Abstract
African trypanosomes cause sleeping sickness in humans, a disease that is typically fatal without chemotherapy. Unfortunately, drug resistance is common and melarsoprol-resistant trypanosomes often display cross-resistance to pentamidine. Although melarsoprol/pentamidine cross-resistance (MPXR) has been an area of intense interest for several decades, our understanding of the underlying mechanisms remains incomplete. Recently, a locus encoding two closely related aquaglyceroporins, AQP2 and AQP3, was linked to MPXR in a high-throughput loss-of-function screen. Here, we show that AQP2 has an unconventional “selectivity filter.” AQP2-specific gene knockout generated MPXR trypanosomes but did not affect resistance to a lipophilic arsenical, whereas recombinant AQP2 reversed MPXR in cells lacking native AQP2 and AQP3. AQP2 was also shown to be disrupted in a laboratory-selected MPXR strain. Both AQP2 and AQP3 gained access to the surface plasma membrane in insect life-cycle–stage trypanosomes but, remarkably, AQP2 was specifically restricted to the flagellar pocket in the bloodstream stage. We conclude that the unconventional aquaglyceroporin, AQP2, renders cells sensitive to both melarsoprol and pentamidine and that loss of AQP2 function could explain cases of innate and acquired MPXR.
Keywords: aquaporin, major instrinsic proteins, transporter, Trypanosoma brucei, trypanosomiasis
African trypanosomes are tsetse fly-transmitted protozoan parasites that cause a range of important human and animal diseases. Human African trypanosomiasis (HAT), also known as sleeping sickness, is caused by Trypanosoma brucei gambiense or Trypanosoma brucei rhodesiense and is typically fatal without chemotherapy (1, 2). The closely related, but human-serum sensitive, Trypanosoma brucei brucei, causes a veterinary disease known as nagana. T. b. gambiense is responsible for the vast majority of HAT cases and occurs in West and Central Africa. Chemotherapy is of major importance due to the absence of a vaccine. During the early stages of HAT, suramin and pentamidine, an aromatic diamidine, are the only drugs available but, unfortunately, suramin cannot be used in West Africa because of the risk of severe allergic reactions if Onchocerca infection is also present (3).
As early-stage symptoms are relatively mild and nonspecific, diagnosis is often late and reveals advanced infections, after trypanosomes have invaded the central nervous system (CNS). In these cases, eflornithine (alone or in combination with nifurtimox) or the melaminophenyl arsenical melarsoprol are needed to target trypanosomes in the CNS. Among the limited options, eflornithine or nifurtimox–eflornithine combination therapies currently offer the most favorable clinical outcome for late-stage disease therapy. However, because eflornithine is expensive, difficult to administer, and ineffective against T. b. rhodesiense (4), melarsoprol is still widely used, despite the fact that it causes a fatal reactive encephalopathy in around 5% of patients (2). Thus, melarsoprol is the only drug active against CNS infections of both T. b. gambiense and T. b. rhodesiense. This desperate situation is made worse by the current and increasing incidence of melarsoprol treatment failures, thought to be due to the emergence and spread of drug resistance, leading to 20–30% treatment failure rates in Uganda, the Democratic Republic of Congo, and the Sudan (2).
Melarsoprol has been suggested to act primarily by forming a toxic adduct with trypanothione, known as Mel T (5, 6), whereas pentamidine is a DNA-binding drug that becomes highly concentrated in trypanosomes and collapses the mitochondrial membrane potential (7). Arsenical/diamidine cross-resistance was first reported over 60 y ago (8). In the 1990s, a trypanosome P2 adenosine transporter (AT1) was then identified that, when expressed in Saccharomyces cerevisiae, conferred susceptibility to melarsoprol and was also found to be mutated and defective in melarsoprol-resistant trypanosomes (9, 10). The same P2/AT1 mediates pentamidine transport in trypanosomes (11, 12) and when expressed in S. cerevisiae (13). AT1 gene knockout generated cells with approximately twofold increased resistance to both melarsoprol and pentamidine (14) and AT1 knockdown, using RNA interference (RNAi), increased resistance to melarsoprol (5, 15) but has not been shown to significantly increase resistance to pentamidine. Additional T. brucei transporters, NT11.1 and NT12.1, can transport pentamidine when expressed in a heterologous system (16) and high and low-affinity pentamidine transporters (HAPT1 and LAPT1, respectively) have been described (12, 17). Indeed, selection for increased resistance to either melarsoprol or pentamidine in cells lacking AT1 leads to loss of HAPT1 activity (18), but a gene encoding this activity has not yet been identified. Mel T efflux also has an impact on melarsoprol accumulation as demonstrated by a 10-fold increased resistance in trypanosomes overexpressing the ATP-binding cassette (ABC) transporter, MRPA (19).
As detailed above, our understanding of the mechanisms underlying MPXR remains incomplete (20). A recent set of high-throughput, loss-of-function RNAi screens linked AT1 and several additional membrane-spanning transporters to melarsoprol or pentamidine resistance (5). In particular, the plasma membrane H+-ATPases, HA1-3, were linked to pentamidine resistance, suggesting that at least one of the relevant pentamidine transporters is a proton symporter. Most notably, however, the screens linked just one locus, encoding a pair of closely related aquaglyceroporins (AQP2 and AQP3) to MPXR; knockout of both the AQP2 and AQP3 genes increased resistance to melarsoprol and pentamidine by >2-fold and >15-fold, respectively (5).
The AQPs are major intrinsic proteins (MIPs) that constitute a superfamily of aquaporins and aquaglyceroporins, channels that facilitate the passive transport of water and small neutral solutes across cell membranes in organisms from bacteria to humans. Although they are absent in many microorganisms, most eukaryotic genomes encode at least 1 of these channels; several plants have >30, mammals have >10, and Plasmodium spp. and other Apicomplexa typically have 1. The reader is referred to reviews dealing with AQPs generally (21), with human AQPs (22), and with AQPs in protozoan parasites (23). The T. brucei aquaglyceroporins, AQP1, AQP2, and AQP3, transport water, glycerol, urea, dihydroxyacetone (24), and ammonia (25), and AQP1 and AQP3 have been shown to be flagellar membrane and plasma membrane proteins, respectively (26).
An improved understanding of the mechanisms underlying MPXR in African trypanosomes will facilitate the development of diagnostic tools and possibly also the development of improved therapies. Here, we show that loss of AQP2, a channel with an unusual selectivity filter, is specifically responsible for MPXR. AQP2 is also specifically restricted to the flagellar pocket in bloodstream-form cells. These unique findings establish a central role for T. brucei AQP2 in MPXR.
Results
T. brucei AQP2 Has an Unusual Selectivity Filter.
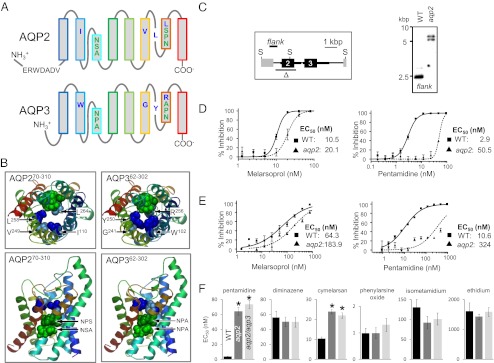
The T. brucei locus recently linked to MPXR encodes the closely related proteins, AQP2 and AQP3 (5). These proteins are also related to the human aquaglyceroporins, AQPs 3, 7, 9, and 10 (22, 24). The structural determinants for gating these channels are the focus of intense interest, and because the selectivity of channels is central to their function, we examined the conserved residues that constitute the AQP2 and AQP3 “selectivity filter” (Fig. 1A). AQPs have six membrane-spanning α-helices and two half helices that fold into the center of the channel, with the termini located on the cytoplasmic face of the membrane. The selectivity filter is typically defined by two prominent constrictions in the channel, one formed by the highly conserved “NPA” motifs within the half helices, and the other, usually narrower, formed by an “aromatic arginine” (ar/R) motif (27–29). Schematic representations of AQP2 and AQP3 indicate the location of the selectivity filter residues relative to the predicted membrane-spanning domains (Fig. 1A). Whereas AQP3 encodes a conventional selectivity filter, as does AQP1, AQP2 lacks the NPA and ar/R motifs. Homology models (Fig. 1B) indicate how the unusual selectivity filter of AQP2 likely impacts the lining of the channel. An analysis of more than 1,000 MIPs from 340 organisms in the MIPModDB database (30) revealed T. brucei AQP2 as the only MIP with NSA/NPS or IVLL motifs; 78% contain NPA/NPA and 89% contain the arginine of the ar/R motif. Thus, AQP2 has a unique selectivity filter.
Fig. 1.
Cells lacking the unusual AQP2 are MPXR. (A) Schematic representation of AQP2 (Tb927.10.14170) and AQP3 (Tb927.10.14160). The selectivity filter residues are indicated; NSA/NPS/IVLL in AQP2 and NPA/NPA/WGYR in AQP3. An AQP2-specific insertion is also indicated. (B) Homology models for AQP2 and AQP3 showing the key amino acids that line the channel (colors as in A). The models were generated using SWISS-MODEL (51) and the Protein Data Bank coordinates 1ldfA. (C) Schematic map (Left) and Southern blot (Right) showing AQP2 gene knockout. Δ indicates the region deleted and “flank” indicates the 671-bp probe. Transcription is from left to right. Thicker black lines indicate the predicted untranslated regions. Genomic DNA was digested with SacII (S) before Southern blotting. (D) Bloodstream-form cells. Dose–response curves for melarsoprol and pentamidine. Wild-type (WT) cells are compared with aqp2 null cells. EC50 values are shown. (E) Insect-stage (procyclic form) cells. Other details as in D above. (F) Bloodstream-form cells. EC50 values for other related drugs. Average and SEM are shown (n ≥ 10). *P < 0.0001 as determined using a one-way ANOVA test.
aqp2 Null Cells Are Melarsoprol and Pentamidine Resistant.
Cells lacking both AQP2 and AQP3 were resistant to melarsoprol and pentamidine (5). Because the analysis above indicated major differences between AQP2 and AQP3 that may be of functional significance, we wanted to determine what contribution each protein makes to the drug susceptibility phenotype. We used a gene knockout approach to test our hypothesis that AQP2 is specifically required for drug susceptibility. An aqp2 null strain was generated by sequential transfection with constructs that replace each AQP2 allele with selectable marker genes, deleting the entire AQP2 protein coding sequence but leaving the AQP3 gene intact (Fig. 1C). AQP2 knockout was confirmed by Southern blotting (Fig. 1C). Neither aqp2 null cells nor aqp2/aqp3 null cells displayed any detectable growth defect relative to wild-type cells. Drug susceptibility analysis revealed that aqp2 null cells were resistant to both melarsoprol and pentamidine (Fig. 1D); the 50% effective growth-inhibitory concentration (EC50) was increased almost 2-fold relative to wild-type cells for melarsoprol and by more than 15-fold for pentamidine. Thus, aqp2 null cells are indistinguishable from aqp2/aqp3 null cells (5) in terms of EC50 for either of these drugs.
Differentiation of African trypanosomes from the bloodstream stage to the insect “procyclic” stage, during the life cycle, involves major changes in surface architecture and energy metabolism (31). To probe the function of AQP2 during and/or following this developmental transition, we differentiated aqp2 and aqp2/aqp3 null strains; triggered in vitro by reduced temperature and by the addition of citric acid cycle intermediates to the culture medium. We saw no evidence for a defect in the process of differentiation in either aqp2 null or aqp2/aqp3 null cells. Dose–response curves revealed that the impact of AQP2 on drug sensitivity was similar in both stages. In aqp2 null insect-stage cells, the EC50 for melarsoprol was increased by approximately 3-fold compared with wild-type cells, whereas the EC50 for pentamidine was increased by more than 30-fold (Fig. 1E) and we obtained similar results for aqp2/aqp3 null insect-stage cells. We conclude that AQP2 is constitutively expressed and renders both major life-cycle stages drug sensitive. However, neither AQP2 nor AQP3 is required for viability of the bloodstream or procyclic stage or for differentiation in vitro.
In bloodstream-form cells, the resistance phenotype was specific for pentamidine and melaminophenyl arsenicals, with little or no effect on sensitivity to the veterinary diamidine, diminazene aceturate (Berenil), or the phenanthridine trypanocides, isometamidium and ethidium (Fig. 1F). In addition, wild-type and aqp null cells were equally sensitive to the lipophilic arsenical, phenylarsine oxide, which is known to diffuse across membranes (18). Thus, the deletion of AQP2 renders T. brucei MPXR but resistance does not extend to all diamidines or arsenicals.
AQP2 Restores Drug Sensitivity to aqp2/aqp3 Null Cells.
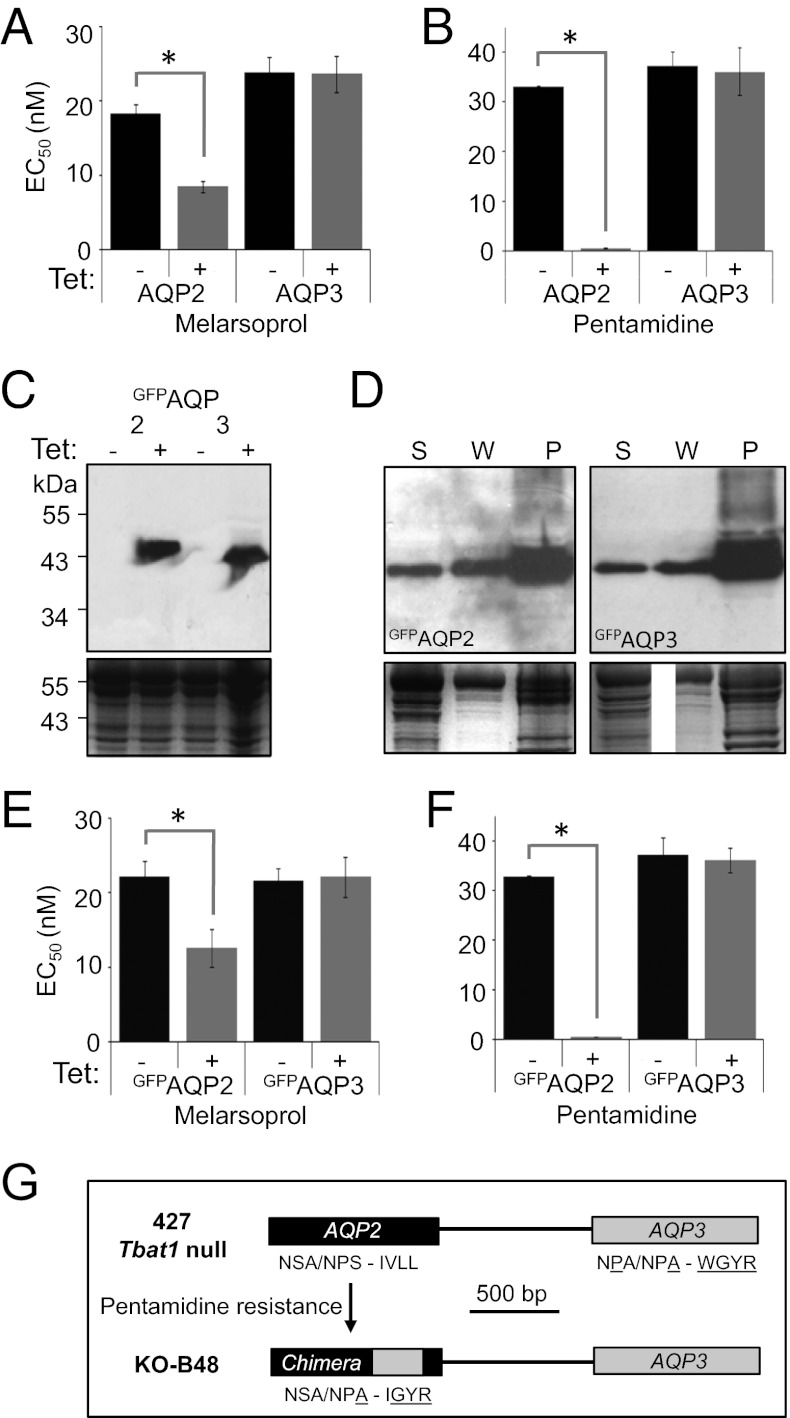
The results above suggest that AQP2 plays an important role in rendering T. brucei sensitive to melarsoprol and pentamidine. To determine whether AQP3 is required for this effect, we reintroduced an inducible copy of AQP2 into aqp2/aqp3 null cells. In the absence of AQP2 induction, these strains displayed a similar drug sensitivity phenotype to aqp2 or aqp2/aqp3 null cells (compare Fig. 1F with Fig. 2 A and B). In striking contrast, induction of AQP2 expression restored sensitivity to both melarsoprol (Fig. 2A) and pentamidine (Fig. 2B). Importantly, drug sensitivity is restored in the absence of AQP3, indicating that AQP3 expression is not required for melarsoprol and pentamidine sensitivity. A similar experiment, but involving the reintroduction of a copy of AQP3, failed to show any impact on MPXR (Fig. 2 A and B), confirming that AQP3 expression does not confer sensitivity to melarsoprol or pentamidine. AQP2 also restored drug sensitivity in an aqp2 null strain with an intact AQP3 gene. We conclude that, whereas AQP2 is required, AQP3 is neither sufficient nor required for drug sensitivity.
Fig. 2.
AQP2 is required for drug sensitivity, whereas AQP3 is neither necessary nor sufficient. (A) EC50 values for melarsoprol following recombinant AQP expression in bloodstream-form cells. aqp2/aqp3 null cells with a tetracycline-regulated (Tet-on) copy of either AQP2 or AQP3 were grown in the absence (black bars) or presence (gray bars) of tetracycline. Error bars, SEM derived from two independent triplicate assays. *P < 0.0001 as determined using nonlinear regression analysis. (B) EC50 values for pentamidine. Other details as in A. (C) Tet-regulated expression of GFP-tagged AQPs. Western blots reveal tightly regulated inducible expression of both GFPAQP2 and GFPAQP3. Both proteins migrated below their predicted molecular mass of ∼60 kDa but proteins with extensive transmembrane regions often display aberrant migration. (D) Both GFPAQP2 and GFPAQP3 are membrane associated. Western blots show supernatant (S), wash (W), and pellet (P; membrane-fraction) following hypotonic lysis. GFPAQPs are expressed in an aqp2/aqp3 null background. (E) EC50 values for melarsoprol. aqp2/aqp3 null cells with a Tet-on copy of GFPAQP2 or GFPAQP3. Other details as in A. (F) EC50 values for pentamidine. Other details as in E. (G) The KO-B48 strain encodes an AQP2/AQP3 chimeric sequence. Schematic illustrates the AQP2/AQP3 locus before and after the emergence of pentamidine resistance. Selectivity filter residues are indicated; six residues are specific to the AQP3 sequence (underlined) and four of these are found in the predicted chimeric protein (Fig. S1).
We also used the approach described above to reintroduce AQPs with green fluorescent protein (GFP) fused to the N termini. This approach allowed us to confirm inducible expression of GFPAQP2 and GFPAQP3 by Western blotting (Fig. 2C) and also to show that both GFPAQP2 and GFPAQP3 partition predominantly into the membrane fraction, as expected (Fig. 2D). GFPAQP2 fully restored drug sensitivity in both aqp2 and aqp2/aqp3 null strains, whereas GFPAQP3 failed to restore drug sensitivity in either strain (Fig. 2 E and F), confirming the results above and indicating that AQP2 remains functional when fused to GFP.
AQP2 Is Disrupted in a Pentamidine-Resistant Strain.
We next asked whether exposure to pentamidine could select for mutations that disrupt AQP2 function, thereby producing drug-resistant strains. To address this question, the AQP2 gene was sequenced in the well-characterized KO-B48 strain, which lacks HAPT1 activity, displays reduced pentamidine uptake, and is MPXR (18). Strikingly, an AQP2/AQP3 chimeric gene was found in place of AQP2 (Fig. 2G); a 272-nucleotide segment was replaced by an AQP3-derived sequence that alters four of the predicted residues (underlined) of the selectivity filter described above, from NSA/NPS/IVLL in AQP2 to NSA/NPA/IGYR in the chimera (Fig. S1).
AQP2 Is Restricted to the Flagellar Pocket Specifically in Bloodstream-Form Cells.
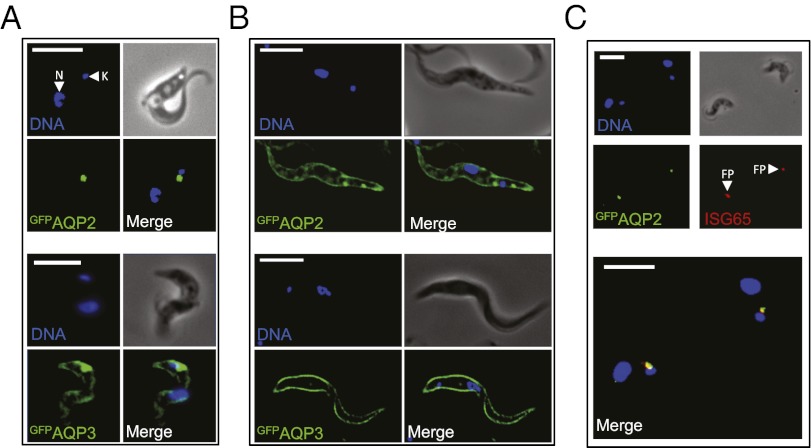
AQP3 is known to be dispersed over the plasma membrane (26) but AQP2 localization has not been determined. We used the strains expressing GFP-tagged AQPs described above to determine AQP subcellular localization and observed GFP fluorescence directly to eliminate potential artifacts associated with immunofluorescence detection of membrane proteins following detergent-based permeabilization. In bloodstream-form cells, the two proteins displayed strikingly different localizations; GFPAQP2 localized to a focal region adjacent to the kinetoplast (mitochondrial DNA) (Fig. 3A, Upper), whereas GFPAQP3 localized to this region and also to the plasma membrane (Fig. 3A, Lower), as demonstrated previously for native AQP3 (26). These results were confirmed by confocal microscopy and were indistinguishable in an aqp2/aqp3 or aqp2 background. Because AQPs typically form tetramers, these results suggest little or no physical interaction between AQP2 and AQP3 and the formation of primarily or exclusively AQP2 and AQP3 homotetramers.
Fig. 3.
AQP2 is restricted to the flagellar pocket specifically in bloodstream-form cells. (A) Fluorescence microscopy reveals the location of GFPAQP2 and GFPAQP3 in bloodstream-form cells. DNA is stained with DAPI and the nucleus (N) and mitochondrial genome (kinetoplast, K) are indicated. (B) Fluorescence microscopy reveals the location of GFPAQP2 and GFPAQP3 in insect-stage cells. (C) Immunofluorescence microscopy reveals the location of GFPAQP2 colocalized with the flagellar pocket (FP) marker, ISG65 in bloodstream-form cells. GFPAQPs are expressed in an aqp2/aqp3 null background. (Scale bars, 10 μm.)
We demonstrated above that AQP2 confers melarsoprol and pentamidine sensitivity to both bloodstream- and insect-stage cells. To determine whether AQP2 localization differs in these two life-cycle stages, the GFPAQP2 and GFPAQP3 strains were differentiated to the insect stage. Both proteins localized to the plasma membrane in insect-stage cells (Fig. 3B), indicating that the mechanism blocking access of AQP2 to the plasma membrane is bloodstream-stage specific and developmentally regulated. The focal structure identified by the AQP2 signal in bloodstream-form cells is adjacent to the kinetoplast and may represent the flagellar pocket, an invagination of the pellicular membrane, which is inaccessible to host innate immune effectors and the exclusive site for endocytosis, exocytosis, and specific receptors (32). To test this hypothesis, we stained invariant surface glycoprotein 65 (ISG65), known to identify the flagellar pocket (33), and visualized both proteins by microscopy. GFPAQP2 displayed almost complete colocalization with the flagellar pocket marker (Fig. 3C). We conclude that all three T. brucei AQPs display distinct subcellular localization in bloodstream-form T. brucei, with AQP2 displaying specific sequestration in the flagellar pocket.
Discussion
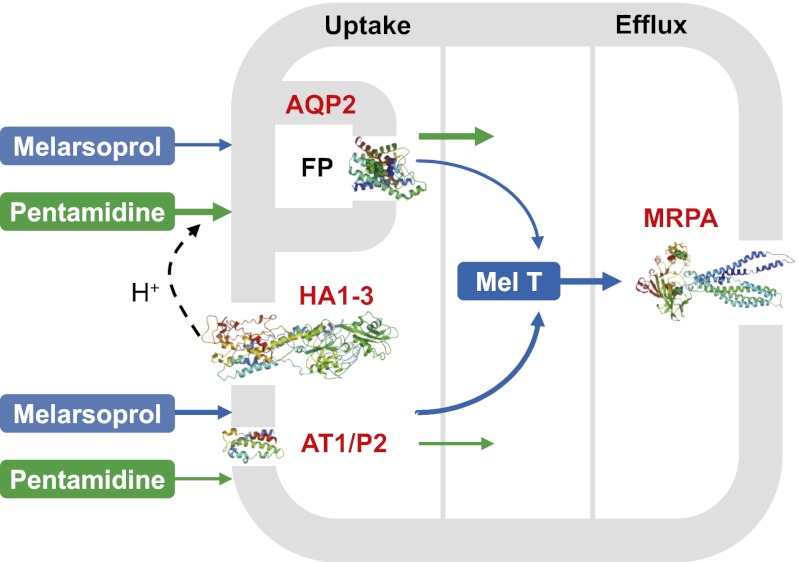
Genome-scale RNAi screening linked a pair of closely related aquaglyceroporins to MPXR (5), suggesting that these channels play the dominant role in controlling susceptibility to these drugs. We have shown here that one of these proteins, AQP2, restricted to the flagellar pocket in bloodstream-form cells, is specifically responsible for MPXR. In support of this conclusion, an aqp2 null strain that retains AQP3 displays MPXR, whereas AQP2 expression in an aqp2/aqp3 null strain restores sensitivity. In addition, AQP2 is shown to be disrupted in the well-characterized MPXR strain, KO-B48. Thus, AQP2 may correspond to the long-sought high-affinity pentamidine transporter (HAPT1) (20). Indeed, pentamidine and cymelarsan were previously identified as HAPT1 substrates (18), whereas diminazene transport is primarily via AT1 (34). The observations that wild-type and aqp2 null strains are equally sensitive to a lipophilic arsenical and that the KO-B48 strain displays reduced pentamidine uptake (18) suggest that melarsoprol and pentamidine uptake are both AQP2 dependent. We present a model (Fig. 4) to explain the role of all four transporters now linked to the control of melarsoprol (MRPA), pentamidine (HA1-3), or melarsoprol and pentamidine (AT1 and AQP2) susceptibility.
Fig. 4.
Model to explain melarsoprol and pentamidine transport in T. brucei. Four T. brucei transporters have been linked to the control of melarsoprol and or pentamidine susceptibility. Both drugs enter the cell through AT1/P2 (Tb927.5.286b) and AQP2. HA1-3 (Tb927.10.12500–10, only two annotated in the reference genome) is specifically linked to pentamidine susceptibility and generates the proton motive force required for pentamidine symport via AQP2. Efflux of the toxic melarsoprol adduct, Mel T, is via MRPA (Tb927.8.2160). The AT1/P2 and AQP2 channels transport the drugs with different efficiencies as indicated by the weighted arrows. AT1/P2 and MRPA have not been localized. FP, flagellar pocket; Mel T, melarsoprol–trypanothione adduct. The homology model for AT1/P2 is restricted to residues 340–454.
AQP1, AQP2, and AQP3 have been reported to transport a similar set of substrates (24, 25) but the roles of AQPs in trypanosomes have not been clearly defined. It seems likely that they play roles in osmoregulation and metabolism, such as in the efflux of glycerol from the bloodstream form (35). So what property of AQP2 allows this porin to specifically contribute to drug susceptibility? With molecular masses of 398 Da and 340 Da, respectively, melarsoprol and pentamidine are substantially larger than glycerol, with a mass of only 92 Da, or other compounds known to be transported by AQPs (36). Inorganic trivalent arsenic and antimony (in the form of arsenite or potassium antimonyl tartrate) are transported by AQPs, such as Leishmania sp. AQP1 (37), whereas AQP-mediated transport of large metalloorganic compounds or diamidines has not been reported. It will therefore be important to consider whether melarsoprol or pentamidine preparations contain or form lower molecular mass toxins. However, both drugs appear to enter trypanosomes intact (6, 38) and it may be the unique selectivity filter of AQP2 that allows the passage of these large molecules. In addition, the arginine (R) of the ar/R filter, not present in AQP2, is thought to repel protons, and AQPs lacking this residue display enhanced proton conductance (29, 39, 40). We therefore propose direct passage of melarsoprol and pentamidine via AQP2 and proton symport in the case of pentamidine (Fig. 4).
AQP3 is dispersed over the plasma membrane, whereas AQP1 is predominantly restricted to the flagellum (26). We have now shown that AQP2 displays a distinct localization that is specific to bloodstream-stage cells, being retained in the flagellar pocket. Thus, all three T. brucei AQPs display distinct subcellular localization in bloodstream-stage cells. Although our analysis of insect-stage cells indicates that pocket sequestration is not strictly required for the protein to confer drug sensitivity, it will be of interest to identify the features of AQP2 that retard transit from the flagellar pocket to the cell surface plasma membrane in a stage-specific manner.
The selective pressure associated with widespread drug use, including mass pentamidine chemoprophylaxis in the 1940s, could have contributed to the selection and spread of MPXR parasites (17, 34, 41). AT1 has been found to be mutated, deleted, or down-regulated in melarsoprol-resistant trypanosomes (10, 42) but AT1 knockout generated cells with only a twofold increased resistance to both melarsoprol and pentamidine (14), and there is evidence for resistance that is independent of AT1 disruption (43, 44). Our results now show that AQP2, when defective, could explain cases of innate MPXR and/or acquired MPXR of clinical relevance.
These unique findings offer potential explanations for many earlier observations and establish a central role for T. brucei AQP2 in MPXR. Our prospects for understanding cases of innate and acquired MPXR are now greatly improved. Aquaglyceroporins typically transport water and small solutes but AQP2 may be an exception in terms of an ability to transport larger cargo. It will now be important to investigate the status of AQP2 in drug-resistant isolates and to establish whether AQP2 status is predictive of clinical outcome.
Experimental Procedures
Strains.
Bloodstream-form T. brucei, Lister 427, MiTat 1.2, clone 221a, and derivatives were maintained as previously described (45); 2T1 (46), KO-B48 (18), and aqp2/aqp3 null strains (5) were described previously. Strains were transfected using a Nucleofector apparatus (Lonza) in conjunction with cytomix or T-cell nucleofection solution. Transformants were selected with blasticidin (10 μg/mL), G418 (2 μg/mL), or hygromycin (2.5 μg/mL). Southern blotting was carried out according to standard protocols. Differentiation to the insect stage was triggered by transferring the cells to glucose-free DTM medium (47) supplemented with citrate and cis-aconitate at 27 °C. AQP and GFPAQP expression was induced by exposing cells to 1 μg/mL tetracycline for 24 h. EC50 assays were carried out using alamarBlue as described (48, 49). The insect-stage cells were analyzed ≥6 d after differentiation was initiated. The AQP2/AQP3 chimera was PCR amplified from KO-B48 genomic DNA using a high-fidelity polymerase; two independent products were sequenced using standard procedures.
Plasmid Construction.
The AQP2 locus was disrupted in 2T1 cells by replacement of a 2,535-bp fragment with NPT and BLA selectable markers (the T. brucei genome is diploid). Briefly, AQP2-flanking sequences were inserted on both sides of the selectable marker cassettes and, before transfection, the plasmids were linearized by cleaving at the distal ends of these AQP2 targeting regions. The pRPaGFPx construct (46) was modified to express native or GFP-tagged AQPs under the control of a Tet-regulated RRNA promoter. Both AQP2 constructs were checked for the presence of the expected SacII cleavage site. Primer sequences are available upon request.
Protein Analysis.
Subcellular fractionation by hypotonic lysis was carried out as described (5). All protein samples were stored in the presence of a protease inhibitor mixture (Roche) and were not boiled. Whole cell lysates and hypotonic lysis fractions were separated by SDS/PAGE and electroblotted according to standard protocols. For immunoblotting, we used an enhanced chemiluminescent kit (GE Healthcare), according to the manufacturer’s instructions. GFP was detected on Western blots and by fluorescence microscopy using polyclonal rabbit α-GFP (Europa; 1:2,000). Immunofluorescence was carried out according to standard protocols. Briefly, cells were permeabilized and ISG65 was detected using rabbit α-ISG65 (1:1,000) primary antibody (50) and a rhodamine-conjugated α-rabbit secondary antibody (1:100). Cells were settled on slides and mounted in Vectashield (Vector Laboratories) containing the DNA counterstain, 4,6-diamidino-2-phenylindole (DAPI). Images were captured using a Nikon Eclipse E600 epifluorescence microscope in conjunction with a Coolsnap FX (Photometrics) charge-coupled device (CCD) camera and processed in Metamorph 5.0 (Photometrics).
Supplementary Material
Acknowledgments
We thank Mark Carrington (University of Cambridge) for α-ISG65 sera and Sam Alsford for technical advice. This work was funded by Grant 093010/Z/10/Z (to D.H.) from The Wellcome Trust and Grant 84733 (to H.P.d.K. and M.P.B.) from the Medical Research Council. N.B. was supported by a Bloomsbury Colleges doctoral studentship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. JX026928).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202885109/-/DCSupplemental.
References
- 1.Malvy D, Chappuis F. Sleeping sickness. Clin Microbiol Infect. 2011;17:986–995. doi: 10.1111/j.1469-0691.2011.03536.x. [DOI] [PubMed] [Google Scholar]
- 2.Pépin J, Milord F. The treatment of human African trypanosomiasis. Adv Parasitol. 1994;33:1–47. doi: 10.1016/s0065-308x(08)60410-8. [DOI] [PubMed] [Google Scholar]
- 3.Brun R, Blum J, Chappuis F, Burri C. Human African trypanosomiasis. Lancet. 2010;375:148–159. doi: 10.1016/S0140-6736(09)60829-1. [DOI] [PubMed] [Google Scholar]
- 4.Iten M, et al. Alterations in ornithine decarboxylase characteristics account for tolerance of Trypanosoma brucei rhodesiense to D,L-α-difluoromethylornithine. Antimicrob Agents Chemother. 1997;41:1922–1925. doi: 10.1128/aac.41.9.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alsford S, et al. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature. 2012;482:232–236. doi: 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fairlamb AH, Henderson GB, Cerami A. Trypanothione is the primary target for arsenical drugs against African trypanosomes. Proc Natl Acad Sci USA. 1989;86:2607–2611. doi: 10.1073/pnas.86.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanteri CA, Tidwell RR, Meshnick SR. The mitochondrion is a site of trypanocidal action of the aromatic diamidine DB75 in bloodstream forms of Trypanosoma brucei. Antimicrob Agents Chemother. 2008;52:875–882. doi: 10.1128/AAC.00642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rollo IM, Williamson J. Acquired resistance to ‘Melarsen’, tryparsamide and amidines in pathogenic trypanosomes after treatment with ‘Melarsen’ alone. Nature. 1951;167:147–148. doi: 10.1038/167147a0. [DOI] [PubMed] [Google Scholar]
- 9.Carter NS, Fairlamb AH. Arsenical-resistant trypanosomes lack an unusual adenosine transporter. Nature. 1993;361:173–176. doi: 10.1038/361173a0. [DOI] [PubMed] [Google Scholar]
- 10.Mäser P, Sütterlin C, Kralli A, Kaminsky R. A nucleoside transporter from Trypanosoma brucei involved in drug resistance. Science. 1999;285:242–244. doi: 10.1126/science.285.5425.242. [DOI] [PubMed] [Google Scholar]
- 11.Carter NS, Berger BJ, Fairlamb AH. Uptake of diamidine drugs by the P2 nucleoside transporter in melarsen-sensitive and -resistant Trypanosoma brucei brucei. J Biol Chem. 1995;270:28153–28157. doi: 10.1074/jbc.270.47.28153. [DOI] [PubMed] [Google Scholar]
- 12.De Koning HP. Uptake of pentamidine in Trypanosoma brucei brucei is mediated by three distinct transporters: Implications for cross-resistance with arsenicals. Mol Pharmacol. 2001;59:586–592. doi: 10.1124/mol.59.3.586. [DOI] [PubMed] [Google Scholar]
- 13.de Koning HP, et al. The trypanocide diminazene aceturate is accumulated predominantly through the TbAT1 purine transporter: Additional insights on diamidine resistance in african trypanosomes. Antimicrob Agents Chemother. 2004;48:1515–1519. doi: 10.1128/AAC.48.5.1515-1519.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matovu E, et al. Mechanisms of arsenical and diamidine uptake and resistance in Trypanosoma brucei. Eukaryot Cell. 2003;2:1003–1008. doi: 10.1128/EC.2.5.1003-1008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schumann Burkard G, Jutzi P, Roditi I. Genome-wide RNAi screens in bloodstream form trypanosomes identify drug transporters. Mol Biochem Parasitol. 2011;175:91–94. doi: 10.1016/j.molbiopara.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Ortiz D, Sanchez MA, Quecke P, Landfear SM. Two novel nucleobase/pentamidine transporters from Trypanosoma brucei. Mol Biochem Parasitol. 2009;163:67–76. doi: 10.1016/j.molbiopara.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Koning HP, Jarvis SM. Uptake of pentamidine in Trypanosoma brucei brucei is mediated by the P2 adenosine transporter and at least one novel, unrelated transporter. Acta Trop. 2001;80:245–250. doi: 10.1016/s0001-706x(01)00177-2. [DOI] [PubMed] [Google Scholar]
- 18.Bridges DJ, et al. Loss of the high-affinity pentamidine transporter is responsible for high levels of cross-resistance between arsenical and diamidine drugs in African trypanosomes. Mol Pharmacol. 2007;71:1098–1108. doi: 10.1124/mol.106.031351. [DOI] [PubMed] [Google Scholar]
- 19.Shahi SK, Krauth-Siegel RL, Clayton CE. Overexpression of the putative thiol conjugate transporter TbMRPA causes melarsoprol resistance in Trypanosoma brucei. Mol Microbiol. 2002;43:1129–1138. doi: 10.1046/j.1365-2958.2002.02831.x. [DOI] [PubMed] [Google Scholar]
- 20.de Koning HP. Ever-increasing complexities of diamidine and arsenical crossresistance in African trypanosomes. Trends Parasitol. 2008;24:345–349. doi: 10.1016/j.pt.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Benga G. Water channel proteins (later called aquaporins) and relatives: Past, present, and future. IUBMB Life. 2009;61:112–133. doi: 10.1002/iub.156. [DOI] [PubMed] [Google Scholar]
- 22.King LS, Kozono D, Agre P. From structure to disease: The evolving tale of aquaporin biology. Nat Rev Mol Cell Biol. 2004;5:687–698. doi: 10.1038/nrm1469. [DOI] [PubMed] [Google Scholar]
- 23.Beitz E. Aquaporins from pathogenic protozoan parasites: structure, function and potential for chemotherapy. Biol Cell. 2005;97:373–383. doi: 10.1042/BC20040095. [DOI] [PubMed] [Google Scholar]
- 24.Uzcategui NL, et al. Cloning, heterologous expression, and characterization of three aquaglyceroporins from Trypanosoma brucei. J Biol Chem. 2004;279:42669–42676. doi: 10.1074/jbc.M404518200. [DOI] [PubMed] [Google Scholar]
- 25.Zeuthen T, et al. Ammonia permeability of the aquaglyceroporins from Plasmodium falciparum, Toxoplasma gondii and Trypansoma brucei. Mol Microbiol. 2006;61:1598–1608. doi: 10.1111/j.1365-2958.2006.05325.x. [DOI] [PubMed] [Google Scholar]
- 26.Bassarak B, Uzcátegui NL, Schönfeld C, Duszenko M. Functional characterization of three aquaglyceroporins from Trypanosoma brucei in osmoregulation and glycerol transport. Cell Physiol Biochem. 2011;27:411–420. doi: 10.1159/000327968. [DOI] [PubMed] [Google Scholar]
- 27.de Groot BL, Grubmüller H. Water permeation across biological membranes: Mechanism and dynamics of aquaporin-1 and GlpF. Science. 2001;294:2353–2357. doi: 10.1126/science.1066115. [DOI] [PubMed] [Google Scholar]
- 28.Savage DF, O’Connell JD, 3rd, Miercke LJ, Finer-Moore J, Stroud RM. Structural context shapes the aquaporin selectivity filter. Proc Natl Acad Sci USA. 2010;107:17164–17169. doi: 10.1073/pnas.1009864107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sui H, Han BG, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- 30.Gupta AB, et al. MIPModDB: A central resource for the superfamily of major intrinsic proteins. Nucleic Acids Res. 2012;40(Database issue):D362–D369. doi: 10.1093/nar/gkr914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fenn K, Matthews KR. The cell biology of Trypanosoma brucei differentiation. Curr Opin Microbiol. 2007;10:539–546. doi: 10.1016/j.mib.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Field MC, Carrington M. The trypanosome flagellar pocket. Nat Rev Microbiol. 2009;7:775–786. doi: 10.1038/nrmicro2221. [DOI] [PubMed] [Google Scholar]
- 33.Chung WL, Carrington M, Field MC. Cytoplasmic targeting signals in transmembrane invariant surface glycoproteins of trypanosomes. J Biol Chem. 2004;279:54887–54895. doi: 10.1074/jbc.M409311200. [DOI] [PubMed] [Google Scholar]
- 34.Teka IA, et al. The diamidine diminazene aceturate is a substrate for the high-affinity pentamidine transporter: Implications for the development of high resistance levels in trypanosomes. Mol Pharmacol. 2011;80:110–116. doi: 10.1124/mol.111.071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gruenberg J, Schwendimann B, Sharma PR, Deshusses J. Role of glycerol permeation in the bloodstream form of Trypanosoma brucei. J Protozool. 1980;27:484–491. doi: 10.1111/j.1550-7408.1980.tb05404.x. [DOI] [PubMed] [Google Scholar]
- 36.Tsukaguchi H, Weremowicz S, Morton CC, Hediger MA. Functional and molecular characterization of the human neutral solute channel aquaporin-9. Am J Physiol. 1999;277:F685–F696. doi: 10.1152/ajprenal.1999.277.5.F685. [DOI] [PubMed] [Google Scholar]
- 37.Figarella K, et al. Biochemical characterization of Leishmania major aquaglyceroporin LmAQP1: Possible role in volume regulation and osmotaxis. Mol Microbiol. 2007;65:1006–1017. doi: 10.1111/j.1365-2958.2007.05845.x. [DOI] [PubMed] [Google Scholar]
- 38.Stewart ML, et al. Detection of arsenical drug resistance in Trypanosoma brucei with a simple fluorescence test. Lancet. 2005;366:486–487. doi: 10.1016/S0140-6736(05)66793-1. [DOI] [PubMed] [Google Scholar]
- 39.Beitz E, Wu B, Holm LM, Schultz JE, Zeuthen T. Point mutations in the aromatic/arginine region in aquaporin 1 allow passage of urea, glycerol, ammonia, and protons. Proc Natl Acad Sci USA. 2006;103:269–274. doi: 10.1073/pnas.0507225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, et al. Enhancement of proton conductance by mutations of the selectivity filter of aquaporin-1. J Mol Biol. 2011;407:607–620. doi: 10.1016/j.jmb.2011.01.036. [DOI] [PubMed] [Google Scholar]
- 41.Barrett MP. Veterinary link to drug resistance in human African trypanosomiasis? Lancet. 2001;358:603–604. doi: 10.1016/S0140-6736(01)05817-2. [DOI] [PubMed] [Google Scholar]
- 42.Stewart ML, et al. Multiple genetic mechanisms lead to loss of functional TbAT1 expression in drug-resistant trypanosomes. Eukaryot Cell. 2010;9:336–343. doi: 10.1128/EC.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berger BJ, Carter NS, Fairlamb AH. Characterisation of pentamidine-resistant Trypanosoma brucei brucei. Mol Biochem Parasitol. 1995;69:289–298. doi: 10.1016/0166-6851(94)00215-9. [DOI] [PubMed] [Google Scholar]
- 44.Matovu E, et al. Genetic variants of the TbAT1 adenosine transporter from African trypanosomes in relapse infections following melarsoprol therapy. Mol Biochem Parasitol. 2001;117:73–81. doi: 10.1016/s0166-6851(01)00332-2. [DOI] [PubMed] [Google Scholar]
- 45.Alsford S, Kawahara T, Glover L, Horn D. Tagging a T. brucei RRNA locus improves stable transfection efficiency and circumvents inducible expression position effects. Mol Biochem Parasitol. 2005;144:142–148. doi: 10.1016/j.molbiopara.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alsford S, Horn D. Single-locus targeting constructs for reliable regulated RNAi and transgene expression in Trypanosoma brucei. Mol Biochem Parasitol. 2008;161:76–79. doi: 10.1016/j.molbiopara.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Overath P, Czichos J, Haas C. The effect of citrate/cis-aconitate on oxidative metabolism during transformation of Trypanosoma brucei. Eur J Biochem. 1986;160:175–182. doi: 10.1111/j.1432-1033.1986.tb09955.x. [DOI] [PubMed] [Google Scholar]
- 48.Baker N, Alsford S, Horn D. Genome-wide RNAi screens in African trypanosomes identify the nifurtimox activator NTR and the eflornithine transporter AAT6. Mol Biochem Parasitol. 2011;176:55–57. doi: 10.1016/j.molbiopara.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Räz B, Iten M, Grether-Bühler Y, Kaminsky R, Brun R. The Alamar Blue assay to determine drug sensitivity of African trypanosomes (T.b. rhodesiense and T.b. gambiense) in vitro. Acta Trop. 1997;68:139–147. doi: 10.1016/s0001-706x(97)00079-x. [DOI] [PubMed] [Google Scholar]
- 50.Hanrahan O, et al. The glycosylphosphatidylinositol-PLC in Trypanosoma brucei forms a linear array on the exterior of the flagellar membrane before and after activation. PLoS Pathog. 2009;5:e1000468. doi: 10.1371/journal.ppat.1000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.