Abstract
The human distal gut harbors a vast ensemble of microbes (the microbiota) that provide us with important metabolic capabilities, including the ability to extract energy from otherwise indigestible dietary polysaccharides1–6. Studies of a small number of unrelated, healthy adults have revealed substantial diversity in their gut communities, as measured by sequencing 16S rRNA genes6–8, yet how this diversity relates to function and to the rest of the genes in the collective genomes of the microbiota (the gut microbiome) remains obscure. Studies of lean and obese mice suggest that the gut microbiota affects energy balance by influencing the efficiency of calorie harvest from the diet, and how this harvested energy is utilized and stored3–5. To address the question of how host genotype, environmental exposures, and host adiposity influence the gut microbiome, we have characterized the fecal microbial communities of adult female monozygotic and dizygotic twin pairs concordant for leanness or obesity, and their mothers. Analysis of 154 individuals yielded 9,920 near full-length and 1,937,461 partial bacterial 16S rRNA sequences, plus 2.14 gigabases from their microbiomes. The results reveal that the human gut microbiome is shared among family members, but that each person’s gut microbial community varies in the specific bacterial lineages present, with a comparable degree of co-variation between adult monozygotic and dizygotic twin pairs. However, there was a wide array of shared microbial genes among sampled individuals, comprising an extensive, identifiable ‘core microbiome’ at the gene, rather than at the organismal lineage level. Obesity is associated with phylum-level changes in the microbiota, reduced bacterial diversity, and altered representation of bacterial genes and metabolic pathways. These results demonstrate that a diversity of organismal assemblages can nonetheless yield a core microbiome at a functional level, and that deviations from this core are associated with different physiologic states (obese versus lean).
We characterized gut microbial communities in 31 monozygotic (MZ) twin pairs, 23 dizygotic (DZ) twin pairs, and where available their mothers (n=46) (Supplementary Tables 1–5). MZ and DZ co-twins and parent-offspring pairs provided an attractive paradigm for assessing the impact of genotype and shared early environment exposures on the gut microbiome. Moreover, genetically ‘identical’9 MZ twin pairs gain weight in response to overfeeding in a more reproducible way than do unrelated individuals10 and are more concordant for body mass index (BMI) than DZ twin pairs11.
Twin pairs who had been enrolled in the Missouri Adolescent Female Twin Study (MOAFTS12) were recruited for this study (mean period of enrollment in MOAFTS, 11.7±1.2 years; range, 4.4–13.0 years). All twins were 25–32 years old, of European or African ancestry (EA and AA, respectively), were generally concordant for obesity (BMI≥30 kg/m2) or leanness (BMI=18.5–24.9 kg/m2) [1 twin pair was lean/overweight (overweight defined as BMI ≥25 and <30) and 6 pairs were overweight/obese], and had not taken antibiotics for at least 5.49±0.09 months. Each participant completed a detailed medical, lifestyle, and dietary questionnaire: they were broadly representative of the overall Missouri population with respect to BMI, parity, education, and martial status (see Supplementary Results). Although all were born in Missouri, they currently live throughout the USA: 29% live in the same house, but some live >800 km apart. Since fecal samples are readily attainable and representative of interpersonal differences in gut microbial ecology7, they were collected from each individual and frozen immediately. The collection procedure was repeated again with an average interval between sampling of 57±4 days.
To characterize the bacterial lineages present in the fecal microbiotas of these 154 individuals, we performed 16S rRNA sequencing, targeting the full-length gene with an ABI 3730xl capillary sequencer. Additionally, we performed multiplex pyrosequencing with a 454 FLX instrument to survey the gene’s V2 variable region13 and it’s V6 hypervariable region14 (Supplementary Tables 1–3).
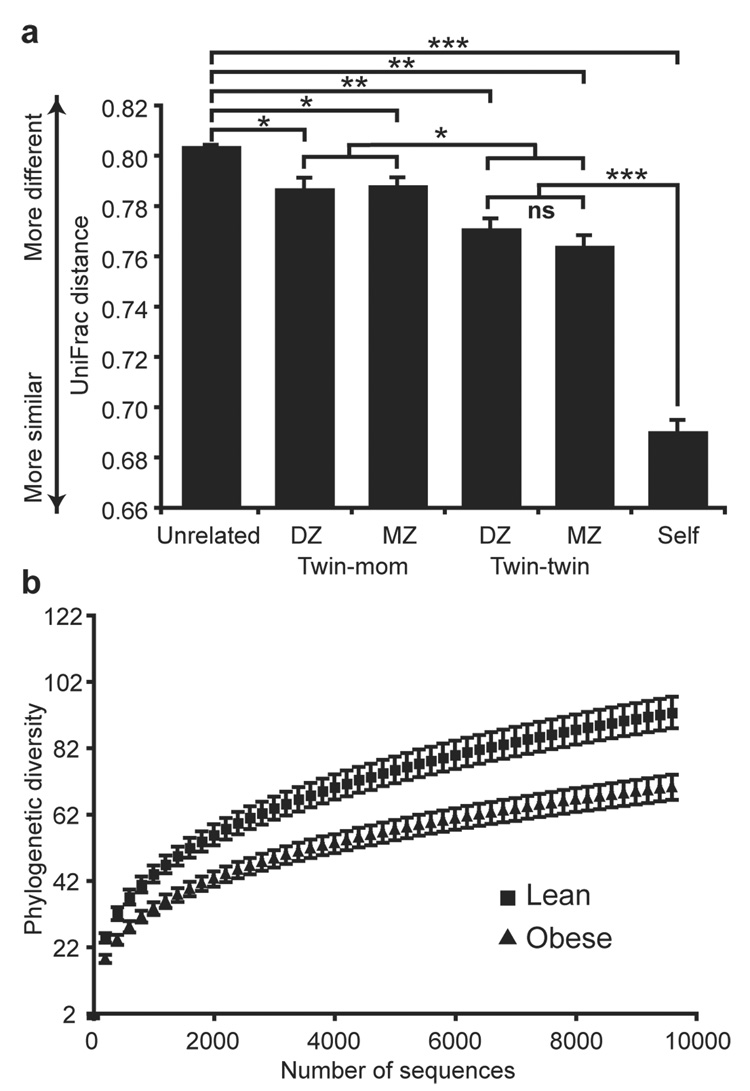
Complementary phylogenetic and taxon-based methods were used to compare 16S rRNA sequences among fecal communities (see Methods). No matter which region of the gene was examined, individuals from the same family (a twin and her co-twin, or twins and their mother) had a more similar bacterial community structure than unrelated individuals (Fig. 1A and Supplementary Fig. 1A,B), and shared significantly more species-level phylotypes (defined as sharing ≥97% identity in their 16S rRNA sequences) [G=55.2, p<10−12 (V2); G=12.3, p<0.001 (V6); G=11.3, p<0.001 (full-length)]. No significant correlation was seen between the degree of physical separation of family members’ current homes and the degree of similarity between their microbial communities (defined by UniFrac15). The observed familial similarity was not due to an indirect effect of the physiologic states of obesity versus leanness; similar results were observed after stratifying twin-pairs and their mothers by BMI category (concordant lean or concordant obese individuals; Supplementary Fig. 2). Surprisingly, there was no significant difference in the degree of similarity in the gut microbiotas of adult MZ versus DZ twin-pairs (Fig. 1A). However, in the present study we could not assess whether MZ and DZ twin pairs had different degrees of similarities at earlier stages of their lives.
Figure 1. 16S rRNA gene surveys reveal familial similarity and reduced diversity of the gut microbiota in obese individuals.
(A) Average unweighted UniFrac distance (a measure of differences in bacterial community structure) between individuals over time (self), twin-pairs, twins and their mother, and unrelated individuals [1,000 sequences per V2 dataset; Student’s t-test with Monte Carlo; *p<10−5; **p<10−14; ***p<10−41; mean±SEM]. (B) Phylogenetic diversity curves for the microbiota of lean and obese individuals (based on 1 to 10,000 sequences per V6 dataset; mean±95%CI shown).
Multiplex pyrosequencing of V2 and V6 amplicons allowed higher levels of coverage compared to what was feasible using Sanger sequencing, reaching on average 3,984±232 (V2) and 24,786±1,403 (V6) sequences per sample. To control for differences in coverage, all analyses were performed on an equal number of randomly selected sequences [200 full-length, 1,000 V2, and 10,000 V6]. At this level of coverage, there was little overlap between the sampled fecal communities. Moreover, the number of 16S rRNA gene sequences belonging to each phylotype varied greatly between fecal microbiotas (Supplementary Tables 6–8).
Since this apparent lack of overlap could reflect the level of coverage (Supplementary Tables 1–3), we subsequently searched all hosts for bacterial phylotypes present at high abundance using a sampling model based on a combination of standard Poisson and binomial sampling statistics. The analysis allowed us to conclude that no phylotype was present at more than ~0.5% abundance in all of the samples in this study (see Supplementary Results). Finally, we subsampled our dataset by randomly selecting 50–3,000 sequences/sample; again, no phylotypes were detectable in all individuals sampled within this range of coverage (Supplementary Fig. 3).
Samples taken from the same individual at the initial collection point and 57±4 days later were consistent with respect to the specific phylotypes found (Supplementary Figs. 4,5), but showed variations in relative abundance of the major gut bacterial phyla (Supplementary Fig. 6). There was no significant association between UniFrac distance and the time between sample collections. Overall, fecal samples from the same individual were much more similar to one another than samples from family members or unrelated individuals (Fig. 1A), demonstrating that short-term temporal changes in community structure within an individual are minor compared to inter-personal differences.
Analysis of 16S rRNA datasets produced by the three PCR-based methods, plus shotgun sequencing of community DNA (see below), revealed a lower proportion of Bacteroidetes and a higher proportion of Actinobacteria in obese versus lean EA and AA individuals (Supplementary Table 9). Combining the individual p-values across these independent analyses using Fisher’s method disclosed significantly less Bacteroidetes (p=0.003), more Actinobacteria (p=0.002), but no significant difference in Firmicutes (p=0.09). These findings are in agreement with previous work showing comparable differences in both taxa in mice2 and a progressive increase the representation of Bacteroidetes when 12 unrelated obese humans lost weight after being placed on one of two reduced calorie diets6.
Across all methods, obesity was associated with a significant decrease in the level of diversity (Fig. 1B plus Supplementary Fig. 1C–F). This reduced diversity suggests an analogy: the obese gut microbiota is not like a rainforest or reef, which are adapted to high energy flux and are highly diverse, but rather may be more like a fertilizer runoff where a reduced diversity microbial community blooms with abnormal energy input16.
We subsequently characterized the microbial lineage and gene content of the fecal microbiomes of 18 individuals representing 6 of the families (3 lean EA MZ twin-pairs and their mothers plus 3 obese EA MZ twin pairs and their mothers) through shotgun pyrosequencing (Supplementary Tables 4,5) and BLASTX comparisons against a number of databases [KEGG17 (v44) and STRING18] plus a custom database of 44 reference human gut microbial genomes (Supplementary Figs. 7–10 and Supplementary Results). Our analysis parameters were validated using control datasets comprised of randomly fragmented microbial genes with annotations in the KEGG database17 (Supplementary Fig. 11 and Supplementary Methods). We also tested how technical advances that produce longer reads might improve these assignments by sequencing fecal community samples from one twin pair using Titanium pyrosequencing methods [average read length of 341±134 nt (SD) versus 208±68 nt for the standard FLX method]. Supplementary Fig. 12 shows that the frequency and quality of sequence assignments is improved as read length increases from 200 to 350 nt.
The 18 microbiomes were searched to identify sequences matching domains from experimentally validated Carbohydrate-Active enZymes (CAZymes). Sequences matching 156 total CAZy families were found within at least one human gut microbiome, including 77 glycoside hydrolase, 21 carbohydrate-binding module, 35 glycosyltransferase, 12 polysaccharide lyase, and 11 carbohydrate-esterase families (Supplementary Table 10). On average 2.62±0.13% of the sequences in the gut microbiome could be assigned to CAZymes (total of 217,615 sequences), a percentage that is greater than the most abundant KEGG pathway (‘Transporters’; 1.20±0.06% of the filtered sequences generated from each sample), and indicative of the abundant and diverse set of microbial genes directed towards accessing a wide range of polysaccharides.
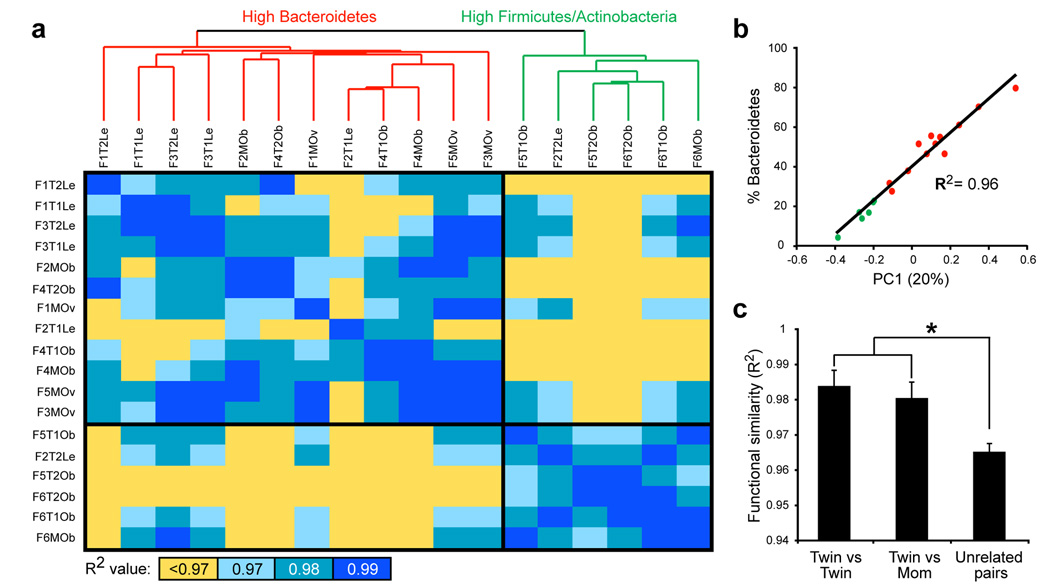
Category-based clustering of the functions from each microbiome was performed using Principal Components Analysis (PCA) and hierarchical clustering19. Two distinct clusters of gut microbiomes were identified based on metabolic profile, corresponding to samples with an increased abundance of Firmicutes and Actinobacteria, and samples with a high abundance of Bacteroidetes (Fig. 2A). A linear regression of the first principal component (PC1, explaining 20% of the functional variance) and the relative abundance of the Bacteroidetes showed a highly significant correlation (R2=0.96, p<10−12; Fig. 2B). Functional profiles stabilized within each individual’s microbiome after ~20,000 sequences had been accumulated (Supplementary Fig. 13). Family members had more similar profiles than unrelated individuals (Fig. 2C), suggesting that shared bacterial community structure (who’s there based on 16S rRNA analyses) also translates into shared community-wide relative abundance of metabolic pathways. Accordingly, a direct comparison of functional and taxonomic similarity (see Supplementary Methods) disclosed a significant association: individuals with similar taxonomic profiles also share similar metabolic profiles (p<0.001; Mantel test).
Figure 2. Metabolic pathway-based clustering and analysis of the human gut microbiome of MZ twins.
(A) Clustering of functional profiles based on the relative abundance of KEGG metabolic pathways. All pairwise comparisons were made of the profiles by calculating each R2 value. Sample ID nomenclature: Family number, Twin number or mom, and BMI category (Le=lean, Ov=overweight, Ob=obese; e.g. F1T1Le stands for family 1, twin 1, lean). (B) The relative abundance of Bacteroidetes as a function of the first principal component derived from an analysis of KEGG metabolic profiles. (C) Comparisons of functional similarity between twin pairs, between twins and their mother, and between unrelated individuals. Asterisks indicate significant differences (Student’s t-test with Monte Carlo; p<0.01; mean±SEM).
Functional clustering of phylum-wide sequence bins representing microbiome reads assigned to 23 human gut Firmicutes and 14 Bacteroidetes reference genomes showed discrete clustering by phylum (Supplementary Figs. 14A,15). Bootstrap analyses of the relative abundance of metabolic pathways in the microbiome-derived Firmicutes and Bacteroidetes sequence bins, disclosed 26 pathways with a significantly different relative abundance (Supplementary Fig. 14A). The Bacteroidetes bins were enriched for a number of carbohydrate metabolism pathways, while the Firmicutes bins were enriched for transport systems. The finding is consistent with our CAZyme analysis, which revealed a significantly higher relative abundance of glycoside hydrolases, carbohydrate-binding modules, glycosyltransferases, polysaccharide lyases, and carbohydrate esterases in the Bacteroidetes sequence bins (Supplementary Fig. 14B).
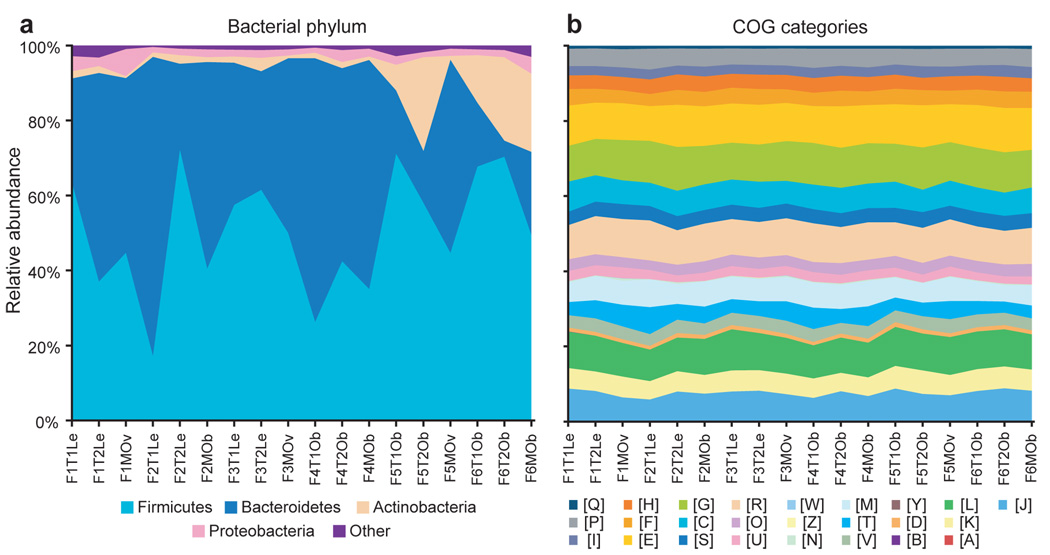
One of the major goals of the International Human Microbiome Project(s) is to determine whether there is an identifiable ‘core microbiome’ of shared organisms, genes, or functional capabilities found in a given body habitat of all or the vast majority of humans1. Although all of the 18 gut microbiomes surveyed showed a high level of beta-diversity with respect to the relative abundance of bacterial phyla (Fig. 3A), analysis of the relative abundance of broad functional categories of genes (COG) and metabolic pathways (KEGG) revealed a generally consistent pattern regardless of the sample surveyed (Fig. 3B and Supplementary Table 11): the pattern is also consistent with results we obtained from an meta-analysis of previously published gut microbiome datasets from nine adults20,21 (Supplementary Fig. 16). This consistency is not simply due to the broad level of these annotations, as a similar analysis of Bacteroidetes and Firmicutes reference genomes revealed substantial variation in the relative abundance of each category (see Supplementary Fig. 17). Furthermore, pair-wise comparisons of metabolic profiles obtained from the 18 microbiomes in this study revealed an average R2 of 0.97±0.002 (Fig. 2A), indicating a high level of functional similarity.
Figure 3. Comparison of taxonomic and functional variations in the human gut microbiome.
(A) Relative abundance of major phyla across 18 fecal microbiomes from MZ twins and their mothers, based on BLASTX comparisons of microbiomes and the NCBI non-redundant database. (B) Relative abundance of COG categories across each sampled gut microbiome.
Overall functional diversity was compared using the Shannon index22, a measurement that combines diversity (the number of different types of metabolic pathways) and evenness (the relative abundance of each pathway). The human gut microbiomes surveyed had a stable and high Shannon index value (4.63±0.01), close to the maximum possible level of functional diversity (5.54; see Supplementary Methods). Despite the presence of a small number of abundant metabolic pathways (listed in Supplementary Table 11), the overall functional profile of each gut microbiome is quite even (Shannon evenness of 0.84±0.001 on a scale of 0 to 1), demonstrating that most metabolic pathways are found at a similar level of abundance. Interestingly, the level of functional diversity in each microbiome was significantly linked to the relative abundance of the Bacteroidetes (R2=0.81, p<10−6); microbiomes enriched for Firmicutes/Actinobacteria had a lower level of functional diversity. This observation is consistent with an analysis of simulated metagenomic reads generated from each of 36 Bacteroidetes and Firmicutes genomes (Supplementary Fig. 18): on average, the Bacteroidetes genomes have a significantly higher level of both functional diversity and evenness (Mann-Whitney, p<0.01).
At a finer level, 26–53% of ‘enzyme’-level functional groups (KEGG/CAZy/STRING) were shared across all 18 microbiomes, while 8–22% of the groups were unique to a single microbiome (Supplementary Fig. 19A–C). The ‘core’ functional groups present in all microbiomes were also highly abundant, representing 93–98% of the total sequences. Given the higher relative abundance of these ‘core’ groups, >95% were found after 26.11±2.02 Mb of sequence was collected from a given microbiome, whereas the ‘variable’ groups continue to increase substantially with each additional Mb of sequence. Of course, any estimate of the total size of the core microbiome will be dependent upon sequencing effort, especially for functional groups found at a low abundance. On average, our survey achieved greater than 450,000 sequences per fecal sample, which, assuming an even distribution, would allow us to sample groups found at a relative abundance of 10−4. To estimate the total size of the core microbiome based on the 18 individuals, we randomly sub-sampled each microbiome in 1,000 sequence intervals (Supplementary Fig. 19D). Based on this analysis, the core microbiome is approaching a total of 2,142 total orthologous groups (one site binding hyperbola curve fit to the resulting rarefaction curve, R2=0.9966), indicating that we have identified 93% of functional groups (defined by STRING) found within the core microbiome of the 18 individuals surveyed. Of these core groups, 71% (CAZy), 64% (KEGG), and 56% (STRING) were also found in the 9 previously published but much lower coverage datasets generated by capillary sequencing of adult fecal DNA20,21 (average of 78,413±2,044 bidirectional reads/sample; see Supplementary Methods).
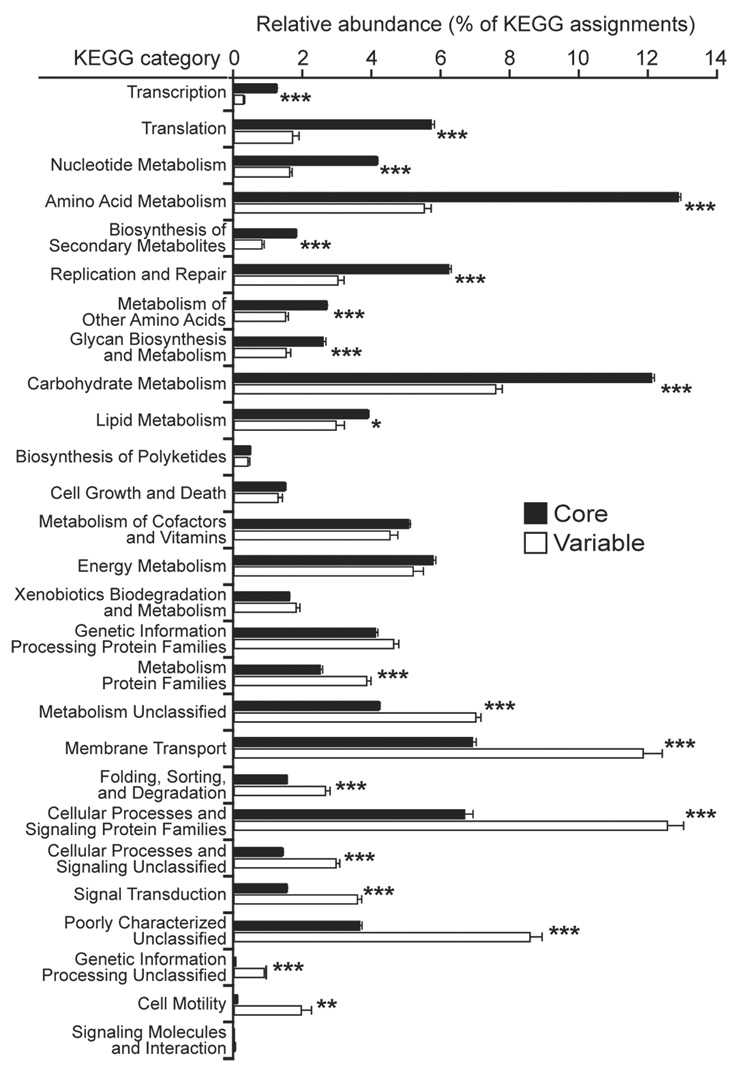
Metabolic reconstructions of the ‘core’ microbiome revealed significant enrichment for a number of expected functional categories, including those involved in transcription and translation (Fig. 4). Metabolic profile-based clustering indicated that the representation of ‘core’ functional groups was highly consistent across samples (Supplementary Fig. 20), and includes a number of pathways likely important for life in the gut, such as those for carbohydrate and amino acid metabolism (e.g. fructose/mannose metabolism, aminosugar metabolism, and N-Glycan degradation). Variably represented pathways and categories include cell motility (only a subset of Firmicutes produce flagella), secretion systems, and membrane transport (e.g. phosphotransferase systems involved in the import of nutrients, including sugars; Fig. 4 and Supplementary Fig. 20).
Figure 4. KEGG categories enriched or depleted in the core versus variable components of the gut microbiome.
Sequences from each of the 18 fecal microbiomes were binned into the ‘core’ or ‘variable’ microbiome based on the co-occurrence of KEGG orthologous groups (core groups were found in all 18 microbiomes while variable groups were present in fewer (<18) microbiomes; see Supplementary Figure 19A). Asterisks indicate significant differences (Student’s t-test, *p<0.05, **p<0.001, ***p<10−5; mean±SEM).
The distribution of CAZy glycoside hydrolase and glycosyltransferase families was compared between each pair of microbiomes (see Supplementary Table 10 for CAZy families with a relative abundance >1%). This analysis revealed that all individuals have a similar profile of glycosyltransferases (R2=0.96±0.003), while the profiles of glycoside hydrolases were significantly more variable, even between family members (R2=0.80±0.01; p<10−30, paired Student’s t-test). This suggests that the number and spectrum of glycoside hydrolases is probably affected by ‘external’ factors such as diet more than the glycosyltransferases.
To identify metabolic pathways associated with obesity, only non-core associated (variable) functional groups were included in a comparison of the gut microbiomes of lean versus obese twin pairs. A bootstrap analysis23 was used to identify metabolic pathways that were enriched or depleted in the variable obese gut microbiome. For example, similar to a mouse model of diet-induced obesity4, the obese human gut microbiome was enriched for phosphotransferase systems involved in microbial processing of carbohydrates (Supplementary Table 12). All gut microbiome sequences were compared against the custom database of 44 human gut genomes: an odds ratio analysis revealed 383 genes that were significantly different between the obese and lean gut microbiome (q-value < 0.05; 273 enriched and 110 depleted in the obese microbiome; Supplementary Tables 13,14). By contrast, only 49 genes were consistently enriched or depleted between all twin-pairs (see Supplementary Methods).
These obesity-associated genes were representative of the taxonomic differences described above: 75% of the obesity-enriched genes were from Actinobacteria (vs. 0% of lean-enriched genes; the other 25% are from Firmicutes) while 42% of the lean-enriched genes were from Bacteroidetes (vs. 0% of the obesity-enriched genes). Their functional annotation indicated that many are involved in carbohydrate, lipid, and amino acid metabolism (Supplementary Tables 13,14). Together, they comprise an initial set of microbial biomarkers of the obese gut microbiome.
Our finding that the gut microbial community structures of adult MZ twin pairs had a degree of similarity that was comparable to that of DZ twin pairs, and only slightly more similar compared to their mothers, is consistent with an earlier fingerprinting study of adult twins24, and with a recent microarray-based analysis, which revealed that gut community assembly during the first year of life followed a more similar pattern in a pair of DZ twins compared to 12 unrelated infants25. Intriguingly, another fingerprinting study of MZ and DZ twins in childhood showed a slightly reduced similarity profile in DZ twins26. Thus, comprehensive time-course studies, comparing MZ and DZ twin pairs from birth through adulthood, as well as intergenerational analyses of their families’ microbiotas, will be key to determining the relative contributions of host genotype and environmental exposures to (gut) microbial ecology.
The hypothesis that there is a core human gut microbiome, definable by a set of abundant microbial organismal lineages that we all share, may be incorrect: by adulthood, no single bacterial phylotype was detectable at an abundant frequency in the guts of all 154 sampled humans. Instead, it appears that a core gut microbiome exists at the level of metabolic functions. This conservation suggests a high degree of redundancy in the gut microbiome and supports an ecological view of each individual as an ‘island’ inhabited by unique collections microbial phylotypes: as in actual islands, different species assemblages converge on shared core functions provided by distinctive components. Our findings raise the question of how core functionality is assembled in this body habitat. Understanding the underlying principles should provide insights about microbial adaptation to, and perhaps mutualistic community assembly within, a wide range of environments.
METHODS SUMMARY
Fecal samples were collected from each individual. Community DNA was prepared and used for pyrosequencing (454 Life Sciences), as well as for PCR and sequencing of bacterial 16S rRNA genes. Shotgun reads were mapped to reference genomes using the NCBI ‘non-redundant’ database, KEGG17, STRING18, CAZy (http://www.cazy.org/), and a 44-member human gut microbial genomes database. Metabolic reconstructions were performed based on CAZy, KEGG, and STRING annotations. The relative abundance of KEGG metabolic pathways is referred to as a ‘metabolic profile.’
METHODS
Community DNA preparation
Fecal samples were frozen immediately after they were produced. De-identified samples were stored at −80°C before processing. 10–20g of each sample was pulverized in liquid nitrogen with a mortar and pestle. An aliquot (~500mg) of each sample was then suspended, while frozen, in a solution containing 500 µl of extraction buffer [200 mM Tris (pH 8.0), 200 mM NaCl, 20 mM EDTA], 210 µl of 20% SDS, 500 µl of a mixture of phenol:chloroform:isoamyl alcohol (25:24:1, pH 7.9), and 500 µl of a slurry of 0.1 mm-diameter zirconia/silica beads (BioSpec Products, Bartlesville, OK). Microbial cells were subsequently lysed by mechanical disruption with a bead beater (BioSpec Products) set on high for 2 min at room temperature, followed by extraction with phenol:chloroform:isoamyl alcohol, and precipitation with isopropanol. DNA obtained from three separate 10 mg frozen aliquots of each fecal sample were pooled (≥200µg DNA) and used for pyrosequencing (see below).
16S rRNA gene sequence-based surveys
Complementary phylogenetic and taxon-based methods were used to compare 16S rRNA sequences among fecal communities. Phylogenetic clustering with UniFrac15 is based on the principle that communities can be compared in terms of their shared evolutionary history, as measured by the degree to which they share branch length on a phylogenetic tree. We complemented this approach with taxon-based methods27, which disregard some of the information contained in the phylogenetic tree of the taxa in question, but have the advantage that specific taxa unique to, or shared among, groups of samples can be identified (e.g., those from lean or obese individuals). Prior to both types of analyses, we grouped 16S rRNA gene sequences into Operational Taxonomic Units (OTUs/phylotypes) using both cd-hit28 and the furthest-neighbor-like (FNL) algorithm, with a sequence identity threshold of 97%, which is commonly used to define ‘species’-level phylotypes. Taxonomy was assigned using the best-BLAST-hit against Greengenes29 (E-value cutoff of 1e-10, minimum 88% coverage, 88% percent identity) and the Hugenholtz taxonomy (downloaded from http://greengenes.lbl.gov/Download/Sequence_Data/Greengenes_format/ on May 12, 2008, excluding sequences annotated as chimeric).
Selection of operational taxonomic units (OTUs)
16S rRNA gene-derived pyrosequencing data were pre-processed to remove sequences with low quality scores, sequences with ambiguous characters, or sequences outside of the length bounds (V6 < 50nt, V2 < 200nt), and binned according to sample-specific barcode (e.g. ref. 13). Similar sequences were identified using Megablast30 and cd-hit, with the following parameters: E-value 1e−10 (Megablast only); minimum coverage, 99%; and minimum pairwise identity, 97%. Candidate OTUs were identified as sets of sequences connected to each other at this level using a maximum of 4000 hits per sequence. Each candidate OTU was considered valid if the average density of connection was above threshold; otherwise it was broken up into smaller connected components27.
Tree building and UniFrac clustering for PCA analysis
A relaxed neighbor-joining tree was built from one representative sequence per OTU using Clearcut31, employing the Kimura correction (the PH Lane mask was applied to V2 and full-length data), but otherwise with default comparisons. Unweighted UniFrac15 was run using the resulting tree. PCA was performed on the resulting matrix of distances between each pair of samples. To determine if the UniFrac distances were on average significantly different for pairs of samples (i.e. between twin-pairs, between twins and their mother, or between unrelated individuals), we performed a t-test on the UniFrac distance matrix, and generated a p-value for the t-statistic by permutation of the rows and columns as in the Mantel test, regenerating the t-statistic for 1,000 random samples, and using the distribution to obtain an empirical p-value.
Rarefaction and phylogenetic diversity (PD) measurements
To determine which individuals had the most diverse communities of gut bacteria, rarefaction plots and Phylogenetic Diversity (PD) measurements, as described by Faith32, were made for each sample. PD is the total amount of branch length in a phylogenetic tree constructed from the combined 16S rRNA datasets, leading to the sequences in a given sample. To account for differences in sampling effort between individuals, and to estimate how far we were from sampling the diversity of each individual completely, we plotted the accumulation of PD (branch length) with sampling effort, in a manner analogous to rarefaction curves. We generated the PD rarefaction curve for each individual by applying custom python code (http://bmf2.colorado.edu/unifrac/about.psp) to the Arb parsimony insertion tree27.
Pyrosequencing of total community DNA
Shotgun sequencing runs were performed on the 454 FLX pyrosequencer from total fecal community DNA. Two samples were also analyzed in a single run employing Titanium extra long read pyrosequencing technology (see Supplementary Table 4,5). Sequencing reads with degenerate bases (“Ns”) were removed along with all duplicate sequences, as sequences of identical length and content are a common artifact of the pyrosequencing methodology. Finally, human sequences were removed by identifying sequences homologous to the H.sapiens reference genome (BLASTN e-value<10−5, %identity>75, and score>50).
CAZyme analysis
Metagenomic sequence reads were searched against a library of modules derived from all entries in the Carbohydrate-Active enZymes (CAZy) database (www.cazy.org using FASTY33 e-value<10−6). This library consists of ~180,000 previously annotated modules (catalytic modules, carbohydrate binding modules (CBMs) and other non-catalytic modules or domains of unknown function) derived from ~80,000 protein sequences. The number of sequencing reads matching each CAZy family was divided by the number of total sequences assigned to CAZymes and multiplied by 100 to calculate a relative abundance. An R2 value was calculated for each pair of CAZy profiles. We then compared the distribution of glycoside hydrolase similarity scores to the distribution of glycosyltransferase similarity scores.
Statistical analyses
Xipe23 (version 2.4) was employed for bootstrap analyses of pathway enrichment and depletion, using the parameters sample size=10,000 and confidence level=0.95. Linear regressions were performed in Excel (version 11.0, Microsoft). Mann-Whitney and Student’s t-tests were utilized to identify statistically significant differences between two groups (Prism v4.0, GraphPad; Excel version 11.0, Microsoft). The Bonferroni correction was used to correct for multiple hypotheses. The Mantel test was used to compare distance matrices: the matrix of each pairwise comparison of the abundance of each reference genome, and the abundance of each metabolic pathway, were compared (Mantel program in Python using PyCogent34; 10,000 replicates). Data are represented as mean±SEM unless otherwise indicated.
Microbiome sequences were compared against the custom database of 44 gut genomes (BLASTX e-value<10−5, bitscore>50, and %identity>50). A gene by sample matrix was then screened to identify genes ‘commonly-enriched’ in either the obese or lean gut microbiome (defined by an odds ratio greater than 2 or less than 0.5 when comparing the pooled obese twin microbiomes to the pooled lean twin microbiomes and when comparing each individual obese twin microbiome to the aggregate lean twin microbiome, or vice versa). The statistical significance of enriched or depleted genes was then calculated using a modified t-test (q-value<0.05; calculated with code kindly supplied by Mihai Pop and J.R. White, University of Maryland). We also searched for genes that were consistently enriched or depleted in all six MZ twin-pairs. A gene-by-sample matrix was generated based on BLASTX comparisons of each microbiome with our custom 44-genome database, in order to calculate an odds ratio based on the frequency of each gene in each twin versus the respective co-twin. The analysis revealed only 49 genes (odds ratio>2 or <0.5): they represent a variety of taxonomic groups, including Firmicutes, Bacteroidetes, and Actinobacteria, and did not show any clear functional trends.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
We thank Sabrina Wagoner and Jill Manchester for technical support; Stacey Marion and Deborah Hopper for recruitment of participants and sample collection, Andrew Goodman, Brian Muegge, and Michael Mahowald for helpful suggestions, plus Sue Huse (Marine Biological Laboratory), Faheem Niazi and Sayid Attiya (454 Life Sciences), Chris Markovic, Lucinda Fulton, Bob Fulton, Elaine Mardis and Richard Wilson (Washington University Genome Sequencing Center), and Simone Macmil, Graham Wiley, Chunmei Qu, and Ping Wang (University of Oklahoma) for their assistance with sequencing, and Pedro M. Coutinho (Université de Provence, France) for help with the CAZy analysis. Deep draft assemblies of reference gut genomes were generated as part of an NHGRI-sponsored human gut microbiome initiative (HGMI, http://genome.wustl.edu/pub/organism/Microbes/Human_Gut_Microbiome/). This work was supported in part by the NIH (DK78669/ES012742/AA09022/HD049024), the NSF (OCE0430724), the W.M. Keck Foundation, and the Crohn’s and Colitis Foundation of America.
Footnotes
Author Information – PJT, ACH, RK, and JIG designed the experiments. PJT, TY, AD, REL, MLS, WJJ, BAR, JPA, and ME generated the data. PJT, MH, MLS, BLC, AD, BH, ACH, RK, and JIG analyzed the data. PJT, ACH, RK, and JIG wrote the manuscript with input from the other members of the team. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under accession number 32089. 454 pyrosequencing reads have been deposited in the NCBI Short Read Archive. Nearly full-length 16S rRNA gene sequences are deposited in GenBank under the accession numbers FJ362604-FJ372382. Annotated sequences are also available for further analysis in MG-RAST (http://metagenomics.nmpdr.org/). The authors declare no competing financial interests.
REFERENCES
- 1.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, et al. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäckhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 7.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruder CE, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am. J. Hum. Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouchard C, et al. The response to long-term overfeeding in identical twins. N. Engl. J. Med. 1990;322:1477–1482. doi: 10.1056/NEJM199005243222101. [DOI] [PubMed] [Google Scholar]
- 11.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav. Genet. 1997;27:325–351. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- 12.Heath AC, et al. Ascertainment of a mid-western US female adolescent twin cohort for alcohol studies: assessment of sample representativeness using birth record data. Twin Research. 2002;5:107–112. doi: 10.1375/1369052022974. [DOI] [PubMed] [Google Scholar]
- 13.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sogin ML, et al. Microbial diversity in the deep sea and the underexplored "rare biosphere". Proc. Natl. Acad. Sci. USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lozupone C, Hamady M, Knight R. UniFrac-an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watson SB, McCauley E, Downing JA. Patterns in phytoplankton taxonomic composition across temperate lakes of differing nutrient status. Limnology and Oceanography. 1997;42:487–495. [Google Scholar]
- 17.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Mering C, et al. STRING 7-recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 20.Gill, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurokawa, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinsdale EA, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez-Brito B, Rohwer F, Edwards RA. An application of statistics to comparative metagenomics. BMC Bioinformatics. 2006;7:162. doi: 10.1186/1471-2105-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zoetandal EG, Akkermans ADL, Akkermans-van Vliet WM, de Visser JA, de Vos WM. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb. Ecol. Health Disease. 2001;13:129–134. [Google Scholar]
- 25.Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the human infant intenstinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart JA, Chadwick VS, Murray A. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J. Med. Microbiol. 2005;54:1239–1242. doi: 10.1099/jmm.0.46189-0. [DOI] [PubMed] [Google Scholar]
METHODS REFERENCES
- 27.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 29.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 31.Sheneman L, Evans J, Foster JA. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics. 2006;22:2823–2824. doi: 10.1093/bioinformatics/btl478. [DOI] [PubMed] [Google Scholar]
- 32.Faith DP. Conservation evaluation and phylogenetic diversity. Biological Conservation. 1992;61:1. [Google Scholar]
- 33.Pearson WR, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- 34.Knight R, et al. PyCogent: a toolkit for making sense from sequence. Genome Biol. 2007;8:R171. doi: 10.1186/gb-2007-8-8-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.