Abstract
Nrf2 is a transcription factor that has emerged as the cell's main defense mechanism against many harmful environmental toxicants and carcinogens. Nrf2 is negatively regulated by Keap1, a substrate adaptor protein for the Cullin3 (Cul3)-containing E3-ligase complex, which targets Nrf2 for ubiquitination and degradation by the ubiquitin proteasome system (UPS). Recent evidence suggests that constitutive activation of Nrf2, due to mutations in Keap1 or Nrf2, is prominent in many cancer types and contributes to chemoresistance. Regulation of Nrf2 by the Cul3–Keap1-E3 ligase provides strong evidence that tight regulation of Cullin-ring ligases (CRLs) is imperative to maintain cellular homeostasis. There are seven known Cullin proteins that form various CRL complexes. They are regulated by neddylation/deneddylation, ubiquitination/deubiquitination, CAND1-assisted complex assembly/disassembly, and subunit dimerization. In this review, we will discuss the regulation of each CRL using the Cul3–Keap1-E3 ligase complex as the primary focus. The substrates of CRLs are involved in many signaling pathways. Therefore, deregulation of CRLs affects several cellular processes, including cell cycle arrest, DNA repair, cell proliferation, senescence, and death, which may lead to many human diseases, including cancer. This makes CRLs a promising target for novel cancer drug therapies. Antioxid. Redox Signal. 13, 1699–1712.
Introduction
Nrf2 (NF-E2-related factor 2) is a transcription factor that has emerged as the cell's main defense mechanism against many harmful environmental toxicants and carcinogens. The main function of Nrf2 is to activate the antioxidant response and induce transcription of a wide array of genes that are able to combat the harmful effects of oxidative stress, thus restoring intracellular homeostasis. These genes include: (i) intracellular redox-balancing proteins [glutamate cysteine ligase (GCL) and heme oxygenase-1 (HMOX-1)]; (ii) phase II detoxifying enzymes [glutathione S-transferase (GST) and NAD(P)H quinine oxidoreductase-1 (NQO1)]; and (iii) transporters (multidrug resistance-associated proteins, MRPs) (26, 27, 40, 46, 59). The promoter regions of Nrf2 target genes contain a specific DNA sequence, called the antioxidant response element (ARE), that is required for Nrf2 binding and gene induction. Other Nrf2 downstream genes play a role in a wide variety of functions such as the inflammatory response, cell growth and apoptosis, DNA repair, and the ubiquitin-mediated degradation pathway (99) The diverse nature of Nrf2 downstream genes demonstrates its vital importance in cell survival and protection. The Nrf2 antioxidant response has been shown to protect against cancer, neurodegenerative diseases, aging, diabetes, photo-oxidative stress, cardiovascular disease, inflammation, pulmonary fibrosis, and acute pulmonary injury (35, 40, 46, 59, 99).
Since its discovery, Nrf2 has been regarded as a regulator of the cell survival response, and as a result it has been found to not only promote survival of normal cells, but also cancer cells. Overexpression of Nrf2 in cancer cells creates an environment conducive for cell growth and protection against oxidative stress and chemotherapeutic agents. This phenomenon has recently been termed “the dark side of Nrf2.” Yamamoto and Biswal's groups have found that Nrf2 is constitutively activated in lung tumors and multiple cancer cell lines. Sequence analysis identified somatic mutations, with many of these mutations disrupting the Keap1(Kelch-like ECH-associated protein 1)-mediated negative regulation of Nrf2 (66, 80). Furthermore, we have shown that Nrf2 status correlates with chemoresistance. High levels of Nrf2 protect cancer cells from the effects of various chemotherapeutic drugs, whereas knockdown of Nrf2, transiently or stably, increases the sensitivity of cancer cells to chemotherapeutic-induced cell death. Moreover, we have shown that Nrf2 is overexpressed at later stages of lung cancer and type II endometrial cancer (89). This discovery has set a new paradigm for treating cancer and has opened up a broad spectrum of research that needs to be conducted in order to translate this research from the bench-top to the clinic. This research includes identifying Nrf2 inhibitors that can be used as chemosensitizers. Additionally, targeting the E3 ubiquitin ligase may prove to be another means of altering Nrf2 expression to combat chemoresistance. The ubiquitin proteasome system (UPS) has been an up and coming target for cancer therapy and has already proved to work in the clinic. This issue will be discussed in greater detail in this review.
Due to the profound effects of Nrf2 on cell survival, tight regulation of the Nrf2-mediated antioxidant response is very important. For over a decade, the regulation of Nrf2 has been investigated extensively and a huge milestone was reached when Keap1 was identified as the key negative regulator of Nrf2 (32). Under unstressed conditions, Nrf2 is regulated by the Cul3-Keap1-E3 ligase and the UPS to maintain low basal levels of Nrf2. Cullin 3 (Cul3) serves as a scaffolding protein that is bound to both Rbx1 and Keap1, which is the substrate adaptor protein that binds to Nrf2 (Fig. 1). Conversely, under stressed conditions, the ability of the Cul3-Keap1-E3 ligase to ubiquitinate Nrf2 is inhibited, allowing Nrf2 to translocate to the nucleus, and initiate the antioxidant response (43, 59, 99, 100).
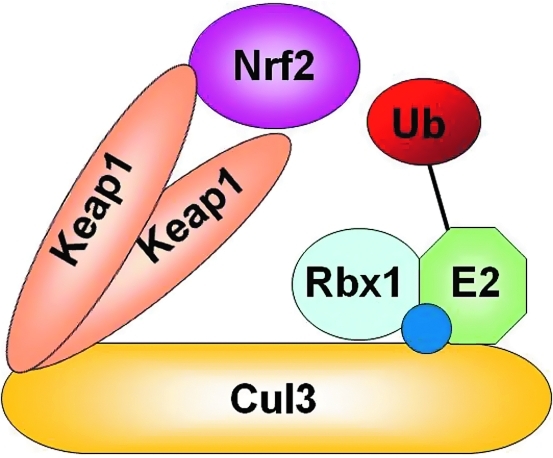
FIG. 1.
A schematic of the Cul3-Keap1-E3 ligase. Cul3 serves as a scaffolding protein that binds the Ub-loaded E2, Rbx1, and Keap1 proteins. Following Nedd8 conjugation (blue circle), a Keap1 homodimer functions as a substrate adaptor to recruit Nrf2 to the Cul3–Keap1–E3 ligase complex for ubiquitination.
There are two main types of E3 ligases, those containing a HECT (Homologous to E6-AP C-Terminus) domain and those containing a RING (Really Interesting New Gene) domain. For the scope of this review, HECT E3s will not be discussed. There are numerous Cullin-ring ligases (CRLs) composed of different Cullin and substrate adaptor proteins (Fig. 2). Their primary function is to regulate the stability of their substrates in a 26S proteasome-dependent manner. The regulation of Nrf2 by the Cul3-Keap1-E3 ligase is a prime example of how CRLs regulate their substrates.
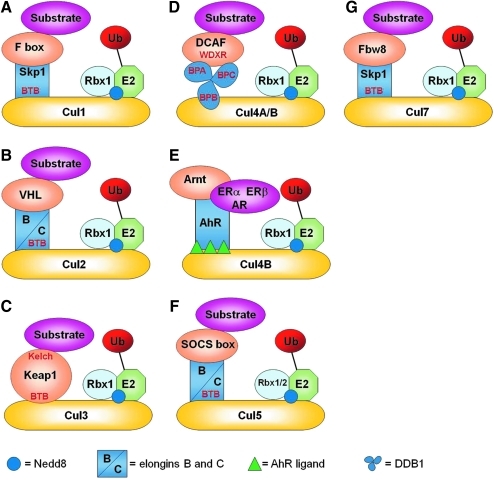
FIG. 2.
A general schematic of the seven Cullin-ring ligases. (A) Cul1, (B) Cul2, (C) Cul3, (D) Cul4A/B = Cul4A and Cul4B, (E) Cul4B, (F) Cul5, and (G) Cul7 Cullin-ring ligases. Words listed in red are all domain names. BPA, BPB, and BPC are the three propeller domains of DDB1.
Ubiquitin (Ub) is a small protein (76 amino acids) that is highly conserved between species. Ubiquitin can modify proteins through an isopeptide bond between the C-terminal glycine of Ub and a lysine residue on the substrate protein. Through a series of thioesterification reactions, Ub is activated and then transferred by the E3 ligase to a lysine residue on the substrate protein. The addition of one molecule of ubiquitin to the substrate is called mono-ubiquitination. Polyubiquitin (poly-Ub) chains can also be generated by repeating this process and adding one ubiquitin onto the previously linked Ub (29). Ubiquitin contains seven lysine residues, including Lys-48 and Lys-63 that are all able to accept Ub to form different poly-Ub linked chains. Proteins conjugated with a poly-Ub Lys-48 linked chain are mainly targeted to the 26S proteasome for degradation; however, those conjugated with a poly-Ub Lys-63 linked chain have been shown to mediate signal transduction and DNA repair. For a comprehensive review please see Reference (50). The ability of CRLs to covalently modify proteins, thus, altering their abundance and function, makes them very important intracellular complexes. CRLs control a variety of cellular processes, such as transcription, cell signaling, and cell cycle progression. Additionally, CRLs regulate biological processes that include immunity, development, glucose sensing, and circadian rhythms (69). This review will discuss in greater detail how Nrf2 is regulated by the Cul3-Keap1-E3 ligase and regulation of IκBα and Hif-1α (hypoxia inducible factor-1α) by their respective CRLs, Cul1-SCF and Cul2-VHL. Furthermore, we will briefly discuss additional CRLs containing the other four Cullin proteins, how CRLs are regulated, and the future in targeting the ubiquitin proteasome system for cancer therapy.
Cullins
Nrf2 regulation by the Cul3–Keap1–E3 ligase complex
Nrf2 is negatively regulated by Keap1, a substrate adaptor for the Cul3–Keap1–E3 ubiquitin ligase complex (Fig. 1). Under basal conditions, the Cul3–Keap1–E3 ligase catalyzes the addition of a Lys-48 linked poly-Ub chain onto Nrf2 and thereby targets it for proteasome-mediated degradation, maintaining low basal levels of Nrf2 (43, 59, 99, 100). Under oxidative stressed conditions, the ability of the E3 ubiquitin ligase complex to ubiquitinate Nrf2 is suppressed and Nrf2 degradation is inhibited, leading to increased protein stability and activation of the antioxidant response. In response to Nrf2 inducers, the activity of the E3 ubiquitin ligase complex is inhibited due to chemical modification of cysteine residues, particularly Cys-151, on Keap1. This modification is thought to alter the conformation of the Cul3–Keap1–E3 ligase, thus impairing the correct assembly of Nrf2 into the complex, which is required for Nrf2 ubiquitination (42, 59, 66, 100). Furthermore, prolonged oxidative stress may result in other modifications of Keap1 that expose lysine residues within Keap1, which then become targets for subsequent ubiquitination (101). Unlike Nrf2, ubiquitinated Keap1 is not targeted to the 26S proteasome for degradation due to the difference in ubiquitin linkage. Nrf2 polyubiquitination is Lys-48 linked, whereas in Keap1, poly-Ub is linked through Lys-63 (101). The function of Lys-63 polyubiquitinated Keap1 still remains elusive. Conceivably, this switch from substrate (Nrf2) to substrate adaptor (Keap1) ubiquitination may be a general mechanism for controlling steady-state levels of CRL substrate proteins. Furthermore, substrate adaptor proteins are known to undergo auto-ubiquitination, destabilizing the CRL complex (92, 104). Therefore, Keap1 auto-ubiquitination may be necessary in order to adequately induce Nrf2 to a level that is sufficient to sustain the antioxidant response over time.
Deregulation of Nrf2 by the Cul3–Keap1–E3 ligase complex in cancer
In normal cells Nrf2 is under constant regulation by the Cul3–Keap1–E3 ligase complex. However, in numerous cancer cell lines this regulation is lost, leading to constitutively active Nrf2, implicating the Nrf2 pathway in cancer promotion. Deregulation can be a result of mutations in Keap1 or in Nrf2. Keap1 mutations or loss of heterozygosity have been found in multiple lung cancer tissues and cell lines, causing inactivation or decreased expression of Keap1. As a result, tight regulation of the Nrf2 pathway becomes disrupted, leading to an increase in Nrf2 expression and activation of its downstream genes (66, 80, 81). Mutations in Keap1 have also been identified in breast cancer cell lines and gall bladder cancer (78), as well as in human patients with lung adenocarcinoma (63). Taken together, these studies imply that Keap1 might function as a tumor suppressor, as loss of function is associated with many types of cancer. More recently, somatic mutations in the coding region of Nrf2 have been identified in multiple human cancers. All mutations were determined to be missense amino acid substitutions and were observed more frequently in patients with squamous cell carcinoma (SCC) or patients with a history of smoking. No synonymous somatic alterations were detected. More specifically, the mutations were altering the amino acids involved in the two-site substrate recognition/hinge and latch model, a model for Nrf2 regulation. In this model, two amino terminal motifs in Nrf2, DLG and ETGE, bind a Keap1 homodimer that positions Nrf2 in the proper orientation to accept Ub, leading to subsequent Nrf2 degradation (56, 86, 87). As a result, these mutations led to constitutively high levels of Nrf2, increased Nrf2 nuclear localization, and increased transcriptional activation of Nrf2 downstream genes (79). Another recent study investigated mutations in exon 2 of the Nrf2 gene, which contains the DLG and ETGE motifs, in a broad range of human cancers. Kim et al. (41) analyzed 1145 cancer carcinoma tissues from skin, lung, esophagus, larynx, breast, colon, stomach, liver, kidney, prostate, urinary bladder, ovary, uterine, cervix, meningiomas, multiple myelomas, and acute leukemias. Their results identified Nrf2 mutations in esophageal SCC (8/70; 11.4%), skin SCC (1/17; 6.3%), broad spectrum lung cancers (10/125; 8.0%), and laryngeal SCC (3/23; 13.0%). Again, these mutations were found within or near the DLG and ETGE motifs and most likely play a role in disrupting the interaction between Nrf2 and Keap1, resulting in constitutively active Nrf2. It is interesting to note that 20/22 (90.9%) patients with Nrf2 mutations were previous or current smokers (41). Collectively, these results implicate Nrf2 in the development of SCC in lung and head/neck cancers, as well as in esophageal and skin cancer. Mounting evidence on the dark side of Nrf2 continues to emerge creating a sense of urgency to develop drugs that inhibit Nrf2 activity in order to enhance chemotherapeutic efficacy. Targeting Keap1 may prove to be a useful tool, thus understanding the mechanisms of Keap1-mediated regulation of Nrf2 is of vital importance.
Cul3–Keap1–E3 ligase regulates the NF-κB signaling pathway
Recently, IκB kinase β (IKKβ) was identified as another substrate for the Keap1–Cul3–E3 ligase complex (48). IKKβ contains an ETGE motif (NQE36TGE39) similar to that of Nrf2 (DEE79TGE82). IKKβ positively regulates the nuclear factor κB (NF-κB) pathway by targeting the negative inhibitory protein, IκBα, for degradation (Fig. 3). IKKβ phosphorylates two serine residues (Ser-32, Ser-36) in human IκBα, which targets it for proteasomal degradation by the SCF (Skp1, S-phase kinase-associated protein 1; Cullin1, Cul1; F-box protein) complex. This enables NF-κB to translocate to the nucleus and activate an array of downstream genes involved in many processes, including the immune response, inflammation, angiogenesis, cell proliferation, cell survival, and tumor invasion and metastasis (48). IKKβ also regulates other proteins involved in the NF-κB pathway (20, 25). Furthermore, IKKβ also regulates NF-κB-independent pathways involved in allergy, inflammation, and immunity by phosphorylating key proteins such as SNAP23 (synaptosomal-associated protein 23) and IRF7 (interferon regulatory factor 7), and also in cancer by phosphorylating proteins such as p53, TSC1 (tuberous sclerosis gene 1), and FOXO3a (forkhead transcription factor). For a more comprehensive list, please refer to Reference (5). The ability of IKKβ to regulate a wide array of processes exemplifies the importance of understanding the mechanisms of its regulation by the UPS. In this recent report, it was shown that knockdown of Keap1 stabilized IKKβ protein expression, which resulted in upregulation of NF-κB-derived tumor angiogenic factors. The Keap1-interacting domain was mapped to the Kelch domain of Keap1, and mutations in the ETGE motif in IKKβ had decreased binding to Keap1 and were resistant to Keap1-mediated ubiquitination and subsequent degradation (48). Although the biochemical data strongly demonstrates that IKKβ is an authentic substrate for the Cul3–Keap1–E3 ligase, the significance of IKKβ ubiquitination in activation of the NF-κB pathway is unclear since Keap1-mediated IKKβ ubiquitination was not affected in response to TNF-α (tumor necrosis factor-α) treatment, a known activator of the NF-κB pathway (48). In addition, many mutations that disrupt Cul3–Keap1–E3 ligase activity have been identified in 26 cancer cell lines and 119 primary tumors (48). However, no mutations were found in the NQE36TGE39 domain of IKKβ, implying that constitutive activation of Nrf2, due to impaired activity of the Cul3–Keap1–E3 ligase, may be the underlying mechanism in cancer promotion rather than activation of the NF-κB pathway due to excessive expression of IKKβ.
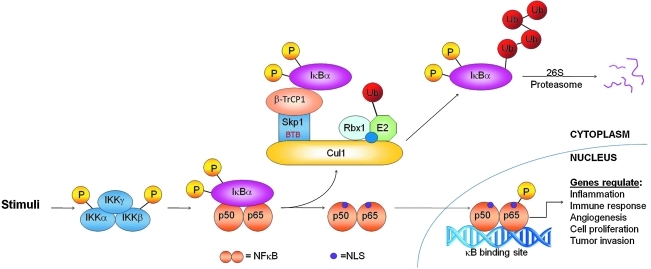
FIG. 3.
Activation of the NF-κB signaling pathway. Various stimuli can lead to phosphorylation and activation the IKK complex composed of three subunits, IKKα, IKKβ, and IKKγ. Upon activation, IKKβ phosphorylates two serine residues in IκBα that targets it for degradation by the Cul1-β-TrCP1–E3 ligase complex. As a result, the NF-κB complex (p50 and p65) is released, exposing a nuclear localization signal (NLS) that allows the NF-κB complex to translocate to the nucleus and bind to κB sites in the promoters of downstream genes to induce their transcription.
The role of other Cullin proteins
The human genome encodes at least seven Cullins including Cul1, Cul2, Cul3, Cul4A, Cul4B, Cul5, and Cul7 (Fig. 2). The Cul1-containing SCF complex represents another well-characterized E3-ligase consisting of Skp1, Cul1, and an F-box protein (Fig. 2A). The F-box protein, which serves as the substrate-recognition component, binds Skp1 via its F-box interacting motif. Skp1 is the linker protein which connects the substrate adaptor F-box protein to Cul1. Most F-box proteins recognize their substrates through various interaction motifs, such as WD40 motifs or Leucine Rich Repeats (LRR). A well-characterized F-box protein, β-TrCP1 (β-transducin repeat-containing protein, also known as Fbw1a or FWD1), functions as a substrate adaptor protein for the SCF-E3-ligase complex to further regulate the NF-κB signaling pathway. Upon IKKβ-mediated phosphorylation of IκBα (S32, S36), the Cul1-β-TrCP1-E3 ligase complex targets IκBα for ubiquitination and subsequent degradation by the 26S proteasome (98). Constitutive activation of the NF-κB pathway in cancer cells often occurs due to the oncogenic activation of IKKβ (as described in the previous section); however, an increase in Cul1-β-TrCP1-E3 ligase activity may also contribute to enhanced NF-κB signaling (19). Furthermore, the Cul1-β-TrCP1-E3 ligase complex also regulates the stability of β-catenin (19), an important transcription factor for the Wnt signaling pathway. Similarly to IκBα, β-catenin requires phosphorylation of two serine residues contained in a similar motif (DSGXXS) before it can be recognized by β-TrCP1 and targeted for proteolysis (54, 98). In the absence of Wnt signaling, β-catenin forms a complex with glycogen synthase kinase 3β (GSK3β), casein kinase 1α (CK1α), Axin and adenomatous polyposis coli (APC), resulting in β-catenin phosphorylation, which targets it for ubiquitination and degradation (Fig. 4). β-catenin is phosphorylated first by CK1α (Ser-45) followed by GSK3β-mediated phosphorylation (Thr-41, Ser-37, Ser33). In response to activators of Wnt signaling, GSK3β-mediated phosphorylation of β-catenin is inhibited, thus preventing β-catenin ubiquitination and degradation. APC is a tumor suppressor gene that is mutated in 80% of all human colorectal cancers (45) and it regulates β-catenin localization, phosphorylation, and ubiquitination (51). APC binds β-catenin and Axin and is necessary for β-catenin phosphorylation and ubiquitination (2, 51). Mutations in APC result in deregulation of β-catenin and excessive expression of β-catenin, which is thought to initiate colorectal neoplasia. High levels of β-catenin are found in colorectal cancer, hepatocellular carcinomas, and malignant melanomas (1). Furthermore, it was shown that β-TrCP1 is not absolutely required for IκBα and β-catenin degradation, suggesting that other E3-ligases play a role in regulating the NF-κB and Wnt signaling pathways (60). In fact, the Cul3–Keap1–E3 ligase and the Siah-SIP-Skp1-Ebi-E3 ligase, have been shown to regulate the NF-κB pathway and β-catenin degradation, respectively (48, 55).
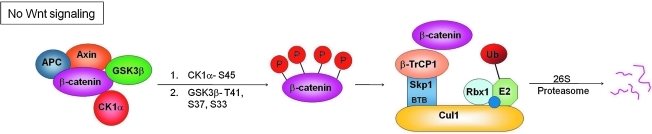
FIG. 4.
Regulation of β-catenin by the Cul1–Skp1–E3 ligase complex. In the absence of Wnt signaling, β-catenin forms a complex with GSK3β, casein kinase 1α (CK1α), Axin, and adenomatous polyposis coli (APC), resulting in CK1α-mediated phosphorylation of β-catenin (S45), followed by GSK3β-mediated phosphorylation of β-catenin at additional residues (T41, S37, S33). Phosphorylation of β-catenin allows it to be recognized by βTrCP1 and targeted for ubiquitination by the Cul1–Skp1–E3 ligase and subsequent degradation by the 26S proteasome.
The von Hippel–Lindau protein (VHL) forms another well-characterized E3 ligase by binding to elongins B and C, and Cul2, creating a multiprotein E3–ligase complex (Fig. 2B) (33, 67). Analogous to the role of β-TrCP1, VHL serves as a substrate adaptor protein which targets Hif-1α for degradation. Under normoxic conditions, proline 564 in Hif-1α becomes hydroxylated and Hif-1α is subsequently targeted to the Cul2-VHL-E3-ligase for degradation (34). Under hypoxic conditions, Hif-1α escapes VHL-mediated degradation and is able to induce transcription of hypoxia-inducible genes, such as vascular endothelial growth factor (VEGF), glucose transporter 1 (GLUT1), and erythropoietin (EPO), and promote vascularization and tumor growth (36). Von Hippel–Lindau disease is a result of inactivation of the VHL gene and is characterized by the VHL syndrome that predisposes individuals to many benign and malignant tumors, including sporadic renal clear cell carcinoma and CNS (central nervous system) hemangioblastomas. Tumors lacking functional VHL have high levels of hypoxia-inducible genes, including VEGF, resulting in hypervascularization (21, 31). To study the effect of VHL mutations, Iwai et al. expressed relevant cancer-related VHL mutants in a cell based model. They found that the VHL mutants were unable to ubiquitinate substrate proteins and this was independent of the VHL mutants forming a complex with elongins B and C, and Cul2 (33). These results suggest that the mutations disrupted the interaction between the substrate proteins and the Cul1–VHL–E3 ligase, providing mechanistic evidence as to how hypoxia-inducible genes are upregulated in patients lacking a functional VHL gene.
More recently characterized Cullins include Cul4A, Cul4B, Cul5, and Cul7. Cul4A and Cul4B share substrate adaptor proteins and have somewhat redundant functions (Fig. 2D). Substrates for Cul4–CRLs include the nucleotide excision repair proteins XPC (Xeroderma Pigmentosum group C) and CSB (Cockayne Syndrome group B), and c-jun (22, 83). Cul4 forms a complex with the linker protein, DDB1 (damage-specific DNA binding protein 1), which binds one or two WDXR motifs in DCAF (DDB1-Cul4-associated factor) substrate adaptors via its unique triple β-propellar configuration (BPA, BPB, BPC), each comprised of 7 WD40-like repeats (Fig. 2D). The BPB propeller is the motif required for the interaction between the linker protein, DDB1, and Cul4, representing a novel mechanism of Cullin-substrate adaptor binding (3). As shown in Figure 2, all other substrate adaptor proteins contain a bric-a-brac, tramtrack, broad complex (BTB) domain that is necessary for binding to their respective Cullin protein, except the Cul4B–AhR/Arnt–E3 ligase complex (Fig. 2E), where Ahr functions as an atypical component of the CRL. The Cul4–DDB1–DCAF complex regulates substrates involved in DNA repair and chromatin regulation (24, 49). Interestingly, the HIV1 viral protein R (Vpr) hijacks the Cul4 complex to arrest cell cycle and mediate Vpr cytostatic activity. Vpr binds DCAF1 and recruits the Cul4A–DDB1 CRL, resulting in degradation of atypical substrates required for the G2--M transition (47). In addition, a fat-soluble ligand-dependent CRL complex has recently been characterized, in which the dioxin receptor (AhR) and Arnt are components of a novel Cul4B–E3 ligase complex that requires the AhR ligand for complex assembly and catalytic activity. This Cul4B– AhR/Arnt–E3-ligase complex targets sex steroid receptors, including estrogen receptor-α (ERα), estrogen receptor-β (ERβ), and androgen receptor (AR), for degradation (Fig. 2E) (64). Similarly to Cul2, Cul5 uses elongins B and C as linker proteins to form a complex between a SOCS box-containing protein and the Cullin protein (Fig. 2F). The substrate adaptor protein determines whether Cul2 or Cul5 will be recruited based on its Cullin interacting motifs, suggesting that the substrate adaptor protein directly interacts with the Cullin protein. Cul5 has also been shown to interact with both Rbx1 and Rbx2 (37). Recently, the Cul5–E3 ligase has been shown to interact with the Hsp90 (heat shock protein 90) chaperone complex and ErbB2, an Hsp90 substrate. The Cul5–E3 ligase is recruited to the plasma membrane where it ubiquitinates ErbB2 receptors and targets them for degradation. Surprisingly, ErbB2 ubiquitination and degradation occurs in the absence of a SOCS box substrate adaptor protein and does not require elongins B and C, suggesting that Cul5 may directly interact with the Hsp90 chaperone complex to form a functional CRL complex. Hif-1α has also been shown to interact with Hsp90. Although Hif-1α is normally degraded by the Cul2–VHL–E3-ligase complex under normoxic conditions, it is also sensitive to geldanamycin (GA)-induced degradation which is independent of oxygen and the Cul2 complex. GA is a benzoquinone ansamysin antiobiotic, currently in human clinical trials as an anti-cancer treatment. GA-induced degradation of Hif-1α was found to be Cul5 dependent, suggesting that Cul5 may regulate multiple Hsp90 substrate proteins (17). Cul7 also serves as a scaffolding protein and binds the F-box protein, Fbw8, as well as Skp1 and Rbx1 to form a CRL complex (Fig. 2G). Unlike Cul1, which can bind Skp1 itself, Cul7 interacts with the Skp1–Fbw8 complex, not Skp1 alone (12). Interestingly, to date, only one F-box substrate adaptor protein has been identified for the Cul7-CRL. The Cul7–Fbw8–E3-ligase complex targets cyclin D1 and insulin receptor substrate-1 (IRS-1) for ubiquitination and degradation and is involved in growth regulation. Cul7 may also have multiple nonproteolytic functions. For a review, please see reference (75).
Interestingly, the majority of CRL substrates are regulated at the level of substrate-CRL binding by post-translational modification of the substrate proteins. Only modified substrates are recognized and targeted to the CRL complex. IκBα, β-catenin, Hif-1α, and cyclin D1 are a few examples. As discussed previously, under induced conditions IκBα is phosphorylated by IKKβ, enabling recognition by the SCF complex. In the absence of Wnt signaling, GSK3β phosphorylates β-catenin, which significantly enhances its binding affinity to the SCF complex (96). Under normoxic conditions, Hif-1α is hydroxylated on proline 564, which targets it to the Cul2–VHL–E3 ligase (34). Lastly, following phosphorylation of T286 by the Erk2 MAP kinase, cyclin D1 is targeted for ubiquitination by the Cul7–Fbw8–E3 ligase (65). However, regulation of Nrf2 by the Cul3–Keap1–E3-ligase occurs in a different manner. The substrate adaptor protein (Keap1) is modified, not the substrate (Nrf2) itself. Furthermore, modification of Keap1 results in activation of Nrf2. Under oxidative stressed or induced conditions, cysteine residues on Keap1 become modified, which may alter the conformation of the Cul3–Keap1–E3 ligase complex. As a result, Nrf2 binding is unfavorable for the addition of ubiquitin to the seven lysine-accepting residues, allowing free Nrf2 to translocate to the nucleus and activate the antioxidant response (99).
Cullin Regulation
Neddylation/Deneddylation
Despite the variance between the CRLs, they are all regulated by similar mechanisms. Neddylation, is the process by which Nedd8 (neural precursor cell expressed developmentally downregulated protein 8), a small ubiquitin-like protein, is conjugated onto a conserved lysine residue of proteins. Nedd8 and Ub have high sequence similarity (57% identical and 76% similar) and are very similar structurally. Nedd8 is conjugated onto the C-terminal end of Cullin proteins (human: K720-Cul1, K689-Cul2, K712-Cul3, K605-Cul4A, K859-Cul4B) (52, 88, 95). Neddylation occurs by a similar process as ubiquitination, involving an E1-like heterodimer (ULA1/UBA3), an E2 enzyme (UBC12), and an E3 ligase (DCN1 or SCCRO) (Fig. 5) (57). It is well known that neddylation activates CRL function. Using purified in vitro systems, several groups have shown that neddylated SCF complexes are more active than deneddylated-complexes, leading to increased ubiquitination of IκBα (74) or p27 (71), a cell cycle inhibitor. Neddylation was also shown to increase ligase activity in Cul2–VHL and Cul3–Keap1 E3 ligase complexes, leading to increased ubiquitination of HIF-1α and Nrf2, respectively (52, 62). It was originally thought that neddylation functions to recruit Ub-activated E2 to the Cullin protein (39), resulting in increased CRL activity; however, recent evidence suggests that the role of Nedd8 in the UPS may be more complex. In fact, neddylation promotes a conformational change in the Cullin protein that facilitates the recruitment of the activated E2 enzyme and releases the auto-inhibition of the C-terminal end of the Cullin protein. This allows Rbx1 to move freely, bringing the Ub-loaded E2 closer to the acceptor lysine residue of the substrate protein. Thus, Nedd8 not only stimulates the transfer of Ub from E2 onto the substrate, but also facilitates Ub chain elongation (57).
FIG. 5.
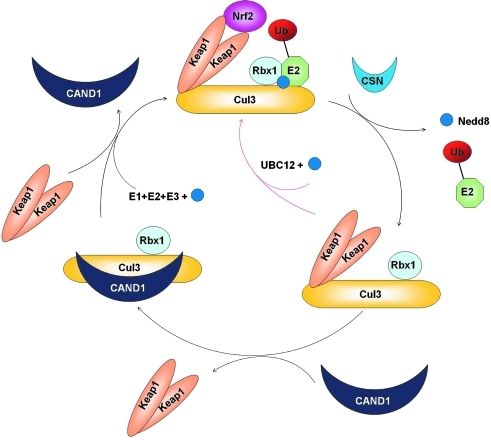
Regulation of the Cul3–Keap1–E3 ligase complex. The Cul3–Keap1–E3 ligase complex is active when it is in the neddylated state, which facilitates the docking of a Keap1 homodimer-bound to Nrf2 into the complex, resulting in ubiquitination and degradation of Nrf2. The complex is inactivated by deneddylation. The CSN complex removes Nedd8 from Cul3 enhancing the association of Cul3 and CAND1, which triggers dissociation of Keap1-Nrf2 from Cul3. Finally, neddylation can reactivate the E3 ligase complex. The complex can also be reactivated following deneddylation without disassembly of the complex. UBC12 can conjugate Nedd8 back onto Cul3 without dissociation of Keap1 from the Cul3–Rbx1 core complex (red arrow).
Neddylation of Cullins is reversible by deneddylation, which involves the COP9-signalosome (CSN), a large complex similar to that of the proteasome lid. The CSN is an evolutionary conserved eight-subunit complex (CSN1-8) responsible for removing Nedd8 from Cullin proteins. The eight subunits contain either a MPN (MPR-PAD1-Nterm) domain (CSN5 and CSN6) or a PCI (Proteasome; COP9-signalosome; elongation initiation factor 3, eIF-3) domain (CSN1-4 and CSN7-8). These domains are also found in the proteasome lid and the eIF3 translation initiation factor complex. Interestingly, each of the eight subunits of the CSN has an analogous counterpart in the proteasome lid that assembles into a similar ring structure (38). It is well known that CSN5 is the catalytic subunit of the CSN complex due to the metalloprotease activity of its JAMM (Jab1/MPN domain metalloenzyme) motif (16). Specific CSN5 mutations, that eliminate the isopeptidase activity of the CSN, led to an accumulation of Nedd8-Cul1 in S. pombe (11), A. thaliana (14, 23), and D. melanogaster (15), and accumulation of Nedd8-Cul3 in early C. elegans embryos (70). Along with CSN5, it is also believed that CSN2 is essential for deneddylation. Polyclonal antibodies raised against all the CSN subunits were tested for their ability to inhibit deneddylation. Only the polyclonal antibodies raised against CSN2 resulted in an accumulation of neddylated Cul1 and Cul2 in cytosolic or total HeLa cell extract (97).
The CSN is recruited to the E3–ligase complex and interacts directly with the Cullin protein and Rbx1, which is necessary for deneddylation (53, 70). Upon knockdown of Rbx1 (ZK287.5 in C. elegans) by RNAi, Pintard et al. observed an increase in Nedd8–Cul3 (70). Additionally, the CSN also functions to stabilize Rbx1. When cells were treated with siRNA directed against CSN1 or CSN3, Rbx1 was degraded at a faster rate than in control cells. Rbx1 was also degraded faster when CSN5 was knocked down using siRNA, but to a lesser extent than CSN1-siRNA or CSN3-siRNA (68). These results provide evidence that the CSN is not the sole protein necessary for deneddylation, but Rbx1 also plays an essential role. Furthermore, the CSN does not only function in deneddylation, but it is important for stabilizing Rbx1, a protein necessary for CRL activity.
Cycling
Since neddylation functions to activate CRL activity, it was speculated that deneddylation opposes this function. Surprisingly, it's been shown that both neddylation and deneddylation are required for CRL activity in vivo (9). Following is a model explaining the apparent paradox between these two opposing functions. The function of neddylation is to recruit ubiquitin-loaded E2 to the Cullin protein and promote substrate ubiquitination and Ub-chain elongation. Subsequently, deneddylation promotes the release of the empty E2, followed by another round of neddylation, Ub-loaded E2 recruitment, and substrate ubiquitination. Cycling between neddylation and deneddylation can be hindered by two mechanisms: (i) impairing neddylation and recruitment of Ub-E2 or (ii) inhibiting deneddylation through mutation of the CSN, thus locking empty-E2 in the E3–ligase complex. Both mechanisms inhibit the activity of the E3–ligase complex and prevent substrate ubiquitination (70).
Furthermore, whether neddylation and deneddylation regulate Cullin protein stability still remains unclear. Wu et al. (94) examined the effect of CSN complex disruption in Drosophila. Cells deficient in CSN activity resulted in increased levels of the neddylated form of the Cullin and enhanced CRL activity. However, Nedd8 conjugation to Cul1 or Cul3 rendered the protein unstable. For instance, in CSN5-knockout cells, there was an increase in neddylated Cul1 and Cul3 that resulted in decreased protein stability compared to control cells as demonstrated by a cycloheximide chase experiment. In a similar cycloheximide chase experiment, knockdown of Nedd8 inhibited Cul1 and Cul3 neddylation and stabilized the proteins (94). Conversely, deneddylation is required to remove Nedd8 and return the Cullin proteins back to a stable state. Another study confirmed that disruption of the CSN complex decreased Cul1 protein stability in Neurospora (28). On the other hand, a recent study determined that Nedd8 modification of Cullin proteins does not regulate their stability in mammalian cells. In this study, HEK293 cells were transfected with either dominant negative-UBC12 (dnUbc12), a Nedd8 conjugating enzyme, or with a Cul1 mutant (L720R), to inhibit Cul1 neddylation. When Cul1 protein levels were compared between the neddylation deficient conditions and wild-type conditions, no change in the steady state level of the Cul1 protein was observed. Similar results were seen when Cul2 and Cul3 proteins levels were examined under the same conditions (7). The discrepancy between these studies may be due to the difference in species examined. Further studies should be conducted to clarify this issue.
Cycling between neddylated and deneddylated states is also important to prevent substrate adaptor autoubiquitination. If the E3–ligase complex is in the neddylated state, it remains active. Therefore, for the CRL to switch to a different substrate two processes can occur, (i) substrate adaptor proteins can undergo autoubiquitination and autocatalytic destruction through the 26S proteasome, or (ii) the complex can cycle through assembly/disassembly. When there is no more substrate or the substrate no longer needs to be ubiquitinated, the substrate adaptor can undergo autocatalytic destruction, allowing the E3-complex to recruit a new substrate adaptor and substrate. Even though substrate adaptor degradation is important for targeting new substrates, it can also be detrimental for E3-ligase activity. It's been shown that CSN-dependent deneddylation of Cullins is necessary to prevent autoubiquitination and adaptor instability (10, 90). In fission yeast, various CSN mutants led to an increase in Pop1p (substrate adaptor protein in SCF-CRL) and Btb3p (substrate adaptor protein in Cul3-CRL) autoubiquitination and subsequent degradation (90). Preventing substrate adaptor degradation is important to maintain a functional E3-ligase complex capable of ubiquitinating newly recruited substrates without having to synthesize new proteins and reassemble the complex. Substrate adaptor instability is also regulated by the CSN associated deubiquitinating enzyme, Ubp12/USP15. Ubp12 has been shown to remove ubiquitin from CRL substrates or substrate adaptor proteins, including BTB containing proteins, and protect CRL components from cellular depletion by preventing autoubiquitination and subsequent degradation. Ubp12 has also been shown to stabilize the adapter proteins, Pop1p and Btb3p, through its deubiquitinating activity (103). Conversely, inhibition of Ubp12 activity led to an increase in Rum1p, a SCF-Pop1p substrate. However, new evidence suggests that the CSN complex and Ubp12 do not stabilize all substrate adaptor proteins in S. pombe. Only a subset of F-box proteins, which contain a critical proline residue in their F-box that enhances their interaction with Cul1-Skp1, required the CSN complex for stabilization. Moreover, none of the proline-containing F-box proteins required Ubp12 for stabilization, unlike the Cul3-E3 ligase substrate adaptor protein, Btb3p (76). Further research is required to determine whether this phenomenon occurs in all species. In conclusion, generally, the CSN along with Ubp12 act synergistically to protect the CRLs from degradation and maintain basal levels of CRL activity. These results demonstrate that cycling is essential to stabilize Cullin protein levels, prevent substrate adaptor autoubiquitination, and promote E3 ligase activity.
CAND1
The CRL not only cycles between neddylation and deneddylation states, but also cycles through assembly and disassembly of the ligase complex, which is dependent on the neddylation state of the Cullin protein. Cycling between neddylation/deneddylation increases the activity of the E3-ligase. Conversely, the activity of the E3-ligase is inhibited by a two step process involving disassembly of the complex. The first step involves removal of Nedd8 from the Cullin protein. The second step is the association of the protein, CAND1 (cullin-associated and neddylation-dissociated 1), with the deneddylated Cullin complex. CAND1 preferentially binds deneddylated-Cullin/Rbx1 complexes, preventing their assembly into new E3-ligase complexes. CAND1 binds directly to both ends of the Cullin, blocking the binding of substrate adaptor proteins at the N-terminal end and the NEDD8-lysine accepting residue at the C-terminal end (Fig. 5) (102). Blockage of substrate adaptor binding and neddylation of Cullin proteins has been shown to inhibit ubiquitination of SCF substrates, IκBα and p27, and stabilize them in vitro. Interestingly, CAND1 mutants have also been shown to stabilize SCF substrates in vivo (6, 8, 18). Furthermore, both overexpression and siRNA-mediated knockdown of CAND1 decreased the ability of the Cul3–Keap1 E3 ubiquitin ligase to ubiquitinate and target Nrf2 for degradation resulting in Nrf2 stabilization (52). This paradox is similar to that seen with the CSN/deneddylation in vivo, indicating a requirement for CAND1 in mediating complex assembly and disassembly. However, the role of CAND1 is more complicated. CAND1 does not bind all Cullin proteins and does not bind the same Cullin proteins in all cell types, indicating the role of CAND1 may only be essential under certain physiological conditions and be cell type dependent (57). It has also been shown that F-box proteins, substrate adaptors for the SCF complex, are destabilized when they are incorporated into the E3 ligase complex. However, binding of CAND1 to the Cullin ligase complex causes them to be displaced and leads to their stabilization (92, 104). The function of CAND1 in stabilizing substrate adaptor proteins has held true until recently. Schmidt et al. have discovered a new role for CAND1. Contrary to the current model, they discovered that CAND1 is not required to maintain the stability of CSN-regulated F-box proteins in S. pombe. Instead, CAND1 maintains the balance of Cul1-E3 ligases, allowing less abundant F-box proteins to bind Cul1 and not be out-competed by higher abundant substrate adaptor proteins (76).
Dimerization
It is well established that Keap1 functions as a homodimer to regulate Nrf2. Nrf2 contains two binding sites for Keap1 in its Neh2 domain, a weak binding site (DLG) and a stronger binding site (ETGE). The Keap1-homodimer recognizes and binds both motifs, positioning the seven ubiquitin-accepting lysine residues that are contained between these two motifs in a favorable position for polyubiquitination. This two-site substrate recognition model is also known as the “hinge-latch model.” The high affinity ETGE motif functions as the “hinge” and the lower affinity DLG motif functions as the “latch”. Under oxidative stressed or inducible conditions, cysteine residues in Keap1 become modified, which may alter the structural confirmation and “unlatch” the weak-binding DLG motif from Keap1 (56, 86, 87). Both the DLG and the ETGE motifs are necessary for Nrf2 ubiquitination. Mutation or deletion of the DLG or the ETGE domains in Nrf2 (79, 80, 87) or disruption of the BTB domain in Keap1, required for its homodimerization (105), inhibited Keap1-mediated Nrf2 ubiquitination. Along with Keap1, many other substrate adaptor proteins have been shown to dimerize, including human (β-TrCP1, β-TrCP2, Skp2), budding yeast (Cdc4p and Met30p), and fission yeast (Pop1p and Pop2p) F-box proteins, as well as the BTB domain-containing protein, promyelocytic leukemia zinc finger (PLZF) (13, 44, 77, 85, 93). Skp2, a substrate adaptor protein for the SCF-E3 ligase, also homodimerizes; however, this interaction is independent of its F-box domain, indicating that Skp2 dimerization is independent of its direct binding to Skp1. Similarly, other F-box proteins, including β-TrCP, Pop1p, and Pop2p, do not rely on their F-box domain for dimerization. Instead, homodimerization is mediated by the N-terminal domain (7).
More recently, there is evidence to suggest Cullin dimerization. Cul1, Cul3, and Cul4a, but not Cul2 or Cul5, have been shown to homodimerize. It has been shown that a Cul3 protein with a mutated BTB binding domain (S53A/F54A/E55A) that prevents Keap1 binding was no longer capable of dimerization when an immunoprecipitation analysis was conducted between wild type Cul3-FLAG and the Cul3-S53A/F54A/E55A mutant protein. These results imply that the BTB binding domain is indispensable for this Cul3–Cul3 interaction and Cul3 dimerization depends on substrate adaptor dimerization (7). Unlike Cul3, a Cul1 mutant (Y42A/M43A/E44A), incapable of binding Skp1 and Skp2, was still able to dimerize indicating that the Cul1–Cul1 interaction is independent of substrate adaptor dimerization (7). Conversely, another group concluded that the BTB binding domain is unnecessary for Cul3 dimerization since the interaction between two Cul3-L52A/E55A mutants, FLAG- or Myc-tagged that are unable to bind any BTB domain-containing protein, was confirmed using immunoprecipitation analysis in HEK293 cells (91). The discrepancy between these two studies is not clear. In short, Skp2 and Cul1 homodimerize independently of each other, whereas Cul3 dimerization may be dependent on Keap1 homodimerization through its BTB domain. Further studies need to be conducted to clarify these results.
In summary, the Cul3–Keap1–E3 ligase complex provides an ideal example of CRL regulation. The Cul3–Keap1–E3 ligase complex is tightly regulated by neddylation of Cul3 at lysine-712, the CSN complex (deneddylation), CAND1, and UBC12 (a Nedd8 conjugating enzyme) (Fig. 5) (52, 58, 99, 102). It was proposed that the E3 ligase complex undergoes dynamic cycles of assembly and disassembly. When the ligase complex is in the active state, Cul3 is neddylated which facilitates the docking of a Keap1 homodimer-bound to Nrf2 into the complex, resulting in ubiquitination and degradation of Nrf2. On the other hand, the CSN complex removes Nedd8 from Cul3 enhancing the association of Cul3 and CAND1, which triggers dissociation of Keap1 from Cul3. Thus, deneddylated Cul3 is the inactive form. The precise regulation of the assembly/disassembly cycle is crucial in controlling the E3 ligase activity and thus the stability of Nrf2, as both overexpression and knockdown of CAND1 have been shown to decrease the activity of the Cul3–Keap1–E3 ligase, resulting in stabilization of Nrf2 (52). Furthermore, the complex can be reactivated following deneddylation, without disassembling as a result of CAND1 binding. This can occur if UBC12 conjugates Nedd8 back onto Cul3 without disassociation of Keap1 from the Cul3-Rbx1 core complex (Fig. 5, red arrow). This allows for repetitive cycles of ubiquitination without having to reassemble the complex. Lastly, neddylation re-activates the E3 ligase complex, allowing Nrf2 to be recruited for ubiquitination.
Future Direction
The ubiquitin proteasome system and its implication in cancer
The UPS tightly regulates many signaling pathways, thus affecting many cellular processes including cell cycle arrest, DNA repair, cell proliferation, senescence, and death. Consequently, disruption of the UPS can severely affect these processes which may lead to many human diseases, including cancer. Many E3 ligases have been linked to the development of cancer, based on their ability to target oncogenes or tumor suppressor genes. For example, Mdm2 (murine double minute 2) and Skp2-SCF are E3 ligases that negatively regulate p53, a tumor suppressor protein, and p27, respectively. Moreover, genes involved in vascularization, a hallmark of cancer, are regulated by the UPS (30). For example, the Cul2–VHL–E3 ligase regulates the stability of Hif-1α in an oxygen-dependent manner. Subsequently, Hif-1α regulates a battery of downstream genes involved in vascularization, including VEGF. As a result, tumor cells deficient in, or containing mutated VHL, have high levels of these downstream genes even under normoxic conditions enhancing vascularization and tumor growth (34, 36). As discussed earlier, mutations in Keap1 or Nrf2 can result in deregulation of the antioxidant response pathway. When Nrf2 is not under the negative control of Keap1, it remains constitutively active and creates an environment conducive for cancer cell growth and is also responsible for chemoresistance (41, 63, 78, 79). These are just a few examples of how the UPS can be involved in cancer development and progression.
Targeting the UPS in cancer therapy
The UPS is a promising drug target for cancer therapy. In fact, Bortezomib, a highly selective and reversible proteasome inhibitor, is currently used as an anticancer drug to treat patients with refractory multiple myeloma and mantle cell lymphoma (4). The fact that many E3-ligases regulate proteins involved in cancer development and progression demonstrates the importance of the UPS as a drug target. p53 and p27 are both negatively regulated by their respective E3-ligases, Mdm2 and Skp2-SCF. Therefore, disrupting the activity of the E3-ligases seems like a promising drug target, which would result in increased levels of p53 and p27 causing cell cycle arrest and apoptosis. This could be accomplished in several ways, (i) inhibit the expression of Mdm2 or Skp2, (ii) block ubiquitin ligase activity, or (iii) prevent the interaction between the E3-ligase and the substrate protein. Current research targeting the UPS system to improve human diseases has been focused on the ubiquitin ligases, since they bind to the target protein, and thus confer substrate specificity. However, in some cases, inhibiting E3-ligase activity may be detrimental. For example, it is undesirable to inhibit E3-ligases that target oncogenes for degradation. Instead, one would want to develop agonists that promote the activity of the ligase (30).
There have been several set-backs in translating these approaches into successful therapies. The small-molecule inhibitors developed have either (i) low potency, (ii) severe off-target effects, (iii) do not work when the target is mutated, (iv) affect other substrates from binding the E3-ligase, or (v) have low bioavailability. Another important thing to consider is that some of the E3-ligases target multiple substrates, such as tumor suppressor genes and oncogenes, which makes developing these types of target specific drugs extremely difficult (30).
Deubiquitinating enzymes (DUBs) have also become recent targets for cancer therapy. USP1, USP28, and USP44 are of most importance because through their deubiquitinating activities they modulate substrates that regulate processes relevant to cancer including, modulation of DNA-repair checkpoints, stabilization of cyclin E1 and c-Myc, and preventing premature silencing of the spindle checkpoint, respectively (61, 72, 73, 82). Another cancer related DUB, CYLD (cylindromatosis), regulates the NF-κB pathway and is a tumor suppressor gene that is mutated in skin tumors and also plays a role in kidney, liver, and cervical cancers (84). This demonstrates the importance of having tightly regulated control mechanisms for the UPS. Other aspects of the UPS that are being investigated as drug targets for cancer therapy include targeting (i) the early steps in the pathway like the E1 and E2 enzymes that activate Ub, (ii) the interaction between the substrate adaptor and substrates, (iii) the ubiquitin-binding domains of specific effector proteins, and (iv) inhibiting the catalytic activity of the proteasome (30).
Abbreviations Used
- APC
adenomatous polyposis coli
- AR
androgen receptor
- ARE
antioxidant response element
- BTB
bric-a-brac, tramtrack, broad complex
- β-TrCP1
β-transducin repeat-containing protein
- CAND1
Cullin-associated and neddylation-dissociated 1
- CK1α
casein kinase 1α
- CNS
central nervous system
- CRL
Cullin ring ligase
- CSB
cockayne syndrome group B
- CSN
COP9-signalosome
- Cul
Cullin
- CYLD
cylindromatosis
- DCAF
DDB1-Cul4-associated factor
- DDB1
damage-specific DNA binding protein 1
- Dn
dominant negative
- DUB
deubiquitinating enzyme
- EPO
erythropoietin
- ERα
estrogen receptor α
- ERβ
estrogen receptor β
- GA
geldanamycin
- GCL
glutamate cysteine ligase
- GLUT1
glucose transporter 1
- GSK3β
glycogen synthase kinase 3β
- HECT
homologous to E6-AP C-terminus
- Hif-1α
hypoxia inducible factor-1α
- HMOX-1
heme oxygenase-1
- Hsp90
heat shock protein 90
- IKKβ
IκB kinase β
- IRF7
interferon regulatory factor 7
- IRS-1
insulin receptor substrate-1
- JAMM
Jab1/MPN domain metalloenzyme
- Keap1
Kelch-like ECH-associated protein 1
- LRR
leucine rich repeats
- Mdm2
murine double minute 2
- MPN
MPR-PAD1-Nterm
- MRP
multidrug resistance-associated protein
- Nedd8
neural precursor cell expressed developmentally down-regulated protein 8
- NF-κB
nuclear factor κB
- NQO1
NAD(P)H quinine oxidoreducatase-1
- Nrf2
NF-E2-related factor 2
- PCI
proteasome, COP9-signalosome, initiation factor 3
- PLZF
promyelocytic leukemia zinc finger
- Poly-ub
polyubiquitin
- RING
really interesting new gene
- SCC
squamous cell carcinoma
- SCF
Skp1, Cul1, F-box protein
- Skp1
S-phase kinase-associated protein 1
- Skp2
S-phase kinase-associated protein 2
- SNAP23
synaptosomal-associated protein 23
- TNFα
tumor necrosis factor α
- TSC1
tuberous sclerosis gene 1
- Ub
ubiquitin
- UPS
ubiquitin proteasome system
- VEGF
vascular endothelial growth factor
- VHL
von Hippel–Lindau protein
- Vpr
viral protein R
- XPC
xeroderma pigmentosum group C
Acknowledgments
This study was supported by the NIH Grant ES015010 and American Cancer Society RSG-07-154-CNE to DD Zhang and the NIEHS training grant ES007091 to NF Villeneuve.
References
- 1.Aberle H. Bauer A. Stappert J. Kispert A. Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBOJ. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amit S. Hatzubai A. Birman Y. Andersen JS. Ben–Shushan E. Mann M. Ben–Neriah Y. Alkalay I. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: A molecular switch for the Wnt pathway. Genes Dev. 2002;16:1066–1076. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angers S. Li T. Yi X. MacCoss MJ. Moon RT. Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 4.Cardoso F. Ross JS. Picart MJ. Sotiriou C. Durbecq V. Targeting the ubiquitin-proteasome pathway in breast cancer. Clin Breast Cancer. 2004;5:148–157. doi: 10.3816/cbc.2004.n.020. [DOI] [PubMed] [Google Scholar]
- 5.Chariot A. The NF-kappaB-independent functions of IKK subunits in immunity and cancer. Trends Cell Biol. 2009;19:404–413. doi: 10.1016/j.tcb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Y. Dai X. Zhao Y. AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis. Plant Physiol. 2004;135:1020–1026. doi: 10.1104/pp.104.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chew EH. Poobalasingam T. Hawkey CJ. Hagen T. Characterization of cullin-based E3 ubiquitin ligases in intact mammalian cells—Evidence for cullin dimerization. Cell Signal. 2007;19:1071–1080. doi: 10.1016/j.cellsig.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Chuang LY. Guh JY. Liu SF. Hung MY. Liao TN. Chiang TA. Huang JS. Huang YL. Lin CF. Yang YL. Regulation of type II transforming-growth-factor-beta receptors by protein kinase C iota. Biochem J. 2003;375:385–393. doi: 10.1042/BJ20030522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cope GA. Deshaies RJ. COP9 signalosome: A multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- 10.Cope GA. Deshaies RJ. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006;7:1. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cope GA. Suh GS. Aravind L. Schwarz SE. Zipursky SL. Koonin EV. Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 12.Dias DC. Dolios G. Wang R. Pan ZQ. CUL7: A DOC domain-containing cullin selectively binds Skp1.Fbx29 to form an SCF-like complex. Proc Natl Acad Sci USA. 2002;99:16601–1666. doi: 10.1073/pnas.252646399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon C. Brunson LE. Roy MM. Smothers D. Sehorn MG. Mathias N. Overproduction of polypeptides corresponding to the amino terminus of the F-box proteins Cdc4p and Met30p inhibits ubiquitin ligase activities of their SCF complexes. Eukaryot Cell. 2003;2:123–133. doi: 10.1128/EC.2.1.123-133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dohmann EM. Kuhnle C. Schwechheimer C. Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell. 2005;17:1967–1978. doi: 10.1105/tpc.105.032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doronkin S. Djagaeva I. Beckendorf SK. The COP9 signalosome promotes degradation of Cyclin E during early Drosophila oogenesis. Dev Cell. 2003;4:699–710. doi: 10.1016/s1534-5807(03)00121-7. [DOI] [PubMed] [Google Scholar]
- 16.Dubiel W. Resolving the CSN and CAND1 paradoxes. Mol Cell. 2009;35:547–549. doi: 10.1016/j.molcel.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Ehrlich ES. Wang T. Luo K. Xiao Z. Niewiadomska AM. Martinez T. Xu W. Neckers L. Yu XF. Regulation of Hsp90 client proteins by a Cullin5-RING E3 ubiquitin ligase. Proc Natl Acad Sci USA. 2009;106:20330–20335. doi: 10.1073/pnas.0810571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng S. Shen Y. Sullivan JA. Rubio V. Xiong Y. Sun TP. Deng XW. Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell. 2004;16:1870–1882. doi: 10.1105/tpc.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs SY. Spiegelman VS. Kumar KG. The many faces of beta-TrCP E3 ubiquitin ligases: Reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh S. Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 21.Gnarra JR. Zhou S. Merrill MJ. Wagner JR. Krumm A. Papavassiliou E. Oldfield EH. Klausner RD. Linehan WM. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc Natl Acad Sci USA. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groisman R. Kuraoka I. Chevallier O. Gaye N. Magnaldo T. Tanaka K. Kisselev AF. Harel–Bellan A. Nakatani Y. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 2006;20:1429–1434. doi: 10.1101/gad.378206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gusmaroli G. Feng S. Deng XW. The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell. 2004;16:2984–3001. doi: 10.1105/tpc.104.025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannah J. Zhou P. Regulation of DNA damage response pathways by the cullin-RING ubiquitin ligases. DNA Repair (Amst) 2009;8:536–543. doi: 10.1016/j.dnarep.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayden MS. Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 26.Hayes JD. McMahon M. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer Lett. 2001;174:103–113. doi: 10.1016/s0304-3835(01)00695-4. [DOI] [PubMed] [Google Scholar]
- 27.Hayes JD. McMahon M. NRF2 and KEAP1 mutations: Permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 28.He Q. Cheng P. He Q. Liu Y. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 2005;19:1518–1531. doi: 10.1101/gad.1322205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 30.Hoeller D. Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 31.Iliopoulos O. Levy AP. Jiang C. Kaelin WG., Jr. Goldberg MA. Negative regulation of hypoxia-inducible genes by the von Hippel–Lindau protein. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itoh K. Wakabayashi N. Katoh Y. Ishii T. Igarashi K. Engel JD. Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwai K. Yamanaka K. Kamura T. Minato N. Conaway RC. Conaway JW. Klausner RD. Pause A. Identification of the von Hippel–Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc Natl Acad Sci USA. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaakkola P. Mole DR. Tian YM. Wilson MI. Gielbert J. Gaskell SJ. Kriegsheim A. Hebestreit HF. Mukherji M. Schofield CJ. Maxwell PH. Pugh CW. Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 35.Jeong WS. Jun M. Kong AN. Nrf2: A potential molecular target for cancer chemoprevention by natural compounds. Antioxid Redox Signal. 2006;8:99–106. doi: 10.1089/ars.2006.8.99. [DOI] [PubMed] [Google Scholar]
- 36.Kaelin WG., Jr. Cancer. Many vessels, faulty gene. Nature. 1999;399:203–204. doi: 10.1038/20309. [DOI] [PubMed] [Google Scholar]
- 37.Kamura T. Maenaka K. Kotoshiba S. Matsumoto M. Kohda D. Conaway RC. Conaway JW. Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapelari B. Bech–Otschir D. Hegerl R. Schade R. Dumdey R. Dubiel W. Electron microscopy and subunit-subunit interaction studies reveal a first architecture of COP9 signalosome. J Mol Biol. 2000;300:1169–1178. doi: 10.1006/jmbi.2000.3912. [DOI] [PubMed] [Google Scholar]
- 39.Kawakami T. Chiba T. Suzuki T. Iwai K. Yamanaka K. Minato N. Suzuki H. Shimbara N. Hidaka Y. Osaka F. Omata M. Tanaka K. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20:4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kensler TW. Wakabayashi N. Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 41.Kim YR. Oh JE. Kim MS. Kang MR. Park SW. Han JY. Eom HS. Yoo NJ. Lee SH. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol. 2010;220:446–451. doi: 10.1002/path.2653. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi A. Kang MI. Okawa H. Ohtsuji M. Zenke Y. Chiba T. Igarashi K. Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi M. Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Kominami K. Ochotorena I. Toda T. Two F-box/WD-repeat proteins Pop1 and Pop2 form hetero- and homo-complexes together with cullin-1 in the fission yeast SCF (Skp1-Cullin-1-F-box) ubiquitin ligase. Genes Cells. 1998;3:721–735. doi: 10.1046/j.1365-2443.1998.00225.x. [DOI] [PubMed] [Google Scholar]
- 45.Kwong LN. Dove WF. APC and its modifiers in colon cancer. Adv Exp Med Biol. 2009;656:85–106. doi: 10.1007/978-1-4419-1145-2_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lau A. Villeneuve NF. Sun Z. Wong PK. Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Rouzic E. Belaidouni N. Estrabaud E. Morel M. Rain JC. Transy C. Margottin– Goguet F. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle. 2007;6:182–188. doi: 10.4161/cc.6.2.3732. [DOI] [PubMed] [Google Scholar]
- 48.Lee DF. Kuo HP. Liu M. Chou CK. Xia W. Du Y. Shen J. Chen CT. Huo L. Hsu MC. Li CW. Ding Q. Liao TL. Lai CC. Lin AC. Chang YH. Tsai SF. Li LY. Hung MC. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell. 2009;36:131–140. doi: 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J. Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell. 2007;26:775–780. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Li W. Ye Y. Polyubiquitin chains: Functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu C. Li Y. Semenov M. Han C. Baeg GH. Tan Y. Zhang Z. Lin X. He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 52.Lo SC. Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol Cell Biol. 2006;26:1235–1244. doi: 10.1128/MCB.26.4.1235-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyapina S. Cope G. Shevchenko A. Serino G. Tsuge T. Zhou C. Wolf DA. Wei N. Shevchenko A. Deshaies RJ. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 54.Margottin F. Bour SP. Durand H. Selig L. Benichou S. Richard V. Thomas D. Strebel K. Benarous R. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 55.Matsuzawa SI. Reed JC. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol Cell. 2001;7:915–926. doi: 10.1016/s1097-2765(01)00242-8. [DOI] [PubMed] [Google Scholar]
- 56.McMahon M. Thomas N. Itoh K. Yamamoto M. Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a "tethering" mechanism: A two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 57.Merlet J. Burger J. Gomes JE. Pintard L. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci. 2009;66:1924–1938. doi: 10.1007/s00018-009-8712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Min KW. Kwon MJ. Park HS. Park Y. Yoon SK. Yoon JB. CAND1 enhances deneddylation of CUL1 by COP9 signalosome. Biochem Biophys Res Commun. 2005;334:867–874. doi: 10.1016/j.bbrc.2005.06.188. [DOI] [PubMed] [Google Scholar]
- 59.Motohashi H. Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 60.Nakayama K. Hatakeyama S. Maruyama S. Kikuchi A. Onoe K. Good RA. Nakayama KI. Impaired degradation of inhibitory subunit of NF-kappa B (I kappa B) and beta-catenin as a result of targeted disruption of the beta-TrCP1 gene. Proc Natl Acad Sci USA. 2003;100:8752–8757. doi: 10.1073/pnas.1133216100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nijman SM. Luna–Vargas MP. Velds A. Brummelkamp TR. Dirac AM. Sixma TK. Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 62.Ohh M. Kim WY. Moslehi JJ. Chen Y. Chau V. Read MA. Kaelin WG., Jr. An intact NEDD8 pathway is required for Cullin-dependent ubiquitylation in mammalian cells. EMBO Rep. 2002;3:177–182. doi: 10.1093/embo-reports/kvf028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohta T. Iijima K. Miyamoto M. Nakahara I. Tanaka H. Ohtsuji M. Suzuki T. Kobayashi A. Yokota J. Sakiyama T. Shibata T. Yamamoto M. Hirohashi S. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 64.Ohtake F. Baba A. Takada I. Okada M. Iwasaki K. Miki H. Takahashi S. Kouzmenko A. Nohara K. Chiba T. Fujii–Kuriyama Y. Kato S. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature. 2007;446:562–566. doi: 10.1038/nature05683. [DOI] [PubMed] [Google Scholar]
- 65.Okabe H. Lee SH. Phuchareon J. Albertson DG. McCormick F. Tetsu O. A critical role for FBXW8 and MAPK in cyclin D1 degradation and cancer cell proliferation. PLoS One. 2006;1:e128. doi: 10.1371/journal.pone.0000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Padmanabhan B. Tong KI. Ohta T. Nakamura Y. Scharlock M. Ohtsuji M. Kang MI. Kobayashi A. Yokoyama S. Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 67.Pause A. Lee S. Worrell RA. Chen DY. Burgess WH. Linehan WM. Klausner RD. The von Hippel–Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc Natl Acad Sci USA. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peth A. Berndt C. Henke W. Dubiel W. Downregulation of COP9 signalosome subunits differentially affects the CSN complex and target protein stability. BMC Biochem. 2007;8:27. doi: 10.1186/1471-2091-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petroski MD. Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 70.Pintard L. Kurz T. Glaser S. Willis JH. Peter M. Bowerman B. Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr Biol. 2003;13:911–921. doi: 10.1016/s0960-9822(03)00336-1. [DOI] [PubMed] [Google Scholar]
- 71.Podust VN. Brownell JE. Gladysheva TB. Luo RS. Wang C. Coggins MB. Pierce JW. Lightcap ES. Chau V. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci USA. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Popov N. Herold S. Llamazares M. Schulein C. Eilers M. Fbw7 and Usp28 regulate myc protein stability in response to DNA damage. Cell Cycle. 2007;6:2327–2331. doi: 10.4161/cc.6.19.4804. [DOI] [PubMed] [Google Scholar]
- 73.Popov N. Wanzel M. Madiredjo M. Zhang D. Beijersbergen R. Bernards R. Moll R. Elledge SJ. Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 74.Read MA. Brownell JE. Gladysheva TB. Hottelet M. Parent LA. Coggins MB. Pierce JW. Podust VN. Luo RS. Chau V. Palombella VJ. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sarikas A. Xu X. Field LJ. Pan ZQ. The cullin7 E3 ubiquitin ligase: A novel player in growth control. Cell Cycle. 2008;7:3154–3161. doi: 10.4161/cc.7.20.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmidt MW. McQuary PR. Wee S. Hofmann K. Wolf DA. F-box-directed CRL complex assembly and regulation by the CSN and CAND1. Mol Cell. 2009;35:586–597. doi: 10.1016/j.molcel.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seibert V. Prohl C. Schoultz I. Rhee E. Lopez R. Abderazzaq K. Zhou C. Wolf DA. Combinatorial diversity of fission yeast SCF ubiquitin ligases by homo- and heterooligomeric assemblies of the F-box proteins Pop1p and Pop2p. BMC Biochem. 2002;3:22. doi: 10.1186/1471-2091-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shibata T. Kokubu A. Gotoh M. Ojima H. Ohta T. Yamamoto M. Hirohashi S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–1368. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 79.Shibata T. Ohta T. Tong KI. Kokubu A. Odogawa R. Tsuta K. Asamura H. Yamamoto M. Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh A. Boldin–Adamsky S. Thimmulappa RK. Rath SK. Ashush H. Coulter J. Blackford A. Goodman SN. Bunz F. Watson WH. Gabrielson E. Feinstein E. Biswal S. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singh A. Misra V. Thimmulappa RK. Lee H. Ames S. Hoque MO. Herman JG. Baylin SB. Sidransky D. Gabrielson E. Brock MV. Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stegmeier F. Rape M. Draviam VM. Nalepa G. Sowa ME. Ang XL. McDonald ER., 3rd Li MZ. Hannon GJ. Sorger PK. Kirschner MW. Harper JW. Elledge SJ. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 83.Sugasawa K. Okuda Y. Saijo M. Nishi R. Matsuda N. Chu G. Mori T. Iwai S. Tanaka K. Tanaka K. Hanaoka F. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 84.Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol. 2008;8:501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suzuki H. Chiba T. Suzuki T. Fujita T. Ikenoue T. Omata M. Furuichi K. Shikama H. Tanaka K. Homodimer of two F-box proteins betaTrCP1 or betaTrCP2 binds to IkappaBalpha for signal-dependent ubiquitination. J Biol Chem. 2000;275:2877–2884. doi: 10.1074/jbc.275.4.2877. [DOI] [PubMed] [Google Scholar]
- 86.Tong KI. Katoh Y. Kusunoki H. Itoh K. Tanaka T. Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: Characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tong KI. Kobayashi A. Katsuoka F. Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: A hinge and latch mechanism. Biol Chem. 2006;387:1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 88.Wada H. Yeh ET. Kamitani T. Identification of NEDD8-conjugation site in human cullin-2. Biochem Biophys Res Commun. 1999;257:100–105. doi: 10.1006/bbrc.1999.0339. [DOI] [PubMed] [Google Scholar]
- 89.Wang XJ. Sun Z. Villeneuve NF. Zhang S. Zhao F. Li Y. Chen W. Yi X. Zheng W. Wondrak GT. Wong PK. Zhang DD. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wee S. Geyer RK. Toda T. Wolf DA. CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat Cell Biol. 2005;7:387–391. doi: 10.1038/ncb1241. [DOI] [PubMed] [Google Scholar]
- 91.Wimuttisuk W. Singer JD. The Cullin3 ubiquitin ligase functions as a Nedd8-bound heterodimer. Mol Biol Cell. 2007;18:899–909. doi: 10.1091/mbc.E06-06-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wirbelauer C. Sutterluty H. Blondel M. Gstaiger M. Peter M. Reymond F. Krek W. The F-box protein Skp2 is a ubiquitylation target of a Cul1-based core ubiquitin ligase complex: Evidence for a role of Cul1 in the suppression of Skp2 expression in quiescent fibroblasts. EMBO J. 2000;19:5362–5375. doi: 10.1093/emboj/19.20.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wolf DA. McKeon F. Jackson PK. F-box/WD-repeat proteins pop1p and Sud1p/Pop2p form complexes that bind and direct the proteolysis of cdc18p. Curr Biol. 1999;9:373–376. doi: 10.1016/s0960-9822(99)80165-1. [DOI] [PubMed] [Google Scholar]
- 94.Wu JT. Lin HC. Hu YC. Chien CT. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat Cell Biol. 2005;7:1014–1020. doi: 10.1038/ncb1301. [DOI] [PubMed] [Google Scholar]
- 95.Wu K. Chen A. Pan ZQ. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J Biol Chem. 2000;275:32317–32324. doi: 10.1074/jbc.M004847200. [DOI] [PubMed] [Google Scholar]
- 96.Xu C. Kim NG. Gumbiner BM. Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle. 2009;8:4032–4039. doi: 10.4161/cc.8.24.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang X. Menon S. Lykke–Andersen K. Tsuge T. Di X. Wang X. Rodriguez–Suarez RJ. Zhang H. Wei N. The COP9 signalosome inhibits p27(kip1) degradation and impedes G1-S phase progression via deneddylation of SCF Cul1. Curr Biol. 2002;12:667–672. doi: 10.1016/s0960-9822(02)00791-1. [DOI] [PubMed] [Google Scholar]
- 98.Yaron A. Gonen H. Alkalay I. Hatzubai A. Jung S. Beyth S. Mercurio F. Manning AM. Ciechanover A. Ben–Neriah Y. Inhibition of NF-kappa-B cellular function via specific targeting of the I-kappa-B-ubiquitin ligase. EMBO J. 1997;16:6486–6494. doi: 10.1093/emboj/16.21.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 100.Zhang DD. Lo SC. Cross JV. Templeton DJ. Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang DD. Lo SC. Sun Z. Habib GM. Lieberman MW. Hannink M. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem. 2005;280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 102.Zheng J. Yang X. Harrell JM. Ryzhikov S. Shim EH. Lykke–Andersen K. Wei N. Sun H. Kobayashi R. Zhang H. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002;10:1519–1526. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- 103.Zhou C. Wee S. Rhee E. Naumann M. Dubiel W. Wolf DA. Fission yeast COP9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Mol Cell. 2003;11:927–938. doi: 10.1016/s1097-2765(03)00136-9. [DOI] [PubMed] [Google Scholar]
- 104.Zhou P. Howley PM. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol Cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- 105.Zipper LM. Mulcahy RT. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem. 2002;277:36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]