Abstract
Circadian clocks align behavioral and biochemical processes with the day/night cycle. Nearly all vertebrate cells possess self-sustained clocks that couple endogenous rhythms with changes in cellular environment. Genetic disruption of clock genes in mice perturbs metabolic functions of specific tissues at distinct phases of the sleep/wake cycle. Circadian desynchrony, a characteristic of shift work and sleep disruption in humans, also leads to metabolic pathologies. Here we review advances in understanding the interrelationship among circadian disruption, sleep deprivation, obesity and diabetes, and implications for rational therapeutics for these conditions.
2-Introduction
The rising and setting of the sun has captivated naturalists interested in the cause of daily rhythmic phenomenon, prompting de Marian to demonstrate the existence of an internal clock in the 18th Century by placing the Mimosa plant in a dark box and showing that its leaves continued to open and close every 24 hrs. Over two centuries later, genetic studies in fruit flies paved the way for discovery that the circadian clock is encoded by a set of transcriptional activators and repressors that comprise an autoregulatory transcription-translation feedback loop. The circadian clock in mammals is expressed within pacemaker neurons of the suprachiasmatic nucleus (SCN) that in turn maintain proper phase alignment of peripheral tissue clocks present in nearly all cells. Thus the brain SCN clock provides ‘standard time’ for all peripheral tissue clocks. In experimental models, clock disruption leads to disorders in glucose metabolism, confirming a role for these genes as key regulators of metabolism, and supporting the hypothesis originally proposed by McKnight and colleagues that circadian cycles are intimately interconnected with metabolic cycles (1). Accumulating evidence has revealed that multiple clock genes participate in metabolic homeostasis, suggesting that these proteins have evolved overlapping (or convergent) functions both as intrinsic “hands” of the clock and as regulators of metabolism. While still at an early stage, emerging studies in humans suggest parallels in the role of circadian genes and metabolic homeostasis. At the epidemiological level, it has been suggested that increased activity during what was ‘rest’ time in the pre-modern world, together with sleep disruption, have been associated with increased prevalence of obesity, diabetes and cardiovascular disease, in addition to certain cancers and inflammatory disorders. This review highlights advances in understanding the molecular coupling between metabolic and clock networks, and its relevance to gene-environment and brain-behavioral systems important in energy balance and metabolic disease.
3-Core transcriptional components of the clock and posttranslational regulation
Features of the circadian clock in all organisms include its persistence under constant conditions, a periodicity that is temperature compensated, and its entrainment to light from the sun (Fig 1). In mammals, cell autonomous circadian clocks are generated by a transcriptional autoregulatory feedback loop composed of the transcriptional activators, CLOCK and BMAL1, and their target genes, Period and Cryptochrome, which rhythmically accumulate and form a repressor complex that interacts with CLOCK-BMAL1, to inhibit their own transcription (2). This autoregulatory loop is post-transcriptionally regulated by casein kinases (CK1ε and CK1δ), which target the PER proteins for degradation via the SCF/β-TrCP ubiquitin ligase complex, and by AMP kinase, which targets the CRY proteins for degradation via the SCF/FBXL3 ubiquitin ligase complex by the 26S proteosome (reviewed in (2), Fig 2).
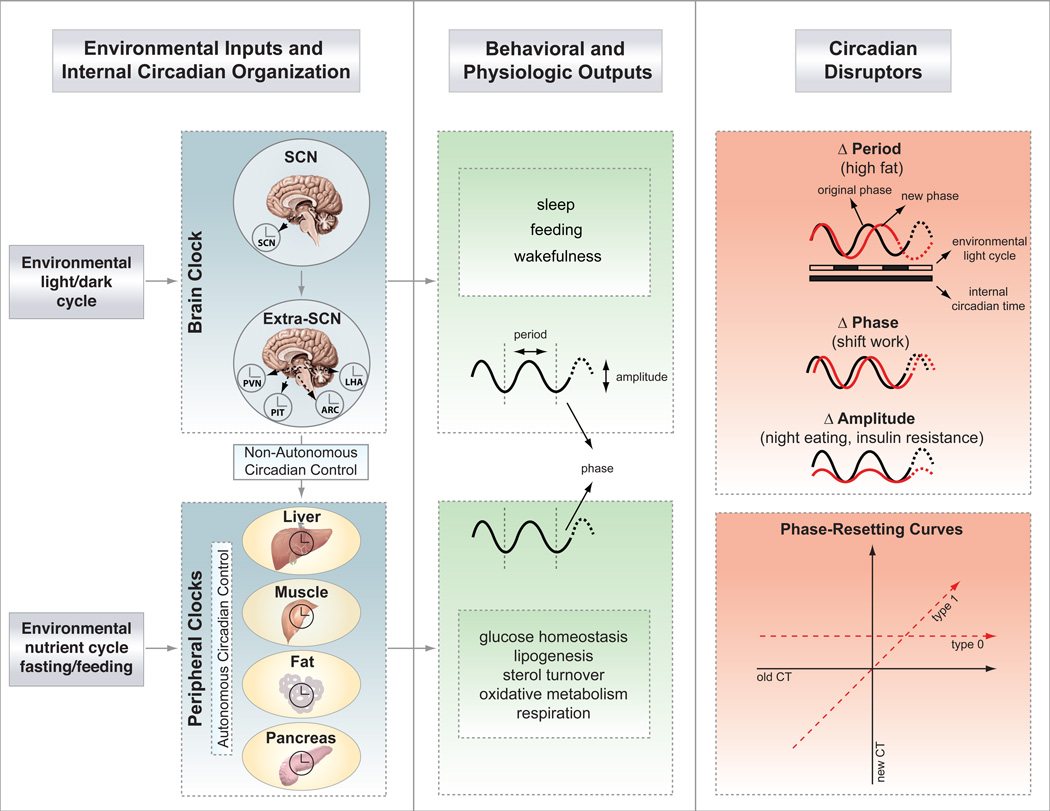
Figure 1. Central and peripheral clocks coordinate external cues with behavior and metabolic outputs.
Light entrains the master pacemaker in the suprachiasmatic nucleus (SCN), which in turn synchronizes extra-SCN and peripheral clocks. Brain clock outputs include behavioral rhythms (i.e., sleep, feeding), while peripheral clock outputs include metabolic rhythms (i.e., glucose and lipid homeostasis, etc). The hierarchical organization of the mammalian clock is highlighted, with “non-autonomous” regulation of peripheral tissue clocks denoting the regulation of peripheral tissue oscillators through direct neural and humoral signals, and “autonomous” regulation indicating the intrinsic regulation of local cellular oscillators independently of the brain clock. Highlighted to the right are the three possible ways to disrupt the clock, by changing either period, phase, or amplitude, each of which can trigger disorders of metabolism. Phase resetting can be broadly classified into two groups based upon phase response following delivery of the agent at sequential time points across the 24 hr cycle. Type 1 resetting indicates that the slope of the plot relating the new to old circadian phase is 1 (interventions that cause different phase shifts at different circadian times). Type 0 resetting indicates that the slope of the new to old circadian phase is 0 (i.e., interventions that cause the same phase at all circadian times). Abbreviations: PVN, paraventricular nucleus; PIT, pituitary; ARC, arcuate nucleus; LHA, lateral hypothalamic area.
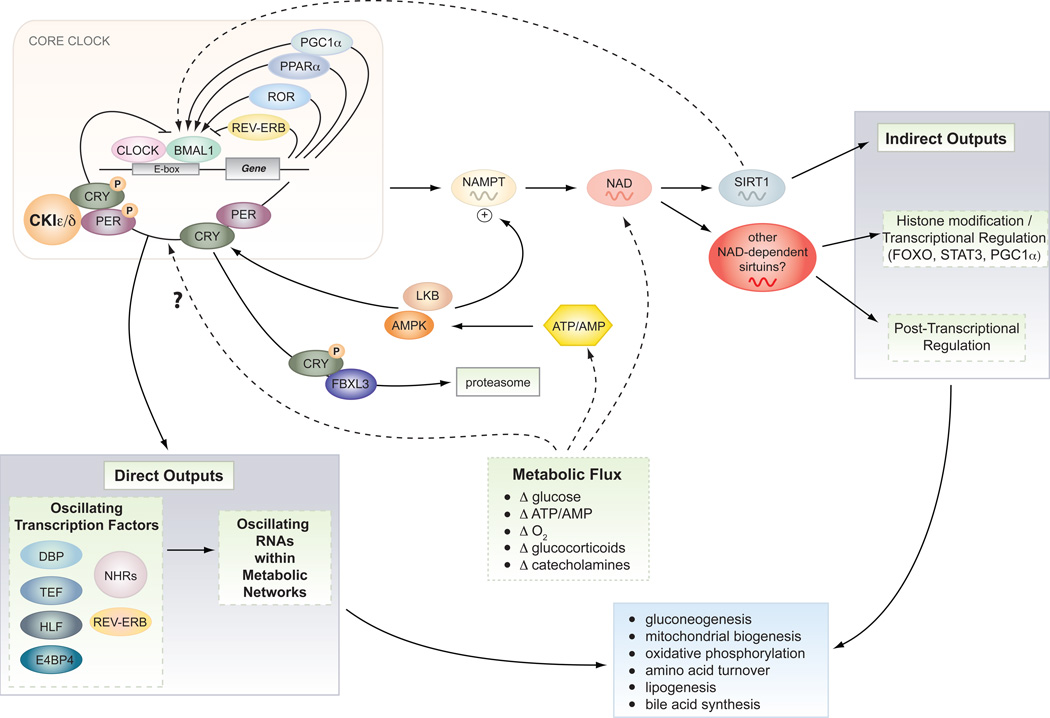
Figure 2. Direct and indirect outputs of the core clock mechanism.
The core clock consists of a series of transcription/translation feedback loops that synchronize diverse metabolic processes through both direct and indirect outputs including gluconeogenesis and oxidative metabolism (see text for details). The clock also receives reciprocal input from nutrient signaling pathways (including SIRT1 and AMPK), which function as rheostats to couple circadian cycles to metabolic flux especially in peripheral tissues.
The prevailing model of the circadian clock involves the transcription-translation feedback loop, but less is known concerning non-transcriptional mechanisms that may generate circadian oscillations. In cyanobacteria, cycles of protein phosphorylation are sufficient to generate biological rhythms in the absence of transcription (3). In the mammalian SCN, changes in cyclic AMP levels alter period length, an additional example of post-translational signaling as a mechanism controlling of circadian cycles (4); and, recent work has shown that the SCN neuronal coupling network itself has intrinsic oscillatory function that can emerge in the absence of cell autonomous oscillators (5). RNAi screening of mammalian cells also indicates coupling of the peripheral clock to PI-3 kinase signaling (6). Further research is warranted to elucidate the impact of post-translational signaling pathways on the core clock and its physiological outputs.
Fibroblast cell lines display ~24 hr oscillation of core clock genes, demonstrating that the clock is not only expressed in neurons, but also in peripheral tissues (7). Intrinsic oscillation of clocks in liver cells can be entrained by food, whereas oscillation of the brain clock is resilient and entrained primarily by light (8). A recurring theme in understanding the coupling between circadian and metabolic systems is the recognition that the two systems are reciprocally regulated; food entrains the liver clock, whereas light acts through the brain clock to control feeding time.
4-Crosstalk between the clock and metabolic transcription networks
a-Nuclear hormone receptors and the phase alignment of metabolic gene expression cycles
Direct evidence for metabolic input into the core clock includes the finding that the orphan nuclear hormone receptor (NHR) reverse-erb alpha (REV-ERBα) (a repressor), and the opposing retinoic acid orphan receptors (RORα and β) (activators), constitute a short feedback loop controlling Bmal1 transcription (9) (10) (Fig 2). PPARα and the coactivator PGC1α also modulate Bmal1 transcription through this feedback loop (11), indicating that REV-ERBα is a nodal point for metabolic input into the clock (12). NHR profiling has revealed rhythmic clustering of these factors in metabolic tissues across the day/night cycle, suggesting extensive coupling between circadian and nuclear receptor signaling networks (13). These findings raise the possibility that disruption of NHR cycles may perturb the clock, and conversely, that delay, advance or reduced amplitude of circadian oscillations may impair NHR function. Knockin mice of the NHR co-repressor NCor display increased energy expenditure, and a shift in the oscillation in the abundance of RNAs encoding oxidative, glycolytic and respiratory genes, indicating that disruption of the phase of expression of NHRs contributes to metabolic dysregulation. Mistiming of gene expression rhythms as a cause of metabolic dysregulation has also been suggested by studies in Rev-erbα mutant animals, in which a phase shift in oscillating rhythms of metabolic gene transcription, rather than changes in total abundance of RNA, correspond with altered energy balance (14). Misalignment between gene transcription cycles within metabolic tissues and the behavioral cycle (of fasting and feeding) may be sufficient to alter energy homeostasis. For instance, high fat feeding provided at the incorrect circadian time leads to greater weight gain in mice than isocaloric feeding at the normal circadian time (15).
b-Direct versus indirect role of clock transcription factors in metabolic gene regulation
It is likely that disruption within the core clock may be transmitted to metabolic outputs through alterations in NHRs or directly by actions of clock activators or repressors. For example, PER2 directly occupies promoters of certain metabolic genes (16). Alternatively, the clock activator loop drives D-element-binding-protein expression, providing indirect regulation of gluconeogenic genes (17). This raises the question as to whether the effects of clock gene disruption relate to direct alterations in “timing” per se, or to indirect effects arising due to independent activity of the clock factors on metabolic networks. The dichotomy between circadian versus non-circadian actions of clock proteins may not be fully valid, since the rhythmic abundance in the expression level of these proteins in turn may produce rhythmic changes in metabolism. For example, CRY, a rhythmically expressed clock repressor, modulates gluconeogenesis through interference with glucagon signaling and inhibition of the phosphorylation of cyclic response element AMP binding protein (CREB) (18). Conceptually, the question of timing versus expression as a cause of metabolic disorders following disruption of clock genes is akin to the difference between a musical performer playing the wrong notes or playing the right notes at the wrong time. One experimental approach to tease apart the role of circadian timing per se in physiology would be to test whether physiological defects could be corrected by alignment of the internal period with the external light cycle (i.e., a test of “resonance”). In plants, various period-length mutants have improved photosynthesis and growth when exposed to external light cycles that matched the endogenous circadian period (19).
c-Reciprocal control of clock by metabolic signaling
An intriguing question remains the extent to which NHRs modulate circadian systems according to changing environmental conditions, such as humoral or nutritional factors. For example, variation in the concentration of glucocorticoid hormone, retinoic acid, heme and fatty acids impact GRs, RAR, REV-ERBα, NPAS2, and PPARs, respectively. So variation in cellular concentrations of any one of these ligands might influence Bmal1 transcription and thereby modulate local cellular circadian rhythms. Within brain, heme and carbon monoxide may modulate NPAS2 activity (20), whereas within vascular cells, retinoic acid influences circadian oscillations through activation of RARα and RXRα (21). Likewise, rhythmic variation in NHR ligands may exert distinct effects on local tissue clock function at different times in the day-night cycle.
Local differences in NHR expression may give rise to tissue-specific differences in coupling of circadian and metabolic cycles, although this remains to be tested. For example, PER2 forms physical interactions with PPARα and REV-ERBα (16), in turn modulating transcription of the gluconeogenic factor G6pase. Conversely, oscillation in NHR ligands may not only impact the phase and amplitude of circadian rhythms, but also physiological outputs of the circadian system. For example, glucocorticoid receptor binding to the promoter of PER2 modulates leptin production and glucose tolerance (22). Non-autonomous signals, such as glucocorticoids or other systemic cues, may have an especially important role in sustaining oscillations of PER2 even in the absence of rhythmic oscillation of CLOCK-BMAL1 (23). It may be possible to exploit tissue-specific differences in NHR-clock interactions for therapeutic purposes.
NHRs may also participate in entrainment of central and peripheral clocks. In vertebrates, a hierarchy of signals within suprachiasmatic nucleus (SCN) pacemaker neurons in the brain and downstream extra-SCN neurons generates entraining cues to maintain phase alignment between oscillators in multiple peripheral tissues. An unresolved question is whether peripheral organ clocks are principally entrained through direct neural wiring, or through circulating hormones, such as glucocorticoids (24). The impact of glucocorticoids on hepatic entrainment has important implications for health under conditions of circadian misalignment, such as phase resetting during jet lag or shift work (25). The finding that liver and brain respond to different entraining signals points towards a possible weak point in the system; conditions such as insulin resistance, where signaling through glucocorticoid, catecholiminergic or peptideric hormones may be attenuated, may cause misalignment between the phase of central and peripheral oscillators. A related phenomenon is food anticipatory activity (FAA) caused by food presentation at the incorrect circadian time. Although clock gene function in FAA behavior has been debated, abrogation of melanocortinergic signaling influences this behavior, consistent with a non-circadian timing mechanism (26). Finally, body temperature has been shown to be a powerful entraining agent for peripheral oscillators. Indeed most signals that synchronize peripheral oscillators affect either body temperature or the heat shock pathway so this may be a final common pathway for resetting clocks in mammals (27).
5-How circadian disruption causes metabolic pathologies: experimental genetic models and human clinical studies
a-REV-ERB & bile acid synthesis
Further studies on the repressor REV-ERBα have uncovered a connection between the clock and the master pathway of hepatic lipid metabolism involving the sterol regulatory element-binding protein (SREBP). SREBPs control both fatty acid and sterol biosynthesis through modulation of rate-limiting enzymes in these pathways. Using a combination of genetic loss and gain of function approaches, Le Martelot et al. observed that the nuclear receptor REV-ERBα controls oscillation in the abundance and activation of the sterol regulatory element-binding proteins, through modulation of the enzyme INSIG2 (insulin-induced gene 2), an insulin-responsive factor that regulates SREBP release from the endoplasmic reticulum. REV-ERBα knockout mice develop increased lipogenesis through upregulation of SREBP1c and SREBP2 target genes independently of nutrient state. In contrast, REV-ERBα-overexpressing mice have decreased SREBP target gene transcription and correspondingly reduced circulating lipid concentrations. The effects of REV-ERBα on bile acid metabolism are mediated through alteration of oxysterol synthesis and liver X receptor (LXR) activity (14), though effects the transcription factor the small heterodimeric protein (SHP) and E4BP4 (adenoviral E4 protein binding protein) have also been implicated in this process (28). Because REV-ERBα expression is controlled by CLOCK-BMAL1, the rhythmic regulation of bile acid production may represent one of the first direct molecular outputs of circadian clock on metabolic physiology.
b-Clock network and lipogenesis
In addition to clock control of bile acid synthesis, mounting evidence has implicated both a direct and indirect effect of clock transcription factors on other aspects of lipogenesis. These observations stem from the finding that ClockΔ19 mutant mice develop hypertriglyceridemia (29), due to effects both within enterocytes and liver (30). At the level of intestine, the clock regulates triglyceride packaging into chylomicron (globules that transport dietary lipids), whereas in liver, clock disruption triggers lipid accumulation (30). The clock controlled gene Nocturnin also impacts the interrelated processes of lipogenesis, osteogenesis (bone formation), and energy homeostasis (31) (32) (33). Effects of circadian gene mutations on lipid absorption are strain-dependent in mice, with severe steatorrhea (excess fecal fat) and malabsorption occurring in the ICR (Institute for Cancer Research) strain, thereby masking the effects of the ClockΔ19 mutation on hepatic triglyceride production, and diet-induced obesity (34).
The clock also functions in ultradian variation (cycles occurring multiple times in 24-hrs) of endoplasmic reticulum (ER) stress signaling, which in turn modulates SREBP activation through a post-transcriptional pathway (35). Rhythmic oscillation of phosphorylation of inositol-requiring enzyme 1α (IRE1α), a transducer of the ER stress response, triggers rhythmic cleavage and translocation of SREBP into the nucleus. The ER stress response detects unfolded or improperly folded proteins, thus rhythmic activation of IRE1α integrates circadian, stress signaling, and lipogenic pathways. Indeed, ultradian rhythms within liver appear with a 12 hr periodicity in the expression of many clock-controlled RNAs (36), and even in rhythmic oscillation of the metabolite NAD+. These shorter cycles are harmonics, of the 24 hr cycle, and may in turn produce rhythmic patterns in physiologic pathways such as lipogenesis. In Cry1/Cry2 double knockout mice, which harbor disruption within the repressor limb of the core clock, loss of circadian oscillation corresponded with constitutive IRE1α activation and accumulation of hepatic lipids. In contrast, in ClockΔ19 mutant mice, which are deficient in the clock activation limb, there were opposite effects on SREBP activation. The finding that ablation of activators and repressors each produce physiologic effects builds evidence that lipogenesis is driven by the circadian clock, rather than an epiphenomenon of clock gene disruption.
The aforementioned studies in mice also have implications for metabolic functions of the clock in humans, as clock genes oscillate within human adipocytes (37), and alterations in clock gene expression are correlated with obesity (38). These findings increase the need to delineate the relationship between chronotype (e.g., whether one is a ‘lark’ or a ‘night owl’), clock genotype and metabolic physiology in humans.
c-Circadian regulation of cardiovascular function, inflammation and thrombosis
It is axiomatic in clinical medicine that certain cardiovascular catastrophes, including myocardial infarction and thrombosis, cluster early in the morning. Yet, the mechanistic underpinnings of timing in cardiovascular disease are not understood. Many aspects of fatty acid metabolism, a key fuel for cardiac muscle, exhibit strong circadian variation, and clock disruption impacts chronotropic function (39). Ablation of Bmal1 also increases the extent of arterial wall lesions following endothelial injury (40), suggesting multiple ways through which clock genes may influence susceptibility to myocardial damage. Similarly, autonomic and mineralocorticoid control of vascular tone, factors in cardiovascular disease risk, have been tied to the clock (41) (42). In individuals with metabolic syndrome, one predictor of cardiovascular risk is the absence of normal nocturnal variation in blood pressure, so-called “non-dippers” (43). Production of the prothrombotic molecule plasminogen activation-inhibitor-1 (PAI-1) has been shown to exhibit circadian regulation (44) (45). Thus, inflammation, thrombosis, cardiomyocyte metabolism, vascular tone and response to vascular injury each represent phenotypes affected by circadian clock function.
d-Circadian systems in glucose homeostasis and diabetes
Glucose concentrations in the blood are highly rhythmic because of changes in insulin sensitivity and insulin secretory capacity of endocrine pancreas (46). Individuals with type 2 diabetes, and even their first-degree relatives not yet affected with the disease, display altered rhythmicity in glucose tolerance (47). Although early morning insulin resistance has been ascribed to the surge in growth hormone during slow wave sleep, rhythmic variation in insulin sensitivity is in part due to autonomic rhythms generated by afferent input from hypothalamus to liver, downstream of the circadian clock (48). Ever since the inception of insulin use in clinical practice, recapitulating the endogenous rhythm of insulin production, and achieving a proper match in the variation in insulin requirement throughout the day/night cycle, has been a pragmatic clinical goal.
Rhythmic production of insulin regulated by peripheral β-cell clocks was revealed by continuous perifusion of isolated islets from the rat, which have a 24-hr rhythm (49). Live-cell imaging in islets isolated from Per2-Luciferase mice shows that they express a self-sustained oscillator with period length matching that of the liver and pituitary (50). ClockΔ19 mutant mice develop age-dependent hyperglycemia in both the light and dark phase of the cycle, corresponding with periods of fasting and feeding (29). These animals also develop susceptibility to diet induced obesity; however, rather than displaying the anticipated hyperinsulinemia, they instead have inappropriately low concentrations of insulin. ClockΔ19 mutant mice display a steeper drop in blood sugar in response to treatment with insulin, a sign that these animals have enhanced insulin sensitivity, thereby masking their β-cell deficiency (50). Bmal1 mutant mice also have impaired glucose tolerance (51), increased insulin sensitivity, and a progressive myopathy with aging that causes cachexia, which limits interpretation of glucose turnover studies. Studies in isolated islets revealed first that the clock oscillator is expressed and self-sustained in this tissue, and secondly, that glucose responsiveness in islets is diminished when the clock is disrupted. After middle age, the mutants also have smaller islet size, reduced proliferation, and transcriptome-wide decreases in proliferative gene expression. Studies in tissue-specific knockout mice have supported the hypothesis that function of the clock activators in liver opposes their function in pancreas (51). Whereas ablation of Bmal1 exclusively within the islet does not impact activity behavior, feeding, or body weight, these mice display a much greater impairment of glucose tolerance than the global knockout, as predicted. Islets from both global and pancreas-specific knockouts have normal insulin content, and influx of calcium in response to glucose is intact. However, exocytosis is impaired, suggesting that clock controls the latest stage in stimulus-secretion coupling.
Findings in experimental genetic models of clock gene ablation may also have implications for understanding emerging evidence that the circadian system participates in human glucose metabolism. For instance, in genome-wide association studies variation in the Melatonin 1b Receptor (MTNR1B) and in Cryptochrome2 (Cry2) are both associated with blood glucose concentrations ((52) and reviewed in (53)). MTNR1B, the cognate receptor of the circadian-regulated hormone melatonin, is expressed in many metabolic tissues, whereas Cry2 encodes a clock repressor. These findings underscore the need to incorporate temporal considerations at the planning stages in future studies to account for circadian variation. Similarly, temporal considerations may aid in analysis of experimental genetic models since testing at different times and under different environmental light cycles may uncover unanticipated effects.
e-Sleep and forced circadian misalignment: genetic models and human studies
Ties between circadian disruption and metabolic disturbance have garnered attention, including large cross sectional sampling of populations subjected to shift work. Extensive studies also indicate a correlation between sleep time and body mass index (BMI). Disruption in specific phases of sleep may be connected to metabolic function. Subtle tones sufficient to selectively deprive subjects of slow wave sleep without producing conscious wakefulness were sufficient to impair glucose tolerance (54). Neuroanatomic studies also indicate interconnections between regions of hypothalamus important in circadian signaling, energetics, and sleep (55) (56). At the molecular level, orexin (also termed hypocretin), originally discovered as a neuropeptide produced in the feeding-stimulatory neurons of lateral hypothalamus, is positioned at the intersection of neuronal systems controlling sleep, circadian output, and metabolism (56). Analysis of orexin receptor 2 knockout mice indicates that lack of orexin signaling increases susceptibility to obesity (rather than the original expectation that orexin, a potent wakefulness-inducing peptide, would induce adiposity) (57). Orexin receptor 2 mutations also account for canine narcolepsy, and orexin deficiency is a hallmark of the disease in humans (58). Activity of the orexin neuron is modulated by glucose and integrates signals downstream of leptin-responsive neurons within the arcuate nucleus. Leptin also impacts sleep, possibly independently of effects on body weight, raising the need to further define leptin actions in this process (59). Manipulation of orexin signaling, an integrator of energetic and circadian signals, may thus provide opportunities to intervene not only in disorders of sleep, but also related metabolic complications.
In humans exposed to a light-dark cycle lengthened to 28 hrs, out of synchrony with the endogenous clock, the sleep-wake cycle is driven at 28-hrs, whereas the melatonin and body temperature rhythm free-runs with a ~24-hr period (60). Such “forced desynchrony”, a manipulation that is intended to simulate deleterious effects of jet-lag or shift work, caused impaired glucose tolerance and hypoleptinemia. Whether circadian disruption might also effect endocrine pancreas insulin secretion, hepatic gluconeogenesis, and glucose disposal in skeletal muscle in humans awaits further study; however these results emphasize the clinical linkages between circadian function and metabolic homeostasis.
6-Coupling and Outputs: How do clocks sense and respond to nutrient signals?
Under homeostatic conditions, the clock acts as a driver of metabolic physiology (Fig 3). However, with perturbations in either circadian or metabolic systems, such as forced behavioral misalignment with shift work, or conversely, high-fat feeding, a vicious cycle ensues in which disruption of metabolic pathways damp and lengthen circadian oscillations (61). The identity of metabolic sensors that may act as intermediates in coupling circadian cycles with physiologic systems remains to be identified. For instance, do changes in cell nutrient signaling in turn produce changes in circadian clock function? Does metabolic disease lead to altered amplitude or phase of circadian cycles within brain or peripheral organs? Two lines of research have begun to address these questions: first, involving the cellular pathway of adenosine monophosphate concentrations, and second, involving NAD+ metabolism. Using phosphopeptide mapping, Lamia et al. identified a consensus motif for phosphorylation by adenosine monophosphate-activated protein kinase (AMP-kinase), a sensor of AMP/ATP ratio, within the CRY protein (62). AMP kinase activator 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR) promoted degradation of CRY, which was abrogated by mutation of the AMPK consensus motif. AMPK knockout mouse embryonic fibroblasts (MEFs) also displayed altered rhythmicity, leading to the proposal that AMP concentration directly couples circadian rhythms to nutrient state in peripheral cells.
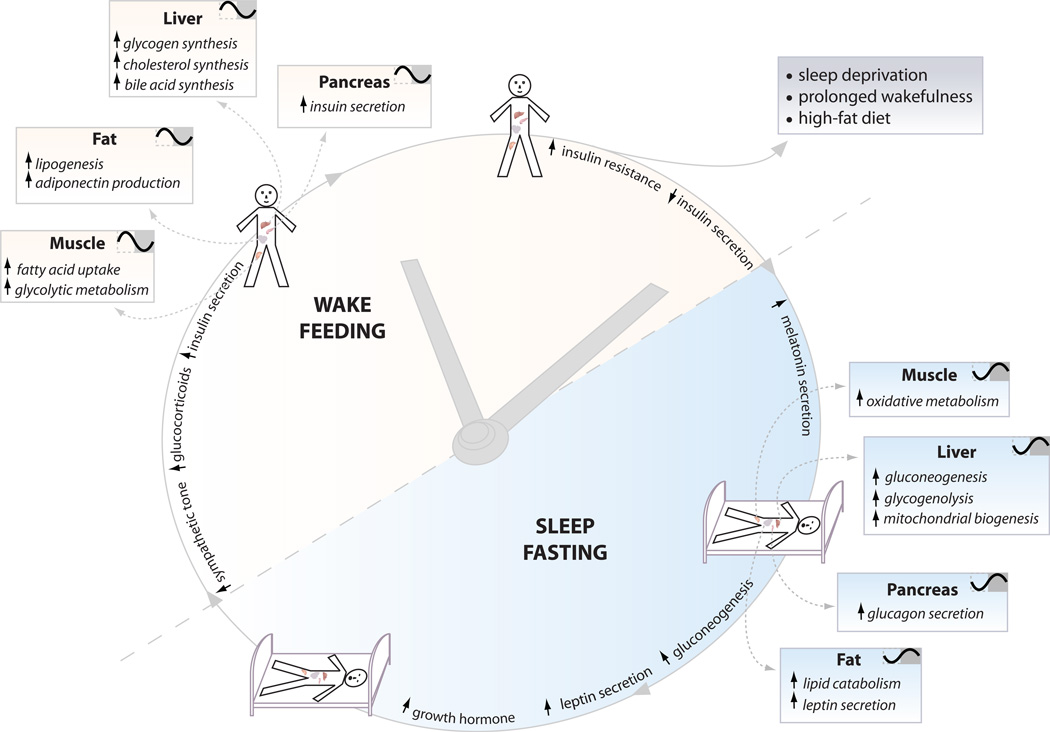
Figure 3. The clock partitions behavioral and metabolic processes according to time of day.
The clock coordinates appropriate metabolic responses within peripheral tissues with the light-dark cycle. For example, the liver clock promotes gluconeogenesis and glycogenolysis during the sleep/fasting period, while it promotes glycogen and cholesterol synthesis during the wake/feeding period. Proper functioning of peripheral clocks keeps metabolic processes in sync with the environment, which is critical for maintaining health of the organism. Different tissues exhibit distinct clock-controlled properties thus ablation of the clock in certain tissues will cause opposing effects on metabolic function as uncovered through dynamic challenges at different times in the cycle under different nutrient conditions. Aging, diet, and environmental disruption such as shift-work may also impact the integration of circadian and metabolic systems.
A second example in metabolic coupling with the core clock originated with the finding that the mammalian ortholog of the yeast sirtuin deacetylases (for silent information regulator), proteins that activate or silence chromatin according to availability of fuel, comprises part of an additional feedback loop with the core clock (63) (64). Sirtuins are present in transcription complexes with the clock, and in turn modulate activity of clock transcription factors. CLOCK-BMAL1 activates the major pathway for mammalian NAD+ synthesis, involving its regeneration from nicotinamide mediated by nicotinamide phosphoribosyltransferase (NAMPT) (65) (66). NAD+ concentration in cells varies across the light-dark cycle, consistent with a role for NAD+ as an oscillating metabolite linking metabolic cycles with the clock. Both NAMPT and SIRT1, similar to the NHRs and AMPK, are regulated not only by the clock, but also by the nutritional status of the organism. For example, fasting increases NAMPT expression in an AMPK-dependent manner in skeletal muscle, whereas fasting and caloric restriction increase SIRT1 activity across multiple tissues. Thus, regulation of the clock by NAD+ and SIRT1 allows for fine-tuning and synchronization of the core molecular clock with the environment. Because NAD+-dependent deacetylases regulate gluconeogenesis and many other pathways (67), it will be important to further delineate the role of clock in NAD+-driven metabolism (Fig 2). A second NAD+-regulated pathway has recently been linked to circadian feeding cycles: PARP-1 activity is circadian in the liver and interacts with the CLOCK protein to poly-ADP-ribosylate it. Since PARP-1 is regulated by NAD+, this provides yet another pathway for metabolic signals to regulate the core clock pathway (68). Collectively, these findings identify incoming (AMPK, PARP-1) and outgoing (NAD+/Sirtuin) sensors that couple nutrient availability, metabolism, and the clock.
7-Conclusion
In just the past twenty years, the mystery of biological timing has been transformed through genetic discovery. As a consequence, availability of molecular clock genes has now provided tools to understand the physiological functions of the circadian system in unprecedented detail. As we experimentally dismantle the clock, the interdependence of timing and energetics seems inextricable. Major gaps in our understanding include: (1) the connection between brain and peripheral tissue clocks in metabolic homeostasis; (2) the interplay between circadian and sleep disruption in energetics; (3) the relationship between nutrient state and circadian homeostasis; and (4) the impact of circadian clock systems on human physiology. Ultimately, such studies will yield deeper insight into the interconnections between genes, behavior and metabolic disease.
Acknowledgments
We thank Ravi Allada, Fred Turek, Brian Chung and Kate-Moynihan Ramsey for helpful suggestions and Billie Marcheva for figures. This work was supported by NIH (PO1 AG011412 and R01HL097817), Chicago Biomedical Consortium Searle Funds, Islet Biology Core of the University of Chicago DRTC, ADA, and JDRF to J.B., and NIH P50 MH074924 and R01 MH078024 to J.S.T. J.S.T. is an Investigator in the Howard Hughes Medical Institute. J.S.T. is a cofounder of ReSet Therapeutics, Inc., and J.S.T. and J.B. are members of its scientific advisory board. J.B. is also an advisor and receives support from Amylin Pharmaceuticals.
References
- 1.Rutter J, Reick M, McKnight SL. Annu Rev Biochem. 2002;71:307. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi JS, Hong HK, Ko CH, McDearmon EL. Nat Rev Genet. 2008;9:764. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakajima M, et al. Science. 2005;308:414. [Google Scholar]
- 4.O'Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. Science. 2008;320:949. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ko CH, et al. PLoS Biol. 2010;8:e1000513. doi: 10.1371/journal.pbio.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang EE, et al. Cell. 2009;139:199. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balsalobre A, Damiola F, Schibler U. Cell. 1998;93:929. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 8.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Science. 2001;291:490. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 9.Preitner N, et al. Cell. 2002;110:251. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 10.Duez H, Staels B. Diab Vasc Dis Res. 2008;5:82. doi: 10.3132/dvdr.2008.0014. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Li S, Liu T, Borjigin J, Lin JD. Nature. 2007;447:477. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 12.Chawla A, Lazar MA. J Biol Chem. 1993;268:16265. [PubMed] [Google Scholar]
- 13.Yang X, et al. Cell. 2006;126:801. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 14.Le Martelot G, et al. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Obesity. 2009;17:2100. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. Genes Dev. 2010;24:345. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roesler WJ, McFie PJ, Dauvin C. J Biol Chem. 1992;267:21235. [PubMed] [Google Scholar]
- 18.Zhang EE, et al. Nat Med. 2010;16:1152. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodd AN, et al. Science. 2005;309:630. [Google Scholar]
- 20.Dioum EM, et al. Science. 2002;298:2385. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 21.McNamara P, et al. Cell. 2001;105:877. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- 22.So AY, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ. Proc Natl Acad Sci U S A. 2009;106:17582. doi: 10.1073/pnas.0909733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balsalobre A, et al. Science. 2000;289:2344. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 25.Kiessling S, Eichele G, Oster H. J Clin Invest. 2010;120:2600. doi: 10.1172/JCI41192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutton GM, et al. J Neurosci. 2008;28:12946. doi: 10.1523/JNEUROSCI.3615-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buhr ED, Yoo SH, Takahashi JS. Science. 2010;330:379. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duez H, et al. Gastroenterology. 2008;135:689. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 29.Turek FW, et al. Science. 2005;308:1043. [Google Scholar]
- 30.Baggs JE, et al. PLoS Biol. 2009;7:e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawai M, et al. Proc Natl Acad Sci U S A. 2010;107:10508. doi: 10.1073/pnas.1000788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green CB, et al. Proc Natl Acad Sci U S A. 2007;104:9888. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai M, Delany AM, Green CB, Adamo ML, Rosen CJ. Endocrinology. 2010;151:4861. doi: 10.1210/en.2010-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kudo T, Tamagawa T, Kawashima M, Mito N, Shibata S. J Biol Rhythms. 2007;22:312. doi: 10.1177/0748730407302625. [DOI] [PubMed] [Google Scholar]
- 35.Cretenet G, Le Clech M, Gachon F. Cell Metab. 2010;11:47. doi: 10.1016/j.cmet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 36.Hughes ME, et al. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu X, et al. Obesity (Silver Spring) 2007;15:2560. doi: 10.1038/oby.2007.308. [DOI] [PubMed] [Google Scholar]
- 38.Wu X, et al. Int J Obes (Lond) 2009;33:971. doi: 10.1038/ijo.2009.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durgan DJ, Young ME. Circ Res. 2010;106:647. doi: 10.1161/CIRCRESAHA.109.209957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anea CB, et al. Circulation. 2009;119:1510. doi: 10.1161/CIRCULATIONAHA.108.827477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curtis AM, et al. Proc Natl Acad Sci U S A. 2007;104:3450. doi: 10.1073/pnas.0611680104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allaman-Pillet N, et al. Mol Cell Endocrinol. 2004;226:59. doi: 10.1016/j.mce.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Ayala DE, et al. Chronobiol Int. 2009;26:1189. doi: 10.3109/07420520903206294. [DOI] [PubMed] [Google Scholar]
- 44.Westgate EJ, et al. Circulation. 2008;117:2087. doi: 10.1161/CIRCULATIONAHA.107.739227. [DOI] [PubMed] [Google Scholar]
- 45.Schoenhard JA, et al. J Mol Cell Cardiol. 2003;35:473. doi: 10.1016/s0022-2828(03)00051-8. [DOI] [PubMed] [Google Scholar]
- 46.Polonsky KS, et al. N Engl J Med. 1988;318:1231. doi: 10.1056/NEJM198805123181903. [DOI] [PubMed] [Google Scholar]
- 47.Boden G, Chen X, Polansky M. Diabetes. 1999;48:2182. doi: 10.2337/diabetes.48.11.2182. [DOI] [PubMed] [Google Scholar]
- 48.Kalsbeek A, et al. PLoS One. 2008;3:e3194. doi: 10.1371/journal.pone.0003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peschke E, Peschke D. Diabetologia. 1998;41:1085. doi: 10.1007/s001250051034. [DOI] [PubMed] [Google Scholar]
- 50.Marcheva B, et al. Nature. 2010;466:571. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lamia KA, Storch KF, Weitz CJ. Proc Natl Acad Sci U S A. 2008;105:15172. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dupuis J, et al. Nat Genet. 2010;42:105. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mulder H, Nagorny CL, Lyssenko V, Groop L. Diabetologia. 2009;52:1240. doi: 10.1007/s00125-009-1359-y. [DOI] [PubMed] [Google Scholar]
- 54.Tasali E, Leproult R, Ehrmann DA, Van Cauter E. Proc Natl Acad Sci U S A. 2008;105:1044. doi: 10.1073/pnas.0706446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saper CB, Scammell TE, Lu J. Nature. 2005;437:1257. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- 56.Adamantidis A, de Lecea L. Trends Endocrinol Metab. 2008;19:362. doi: 10.1016/j.tem.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 57.Funato H, et al. Cell Metab. 2009;9:64. doi: 10.1016/j.cmet.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taheri S, Zeitzer JM, Mignot E. Annu Rev Neurosci. 2002;25:283. doi: 10.1146/annurev.neuro.25.112701.142826. [DOI] [PubMed] [Google Scholar]
- 59.Laposky AD, Bradley MA, Williams DL, Bass J, Turek FW. Am J Physiol Regul Integr Comp Physiol. 2008;295:R2059. doi: 10.1152/ajpregu.00026.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Proc Natl Acad Sci U S A. 2009;106:4453. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kohsaka A, et al. Cell Metab. 2007;6:414. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 62.Lamia KA, et al. Science. 2009;326:437. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asher G, et al. Cell. 2008;134:317. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 64.Nakahata Y, et al. Cell. 2008;134:329. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramsey KM, et al. Science. 2009 [Google Scholar]
- 66.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Science. 2009;324:654. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodgers JT, Puigserver P. Proc Natl Acad Sci U S A. 2007;104:12861. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Asher G, et al. Cell. 2010;142:943. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]