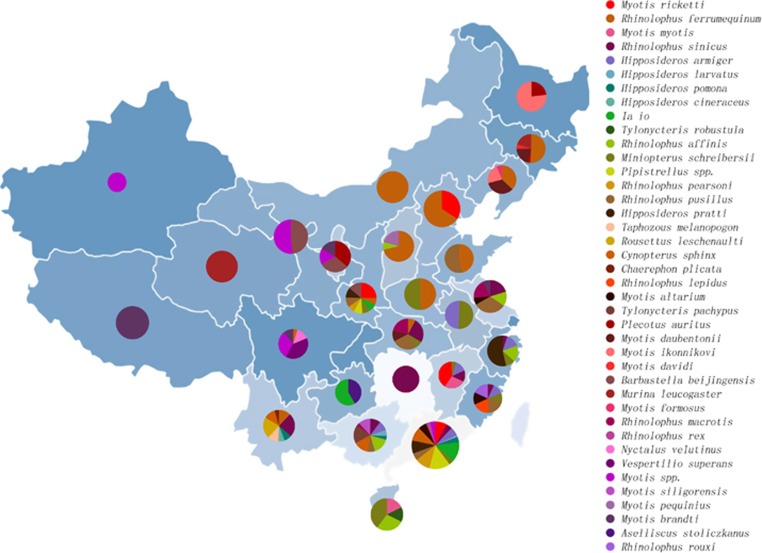
A total of 4440 bats were sampled by obtaining both pharyngeal and anal swabs from October 2010 through October 2013 in 29 provinces throughout China (Figure 1, Supplementary Table S1 and Supplementary Figure S1). These represent the 40 main bat species, the 17 most common genera and 6 families of both frugivorous and insectivorous bats that reside in urban, rural and wild areas throughout China.
Figure 1.
Numbers of bat samples from various provinces. The numbers of the 4440 bat samples belonging to the 40 bat species identified are indicated by a pie chart for each province. The numbers of samples from the 40 bat species and the provinces and dates of collection are detailed in Supplementary Table S1.
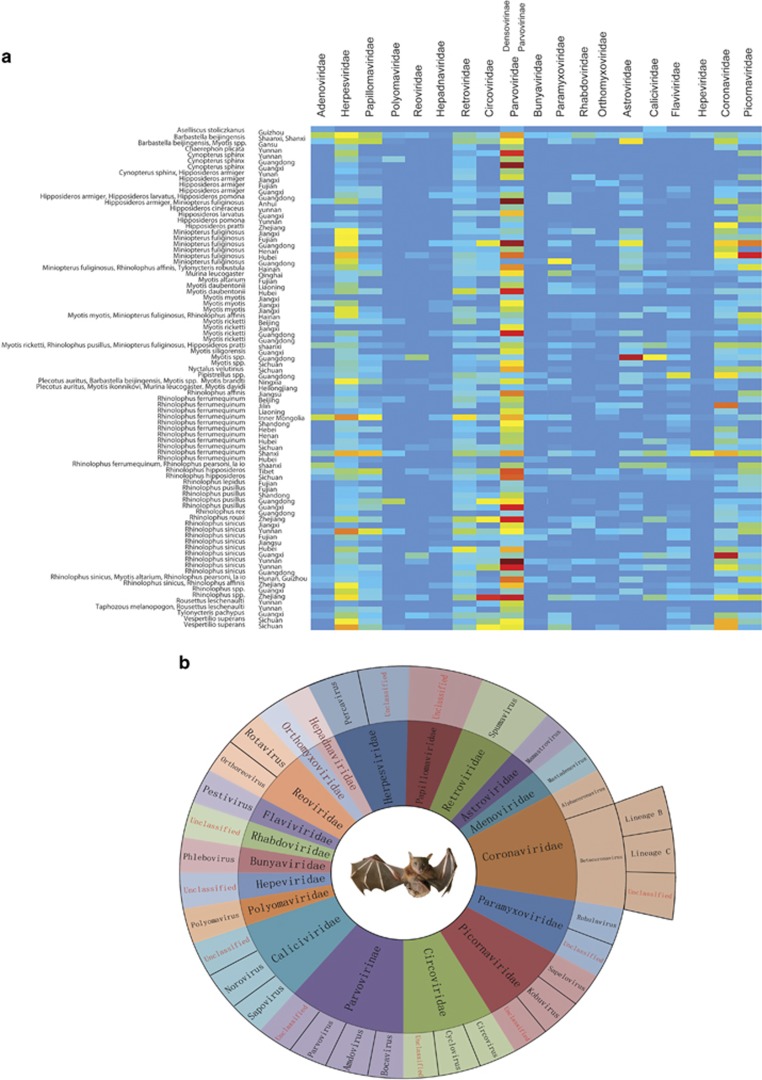
All pharyngeal and anal swab samples were classified and combined into 84 pools and then subjected to virome analysis (Wu et al., 2012). Finally, a total of 179 GB of nt data (2 241 761 959 valid reads, 81 bp in length) was obtained. In total, 20 632 698 reads were best matched with viral proteins available in the NCBI NR database (~0.8% of the total sequence reads). The number of virus-associated reads in each lane varied between 8283 and 2 919 423. In total, 79 families of phages, insect viruses, fungal viruses, mammalian viruses and plant viruses were parsed. After excluding bat habit-related non-mammalian viruses, including insect viruses (mainly of the families Baculoviridae, Iflaviridae, Dicistroviridae and Tetraviridae; subfamily Densovirinae), fungal viruses (mainly of the families Chrysoviridae, Hypoviridae, Partitiviridae and Totiviridae), phages (the order Caudovirales and families Inoviridae and Microviridae) and plant viruses (matched reads of the 60 non-mammalian virus families are provided in Supplementary Table S23), overview of the reads of 19 families of mammalian viruses in each pooled sample is shown in Figure 2a and Supplementary Table S24. In addition, an overview of the classification from family to genus of the identified bat viruses is shown in Figure 2b. The reads related to the family Parvoviridae comprised the largest proportion of viruses as shown in Figure 2, most of which were classified into the subfamily Densovirinae. The dominating abundance of insect densoviruses was associated with the insectivorous habits of bats.
Figure 2.
Overall view of the reads from 19 families of mammalian viruses in each pooled sample. (a) Heatmap based on the normalized sequence reads of 19 families of mammalian viruses in each pooled sample. The bat species are listed in the left text column. Location information is provided in the right text column. The names of the mammalian viral families are presented in the top text row. The boxes colored from blue to red represent the metagenomic sequencing reads observed (reads varied between 8283 and 2 919 423, respectively). (b) Overview of classifications from family to genus of the viruses of the 19 families identified in the bats in this study. Different families are labeled in different colors. The viruses that could not be assigned to any known genus are labeled in red font.
The existence and prevalence of virus strains in 17 of the 19 families were confirmed. In total, 692 positive results were confirmed by PCR screening (positive rates are shown in Supplementary Table S2) and 144 viruses of representative positive samples were selected for genomic sequencing as quasi-species of these viruses (Supplementary Table S2; the 94 full-length sequenced viruses are labeled in red). The most widely distributed families of mammalian viruses were Herpesviridae, Papillomaviridae, Retroviridae, Adenoviridae and Astroviridae. The diverse reads related to these families occupied ~61% of the total viral sequence reads (Supplementary Table S24). The assessment of diverse reads of bat herpesviruses, bat papillomaviruses, bat retroviruses, bat astroviruses and bat adenoviruses (Supplementary Figure S2), and full-length sequenced representative strains of these viruses (Wu et al., 2012) revealed that most of these viruses were distinct from each other within each family (30%–65% nt identities). In addition to the above families, many reads related to the families Circoviridae, Paramyxoviridae, Coronaviridae, Caliciviridae, Polyomaviridae, Rhabdoviridae, Hepeviridae, Bunyaviridae, Reoviridae, Flaviviridae and Picornaviridae, and the subfamily Parvovirinae, exhibited low nt or aa sequence identities with known viruses. Although sequence reads related to the families Orthomyxoviridae and Hepadnaviridae were occasionally present in some of the samples, we failed to amplify any sequences of viruses in these families, which may have been a result of very low viral loads.
Although samples from 307 frugivorous bats of two species (Rousettus leschenaultia and Cynopterus sphinx) were collected for virome analysis, only a few reads related to herpesvirus, papillomavirus and retrovirus were found. This finding revealed that the virome of frugivorous bats is far less abundant than that of insectivorous bats in this study.
Bat main single-stranded RNA viruses (Coronaviridae, Paramyxoviridae and Picornaviridae)
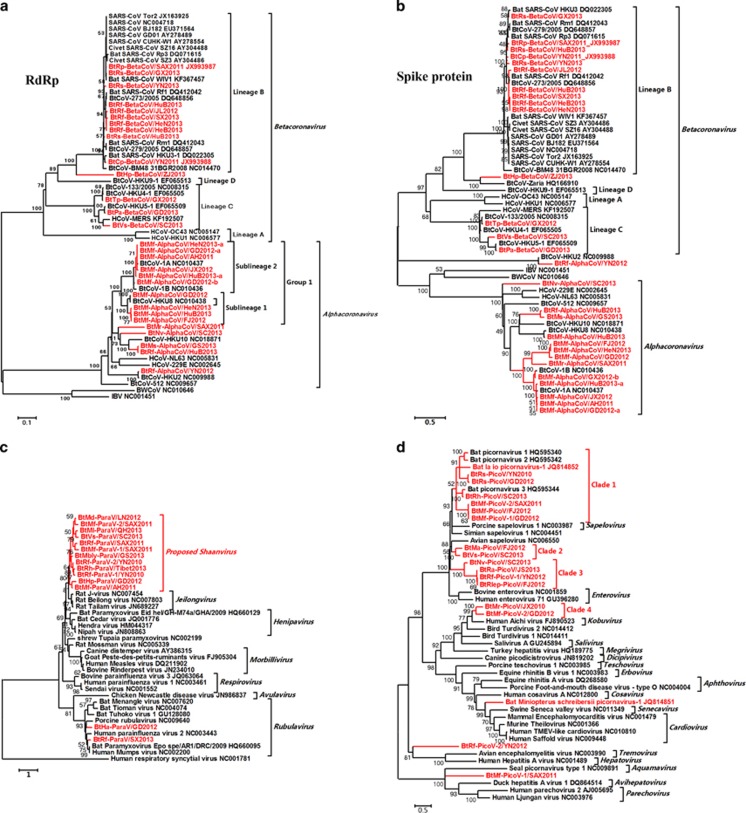
The family Coronaviridae contains two subfamilies, Coronavirinae and Torovirinae. The subfamily Coronavirinae includes four approved genera, Alphacoronavirus, Betacoronavirus, Deltacoronavirus and Gammacoronavirus. It is a group of large enveloped viruses with a positive single-stranded RNA genome (~26 to 32 kb in length). Six α- and β-CoVs (HKU1, OC43, NL63, 229E, SARS-CoV and MERS-CoV) are human pathogens that cause mild-to-severe disease (Corman et al., 2014b; Li et al., 2005; Cui et al., 2007; O'Shea et al., 2014). Here, 30 novel bat CoVs (BtCoVs) were identified separately in 12 bat species of 9 genera from 15 provinces (Supplementary Table S2 and Supplementary Figure S1). CoVs from seven bat species are reported for the first time. Phylogenetic trees were constructed based on the deduced RNA-dependent RNA polymerase (NSP12) (Figure 3a Spike (S) and Figure 3b proteins). The sequence identities of these BtCoVs are shown in Supplementary Table S4.
Figure 3.
Phylogenetic analysis of bat main single-stranded RNA (ssRNA) viruses. (a) Phylogenetic tree based on the complete RNA-dependent RNA polymerase (RdRp, NSP12) proteins of CoVs. (b) Phylogenetic tree based on the complete Spike (S) proteins of CoVs. (c) Phylogenetic tree based on the L proteins of ParaVs. (d) Phylogenetic tree based on the complete RNA-dependent RNA polymerase proteins of PicoVs.
Eleven BtCoVs were assigned to a group with lineage-B beta-CoVs and three BtCoVs were assigned to a group with lineage-C beta-CoVs. The BtVs-BetaCoV/SC2013 identified in Vespertilio superans bats had a closer genetic relationship with MERS-CoV than with other BtCoVs (Supplementary Figure S3), as well as with NeoCoV identified in African Neoromicia capensis bats (Corman et al., 2014a). One BtCoV, BtHp-BetaCoV/ZJ2013, represented a separate clade related to lineage B. The overall nt identity of this CoV genome with lineage-B CoVs was only 58% and the RNA-dependent RNA polymerase and S proteins shared only 78% and 41% aa identities with those of lineage-B CoVs. This CoV contained an unusual putative S-related ORF2 between ORF1ab and the S gene (Supplementary Figure S4 and Supplementary Table S3) (Quan et al., 2010). A signal peptide (1–13 aa) and a transmembrane region (413–435 aa) were identified in the S-related ORF2, suggesting that this protein might be a surface protein.
The remaining 15 BtCoVs were all assigned to the genus Alphacoronavirus and formed many novel separate clades. For BtMr-AlphaCoV/SAX2011 and BtNv-AlphaCoV/SC2013, although the phylogenetic tree constructed based on RNA-dependent RNA polymerase indicated that these two viruses were clustered with HKU10, the phylogenetic tree based on the S protein indicated that these two viruses represented two separate clades far from other CoVs, suggesting that recombination may occur in their genomes. Although the ORF1ab, E, M and N genes of BtRf-AlphaCoV/HuB2013 and BtMs-AlphaCoV/GS2013 shared very high sequence identities (higher than 98%), the S genes of these two viruses shared only 85% nt identity. A similar phenomenon was observed between BtRf-AlphaCoV/YN2012 and HKU2.
The results of sample-by-sample screening of bat samples from the same gathering place revealed that BtCoV strains of the same species identified in the same cave had significantly diverse features. Some key gene segments, such as the S and ORF8 genes, presented great diversity with low sequence identities (Supplementary Figure S5), indicating that these two genes are hypervariable regions within the BtCoV genome.
Similar to recently reported SARS-like CoVs (SL-CoVs) (WIV1, Rs3367 and LYRa11) (Ge et al., 2013; He et al., 2014), two BtCoVs, BtRs-betaCoV/YN2013 and BtRs-betaCoV/GX2013, identified in Rhinolophus sinicus from the Yunnan and Guangxi provinces shared the highest similarities with SARS-CoVs in the backbone (including ORF1ab, E, M and N genes), ORF6, ORF7a, ORF7b and ORF8 genes compared with other bat lineage-B β-CoVs (Supplementary Tables S11–S18 and Supplementary Figures S6A and B and S7). Furthermore, the S genes of lineage-B β-CoVs from R. sinicus had much higher genetic diversity and were scattered among the phylogenetic clades of SARS-CoVs and lineage-B CoVs from other bat species (Supplementary Figure S6C).
Co-infections of CoV strains of sublineages 1 and 2 of group 1 in Miniopterus fuliginosus were detected in two anal specimens collected in Guangdong and Henan. The CoVs of sublineage 1 with highly similar backbone sequences presented differing degrees of variation in the S region. Recombination was confirmed by similarity plots, bootscan analysis and detection of putative breakpoints around the S regions in the genomes of lineage 1 CoVs in M. fuliginosus and M. pusillus. (Supplementary Table S21 and Supplementary Figure S8).
The family Paramyxoviridae is a group of large enveloped viruses with negative-sense single-stranded RNA genomes (~15 to 19 kb in length) that are responsible for a variety of mild-to-severe human and animal diseases (Mayo, 2002; Smith and Wang, 2013). Bat paramyxoviruses (BtParaVs) were separately identified in 9 bat species from 10 provinces of China (Supplementary Table S2 and Supplementary Figure S1). The full-length sequence of 3 viruses (BtMf-ParaV/AH2011, BtMl-ParaV/QH2013 and BtHa-ParaV/GD2012) were almost completely determined (Supplementary Figure S9 and Supplementary Table S6) and 11 other viruses were partially or completely sequenced in the L gene. Twelve of the 14 novel BtParaVs identified in the different bat species could be clustered together and formed a separate phylogenetic clade. Alternatively, these BtParaVs could be classified into a potential separate genus, Shaanvirus, with BtMf-ParaV/AH2011 and BtMl-ParaV/QH2013 as prototypes. The genomic organizations of these two viruses were similar to those of three previously reported members of the genus Jeilongvirus. However, the genomes of BtMs-ParaV/Anhui2011 and BtMl-ParaV/QH2013 were shorter in length than those of the three rodent viruses and the sequence identities were low (Supplementary Table S5). The remaining two novel BtParaVs could be clustered together with Rubulaviruses as new species (Figure 3c. The central domain of the N protein contained three conserved motifs common to all paramyxoviruses, and the six conserved domains within the L proteins of the order Mononegavirales (Lau et al., 2010) could be found in all three full-length sequenced viruses.
Picornaviruses (PicoVs) of the family Picornaviridae are small, non-enveloped, positive single-stranded RNA viruses with a genome of 7–9 kb in size. The members of the family Picornaviridae cause mucocutaneous, encephalic, cardiac, hepatic, neurological and respiratory diseases in a wide variety of vertebrate hosts (Tracy et al., 2006; Wang et al., 2015). Nineteen bat PicoVs (BtPicoVs) were identified in 11 bat species from 9 provinces (Supplementary Table S2 and Supplementary Figure S1). Phylogenetic analysis of the RNA-dependent RNA polymerase genes was conducted (Figure 3d. Seven BtPicoVs could be clustered with three previously reported BtPicoVs (clade 1) and could then be divided into separate sub-clades according to their host genera. Clade 3 contained two sub-clades formed by four BtPicoVs of two bat genera. Clade 2 was formed by two BtPicoVs identified in different bat genera and clustered in a sister relationship with the genus Sapelovirus. Clade 4 contained two BtPicoVs identified in the same bat genus that were closely related to the genus Kobuvirus. The previously reported Miniopterus schreibersii PicoV-1 was closely related to the genera Cardiovirus and Senecavirus. Two BtPicoVs, BtRf-PicoV-2/YN2012 and BtMf-PicoV-1/SAX2011, showed lower aa identities with other known PicoVs. Different BtPicoVs identified from the same bat genus, such as Rhinolophus or Miniopterus, in different locations showed very close genetic relationships. The aa identities of the predicted RNA-dependent RNA polymerase proteins of these novel BtPicoVs were low compared with known PicoVs (Supplementary Table S7). The predicted P1, P2 and P3 regions and cleavage sites of these viruses showed typical features of PicoVs (Supplementary Table S22).
Bat main DNA viruses (Circoviridae and Parvoviridae)
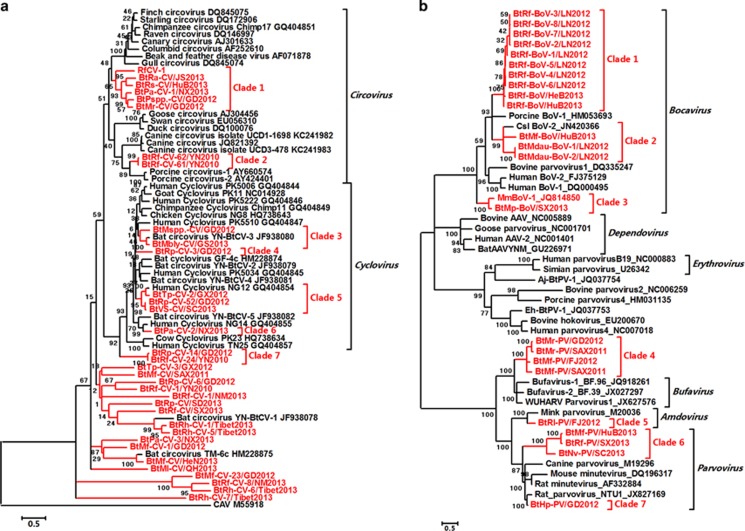
The family Circoviridae is a group of viruses with small, non-enveloped, circular single-strand DNA genomes of 1.7–3 kb in length (Fauquet and Fargette, 2005). Porcine circovirus (CV)-2 is the main swine pathogen (Chae, 2005). Thirty-four novel bat CVs (BtCVs) were identified in 13 bat species from 16 provinces (Supplementary Table S2 and Supplementary Figure S1). The genome sizes of these viruses varied from 1643 to 2979 nts. Seven new clades of BtCVs in the genera Circovirus and Cyclovirus were found. Clade 2 was formed by two viruses clustered in a sister relationship with two pathogenic viruses, porcine and dog CVs (Chae, 2005; Li et al., 2013), at the same root with a short branch length. Three BtCVs were closely related to human cycloviruses. Seven BtCVs were assigned to the proposed genus Cyclovirus and formed four separate clades, three of which were closely related to human cycloviruses. A separate clade, clade 7, was constructed from two BtCVs and was closely related to the genus Cyclovirus. In addition to these viruses, 17 novel BtCVs branched out of the root of CVs and cycloviruses revealed the presence of new genera different from the known genera (Figure 4a).
Figure 4.
Phylogenetic analysis of bat main DNA viruses. (a) Phylogenetic tree based on the complete replicase (Rep) proteins of CVs. (b) Phylogenetic tree based on the VP1 proteins of members of the subfamily Parvovirinae. The bat viruses identified in this study are labeled in red.
Viruses of the family Parvoviridae comprise a group of small, non-enveloped viruses with linear positive-sense single-stranded DNA (~5 kb genomes) that infect vertebrate animals and cause mild-to-severe diseases (Brown, 2010). Bat parvoviruses (BtPVs) and bat bocaviruses were identified in nine bat species from eight provinces (Supplementary Table S2 and Supplementary Figure S1). In addition to viruses in the genera Bocavirus, Parvovirus and Amdovirus, four BtPVs were most similar to the recently reported human Bufavirus members and the bufavirus-related WUHARV parvovirus, with similar NS1 and VP1 proteins (higher than 60% aa identities) (Phan et al., 2012; Yahiro et al., 2014). BtHp-PV/GD2012, in the genus Parvovirus, was very closely related to a recently identified rat PV, with 89% aa identity (Figure 4b). The aa identities among the predicted VP1 proteins of these BtPVs and bat bocaviruses, and other known members of the subfamily Parvovirinae, are shown in Supplementary Table S8. Bat adeno-associated viruses have been described previously (Li et al., 2010b); however, considering the nonpathogenic nature of adeno-associated viruses, we did not perform further verification of these viruses.
Other rare bat viruses
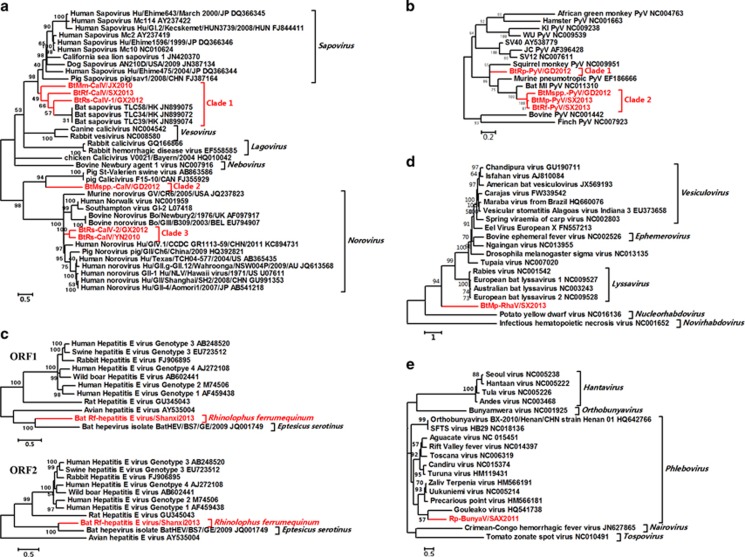
Six bat caliciviruses (BtCalVs), four bat polyomaviruses, one bat hepatitis E virus, one bat rhabdovirus, one bat bunyavirus, one bat orthoreovirus and one bat rotavirus were identified. Phylogenetic analysis (Figure 5, Supplementary Table S2 and Supplementary Figure S1) revealed that these bat viruses represented separate evolutionary lineages within each family (Supplementary Results and Supplementary Tables S9). In addition to the two first identified human and swine norovirus-related BtCalVs (BtRs-CalV-2/GX2012 and BtRs-CalV/YN2010) and one group A rotavirus-related bat rotavirus identified in R. sinicus, the other viruses were evolutionarily distant from known human or animal pathogens, including human hepatitis E viruses, rabies viruses and pathogenic bunyaviruses.
Figure 5.
Phylogenetic analysis of sporadically identified viruses from bats. (a) Phylogenetic tree based on the polyproteins of CalVs. (b) Phylogenetic tree based on the large T proteins of polyomaviruses (PyVs). (c) Phylogenetic tree based on the complete ORF1 and ORF2 sequences of hepatitis E viruses (HEVs). (d) Phylogenetic tree based on the partial L (RNA-dependent RNA polymerase (RdRp)) proteins of rhabdoviruses (RhaVs). (e) Phylogenetic tree based on the partial L proteins of bunyaviruses (BunyaVs). The bat viruses identified in this study are labeled in red.
Discussion
In previous studies, zoonotic viruses in more than 15 virus families have been identified in bats around the world (Chen et al., 2014; O'Shea et al., 2014). Two bat virome analyses conducted by Li et al. (2010a) and Donaldson et al. (2010) have revealed the presence of CoVs, herpesviruses, PicoVs, CVs, adenoviruses, adeno-associated viruses and astroviruses in some bat species of North America. One bat virome analysis conducted by Ge et al. (2012) mainly described insect viruses in some bat species of China. One bat virome analysis conducted by Ng et al. (2013) has revealed the presence of a novel rhabdovirus in big brown bats, and one bat virome analysis conducted by He et al. (2013) has described the spectrum of viruses harbored by several bat species in Myanmar. Different from these previous reports, this study was the first to characterize the pharyngeal and anal virome of representative bat samples in China. We did not perform additional verification of non-mammalian viruses because of the association of the abundance of these viruses (not initially harbored in bats) with their life habits.
This report suggests that bats harbor a large spectrum of mammalian viruses. Except for a few viruses, such as BtCalVs, BtCVs and BtPVs, which are closely related to known viruses, most of the bat viruses identified here that were widely distributed among the bat species were novel and showed no strict geographical restrictions. Large numbers of viruses were identified from bats of Rhinolophus spp. (viruses from 16 families were identified), Miniopterus spp. (viruses from 10 families were identified) and Myotis spp. (viruses from 13 families were identified). These findings reveal that these three bat genera may act as major reservoirs for diverse mammalian viruses in China. Notably, all bats collected in this study were considered to be apparently healthy and showed no overt signs of disease, further confirming that bats can tolerate diverse viruses through their unique metabolic and immune systems (O'Shea et al., 2014).
This study extends the host range for members of each viral family and reveals unique ecological and evolutionary characteristics of bat-borne viruses. The diverse BtCoVs were grouped into several novel evolutionary clades that significantly differed from those of all known α- and β-CoVs, providing additional evidence to support investigations of the evolution of bat-originated CoVs. With regard to BtParaVs, a previous study has revealed that bats host major mammalian ParaVs in the genera Rubulavirus, Morbillivirus, Henipavirus and the subfamily Pneumovirinae (Drexler et al., 2012). However, in this study, except for 2 viruses assigned to the genus Rubulavirus, the remaining 12 viruses formed a new genus distant from the known genera and the 14 identified BtParaVs showed no direct relationship with the known human or animal pathogens of the family Paramyxoviridae. These results suggest an entirely different distribution of BtParaVs in China than previously reported. Although the classifications of bat herpesviruses, bat papillomaviruses, bat retroviruses, bat astroviruses, bat adenoviruses, BtPicoVs, BtCVs, BtPVs and bat bocaviruses were extended according to the current virus taxonomy file released by the International Committee on Taxonomy of Viruses, the large number of novel viruses grouped into the various evolutionary clades identified in this study further expand the taxa to include many new viral genera and species. Many new clades formed by BtCVs distinct from all known members of the genera Circovirus and Cyclovirus could be candidates for many new genera. Viruses related to henipaviruses, Ebola virus, rabies virus and pathogenic bunyaviruses were not detected in the Chinese bat species examined in this study.
Diverse herpesviruses and papillomaviruses identified countrywide support the hypothesis that these DNA viruses from different bat species are located in different phylogenetic positions within each family without strict host or geographic specificity (Garcia-Perez et al., 2014); however, many other DNA or RNA viruses, such as BtCoVs, BtParaVs, BtPicoVs and BtPVs or bat bocaviruses, identified from the same or different bat species from different locations shared high sequence identities and close genetic relationships. These phenomena indicate that certain bat-originated DNA and RNA viruses have the potential for intra- or cross-species transmission concomitant with the migration, co-roosting and intra- or inter-species contact of their bat hosts. The identification of some viruses, such as certain rat PV-related BtPV, norovirus-related BtCalVs, human or swine CV-related BtCVs and bat rotavirus in the rotavirus A group, also provides a new understanding of the evolution of these viruses in different mammalian hosts and possible transmission events that occur between bats and other hosts. Furthermore, BtCoVs had more distinctive features than the other bat viruses. Highly diverse S genes or ORF8s were present in particular CoVs carried by bats of the same species from different locations or even the same gathering place. Considering the diversity of CoVs, co-infections may create opportunities for recombination and the emergence of new CoVs that are able to adapt to new hosts. These findings may explain why tracing the potential CoV-related EIDs in insectivorous bats is often complicated by the presence of diverse key genomic segments and no virus with an identical genome sequence related to the pathogens causing human or animal EIDs has been identified in insectivorous bats. Instead, the origin of the Ebola virus and henipaviruses could be relatively easily confirmed by the identification of identical viruses in frugivorous bats (Calisher et al., 2006; Leroy et al., 2005; Smith and Wang, 2013).
Only lineage-B β-CoVs of six bat species (R. ferrumequinum, R. sinicus, R. pusillus, R. macrotis, R. affinis and Chaerephon plicata) from China (SL-CoVs) are closely related to SARS-CoVs, as similar ORF1ab, E, M and N genes, and the presence of a unique structural ORFs (including ORF6, 7a, 7b and 8) have been identified (Supplementary Figure S10) (Holmes and Enjuanes, 2003; Li et al., 2005; Woo et al., 2009; Quan et al., 2010; Woo et al., 2012; Yang et al., 2013). Recently, a functionally similar S gene has been identified in SL-CoVs of R. sinicus and R. affinis (WIV1, Rs3367 and LYRa11) with less sequence identity to the S gene of SARS-CoV, but which is capable of using the human ACE2 as a receptor for virus entry (Ge et al., 2013; He et al., 2014). However, knowledge gaps exist between bat SL-CoVs and SARS-CoVs with regard to the S gene and the unique structural ORF that prevent the determination of which bat virus species is the direct ancestor of SARS-CoV. In this study, two BtCoVs (BtRs-betaCoV/YN2013 and BtRs-betaCoV/GX2013) in Chinese horseshoe bats (R. sinicus) provided bat-originated unique structural ORFs that were nearly identical to the original SARS-CoV isolated during the earliest phase of the SARS pandemic (after transmission to humans, this region of SARS-CoV experiences ongoing adaptive evolution in humans, with gradual deletion (2004)), providing some information to fill the knowledge gap with regard to the origin of human SARS-CoVs. In addition, the higher similarities of the backbones of these two BtCoVs to SARS-CoVs, and the highly diverse S genes present in only R. sinicus imply that frequent recombination events occur among SL-CoVs of R. sinicus and other hosts, suggesting that the transmission of SL-CoVs from R. sinicus to other mammals is a result of the viruses obtaining novel S genes. These data indicate that human SARS-CoVs most likely originated through zoonotic transfer, either directly or indirectly, from Chinese horseshoe bats via a complicated adaptation process and a series of rare recombination events.
Although the transmission of MERS-CoV has been confirmed by the detection of identical MERS-CoV sequences in dromedary camels and humans (Azhar et al., 2014; Chu et al., 2014; Meyer et al., 2014), the zoonotic transmission of this virus from dromedaries to humans is still considered to be rare (Hemida et al., 2015) and the wildlife source of MERS-CoVs remains unknown. The data obtained here and a recently reported lineage-C BtCoV, NeoCoV, identified by another group (Corman et al., 2014a; Yang et al., 2014a) provide new clues about the sources and pathways of human- and camel-derived MERS-CoVs in bats of the family Vespertilionidae. Bat species of this family may have important roles in MERS-CoV evolution (Yang et al., 2014b). Furthermore, the diverse S genes of these MERS-related CoVs may provide an opportunity for their recombination to ultimately generate new CoVs.
In conclusion, the understanding of the viral community characteristics, genetics and ecological distribution of bat viruses could enable the rapid identification of novel viruses with variant genomes and could thus facilitate the tracing of EIDs in bats. Furthermore, this strategy could be extended to other wildlife or livestock worldwide, ultimately increasing knowledge of the viral population and ecological community, thus minimizing the impact of potential wildlife-originated EIDs on public health by providing meaningful basic data.
Acknowledgments
This study was supported by the Program for Changjiang Scholars and Innovative Research Team in University of China (IRT13007), the National S&T Major Project, ‘China Mega-Project for Infectious Disease' (grant number 2011ZX10004-001 and 2014ZX10004001) from China and the PUMC Youth Fund and the Fundamental Research Funds for the Central Universities (grant number 3332015095).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A et al. (2013). A strategy to estimate unknown viral diversity in mammals. MBio 4: e00598–e00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, Hashem AM et al. (2014). Evidence for Camel-to-Human Transmission of MERS Coronavirus. N Engl J Med 370: 2499–2505. [DOI] [PubMed] [Google Scholar]
- Brown KE. (2010). The expanding range of parvoviruses which infect humans. Rev Med Virol 20: 231–244. [DOI] [PubMed] [Google Scholar]
- Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. (2006) Bats: important reservoir hosts of emerging viruses Clin Microbiol Rev 19: 531–545. [DOI] [PMC free article] [PubMed]
- Chae C. (2005). A review of porcine circovirus 2-associated syndromes and diseases. Vet J 169: 326–336. [DOI] [PubMed] [Google Scholar]
- Chen L, Liu B, Yang J, Jin Q. (2014). DBatVir: the database of bat-associated viruses. Database (Oxford) 2014: bau021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese SARS Molecular Epidemiology Consortium.. (2004). Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303: 1666–1669. [DOI] [PubMed] [Google Scholar]
- Chu DK, Poon LL, Gomaa MM, Shehata MM, Perera RA, Abu Zeid D et al. (2014). MERS Coronaviruses in Dromedary Camels, Egypt. Emerg Infect Dis 20: 1049–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Ithete NL, Richards LR, Schoeman MC, Preiser W, Drosten C et al. (2014. a). Rooting the phylogenetic tree of MERS-Coronavirus by characterization of a conspecific virus from an African Bat. J Virol 88: 11297–11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Kallies R, Philipps H, Gopner G, Muller MA, Eckerle I et al. (2014. b). Characterization of a novel betacoronavirus related to middle East respiratory syndrome coronavirus in European hedgehogs. J Virol 88: 717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Han N, Streicker D, Li G, Tang X, Shi Z et al. (2007). Evolutionary relationships between bat coronaviruses and their hosts. Emerg Infect Dis 13: 1526–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD. (2000). Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science 287: 443–449. [DOI] [PubMed] [Google Scholar]
- Donaldson EF, Haskew AN, Gates JE, Huynh J, Moore CJ, Frieman MB. (2010). Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J Virol 84: 13004–13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Muller MA, Maganga GD, Vallo P, Binger T et al. (2012). Bats host major mammalian paramyxoviruses. Nat Commun 3: 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquet CM, Fargette D. (2005). International Committee on Taxonomy of Viruses and the 3,142 unassigned species. Virol J 2: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez R, Ibanez C, Godinez JM, Arechiga N, Garin I, Perez-Suarez G et al. (2014). Novel papillomaviruses in free-ranging Iberian bats: no virus-host co-evolution, no strict host specificity, and hints for recombination. Genome Biol Evol 6: 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Li Y, Yang X, Zhang H, Zhou P, Zhang Y et al. (2012). Metagenomic analysis of viruses from the bat fecal samples reveals many novel viruses in insectivorous bats in China. J Virol 88: 4620–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH et al. (2013). Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503: 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Li Z, Yang F, Zheng J, Feng Y, Guo H et al. (2013). Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel Mammalian viruses. PLoS One 8: e61950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Zhang Y, Xu L, Yang W, Yang F, Feng Y et al. (2014). Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. J Virol 88: 7070–7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida MG, Al-Naeem A, Perera RA, Chin AW, Poon LL, Peiris M. (2015). Lack of Middle East respiratory syndrome coronavirus transmission from infected camels. Emerg Infect Dis 21: 699–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KV, Enjuanes L. (2003). Virology. The SARS coronavirus: a postgenomic era. Science 300: 1377–1378. [DOI] [PubMed] [Google Scholar]
- Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL et al. (2008). Global trends in emerging infectious diseases. Nature 451: 990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Wong BH, Wong AY, Tsoi HW, Wang M et al. (2010). Identification and complete genome analysis of three novel paramyxoviruses, Tuhoko virus 1, 2 and 3, in fruit bats from China. Virology 404: 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P et al. (2005). Fruit bats as reservoirs of Ebola virus. Nature 438: 575–576. [DOI] [PubMed] [Google Scholar]
- Li L, Victoria JG, Wang C, Jones M, Fellers GM, Kunz TH et al. (2010. a). Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol 84: 6955–6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, McGraw S, Zhu K, Leutenegger CM, Marks SL, Kubiski S et al. (2013). Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerg Infect Dis 19: 534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH et al. (2005). Bats are natural reservoirs of SARS-like coronaviruses. Science 310: 676–679. [DOI] [PubMed] [Google Scholar]
- Li Y, Ge X, Hon CC, Zhang H, Zhou P, Zhang Y et al. (2010. b). Prevalence and genetic diversity of adeno-associated viruses in bats from China. J Gen Virol 91: 2601–2609. [DOI] [PubMed] [Google Scholar]
- Lloyd-Smith JO, George D, Pepin KM, Pitzer VE, Pulliam JR, Dobson AP et al. (2009). Epidemic dynamics at the human-animal interface. Science 326: 1362–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis AD, Hayman DT, O'Shea TJ, Cryan PM, Gilbert AT, Pulliam JR et al. (2013). A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc Biol Sci 280: 20122753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh GA, Wang LF. (2012). Hendra and Nipah viruses: why are they so deadly? Curr Opin Virol 2: 242–247. [DOI] [PubMed] [Google Scholar]
- Mayo MA. (2002). A summary of taxonomic changes recently approved by ICTV. Arch Virol 147: 1655–1663. [DOI] [PubMed] [Google Scholar]
- Meyer B, Muller MA, Corman VM, Reusken CB, Ritz D, Godeke GJ et al. (2014). Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis 20: 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namiki T, Hachiya T, Tanaka H, Sakakibara Y. (2012). MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res 40: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TF, Driscoll C, Carlos MP, Prioleau A, Schmieder R, Dwivedi B et al. (2013). Distinct lineage of vesiculovirus from big brown bats, United States. Emerg Infect Dis 19: 1978–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea TJ, Cryan PM, Cunningham AA, Fooks AR, Hayman DT, Luis AD et al. (2014). Bat flight and zoonotic viruses. Emerg Infect Dis 20: 741–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Leung HC, Yiu SM, Chin FY. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28: 1420–1428. [DOI] [PubMed] [Google Scholar]
- Phan TG, Vo NP, Bonkoungou IJ, Kapoor A, Barro N, O'Ryan M et al. (2012). Acute diarrhea in West African children: diverse enteric viruses and a novel parvovirus genus. J Virol 86: 11024–11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan PL, Firth C, Street C, Henriquez JA, Petrosov A, Tashmukhamedova A et al. (2010). Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria. MBio 1: e00208–e00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan PL, Firth C, Conte JM, Williams SH, Zambrana-Torrelio CM, Anthony SJ et al. (2013). Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad Sci USA 110: 8194–8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I, Wang LF. (2013). Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr Opin Virol 3: 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy S, Chapman NM, Drescher KM, Kono K, Tapprich W. (2006). Evolution of virulence in picornaviruses. Curr Top Microbiol Immunol 299: 193–209. [DOI] [PubMed] [Google Scholar]
- Wang X, Ren J, Gao Q, Hu Z, Sun Y, Li X et al. (2015). Hepatitis A virus and the origins of picornaviruses. Nature 517: 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe ND, Dunavan CP, Diamond J. (2007). Origins of major human infectious diseases. Nature 447: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Huang Y, Yuen KY. (2009). Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med (Maywood) 234: 1117–1127. [DOI] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH et al. (2012). Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 86: 3995–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Ren X, Yang L, Hu Y, Yang J, He G et al. (2012). Virome analysis for identification of novel mammalian viruses in bat species from chinese provinces. J Virol 86: 10999–11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang L, Yang F, Ren X, Jiang J, Dong J et al. (2014). Novel henipa-like virus, mojiang paramyxovirus, in rats, China, 2012. Emerg Infect Dis 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahiro T, Wangchuk S, Tshering K, Bandhari P, Zangmo S, Dorji T et al. (2014). Novel human bufavirus genotype 3 in children with severe diarrhea, bhutan. Emerg Infect Dis 20: 1037–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yang F, Ren L, Xiong Z, Wu Z, Dong J et al. (2011). Unbiased parallel detection of viral pathogens in clinical samples by use of a metagenomic approach. J Clin Microbiol 49: 3463–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wu Z, Ren X, Yang F, He G, Zhang J et al. (2013). Novel SARS-like betacoronaviruses in bats, China, 2011. Emerg Infect Dis 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wu Z, Ren X, Yang F, Zhang J, He G et al. (2014. a). MERS-related betacoronavirus in Vespertilio superans bats, China. Emerg Infect Dis 20: 1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Du L, Liu C, Wang L, Ma C, Tang J et al. (2014. b). Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci USA 111: 12516–12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.