SUMMARY
Commensal microbes can have a substantial impact on autoimmune disorders, but the underlying molecular and cellular mechanisms remain largely unexplored. We report that autoimmune arthritis was strongly attenuated in the K/BxN mouse model under germ-free (GF) conditions, accompanied by reductions in serum autoantibody titers, splenic autoantibody-secreting cells, germinal centers and the splenic T helper 17 (Th17) cell population. Neutralization of interleukin-17 prevented arthritis development in specific-pathogen-free K/BxN mice due to a direct effect of this cytokine on B cells to inhibit germinal center formation. The systemic deficiencies of the GF animals reflected a loss of Th17 cells from the small intestinal lamina propria. Introduction of a single gut-residing species, segmented filamentous bacteria, into GF animals reinstated the lamina propria Th17 cell compartment and production of autoantibodies, and arthritis rapidly ensued. Thus, a single commensal microbe, via its ability to promote a specific Th-cell subset, can drive an autoimmune disease.
INTRODUCTION
Mammals host trillions of microbes at diverse locations throughout the body, in particular in the gut (Backhed et al., 2005; Ley et al., 2006; Ley et al., 2008b). The enormity and complexity of these commensal (or mutualistic) communities have been difficult to deal with until recently, when striking advances in “next-generation” sequencing methods, entailing either 16S rRNA or shot-gun cataloguing, rendered this field navigable terrain.
The gut microbiomes of humans and mice are broadly similar (Backhed et al., 2005; Ley et al., 2006; Ley et al., 2008b; Ley et al., 2008a). In both cases, ~1000 different microbial species from ~10 different divisions colonize the gastrointestinal tract, but just two bacterial divisions – the Bacteroidetes and Firmicutes – and one member of the Archaea appear to dominate, together accounting for ~98% of the 16S rRNA sequences obtained from this site. The number and identity of microbial communities vary along the length of the gut, in a proximal to distal gradient of abundance (small intestine < cecum < colon), and across the three dimensions of the lumen and mucous layers. The total number of genes borne by the gastrointestinal microbiome has been estimated to exceed more than a hundred-fold that of the human genome (Ley et al., 2006). The products of these genes are put to good use by the host, for example in digestion, production of nutrients, detoxification, defense against pathogens and development of a competent immune system (Backhed et al., 2005; Ley et al., 2006; Ley et al., 2008b).
The gastrointestinal microbiome and the immune system are closely tied, each influencing and being influenced by the other (Macpherson and Harris, 2004; Mazmanian and Kasper, 2006; Rakoff-Nahoum and Medzhitov, 2008; Vassallo and Walker, 2008; Duerkop et al., 2009)). In general terms, the incomplete state of the immune system in germ-free (GF) conditions and in neonatal individuals argues that its normal maturation is driven by commensal microbes – for example, GF-housed individuals and neonates can have a reduced fraction of peripheral CD4+ T lymphocytes, a systemic tilt toward the T helper 2 (Th2) cell phenotype, defective T and B cell compartments in gut-associated lymphoid tissue, reduced complements of immunoglobulin G (IgG) and IgA antibodies (Abs), etc (Mazmanian et al., 2005; Rakoff-Nahoum et al., 2004; Ivanov et al., 2008; Atarashi et al., 2008; Mazmanian et al., 2008; Grice et al., 2009; Macpherson and Harris, 2004; Vassallo and Walker, 2008). In more specific terms, gut-resident bacteria – sometimes even a single species – can have a strong influence on the emergence and/or maintenance of particular CD4+ T cell subsets. Examples include the effects of specific bacteria on the emergence of Th17 cells in the intestinal lamina propria (LP) (Ivanov et al., 2008; Atarashi et al., 2008; Salzman et al., 2009; Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009) and the impact of Bacteroides fragilis on systemic Th1 cells and local interleukin-10 (IL-10)-producing regulatory T (Treg) cells (Mazmanian et al., 2008; Mazmanian et al., 2005). In both cases, dendritic cells (DCs) are thought to be the initial target of mediators produced either by the culprit microbe or in response to it – adenosine-5′-triphosphate (ATP) or serum amyloid A (SAA) in the former case (Atarashi et al., 2008; Ivanov et al., 2009), the polysaccharide PSA in the latter (Mazmanian et al., 2005).
Given these tight associations, it is not surprising that gut microbiota have been linked to pathologies of the immune system, notably allergies and autoimmune disorders (Strachan, 1989; Wills-Karp et al., 2001) (Kelly et al., 2007). Ties to inflammatory bowel diseases are easy to understand, but the cellular and molecular mechanisms by which intestinal commensals influence autoimmune responses at distal sites remain enigmatic. The time seems ripe to apply new, and rapidly emerging, knowledge about the composition and properties of the gastrointestinal microbiome and about the activities of recently discovered effector and regulatory T cell subsets to dissecting these mechanisms in autoimmune disease models. We chose to study the K/BxN T cell receptor (TCR) transgenic mouse model of inflammatory arthritis because of its easily distinguishable initiation and effector stages (Kouskoff et al., 1996; Korganow et al., 1999; Matsumoto et al., 1999). The initiation phase relies primarily on the adaptive immune system. T lymphocytes displaying the transgene-encoded TCR recognize a self-peptide derived from glucose-6-phosphate isomerase (GPI) presented by the major histocompatibility complex class II molecule, Ag7; these autoreactive T cells provide exceptionally effective help to GPI-specific B cells, resulting in massive production of GPI autoAbs, primarily of the IgG1 isotype. The effector phase, which can be mimicked by transfer of serum from K/BxN into standard mice, is executed primarily by innate immune system players. GPI:anti-GPI immune complexes initiate a self-sustaining inflammatory response that mobilizes mast cells, neutrophils, the alternative pathway of complement, Fcγ receptors, tumor necrosis factor-α (TNF-α), IL-1, etc. Arthritis ensues, rapidly (beginning at about 4 weeks of age) and with high penetrance (close to 100%).
Here we report that arthritis was attenuated in K/BxN mice housed under GF conditions. Disease dampening was traced to a dearth of Th17 cells, which could be reversed by introducing segmented filamentous bacteria (SFB) into the gut of GF-housed mice, provoking rapid onset of arthritis. Thus, we provide an example of an extra-gut autoimmune disease triggered by a single member of the commensal intestinal microbiota through its promotion of a particular Th cell subset.
RESULTS
Germ-free K/BxN mice have reduced GPI autoAb titers and attenuated arthritis
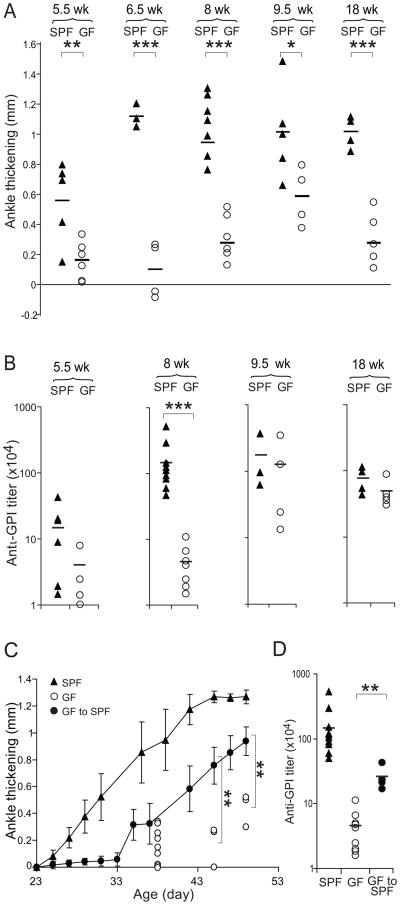
To explore the impact of commensal microbes on the development of autoimmune arthritis, we established GF colonies of KRN/B6 and NOD mice, and mated the two strains to obtain K/BxN experimental animals. As judged by both ankle thickening (Fig. 1A) and clinical index (not shown), GF-housed K/BxN mice developed an attenuated arthritis compared with that of K/BxN animals contemporaneously housed in a specific-pathogen-free (SPF) facility – both delayed in onset and reduced in severity.
Figure 1. Attenuation of arthritis in GF K/xBN mice.
(A) Ankle thickening values and (B) anti-GPI titers for GF and SPF K/BxN mice of the indicated ages. Each symbol represents one animal; bars indicate the group mean. (C) GF K/BxN mice were shipped to our SPF facility and upon weaning (day 21), ankle thickening was measured from day 23. Closed circles: average of ankle thickening ± s.e.m, serially measured on cohorts. Closed triangles: analogous measurements for SPF-housed K/BxN mice. Open circles: values for individual GF-housed K/BxN mice, not measured serially due to experimental contingencies. (D) Sera were collected at the end of the experiment depicted in panel C. The bar indicates the mean. Asterisks indicate statistical significance using the Student s t-Test (*P=0.05, **P<0.05, ***P<0.005)
A key disease landmark in this arthritis model is the production of high titers of serum GPI autoAbs, which separates the initiation phase, dependent on the adaptive immune system, from the effector phase, mostly driven by innate immune system players (Korganow et al., 1999). At 8 weeks of age, serum total IgG (Fig. 1B) as well as IgG1 (Fig. S1), the dominant anti-GPI Ab isotype in the K/BxN model (Korganow et al., 1999), were substantially lower in GF K/BxN mice than in their SPF counterparts. At later timepoints, the difference in anti-GPI titers under the two housing conditions was not so apparent, in contrast to the continued attenuation of ankle thickness in GF mice. Such discordance between autoAb titers and the degree of ankle thickening is frequent at late timepoints in this model. At this stage of disease, ankle thickness reflects primarily bone remodeling and fibrosis, and so is a cumulative index of disease duration and severity rather than an indicator of concomitant inflammation (Kouskoff et al., 1996).
To confirm that the differences in disease parameters really reflected the GF environment – and not, for example, genetic drift – and to evaluate their reversibility, we transferred 21-day-old GF K/BxN mice back into our SPF facility. After 14 days, they had already begun to develop arthritis, which soon surpassed the disease of straight GF K/BxN animals in both its speed and severity, but was still delayed and diminished vis-à-vis straight SPF counterparts (Fig. 1C). Clinical disease severity reflected the titers of GPI autoAb attained at 7 weeks of age under the three housing conditions (Fig. 1D). Thus, commensal microbes are required for the development of high GPI autoAb titers and severe arthritis in the K/BxN model.
The impact of commensal microbes on the adaptive immune system of K/BxN mice
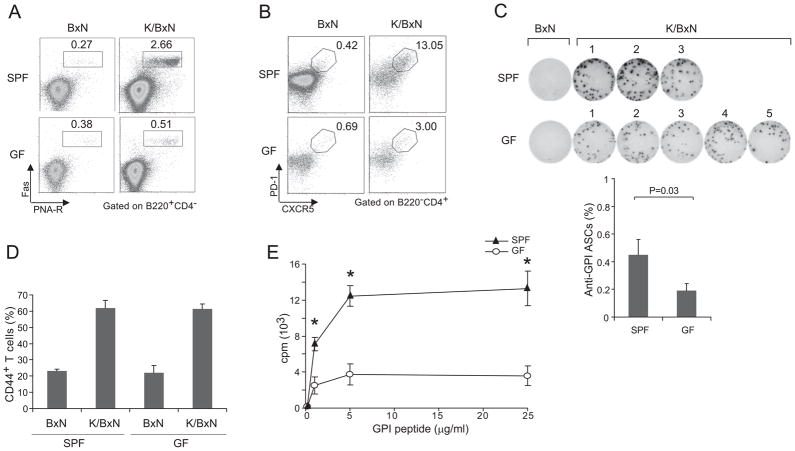
Although influences on effector-phase processes certainly remain possible, the reduced GPI autoAb titers in the absence of commensal microbes suffices, in and of itself, to explain the attenuated arthritis in GF K/BxN mice (Matsumoto et al., 1999). Therefore, we examined the impact of commensals on the adaptive immune system in this model, focusing on the spleen because by far most GPI Ab-secreting cells (ASCs) reside in this organ (Maccioni et al., 2002; Huang et al., 2010). First, we surveyed the B-lymphocyte compartments, performing a four-way comparison of splenocytes from 6- to 8-week-old BxN vs K/BxN mice housed in SPF vs GF conditions. The percentages and numbers of splenic B cells (CD19+) in GF and SPF K/BxN mice were similar (Fig. S2A), as were, more specifically, their: T1 (IgMhiIgDlo), T2 (IgMhiIgDhi), mature (IgMloIgDhi), follicular (CD21loCD23hi) and marginal zone (CD21hiCD23lo) B cell compartments (Fig. S2B and S2C). SPF K/BxN mice showed an increase in the percentage of splenic germinal center (GC) B cells (Fas+PNA-receptor+) vis-à-vis BxN controls, reflecting activation and expansion of the anti-GPI specificities (Fig 2A, upper panels). This augmentation did not occur under GF conditions (Fig 2A, lower panels). Perhaps not surprisingly, then, GF K/BxN mice had a reduced complement of CXCR5+PD1+ T follicular helper (Tfh) cells, which reside primarily in GCs (Fig. 2B). The spleens of GF K/BxN mice also had a smaller fraction of anti-GPI ASCs, as measured by an ELISPOT assay (Fig. 2C). These deficiencies can explain the reduced titer of serum GPI autoAbs in GF K/BxN mice.
Figure 2. Impact of commensal flora on the B and T cell compartments of K/BxN mice.
(A) Splenocytes from GF or SPF BxN or K/BxN mice were stained with Abs recognizing B220, CD4, or FAS or with PNA-R, and were analyzed by flow cytometry, gating as indicated. Values indicate the percentages of Fas+PNA-R+ cells in total B cells. Data are representative of two independent experiments. (B) Splenocytes from GF or SPF BxN or K/BxN mice were stained with mAbs recognizing B220, CD4, CXCR5 and PD-1, and were analyzed by flow cytometry, gating as indicated. Values indicate the percentages of CXCR5+PD-1+ cells in total CD4+ T cells. Data are representative of two independent experiments. (C). Purified B cells of GF or SPF BxN or K/BxN mice were stimulated with GPI protein (10μg/ml) for 6 hrs. An ELISPOT assay revealed GPI-specific ASCs. Fraction of GPI-specific ASCs among total B cells; mean + s.e.m from two independent experiments. (D) Splenocytes were isolated from SPF or GF K/BxN mice, stained and analyzed by flow cytometry. The values indicate percentages of CD44+ cells in CD4+ T cells. Data are representative of two independent experiments [SPF BxN: 23.3 +/− 0.9 s.e.m. (n=2); SPF K/BxN: 61.8 +/− 4.8 s.e.m. (n=3); GF BxN: 22.1 +/− 4.5 s.e.m. (n=2); GF K/BxN: 61.3 +/− 3.2 s.e.m. (n=4)]. (E) Splenocytes were isolated from SPF and GF K/BxN mice and stimulated with GPI282–294 peptide at the indicated concentration. Data are representative of 2 independent experiments. Asterisks indicate statistical significance using the Student s t-Test, *P<0.05.
Given the established T cell dependence of the GPI autoAb response (Kouskoff et al., 1996; Korganow et al., 1999), we also compared the T cell compartments of K/BxN mice kept under the two husbandry conditions. There were only minor changes in the representation of thymic or splenic CD4+ or CD8+ T cells in GF animals, the biggest difference being a 20–50% reduction in the splenic CD4+ T cell compartment compared with that of SPF mice (Fig. S2D and S2E); and the activation state of peripheral cells was similar under the two housing conditions (e.g. Fig. 2D). Also, there was no evident change in either the fraction of CD4+Foxp3+ T regulatory (Treg) cells or in their in vitro suppressive activity (Fig. S2F and G). However, splenocytes from GF K/BxN mice responded less well than those from SPF K/BxN animals when challenged in vitro with the relevant GPI peptide at all doses tested (Fig. 2E). Thus, given the particular B and T cell defects observed, the T helper capabilities of GF K/BxN mice appear to be somehow compromised.
GF K/BxN mice have a dearth of splenic IL-17-producing T cells
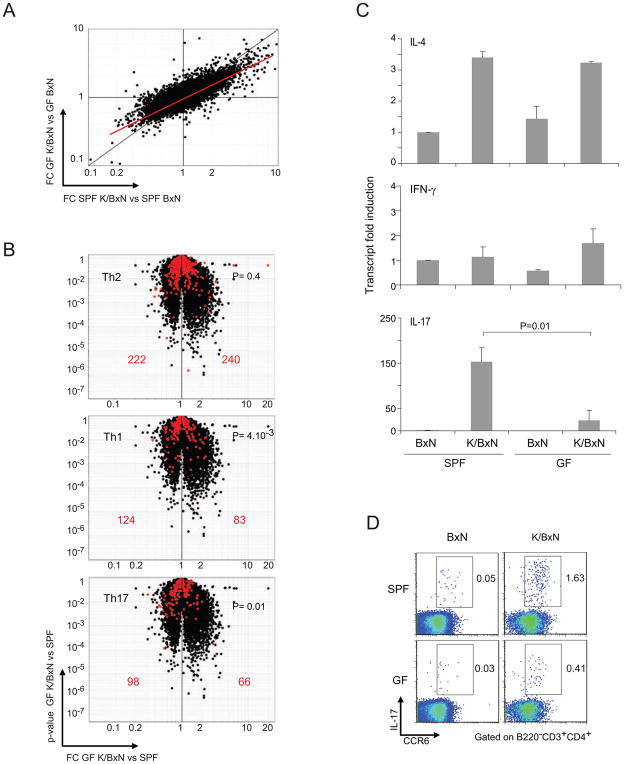
To permit a broad, unbiased comparison of Th cells from mice under the two conditions, we performed microarray-based gene-expression profiling on CD4+ T cells purified from spleens of SPF and GF BxN and K/BxN animals. A FoldChange/FoldChange (FC/FC) plot revealed up-regulation of a large number of transcripts in the K/BxN (vs BxN) T cells; the off-diagonal disposition of the major cloud of dots indicated that induction was muted in GF (vs SPF) mice (Fig. 3A). Another instructive manner to compare gene expression in SPF and GF mice is the “volcano plots” depicted in Fig. 3B, which display for each gene the SPF vs GF FC on the x-axis and the p-value of this FC on the y-axis. Superimposing previously determined Th-cell signatures (Nurieva et al., 2008) onto the plots showed there to be minimal changes in GF CD4+ T cells in transcripts typical of Th2 cells, i.e. no bias to either side of the midline. However, as indicated by their skewed disposition away from the right, the Th1 and Th17 cell signatures were both diminished in GF CD4+ T cells. We did not further pursue the defect in the Th1 cell subset in this study because crossing a null mutation of the gene encoding interferon-γ (IFN-γ), the major Th1 cell cytokine, into the K/BxN model had no impact on the arthritis parameters examined, including ankle thickening, clinical score and histopathology (Fig. S3 and data not shown).
Figure 3. A defective Th17 signature in GF K/BxN mice.
(A) FC (fold change) vs FC plot comparing gene-expression values of SPF K/BxN vs BxN mice (x axis) and GF K/BxN vs BxN mice (y axis). Gene-expression values of each group were the average values of 3 chips from 3 independent experiments. (B) Th cell signatures. Th2, Th1 and Th17 cell signatures were generated from published datasets (Nurieva et al., 2008), using 2 as the cut-off for FC over the expression value of two other cell types. The volcano plots depict the FC for SPF vs GF K/BxN CD4+ T cells versus the p value of the FC. Signature genes are superimposed in red. Values refer to the number of genes upregulated (right) or downregulated (left) in GF vis-à-vis SPF T cells. P values from a X2 test are indicated. (C) IL-4, IFN-γ and IL-17 transcripts in splenic CD4+ T cells for GF or SPF of B/N or K/BxN mice were quantified by RT-PCR. The level in SPF BxN mice was set as 1. Mean + s.e. Results were compiled from three independent experiments with two mice per group. P =0.01 for transcriptional fold-induction of IL-17 (SPF vs. GF K/BxN). (D) Splenocytes of GF or SPF BxN or K/BxN mice were stained with Abs recognizing CCR6 and IL-17, and were analyzed by flow cytometry. Values represent percentages of IL-17+CCR6+ cells in CD3+CD4+B220− cells. Data are representative of two independent experiments.
The transcript profiling pointed to a defect in GF K/BxN Th17 cells that encompassed several of this subset s hallmark proteins: e.g. reductions in RORγt (1/1.8), IL-17A (1/1.3), IL-21 (1/1.3), IL-22 (1/3.2) and CCR6 (1/1.3) The dearth of IL-17A was confirmed by both PCR quantification of splenic CD4+ T cell transcripts (Fig. 3C) and cytofluorimetric evaluation of IL-17 amounts in this population re-stimulated ex-vivo (Fig. 3D). According to both assays, Il17a gene expression was strongly induced in SPF K/BxN vis-à-vis BxN mice, but this induction was minimal under GF conditions.
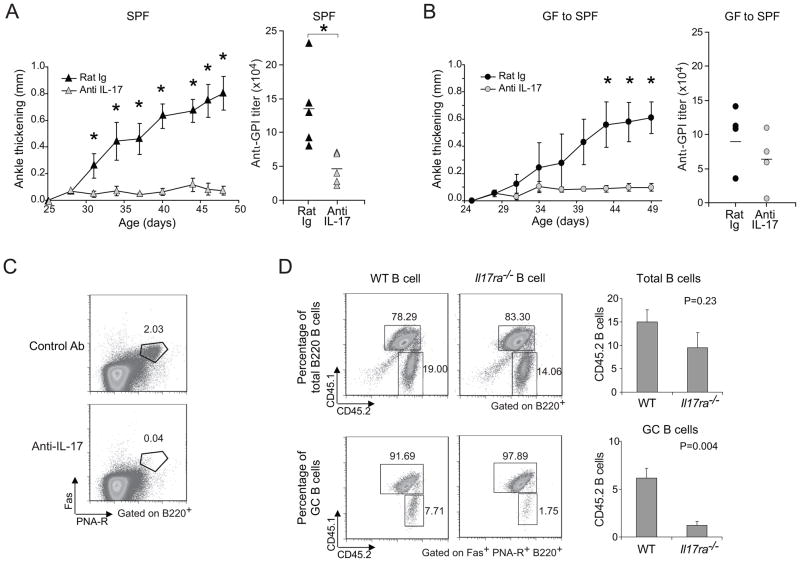
To assess the disease relevance of a defect in the Th17 cell compartment of K/BxN mice, we performed neutralization experiments using an IL-17 monoclonal Ab (mAb). Treatment of 25-day-old SPF-housed K/BxN mice, just at arthritis onset, with anti-IL-17 completely blocked disease progression, which was reflected in low serum GPI autoAb titers (Fig. 4A). In addition, when GF mice were transferred to the SPF facility, they did not succumb to arthritis if anti-IL-17 mAb was administered from the time of transfer (Fig. 4B).
Figure 4. Reduction of arthritis by neutralization of IL-17.
(A) 25-day-old SPF K/BxN mice were treated with 100μg of anti-IL-17 or control rat IgG every 3 days, and ankle thickening was measured over time (left panel). Mean ± s.e of the two groups (n=5 in both groups from two independent experiments is plotted; Asterisks indicate statistical significance using the Student s t-Test, *P<0.05.). Sera were collected at the end of the experiment, and anti-GPI titers quantified (right panel). Symbols represent individual mice; bar indicates the mean. (B) At 23 days of age, GF mice were transferred to our SPF facility, and were treated with 100μg anti-IL-17 or control rat IgG every 3 days from day 24. Otherwise as in the A panels (n=4 in both groups from one experiment; Asterisks indicate statistical significance using the Student s t-Test, *P<0.05). (C) SPF K/BxN mice were treated as in panel A. At the end of treatment, splenocytes were isolated and stained with Abs recognizing B220, CD4 or Fas, or with PNA, and were analyzed by flow cytometry, gating as indicated. Values represent the percentages of Fas+PNA-R+ cells in total B cells. Data are representative of two independent experiments with two mice per group. (D) B cells from either WT or Il17ra−/− mice were combined with splenocytes from arthritic K/BxN mice and transferred into BxN.Rag1−/− recipients. The origins of B cells were identified by expression of the congenic marker: CD45.1+CD45.2+ for K/BxN B cells and CD45.2 for Il17ra−/− or WT B cells. Percentages of K/BxN B cells and Il17ra−/− or WT B cells among total B cell or GC B cell populations are indicated. The quantitative data of Il17ra−/− or WT B cell percentage among total B cells and GC B cells were also shown as mean + s.e. (n=4, data combined from two independent experiments).
As IL-17 has generally been thought of as a pro-inflammatory cytokine, its effect on anti-GPI titers may appear surprising on first consideration. However, Hsu et al recently reported a direct impact of IL-17 on GC formation in the BXD2 mouse strain (Hsu et al., 2008). Indeed, anti-IL-17 blocking studies demonstrated this cytokine to be required for efficient GC formation in the K/BxN model (Fig. 4C). And transfer experiments showed that IL-17 s promotion of GCs was a direct effect on B cells. 1×107 B cells not expressing or expressing IL-17R were purified from spleens of IL-17R-deficient or IL-17R-sufficient B6.H-2g7 (B6g7) littermates; each population was combined with an equal number of splenocytes from arthritic K/BxN mice; each mix was transferred into lightly irradiated (450R) BxN.Rag1−/− recipients; and the recipients splenocytes were analyzed two weeks later by flow cytometry. B cells lacking IL-17R could repopulate the spleen (Fig. 4D, upper panels), but showed a substantially diminished capacity to partake in GCs (Fig. 4D, lower panels). Thus, a paucity of splenic Th17 cells was a critical factor in the diminished arthritis of K/BxN mice; IL-17 promoted GPI autoAb production, enhancing GC formation via a direct effect on B cells.
Linking arthritis to gut commensals
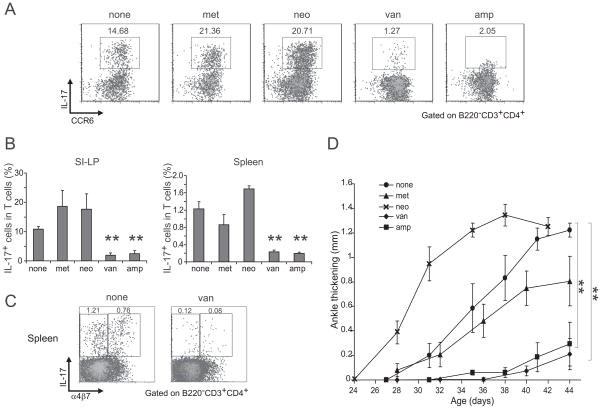
How can commensal microbes impact on the production of IL-17 by splenic T cells? Microbial colonization of the gut promotes Th17 cell differentiation in the small-intestinal lamina-propria (SI-LP), the major site of this subset s differentiation (Ivanov et al., 2008; Atarashi et al., 2008). Indeed, Th17 cells were essentially absent from that site in GF K/BxN animals (Fig. 5A). (In contrast, IL-17-expressing SI-LP CCR6+ γδ T cells were not reduced in K/BxN GF mice – data not shown.) Several experiments were performed to explore a potential link between SI-LP and splenic Th17 cells. First, we compared their appearance through ontogeny: SI-LP Th17 cells arise abruptly between day 16 and day 25 after birth, around the time of weaning (Ivanov et al., 2008), which is just before the window of arthritis development previously reported for the K/BxN model, i.e. day 25–31 (Kouskoff et al., 1996). A direct temporal comparison of the relevant parameters in SPF K/BxN mice revealed that SI-LP Th17 cells appeared in substantial numbers between 2 and 3 weeks of age (Fig. 5B), followed closely by splenic Th17 cells between 3 and 4 weeks (Fig. 5B), and arthritis onset at around 4 weeks (Fig 5B). Second, we looked for the gut homing receptor, α4β7, on splenic Th17 cells. Supporting an intestinal origin, 30–50% of splenic Th17 cells from 5-week-old K/BxN, but not BxN, mice expressed this receptor, imprinted by intestinal-mucosa-associated DCs (Sigmundsdottir and Butcher, 2008) (Fig. 5C). Last, we compared the sensitivities of the SI-LP and splenic Th17 cell compartments and of arthritis development to antibiotic treatments. The differentiation of SI-LP Th17 cells in B6 mice is blocked by ampicillin and vancomycin but not by metronidazole and neomycin, the latter two targeting anaerobes and Gram-negative bacteria, respectively, i.e. >90% of gut microbiota (Ivanov et al., 2008). This pattern of sensitivity was also true of SI-LP and splenic Th17 cells in K/BxN mice (Fig. 6A and B), including those splenic Th17 cells that expressed α4β7 (eg Fig. 6C). Most important, treatment of K/BxN mice from birth with vancomycin or ampicillin, but not metronidazole or neomycin, strongly inhibited the development of arthritis (Fig. 6D). Interestingly, disease was actually exacerbated in the neomycin-treated animals, suggesting an additional negative influence of Gram-negative gut bacteria.
Figure 5. Linking gut and spleen IL-17 cells.
(A) SI-LP lymphocytes were isolated from SPF or GF K/BxN mice. Cells were stained, analyzed by flow cytometry and gated as indicated. Expression of IL-17 versus CCR6 is plotted. The values indicate percentages of IL-17+CCR6+ cells in CD3+CD4+B220− cells. Data are representative of three independent experiments. (B) SI-LP lymphocytes (i) and splenocytes (ii) were isolated from SPF mice of the indicated ages, stained and analyzed by flow cytometry, gated as indicated. Plots displayed IL-17 versus CCR6 expression. Values indicate % of IL-17+CCR6+ cells in total CD4+ T cells (CD3+CD4+B220−). Data are representative of two independent experiments. (iii) Measurement of ankle thickening for the same mice. Each circle represents one animal from two independent experiments. (C) Splenocytes from SPF BxN or K/BxN mice were stained and analyzed by flow cytometry, gated as indicated. Plots depict IL-17 versus α4β7 staining. Values indicate % of IL-17+α4β7+ or IL-17+α4β7− cells in total CD4+ T cells (CD3+CD4+B220−). Data are representative of two independent experiments.
Figure 6. Effects of various antibiotics.
(A) Representative dot plots examining expression of IL-17 and CCR6 by SI-LP lymphocytes in untreated or the indicated antibiotic-treated SPF K/BxN mice, treated from birth to 5 wks of age. Values refer to % of the gated population in total CD4+ T cells. Representative of two independent experiments. (B) SPF K/BxN mice were treated with metronidazole (1g/l), neomycin (1g/l), vancomycin (0.5g/l) or ampicillin (1g/l) in the drinking water from birth. At 5 weeks of age, SI-LP lymphocytes (left) and splenocytes (right) were isolated, stained and analyzed by flow cytometry. Plotted are the % of IL-17+ cells of total CD4+ T cells. Mean + s.e. (data was a combination of two independent experiments with mice treated with metronidazole (n=4), neomycin (n=4), vacomycin (n=4), ampicillin (n=4) or nothing (n=8)) Asterisks indicate statistical significance using the Student s t-Test, **P<0.005. (C) Representative dot plots examining expression of α4β7 on IL-17-producing splenocytes in untreated (left) or vancomycin-treated (right) SPF K/BxN mice, treated from birth to 5 wks of age. Values refer to % of the gated population in total CD4+ T cells. Representative of two independent experiments. (D) K/BxN mice were untreated or were treated with metronidazole (1g/l), neomycin (1g/l), ampicillin (1g/L) or vancomycin (0.5g/l) in the drinking water from birth. Ankle thickening was followed from day 27. Mean ± s.e. (none group: n=5; all other antibiotics treated groups: n=4). Asterisks indicate statistical significance of area under curve using the Student s t-Test, **P<0.005. Representative of two independent experiments.
Monocolonization with SFB triggers arthritis in GF K/BxN mice
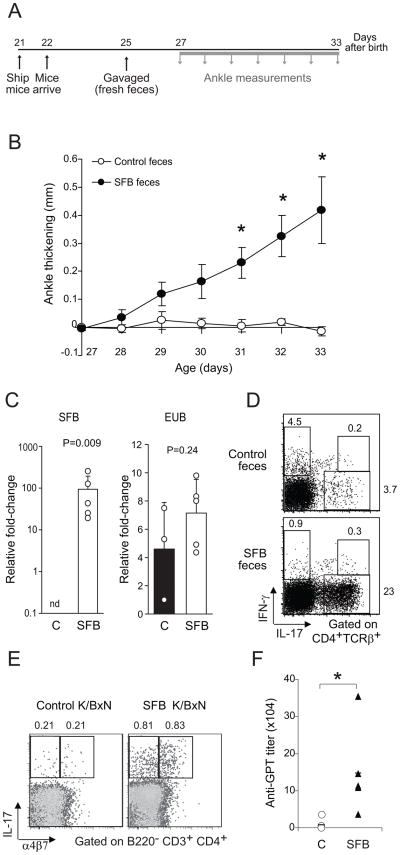
It was recently reported that a single bacterial species that is a component of normal gut microbiota, SFB, was sufficient to induce the development of SI-LP Th17 cells in mice taken from an SPF facility at the Jackson Laboratory, wherein they typically show a dearth of both this bacterium and Th17 cells (Ivanov et al., 2009). This result drew our attention because we had noted lower GPI autoAb titers and attenuated arthritis development when first introducing the K/BxN model into Jackson, vis-à-vis our SPF colonies in Strasbourg and Boston (data not shown). Therefore, we tested whether SFB might be arthritogenic by introducing it, via oral gavage of fecal material from SFB-monocolonized mice (vs feces from control mice), into GF K/BxN mice transferred into an SPF facility at 21 days of age (Fig. 7A). Prior experiments like those illustrated in Fig. 1C had demonstrated that transferred GF mice do develop arthritis, but typically not until 2 weeks after exposure to SPF conditions, i.e. after 35 days of age. Introduction of SFB greatly accelerated arthritis onset in the transferred mice, beginning 3 days after gavage, i.e. after only 28 days of age (Fig. 7B). PCR analysis of fecal material at 6 days after gavage indicated that at this early timepoint only those mice administered SFB-containing feces were colonized with SFB (Fig 7C). Flow cytometry of SI-LP and spleen cells at 33 days of age confirmed the association between SFB colonization, the appearance of Th17 cells in SI-LP, their migration to the spleen, (Fig. 7D and 7E) and the triggering of arthritis (Fig. 7B). As anticipated, introduction of SFB led to an elevation of GPI autoAb titers to amounts that are known to induce arthritis (Fig. 7F; cf Fig. 1B). Thus, a single bacterial species, SFB, is capable of triggering arthritis development in K/BxN mice through promotion of Th17 cell populations in the SI-LP and spleen, leading to high titers of circulating GPI autoAbs, the critical disease driver in this model.
Figure 7. Triggering of arthritis in SFB-colonized GF K/BxN mice.
(A) Experimental scheme. Mice were shipped from the GF Taconic facility to the SPF NYU facility on day 21 after birth and arrived the next day. After a 3-day rest; they were gavaged with SFB mono-feces or control GF feces (the rare animal with already swollen ankles was not used). Ankle thickening was measured every day from day 27 to day 33. (B) Measurement of ankle thickness beginning on day 27. N=9 for SFB-treated and N=5 for controls from 4 independent experiments. Asterisks indicate statistical significance using the Student s t-Test, *P<0.05. (C) Quantitative PCR analysis of SFB and total bacterial (EUB) 16S rRNA genes in mouse feces. GF K/BxN mice were gavaged either with their own feces (C) or with feces from SFB mono-colonized mice (SFB). Genomic DNA was isolated from fecal pellets on day 6 after gavage. Data combined from two separate experiments. (D) SI-LP lymphocytes were isolated from control or SFB- inoculated K/BxN mice. Cells were stained and analyzed by flow cytometry. Expression of IL-17 versus IFN-γ is plotted. Values refer to % of the gated population in total CD4+TCRβ+ cells. (E) Splenocytes were isolated from control or SFB-inoculated K/BxN mice, and were stained and analyzed by flow cytometry, gated as indicated. Plots depict IL-17 versus α4β7 staining. Values indicate % of IL-17+α4β7+ or IL-17+α4β7− cells in total CD4+ T cells (B220− CD3+CD4+). Data are representative of two independent experiments. (F) Sera were collected from control or SFB-inoculated K/BxN animals at the end of the experiment depicted in panel A. The bar indicates the mean. Asterisks indicate statistical significance using the Student s t-Test, *P<0.05.
DISCUSSION
Recent studies have highlighted a critical role for gut microbiota (Niess et al., 2008; Atarashi et al., 2008; Ivanov et al., 2008), in particular SFB (Salzman et al., 2009; Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009), in the differentiation of Th17 cells in the intestinal LP. The data presented herein establish the relevance of these observations for the initiation of autoimmune disease – in particular, a non-gut autoimmune disorder.
SFB are Gram-positive, spore-forming obligate anaerobes that have not yet been successfully cultured in vitro (Klaasen et al., 1992). Most closely related to Clostridia, and provisionally designated Candidatus arthromitus (Snel et al., 1995), they are long and filamentous, comprised of multiple segments with distinct septa (Klaasen et al., 1992). SFB-like bacteria have been detected morphologically in the ileum of all vertebrate species studied to date, including Homo sapiens (Klaasen et al., 1993a). They colonize the gut of mice at weaning (Garland CD et al., 1982), when they adhere tightly to epithelial cells of the ileum (Klaasen et al., 1992). SFB have been known for some time to interact with the immune system, promoting the development of robust LP lymphocyte populations, the secretion of IgA, and the recruitment of intraepithelial lymphocytes (Klaasen et al., 1993b; Talham et al., 1999; Umesaki et al., 1995). Not surprisingly, then, this bacterial species has been reported to impact on intestinal immune-responsiveness (Stepankova et al., 2007; Ivanov et al., 2009).
How does SFB promote the development of joint inflammation in K/BxN mice? K/BxN arthritis relies strongly on IL-17, and the appearance of IL-17-producing Th cells in both the intestinal LP and spleen depends critically on gut microbes, in particular SFB [shown here and (Atarashi et al., 2008; Ivanov et al., 2009; Salzman et al., 2009; Ivanov et al., 2008; Gaboriau-Routhiau et al., 2009; Niess et al., 2008)]. We do not rule out the possibility that other commensals can promote, or can synergize with SFB in promoting, arthritis in this model, but other species, including members of the SFB-related Clostridiaceae family, were not able to induce the accumulation of SI-LP Th17 cells in a previous set of studies (Ivanov et al., 2009).
An early step in K/BxN disease induction is likely to be activation of APCs residing in the intestinal LP, as gut microbes can have an indirect adjuvant effect in pathogen infections [e.g. (Benson et al., 2009)]. It was recently shown that ATP produced by gut bacteria drives a unique population of CD70hiCD11clo cecal LP APCs to produce IL-6, IL-23 and other factors that favor the differentiation of the Th17 cell subset, and that ex vivo co-culture of these APCs with naïve CD4+ T lymphocytes induces the appearance of Th17 cells (Atarashi et al., 2008). However, SFB does not appear to operate via ATP in the SI-LP, instead up-regulating the production of acute-phase isoforms of SAA in the ileum, which can act on DCs from the SI-LP to induce co-cultured naïve CD4+ T cells to differentiate into Th17 cells (Ivanov et al., 2009).
The activation of APCs in the SI-LP should be sufficient to drive an anti-GPI Th17 cell response in the vicinity and, indeed, 6- to 8-week-old SPF K/BxN mice showed a near-doubling of SI-LP Th17 cells compared with SPF BxN animals (data not shown). Given that GPI is expressed in all cell-types, and circulates in low amounts in the blood, and that this is a TCR-transgenic system with a high frequency of self-reactive T cells, there is no need to invoke more complicated scenarios entailing molecular mimicry (Harkiolaki et al., 2009) in this context, i.e. the initial activation of GPI-reactive T cells does not depend on cross-reactivity to a gut-microbe antigen.
Once generated, GPI-reactive SI-LP Th17 cells are competent to exit the gut and re-circulate (Sigmundsdottir and Butcher, 2008). Gut APCs, in particular the CD103+ subset of intestinal LP DCs, produce elevated amounts of retinoic acid, which induces associated T cells to express the gut-homing receptor, the α4β7 integrin. These “gut-imprinted” T cells re-circulate through the intestinal lymphatics, enter the bloodstream and preferentially home back to the LP. In the K/BxN system, a population of α4β7-expressing Th17 cells was retained in the spleen, where they are positioned to provide help for the characteristically massive GPI autoAb response. The alternative explanation that CD103+ DCs migrate from the gut to the spleen and induce α4β7+ Th17 cells there is less likely given reports that gut DCs generally do not migrate beyond the mesenteric lymph nodes (Macpherson and Uhr, 2004; Voedisch et al., 2009). The IL-17 produced by Th17 cells was required for effective GC formation in K/BxN spleens, which was a direct effect of this cytokine on B cells. Although IL-17 is not generally thought of as a “helper” cytokine for B cells, our data are reminiscent of findings on the BXD2 model that argued that this cytokine can act on B cells by suppressing their chemotactic response to CXCL12 (Hsu et al., 2008).
The generation of high titers of GPI autoAbs is a pivotal event in the K/BxN model (Korganow et al., 1999). They combine with circulating GPI to form immune complexes, which are deposited along the non-cellular joint surface where the cartilage meets the articular cavity (Matsumoto et al., 2002). Because of the dearth of inhibitors at this site, the alternative pathway of complement is activated, leading to the recruitment and activation of inflammatory leukocytes. As has been discussed at length (Matsumoto et al., 2002; Binstadt et al., 2006), the joint-specificity of the auto-inflammation in the K/BxN model does not result from joint-specific T or B cell responses, but rather from particularities of joint structure and physiology. Indeed, it is difficult to find any anti-GPI T and B cells in the joint itself (Kouskoff et al., 1996). Although the low titer of anti-GPI in the absence of SFB suffices in and of itself to explain the dampening of arthritis observed in GF-housed mice (Matsumoto et al., 1999), it remains possible that commensal microbes also impact on downstream disease processes. Indeed, a positive influence of Th17 cells on the K/BxN serum-transfer system was recently described (Jacobs JP et al., 2010).
The influence of microbial commensals on arthritis development in other mouse models has been variable, covering the range from inhibition to little effect to augmentation (Bjork et al., 1994; Chervonsky, 2010). However, the significance of these studies is difficult to assess because, in general, they relied on the administration of bacteria or bacterial products (often Complete Freunds Adjuvant) for the induction of disease. Our findings are conceptually different from the observation that fungal infection, likely through the lungs and/or skin of conventionally (compared with SPF-) housed mice, augments arthritis development in the skg model (Yoshitomi et al., 2005).
Because of the relatively high rate of discordance of human rheumatoid arthritis (RA) in monozygotic twins, the role of microbes in this disorder has been of great interest, although the conclusions have often been contentious (Edwards, 2008). Most of the attention has been devoted to disease correlations with infectious microorganisms, resulting in claims of association with a number of them, including Mycobacterium tuberculosis, Proteus mirabilis, Escherichia coli, Epstein-Bar virus, retroviruses, etc. However, none of the associations has emerged as dominating, and mechanistic insights are lacking. Only of late has some of the focus shifted to the potential influence of microbial commensals. Vaahtovuo et al reported differences in the intestinal microbiota of patients with early (<6-month duration) RA vis-à-vis controls with fibromyalgia, as assessed from the 16S rRNA composition of fecal samples (Vaahtovuo et al., 2008), but it is difficult to distinguish cause from effect in such a study. Clearly, this is an area that merits further exploration, which will probably need to partner with studies on animal models to establish causality, permit mechanistic dissection and allow pre-clinical evaluation of suggested therapeutic strategies. Indeed, antibiotics such as sulfasalazine and minocyline have been known for some time to have beneficial effects on RA progression, but underlying mechanisms remain the subject of substantial controversy (Stone et al., 2003).
More generally, commensal microbes can have a variable influence on different spontaneously developing autoimmune diseases (Chervonsky, 2010). For example, introduction of Aire−/− mice into a GF facility had no significant impact on the severity or scope of the multi-organ auto-inflammation that appears under SPF conditions (Gray et al., 2007). And it is well known that the penetrance of type-1 diabetes in the NOD mouse strain increases with cleaner housing conditions, rising to 100% in GF facilities (Pozzilli et al., 1993). It is tempting to speculate that these divergent effects might, at least in part, reflect the various diseases differential dependence on particular Th subsets. In this regard, it may be relevant that for neither of these diseases has there emerged definitive evidence of a critical role for Th17 cells (DeVoss et al., 2008; Martin-Orozco et al., 2009; Bending et al., 2009; Emamaullee et al., 2009).
EXPERIMENTAL PROCEDURES
Mice
K/BxN mice were generated by crossing KRN TCR transgenic mice on the B6 background (KRN/B6) (Kouskoff et al., 1996) with NOD mice in an SPF facility at the Harvard School of Public Health. Pups from KRN/B6 and NOD-background lines were rederived by cesarean section into the GF facility at Taconic Farms (Hudson, NY). Individuals from the two lines were crossed to generate K/BxN experimental animals. All GF mice were given sterilized food (NIH 31M) and water, and were tested weekly to establish that they were free of aerobic and anaerobic bacteria, parasites and fungi. Sentinel mice were also tested routinely and found to be negative for viral serologies. A complete list of excluded organisms is available on request. Mice were shipped in GF containers by Taconic to Boston or New York for measuring arthritis and obtaining experimental organs for analysis. Rag1−/− mice on the B6xNOD background (BxN Rag−/−) were obtained from our colony at the Jackson Laboratory. Il17ra−/− mice (Ye et al., 2001) were obtained from Amgen Washington, and were bred with B6g7 mice at our animal facility at the Harvard School of Public Health. ifng-deficient mice on the B6 genetic background were purchased from the Jackson Laboratory (Dalton et al., 1993), and appropriate crosses were performed to yield K/BxN mice homozygous or heterozygous for the Ifng-null mutation.
IL-17 was neutralized by treatment with a mAb recognizing it (MAB421, R&D Systems). Control Abs were purified polyclonal rat IgG (Jackson Immunoresearch).
For antibiotic treatment, 1 g/L of Ampicillin sodium salt (Sigma), 1g/L of Metronidazole (Acros Organics), 0.5 g/L Vancomycin hydrochloride (Acros Organics), or 1 g/l of Neomycin (Fisher BioReagents), were used as previously described (Atarashi et al., 2008). Antibiotics were added to the drinking water on a weekly basis. Sweetener (Equal) was added to the water (2.5g/L). For the treatment of neonates, antibiotic-supplemented water was provided to lactating mothers.
Ankle thickness was measured with a caliper (J15 Blet micrometer) as described previously (Wu et al., 2007).
ELISAs
Anti-GPI Ab titers were measured as described (Matsumoto et al., 1999). Briefly, ELISA plates were coated with recombinant mouse GPI at 5 mg/ml, and diluted mouse sera added. Subsequently, alkaline-phosphatase (AP)-conjugated anti-mouse IgG, IgG1 or IgG2a followed by AP-conjugated streptavidin were applied. After substrate addition, titers were quantified as optical density (OD) values via an ELISA reader.
Flow cytometry
Cells were collected for flow cytometry by filtering crushed spleen or thymus through a 40 μm nylon membrane. For surface staining: fluorophore-labeled mAbs specific for CD4, CD8, CD3, CD25, CD44, Fas, CD19, IgM, IgD, CD21, CD23, CXCR5, PD-1, CD45.1 or CD45.2 were obtained from BD Pharmingen. Anti-α4β7 and anti-CCR6 were from Biolegend. PNA-R was from Vector Laboratories. For intracellular cytokine staining: immediately after isolation, the cells were incubated for 4 hr with 50 ng/ml phorbol 12-myristate 13-acetate (Sigma), 1μM ionomycin (Sigma) and BD GolgiPlug (1:1000 dilution) at 37°C. Intracellular cytokine staining was performed using Cytofix/Cytoperm (BD Pharmingen) per the manufacturer s instructions. Abs recognizing IL-17 and IFN-γ were obtained from Biolegend and BD Pharmingen, respectively. Foxp3 staining, Foxp3 Staining Buffer Set was obtained from eBioscience, and intracellular staining was performed following the manufacturer s instructions. Cells were run on an LSRII (BD Biosciences), and analysis was performed with FloJo (TreeStar) software.
ELISPOT assay
ELISPOT assays were performed using Multiscreen IP Plates (Millipore). The plates were pre-wet with 15 μl of 35% ethanol (v/v in Milli-Q water) for one minute, rinsed with 150 μl sterile phosphate-buffered saline (PBS) three times, coated with 100 μl (10 μg/ml) recombinant GPI (Matsumoto et al., 2002) in sterile PBS, and incubated overnight at 4°C. The next day, plates were washed with Milli-Q water and blocked with 150 μl per well of tissue-culture medium (RPMI-1640, 10% fetal bovine serum, 1% nonessential amino acids, penicillin, streptomycin, glutamine) for 2 hr at 37°C. B cells from BxN or K/BxN mice were positively isolated with directly conjugated MACS beads (Miltenyi Biotec) according to the manufacturer s instructions. Cells were resupended at 2.5 × 106 cells/ml in medium, and 100 ul of cell suspension was added into the wells. Cells were serially diluted and incubated for 6 hr at 37°C. After washing, alkaline phosphatase-conjugated anti-mouse total IgG was applied, and the plates were incubated for 2 hours at 37°C. Plates were then washed, and 1 step™ NBT/BCIP substrate (Pierce) was added. Spots were developed during 5 min of incubation at room temperature. The plates were rinsed with water, dried overnight in the dark, and analyzed using the CTL-ImmunoSpot® UV Analyzer.
T cell proliferation and T cell suppression assays
For T cell proliferation assays, total splenocytes (2×105) in tissue-culture medium were added to 96-well plates. GPI peptide (GPI282–294) was added to the culture at various concentrations as indicated in the relevant figure. After 2 days of culture, 1μCi of 3H-thymidine was added to each well, the plates were incubated overnight, and cells were harvested and the radioactivity determined by a beta counter. For T cell suppression assays, responder T cells (CD4+CD25−) were sorted from spleens of SPF K/BxN mice and Tregs (CD4+CD25+) were sorted from spleens of either GF or SPF K/BxN mice. Responder T cells were stimulated with anti-CD3/CD28 beads (Dynabeads, Invitrogen) and cultured in complete medium at a density of 2.5×104-5.0×104 cells/well, either alone or with various concentrations of Tregs for 3 days. 3H-thymidine incorporation was examined as described above.
Gene-expression analyses
RNA was prepared as described (Hill et al., 2008). For microarray analysis, RNA was labeled and hybridized to GeneChip Mouse Genome 430 2.0 arrays following the Affymetrix protocols. GF or SPF splenic CD3+CD4+ T cells from BxN or K/BxN mice were isolated via MoFlo sorting (DakoCytomation). Data were analyzed using Multiplot software. The Th1, Th2 and Th17 signatures were derived from the data of Dong and collaborators (Nurieva et al., 2008), each signature generated by using 2 as an arbitrary FC cut-off over the expression value of the other two cell-types. All datasets deposited at NCBI/GEO under accession # XXX.
RNA was isolated from splenocytes via Trizol (6756), and was reverse-transcribed using oligo dT priming and Superscript polymerase (Invitrogen). Quantitative RT-PCR was performed on an Mx3000p instrument (Stratagene), using gene-specific fluorogenic assays (TaqMan, Applied Biosystems). Forward primers (FPs) and reverse primers (RPs) were from MWG Biotech, and probes for IL-4 and IFN-γ were ordered from Applied Biosystems. IL-4 (FP: TCCTCACAGCAACGAAGAACAC, RP: CAAGCATGGAGTTTTCCCATG, probe: TGTAGGGCTTCCAAGGTGCTTCGCATATT), IFN-γ (FP: CAGCAACAGCAAGGCGAAA, RP: CTGGACCTGTGGGTTGTTGAC, probe: TCAAACTTGGCAATACTCATGAATGCATCCT). For IL-17A, a 10 μl final reaction mix containing TaqMan Universal PCR Master Mix (Applied Biosystems) and IL-17A TaqMan Gene Expression Assays (Mm00439619_m1) were used. Cytokine transcripts in spleens were quantified by RT-PCR using hypoxanthine guanine phosphoribosyl transferase mRNA as an internal standard.
Cell Transfers
B cells were positively purified on B220-conjugated MACS beads from B6g7.Il17ra−/− mice or WT B6g7 littermate controls. B cells (1X107) from either WT or Il17ra−/− mice were combined with splenocytes (1.2X107) from arthritic K/BxN mice, and were transferred into lightly irradiated (450R) BxN.Rag1−/− recipients. Recipient mouse splenocytes were isolated after two weeks for flow cytometric analyses of B cell reconstitution.
SI-LP cell isolation and analysis
SI-LP were isolated as described, with some modification (Ivanov et al., 2006; Atarashi et al., 2008). Briefly, the SI was taken, residual mesenteric fat tissue was removed, Peyer’s patches were carefully excised, and the intestine was opened longitudinally. It was then thoroughly washed in ice-cold PBS and cut into 1 cm pieces, which were incubated twice in 25 ml of 5 mM ethylenediaminetetraacetic acid (EDTA) and 0.145 mg/ml DL-Dithiothreitol (DTT) in Dulbecco’s Modified Eagle Medium (DMEM) for 40 min at 37°C at a rotation speed of 200 rpm. After incubation, the epithelial cell layer, containing the intraepithelial lymphocytes, was removed by intensive vortexing and passing through a 100 μm cell strainer. After the second EDTA incubation, the bits were washed in PBS, cut into 1 mm2 pieces using scissors, and placed in 15 ml digestion solution containing 1 mg/ml each of Collagenase D (Roche) and 0.15 mg/ml DNase I (Sigma), and 200 ng/ml liberase Cl (Roche). Digestion was performed by incubating the pieces at 37°C for 20 min with rotation. After the initial 20 min, the solution was vortexed intensely and passed through a 100 μm cell strainer. The supernatants were passed through a 40 μm cell strainer and the cells were resuspended in 10% DMEM medium for stimulation.
Microbiota reconstitution
For inoculation of GF mice with SFB, fecal pellets were collected from SFB-monocolonized mice using sterilized test tubes in the vinyl-isolator, and were preserved frozen under dry ice until immediately before oral administration. Colonizations were performed by oral gavage with 300–400 μl of suspension obtained by homogenizing the fecal pellets in water. Control mice were gavaged with homogenates prepared from their own feces.
16S rRNA gene quantitative PCR analysis
Bacterial genomic DNA was extracted from fresh or frozen fecal samples (within an experiment the samples were treated identically) by phenol-chloroform extraction as previously described (Ivanov et al., 2009).
Statistical Analysis
Asterisks indicate statistical significance. Differences were considered significant at p< 0.05 by the Student s t test (two-tailed, unpaired). Where indicated, p values from chi-square (χ2) tests were used instead. The area under the curve (AUC) was calculated for each animal in an experimental set followed by a Student s t test between groups (Prism 5; Graph-Pad Software, San Diego, CA).
Supplementary Material
Acknowledgments
We thank Dr. Joel Tocker ( Amgen Inc, Seattle, WA) for providing IL-17R−/− mice; Dr A Ortiz-Lopez for RT-PCR primers and assistance with the mouse colonies; Dr. M Kriegel for assistance with part of the SFB quantitative PCR analysis; K. Leatherbee for microarrays; J. Lavecchio and G. Buruzula for flow cytometry and Dr C Laplace for assistance with figure preparation. This work was supported by grants from the NIH to DM and CB (P01 AI065858) or to DRL (RC1AI 087266) and by Joslin s NIDDK-funded DERC cores. JD was funded by National Research Service Award (DFCI/NCI: T32 CA007386) and III by a Pathway to Independence Award from the NIH (1K99DK085329-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Bending D, De La Pena H, Veldhoen M, Phillips JM, Uyttenhove C, Stockinger B, Cooke A. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J Clin Invest. 2009 doi: 10.1172/JCI37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson A, Pifer R, Behrendt CL, Hooper LV, Yarovinsky F. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe. 2009;6:187–196. doi: 10.1016/j.chom.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binstadt BA, Patel PR, Alencar H, Nigrovic PA, Lee DM, Mahmood U, Weissleder R, Mathis D, Benoist C. Particularities of the vasculature can promote the organ specificity of autoimmune attack. Nat Immunol. 2006;7:284–292. doi: 10.1038/ni1306. [DOI] [PubMed] [Google Scholar]
- Bjork J, Kleinau S, Midtvedt T, Klareskog L, Smedegard G. Role of the bowel flora for development of immunity to hsp 65 and arthritis in three experimental models. Scand J Immunol. 1994;40:648–652. doi: 10.1111/j.1365-3083.1994.tb03518.x. [DOI] [PubMed] [Google Scholar]
- Chervonsky AV. Influence of microbial environment on autoimmunity. Nat Immunol. 2010;11:28–35. doi: 10.1038/ni.1801. [DOI] [PubMed] [Google Scholar]
- Dalton DK, Pitts-Meek S, Keshav S, Figari IS, Bradley A, Stewart TA. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- DeVoss JJ, Shum AK, Johannes KP, Lu W, Krawisz AK, Wang P, Yang T, Leclair NP, Austin C, Strauss EC, et al. Effector mechanisms of the autoimmune syndrome in the murine model of autoimmune polyglandular syndrome type 1. J Immunol. 2008;181:4072–4079. doi: 10.4049/jimmunol.181.6.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop BA, Vaishnava S, Hooper LV. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Edwards CJ. Commensal gut bacteria and the etiopathogenesis of rheumatoid arthritis. J Rheumatol. 2008;35:1477–1479. [PubMed] [Google Scholar]
- Emamaullee JA, Davis J, Merani S, Toso C, Elliott JF, Thiesen A, Shapiro AM. Inhibition of Th17 cells regulates autoimmune diabetes in NOD mice. Diabetes. 2009;58:1302–1311. doi: 10.2337/db08-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De PM, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Garland CD, Lee A, Dickson MR. Segmented Filamentous Bacteria in the Rodent Small Intestine: Their Colonization of Growing Animals and Possible Role in Host Resistance to Salmonella. Microb Ecol. 1982;8:181–190. doi: 10.1007/BF02010451. [DOI] [PubMed] [Google Scholar]
- Gray DH, Gavanescu I, Benoist C, Mathis D. Danger-free autoimmune disease in Aire-deficient mice. Proc Natl Acad Sci U S A. 2007;104:18193–18198. doi: 10.1073/pnas.0709160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkiolaki M, Holmes SL, Svendsen P, Gregersen JW, Jensen LT, McMahon R, Friese MA, van BG, Etzensperger R, Tzartos JS, et al. T cell-mediated autoimmune disease due to low-affinity crossreactivity to common microbial peptides. Immunity. 2009;30:348–357. doi: 10.1016/j.immuni.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Hill JA, Hall JA, Sun CM, Cai Q, Ghyselinck N, Chambon P, Belkaid Y, Mathis D, Benoist C. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity. 2008;29:758–770. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HC, Yang P, Wang J, Wu Q, Myers R, Chen J, Yi J, Guentert T, Tousson A, Stanus AL, et al. Interleukin 17-producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat Immunol. 2008;9:166–175. doi: 10.1038/ni1552. [DOI] [PubMed] [Google Scholar]
- Huang H, Benoist C, Mathis D. Rituximab specifically depletes short-lived autoreactive plasma cells in a mouse model of inflammatory arthritis. Proc Natl Acad Sci U S A. 2010;107:4658–4663. doi: 10.1073/pnas.1001074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos RL, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Jacobs JP, Wu H-S, Benoist C, Mathis D. IL-17-producing T cells can augment autoantibody-induced arthritis. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0912152106. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D, King T, Aminov R. Importance of microbial colonization of the gut in early life to the development of immunity. Mutat Res. 2007;622:58–69. doi: 10.1016/j.mrfmmm.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Klaasen HL, Koopman JP, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria. FEMS Microbiol Rev. 1992;8:165–180. doi: 10.1111/j.1574-6968.1992.tb04986.x. [DOI] [PubMed] [Google Scholar]
- Klaasen HL, Koopman JP, Van den Brink ME, Bakker MH, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab Anim. 1993a;27:141–150. doi: 10.1258/002367793780810441. [DOI] [PubMed] [Google Scholar]
- Klaasen HL, Van der Heijden PJ, Stok W, Poelma FG, Koopman JP, Van den Brink ME, Bakker MH, Eling WM, Beynen AC. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect Immun. 1993b;61:303–306. doi: 10.1128/iai.61.1.303-306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korganow AS, Ji H, Mangialaio S, Duchatelle V, Pelanda R, Martin T, Degott C, Kikutani H, Rajewsky K, Pasquali JL, et al. From systemic T cell self-reactivity to organ-specific autoimmune disease via immunoglobulins. Immunity. 1999;10:451–461. doi: 10.1016/s1074-7613(00)80045-x. [DOI] [PubMed] [Google Scholar]
- Kouskoff V, Korganow AS, Duchatelle V, Degott C, Benoist C, Mathis D. Organ-specific disease provoked by systemic autoimmunity. Cell. 1996;87:811–822. doi: 10.1016/s0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. Evolution of mammals and their gut microbes. Science. 2008a;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008b;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Maccioni M, Zeder-Lutz G, Huang H, Ebel C, Gerber P, Hergueux J, Marchal P, Duchatelle V, Degott C, van Regenmortel M, et al. Arthritogenic monoclonal antibodies from K/BxN mice. J Exp Med. 2002;195:1071–1077. doi: 10.1084/jem.20011941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- Martin-Orozco N, Chung Y, Chang SH, Wang YH, Dong C. Th17 cells promote pancreatic inflammation but only induce diabetes efficiently in lymphopenic hosts after conversion into Th1 cells. Eur J Immunol. 2009;39:216–224. doi: 10.1002/eji.200838475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto I, Maccioni M, Lee DM, Maurice M, Simmons B, Brenner M, Mathis D, Benoist C. How antibodies to a ubiquitous cytoplasmic enzyme may provoke joint-specific autoimmune disease. Nat Immunol. 2002;3:360–365. doi: 10.1038/ni772. [DOI] [PubMed] [Google Scholar]
- Matsumoto I, Staub A, Benoist C, Mathis D. Arthritis provoked by linked T and B cell recognition of a glycolytic enzyme. Science. 1999;286:1732–1735. doi: 10.1126/science.286.5445.1732. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat Rev Immunol. 2006;6:849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Niess JH, Leithauser F, Adler G, Reimann J. Commensal gut flora drives the expansion of proinflammatory CD4 T cells in the colonic lamina propria under normal and inflammatory conditions. J Immunol. 2008;180:559–568. doi: 10.4049/jimmunol.180.1.559. [DOI] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzilli P, Signore A, Williams AJK, Beales PE. NOD mouse colonies around the world-recent facts and figures. Immunol Today. 1993;14:193–196. doi: 10.1016/0167-5699(93)90160-M. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Medzhitov R. Innate immune recognition of the indigenous microbial flora. Mucosal Immunol. 2008;1(Suppl 1):S10–S14. doi: 10.1038/mi.2008.49. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2009 doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigmundsdottir H, Butcher EC. Environmental cues, dendritic cells and the programming of tissue-selective lymphocyte trafficking. Nat Immunol. 2008;9:981–987. doi: 10.1038/ni.f.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel J, Heinen PP, Blok HJ, Carman RJ, Duncan AJ, Allen PC, Collins MD. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus”. Int J Syst Bacteriol. 1995;45:780–782. doi: 10.1099/00207713-45-4-780. [DOI] [PubMed] [Google Scholar]
- Stepankova R, Powrie F, Kofronova O, Kozakova H, Hudcovic T, Hrncir T, Uhlig H, Read S, Rehakova Z, Benada O, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm Bowel Dis. 2007;13:1202–1211. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- Stone M, Fortin PR, Pacheco-Tena C, Inman RD. Should tetracycline treatment be used more extensively for rheumatoid arthritis? Metaanalysis demonstrates clinical benefit with reduction in disease activity. J Rheumatol. 2003;30:2112–2122. [PubMed] [Google Scholar]
- Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol. 1995;39:555–562. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol. 2008;35:1500–1505. [PubMed] [Google Scholar]
- Vassallo MF, Walker WA. Neonatal microbial flora and disease outcome. Nestle Nutr Workshop Ser Pediatr Program. 2008;61:211–224. doi: 10.1159/000113496. [DOI] [PubMed] [Google Scholar]
- Voedisch S, Koenecke C, David S, Herbrand H, Forster R, Rhen M, Pabst O. Mesenteric lymph nodes confine dendritic cell-mediated dissemination of Salmonella enterica serovar Typhimurium and limit systemic disease in mice. Infect Immun. 2009;77:3170–3180. doi: 10.1128/IAI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1:69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Sawaya H, Binstadt B, Brickelmaier M, Blasius A, Gorelik L, Mahmood U, Weissleder R, Carulli J, Benoist C, et al. Inflammatory arthritis can be reined in by CpG-induced DC NK cell cross talk. J Exp Med. 2007;204:1911–1922. doi: 10.1084/jem.20070285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang P, Zhang J, et al. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med. 2001;194:519–527. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitomi H, Sakaguchi N, Kobayashi K, Brown GD, Tagami T, Sakihama T, Hirota K, Tanaka S, Nomura T, Miki I, et al. A role for fungal {beta}-glucans and their receptor Dectin-1 in the induction of autoimmune arthritis in genetically susceptible mice. J Exp Med. 2005;201:949–960. doi: 10.1084/jem.20041758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.