Abstract
We sequenced and analyzed the full-length genomes of four coronaviruses (CoVs), each from a distinct wild-ruminant species in Ohio: sambar deer (Cervus unicolor), a waterbuck (Kobus ellipsiprymnus), a sable antelope (Hippotragus niger), and a white-tailed deer (Odocoileus virginianus). The fecal samples from the sambar deer, the waterbuck, and the white-tailed deer were collected during winter dysentery outbreaks and sporadic diarrhea cases in 1993 and 1994 (H. Tsunemitsu, Z. R. el-Kanawati, D. R. Smith, H. H. Reed, and L. J. Saif, J. Clin. Microbiol. 33:3264-3269, 1995). A fecal sample from a sable antelope was collected in 2003 from an Ohio wild-animal habitat during the same outbreak when a bovine-like CoV from a giraffe (GiCoV) was isolated (M. Hasoksuz, K. Alekseev, A. Vlasova, X. Zhang, D. Spiro, R. Halpin, S. Wang, E. Ghedin, and L. J. Saif, J. Virol. 81:4981-4990, 2007). For two of the CoVs (sambar deer and waterbuck), complete genomes from both the cell culture-adapted and gnotobiotic-calf-passaged strains were also sequenced and analyzed. Phylogenetically, wild-ruminant CoVs belong to group 2a CoVs, with the closest relatedness to recent bovine CoV (BCoV) strains. High nucleotide identities (99.4 to 99.6%) among the wild-ruminant strains and recent BCoV strains (BCoV-LUN and BCoV-ENT, isolated in 1998) further confirm the close relatedness. Comparative genetic analysis of CoVs of captive wild ruminants with BCoV strains suggests that no specific genomic markers are present that allow discrimination between the bovine strains and bovine-like CoVs from captive wild ruminants; furthermore, no specific genetic markers were identified that defined cell cultured or calf-passaged strains or the host origin of strains. The results of this study confirm prior reports of biologic and antigenic similarities between bovine and wild-ruminant CoVs and suggest that cattle may be reservoirs for CoVs that infect captive wild ruminants or vice versa and that these CoVs may represent host range variants of an ancestral CoV.
Coronaviruses (CoVs) are the largest known RNA viruses, with a single-stranded positive-sense genome that ranges from 27 to 31 kb in size (2, 55). The enveloped virion of 80 to 160 nm in diameter contains a nucleocapsid (N) and surface spikes (S) resembling a solar corona when imaged by negative-stain electron microscopy (EM) (2, 57). They belong to the Coronaviridae family in the order Nidovirales that includes large RNA viruses sharing common replication strategies—synthesis of the full genomic RNA followed by a nested set of subgenomic RNAs with common 3′ ends (7, 55). The RNA replication machinery shows low fidelity, resulting in a high mutation rate of about one mutation per genome per replication round (18), leading to broad genomic diversity among the virus progeny, referred to as quasispecies (17). Other genomic mechanisms influencing the evolution and ecological properties of CoVs include RNA-RNA recombination (9, 29, 33), with the highest recombination rate among all known nonsegmented, linear, plus-polarity RNA viruses (3). These properties allow CoVs to adapt to environmental conditions and new host species (63, 72). The 2002 to 2003 outbreak of severe acute respiratory syndrome CoV (SARS-CoV), thought to have been due to potential emergence from a bat reservoir (25, 39, 41, 48), resulted in 916 deaths worldwide and had a major international economic impact (24, 73). SARS-like CoVs were isolated from civet cats, raccoon dogs (25), and pigs (11) and were subsequently identified in bat species, now proposed as the host reservoir for SARS CoV (25, 39, 41, 48, 67). Prior to the SARS outbreak, interspecies transmission and genetic recombination of CoVs were suspected for porcine hemagglutinating encephalitis virus (62), human CoV (HCoV) OC-43 (63), and feline infectious peritonitis virus (29). Interspecies transmission of group 2a CoVs was confirmed experimentally: the presence of bovine-like CoVs transmissible to gnotobiotic (Gn) calves from captive wild ruminants (CWRs) was demonstrated by Tsunemitsu and colleagues (59) and Hasoksuz et al. (27), and turkey poults were successfully infected with bovine CoV (BCoV) strain DB2 (30).
Three groups of CoVs are presently recognized on the basis of genomic and antigenic properties. Group 2a CoVs consist of BCoVs, HCoVs (OC-43 and HKU1) (43, 66), mouse hepatitis virus (MHV) (26), and canine respiratory CoVs (21). The SARS-CoV was assigned to a new group, 2b, because of its segregated position in the genus and lack of antigenic relatedness to other, group 2a CoVs (23, 53), although, surprisingly, antigenic cross-reactivity was reported with group 1 CoVs at the level of the N protein (64). BCoVs are commonly associated with winter dysentery (WD) in dairy cattle (16, 49) and with neonatal calf diarrhea (NCD) (45). Besides replicating in the intestine, BCoVs infect the upper and lower respiratory tract (44, 51) and cause respiratory disease in cattle (13, 28, 38), including their recent association with shipping fever in feedlot cattle (56). Interspecies transmission (30, 59) and potential infection of humans with BCoVs has been observed (72). In the late 1970s, bovine-like CoV particles in other ruminant species were first detected by EM. The CoV-like particles were identified in diarrheic feces of Australian sheep (60); sitatunga, waterbuck, and musk oxen (10); and llamas and alpacas (8), and antibodies to BCoV were also identified in caribou (19). BCoV was isolated from infected elk and adapted to cell culture (42), and it was suspected in dromedary (68) calves with enteric disease. In our laboratory, BCoVs from a waterbuck, sambar deer, a white-tailed deer, and a giraffe were isolated in cell culture and characterized more extensively, including biologically, antigenically, and according to their interspecies transmission and pathogenicity in Gn calves (27, 59).
The BCoVs replicate in both the digestive and respiratory tracts and have a broad host range. The availability of Gn-calf models and the closer genetic (group 2) and biologic (pneumoenteric) relatedness of BCoVs to SARS-CoV (12, 40, 52) make BCoVs an attractive model to study CoV genomic properties to define unique phenotypes and the basis for the genetic and biologic diversity of CoVs, including genetic changes associated with adaptation to cell culture or a new host. In the light of the SARS outbreak, the suspected transmission of BCoV to a child, resulting in acute diarrhea (72), also substantiates the importance of studying the genetic diversity of BCoVs circulating in wildlife species. The recent isolation of a human group 2a CoV from the cerebrospinal fluid of a child presumed to have acute disseminated encephalomyelitis further emphasizes the importance of and variability in the clinical manifestations of group 2a CoVs (69). Here we analyze the full-length genomes of four bovine-like CoVs isolated in different years from the feces of CWRs in an Ohio wild-animal habitat and in an Ohio wildlife farm. Analysis of the full genomic sequences demonstrated that they belong to group 2a CoVs, with their closest relatedness to the most-recent BCoV reference strains. No specific genetic markers were identified to discriminate the ruminant bovine-like CoVs from BCoVs, and no consistent genomic changes occurred during calf or cell culture adaptation.
MATERIALS AND METHODS
History of fecal samples collected from CWRs.
The history of the outbreaks of CoV-associated diarrhea and the fecal sampling from sambar deer (Cervus unicolor), a waterbuck (Kobus ellipsiprymnus), a white-tailed deer (Odocoileus virginianus), and a giraffe (Giraffa camelopardalis) were described in our previous reports (27, 59). Briefly, fecal samples were collected from three sambar deer and one waterbuck during a WD outbreak from 1993 to 1994 and from a sable antelope (Hippotragus niger) and a giraffe in 2003 in a wild-animal habitat in Ohio. Fecal samples from a white-tailed deer originated from sporadic diarrhea cases in a wildlife farm in 1994. No cattle were present at either of these premises; cattle herds were within a 2- to 5-mile radius but without direct or any known indirect contact or shared services. All samples were CoV positive by immune EM (IEM) and positive in BCoV antigen capture enzyme-linked immunosorbent assay (ELISA). A single diarrheic fecal sample from a sable antelope was submitted to our laboratory in 2003. The sable antelope was housed in a separate barn about 0.5 miles from the giraffe barn and developed similar signs of diarrhea within 1 or 2 weeks preceding the giraffe diarrhea outbreak. Within the park, the same individuals were responsible for feeding the various animals, and the same dump truck that was used to haul the manure was also used to haul fresh bedding.
CoV strain names and abbreviations.
To be consistent with strain designations for other reported wild-ruminant CoVs (i.e., giraffe CoV [GiCoV]), new or revised strain designations were used for the CWR CoVs. The strains include the original waterbuck CoV (WbCoV) strain OH-WD358 and strains OH-WD358-GnC (GnC indicates that the strain is Gn calf adapted) and OH-WD358-TC (TC indicates that the strain is cell culture adapted); the original sambar deer CoV (SDCoV) strain OH-WD388 and strains OH-WD388-GnC and OH-WD388-TC; white-tailed deer CoV (WtDCoV) strain OH-WD470; and sable antelope CoV (SACoV) strain SACoV-OH1. Different strain names for CWR CoVs were used in our previous paper (59): KI-WB for WbCoV-OH-WD358, KI-D2 for SDCoV-OH-WD388, and WTD for WtDCoV-OH-WD470.
Virus isolation and cell culture passage, plaque induction, and virus purification.
The virus isolation and cell culture passage, plaque induction, and virus purification procedures were performed as described previously (4, 27, 28, 59). Briefly, diluted original fecal samples were clarified by centrifugation and filtered through 0.2-μm filters to remove bacterial contamination. Monolayers of human rectal tumor 18 (HRT-18) cells (59) were inoculated with serial dilutions of the filtered samples, and infection with SDCoV and WbCoV was confirmed by cytopathic effects (observed after the second passage), cell culture immunofluorescence (CCIF) staining, reverse transcription-PCR (RT-PCR), and antigen-capture ELISA. WbCoV-OH-WD358-TC and SDCoV-OH-WD388-TC were plaque purified after seven serial passages on HRT-18 cells and used for RNA extraction and sequencing.
Experimental inoculation of calves.
The preparation of the fecal sample inocula, inoculation methods, and sample collection were described previously (27, 59). Briefly, four 1-week-old Gn calves were inoculated oronasally with 1:10-diluted, filtered original fecal samples from each of the four wildlife species. After necropsy (at day 3 postinoculation for Gn calf inoculated with SDCoV fecal sample and day 6 postinoculation for Gn calf inoculated with WbCoV fecal sample), large intestine contents collected from experimentally infected calves were used as CoV source material for direct RNA extraction and sequencing when appropriate.
RT-PCR.
Total RNA was extracted from clarified and filtered fecal samples (original and from Gn calves) by using an RNeasy minikit (Qiagen, Valencia, CA) according to the manufacturer's instructions. A one-step RT-PCR assay was performed as previously described (9, 27).
IEM.
Briefly, supernatants from 20% fecal samples were mixed with Gn-calf antiserum to the BCoV-Mebus strain, reacted at 4°C for 18 h, and then concentrated by ultracentrifugation and used for IEM as previously described (50).
ELISA and CCIF.
An indirect antigen capture ELISA and CCIF using a pool of three monoclonal antibodies directed against the S, N, and HE structural proteins of BCoV strain DB2 were used to detect CoV antigens and infectious CoV, respectively, in original and Gn-calf fecal suspensions (28).
The sequencing procedures were described in detail previously (27) and applied to all original and Gn-calf fecal suspensions and plaque-purified WbCoV-OH-WD358-TC and SDCoV-OH-WD388-TC.
Sequence analyses.
The reference CoV genome sequences from GenBank compared for phylogenetic analyses are summarized as follows. (i) The group 1 CoVs used included HCoV-229E (NC_002645), HCoV-NL63 (NC_005831), porcine epidemic diarrhea virus CV777 (NC_003436), feline infectious peritonitis virus 79-1146 (NC_007025), and transmissible gastroenteritis virus M6 (DQ811785). (ii) The group 2a CoVs used included HCoV-0C43 ATCC VR-759 (AY391777), HCoV-HKU1 (NC_006577), MHV-A59 (NC_001846), BCoV- Mebus (U00735), BCoV-Quebec (AF220295), BCoV-LUN (AF391542), BCoV-ENT (NC_003045), BCoV-DB2 (DQ811784), and GiCoV (EF424623). (iii) The group 2b CoV used was SARS-Tor2 (NC_004718). (iv) The group 3 CoV used was infectious bronchitis virus Beaudette (NC_001451).
Sequence alignment and phylogenetic analysis were performed as described previously (27).
Nucleotide sequence accession numbers.
The sequences determined in the present study were submitted to GenBank and assigned accession numbers as follows: sable antelope coronavirus US/OH1/2003, EF424621; waterbuck coronavirus US/OH-WD358-TC/1994, FJ425184; waterbuck coronavirus US/OH-WD358-GnC/1994, FJ425185; waterbuck coronavirus US/OH-WD358/1994, FJ425186; white-tailed deer coronavirus US/OH-WD470/1994, FJ425187; sambar deer coronavirus US/OH-WD388-TC/1994, FJ425188; sambar deer coronavirus US/OH-WD388/1994, FJ425189; and sambar deer coronavirus US/OH-WD388-GnC/1994, FJ425190.
RESULTS
Genetic characterization of bovine-like CoVs from CWRs.
Full-length genome sequences of the bovine-like CoVs from four CWR species, waterbuck (Kobus ellipsiprymnus), sambar deer (Cervus unicolor), white-tailed deer (Odocoileus virginianus), and sable antelope (Hippotragus niger) (originating from the 2003 outbreak and used for comparison with our previously sequenced GiCoV originating from the same 2003 outbreak [27]), were obtained and analyzed. The isolation of CoVs from the sambar deer, waterbuck, white-tailed deer, and giraffe (from the same outbreak as SACoV) fecal samples and their antigenic properties were described previously (27, 59). We also performed full-length genome sequencing and genetic characterization for two cell culture-adapted (WbCoV-WD358-TC and SDCoV-WD388-TC) and two Gn-calf-adapted (WbCoV-WD358-GC and SDCoV-WD388-GC) CWR CoV strains. Unlike the GiCoV and other CWR CoVs, we failed to adapt the sable antelope bovine-like CoV (SACoV-OH1) to HRT-18 cell cultures or to successfully passage it in Gn calves, suggesting that this strain was nonviable.
The results of analysis of the eight full-length CoV genome sequences revealed that their organization and structure resembled those of known group 2a CoVs, particularly BCoV (Fig. 1). The genomes were organized into 10 open reading frames (ORFs) and contained the indicated total numbers of nucleotides: WbCoV OH-WD358 (31,010 bp), WbCoV OH-WD358-GnC (30,962 bp), WbCoV OH-358-TC (30,995 bp), SDCoV OH-WD388 (31,011 bp), SDCoV OH-WD388-GnC (31,009 bp), SDCoV OH-WD388-TC (30,995 bp), WtDCoV OH-WD470 (31,020 bp), and SACoV-OH1 (30,995 bp). The observed variations in sequence length among the CWR bovine-like CoVs were mainly due to different numbers of nucleotides at the ends of the 5′-untranslated region (5′-UTR) and the 3′-UTR. All of our sequences were 9 to 16 nucleotides shorter in the 5′-UTR and from 53 nucleotides shorter (OH-WD358-GnC) to 5 nucleotides longer (OH-WD470) in the 3′-UTR; the reason for the differences is unclear, but it could be related to the sequencing strategy.
FIG. 1.
Map of the BCoV genome isolated from CWRs. The overlapping ORF1a and ORF1b encode proteins of the replicative complex; the approximate positions of putative functional domains (identified according to homology predicted from MHV-A59 domains from the sequence NC_001846) are given above the BCoV genome scheme: AC-PL1-PL2-X-TM1 contains the N-terminal acidic (AC) domain, papain-like proteinase domains PL1-PRO and PL2-PRO, ADP-ribose 1′-phosphatase (formerly known as “X-domain”), and transmembrane domain 1 (TM1). The overall identity with the group 2a MHV genome in this region is 66%. For the other domains, the proteins and their identities to MHV (in parentheses) are as follows. TM2 (78.1%) and TM3 (78.5%) are transmembrane-spanning domains, 3CL (76.9%) is the main protease, and SS-RNA binding protein (81.2%) is the last domain before the ribosomal slippage site which leads to −1 frameshift and synthesis of ORF1a and ORF1b as a single polyprotein, ORF1ab. RdRp (85.1%) is the RNA-dependent RNA polymerase, the key and most-conserved protein in the CoV replicative complex; next are the metal-binding and NTPase/helicase domains (83.1%), nuclease ExoN homolog (80.7%), uridylate-specific endoribonuclease NendoU (79.6%), and 2′-O-ribose methyltransferase (79.9%). BCoV genes in the 3′ region of the genome encode the next proteins, abbreviated as follows: 32kDa, 32-kDa nsp; HE, hemagglutinin-esterase protein; S, spike protein; 4.8kDa, 4.9kDa and 12.7kDa, nsp's by size; E, small membrane/envelope protein; M, membrane protein; and N, nucleocapsid protein.
The results of phylogenetic analysis with representatives from all known groups of CoVs demonstrated that the CWR CoVs belong to CoV group 2a, forming a tight cluster together with reference BCoV isolates (data not shown). To define the relatedness between CoVs isolated in our laboratory and known BCoV isolates, we performed phylogenetic analysis with five genomes of reference BCoV strains from different years of isolation and reflecting the evolution of the BCoV genome during the last 35 years. The BCoV strains Mebus and Quebec, isolated in 1972, represent the earliest group; BCoV-DB2, isolated in our laboratory in 1984, represents an intermediate group; and two isolates from 1998, BCoV-LUN and BCoV-ENT, represent the most-recent BCoV reference strains available. The GiCoV recently isolated in our laboratory (27) was the most-recent isolate of a bovine-like CoV and originated from the same outbreak as the SACoV. The resulting phylogenetic tree for these isolates is shown in Fig. 2. Sequences represented on the tree were grouped according to year of isolation, demonstrating the changes accumulated in bovine and bovine-like CoV genomes over time.
FIG. 2.
Phylogenetic analysis of the WbCoV, SDCoV, WtDCoV, and SACoV isolates to identify their relatedness with known reference strains of BCoV or bovine-like CoVs. The range of rates of nucleotide substitution per 100 nucleotides is indicated below. The year of isolation is given for each strain. The Quebec and Mebus BCoV reference strains were attenuated by serial passages in cell culture. The LUN and ENT BCoV reference strains were also adapted to cell culture before sequencing, but their attenuation status is unknown.
The full-length genome identity among the CWR CoVs and BCoV strains was high. The earliest strains (Mebus and Quebec) had the lowest levels of nucleotide identity, 98.4 to 98.8%, to the most-recent BCoV and BCoV-like isolates, including CWR CoVs (Table 1). The nucleotide identity between strain DB2 isolated in the mid-1980s and the early Mebus and Quebec strains was higher (98.8 to 99%), whereas the levels of identity between strain DB2 and the CWR bovine-like CoVs varied from 99.2% for the most-recent GiCoV and SACoV strains to 99.4 to 99.5% for the earlier WbCoV, SDCoV, and WtDCoV strains. The most-recent reference BCoV strains, ENT and LUN (1998), had 99.5 to 99.6% nucleotide identity with the four CWR bovine-like CoVs, whereas they showed only 98.5 to 98.7% identity with the Mebus and Quebec strains. Almost the same percent identity (99.4 to 99.6%) was observed between the CWR CoV strains that originated from different outbreaks in different years. Thus, similar high genomic nucleotide identities were observed among the four CWR bovine-like CoVs and the most-recent reference BCoV strains and similar degrees of divergence were evident between these genomes and the earliest BCoV strains.
TABLE 1.
Nucleotide identity between CWR CoVs and reference BCoV strains isolated during different years
| CWR virus(es),a source, yr of isolation | % Identity of indicated reference strain(s) with CWR strain(s)
|
||
|---|---|---|---|
| BCoV-Quebec and BCoV-Mebus, 1972 | BCoV-DB2, 1984 | BCoV-ENT and BCoV-LUN, 1998 | |
| SDCoV and WbCoV,b diarrheal outbreak, 1994 | 98.6 | 99.5 | 99.5 |
| WtDCoV, sporadic diarrhea, 1994 | 98.6-98.8 | 99.4 | 99.6 |
| SACoV,c diarrheal outbreak, 2003 | 98.4-98.6 | 99.2 | 99.5-99.6 |
Identity between CWR CoVs from the different outbreaks (or the case of sporadic diarrhea) was 99.4 to 99.6%.
The SDCoV and WbCoV isolated from the same outbreak (1994) showed 99.9% nucleotide identity.
The same level of identity with BCoV reference strains was observed for GiCoV originating from the same outbreak as SACoV (27). The GiCoV and SACoV isolated from the same outbreak (2003) showed 99.9% identity.
No substantial nucleotide differences were noted between the CWR CoV and reference BCoV genomes in the noncoding regions and regions of a putative RNA pseudoknot and a ribosomal slippery site. No variable positions were present in the 5′-UTR compared to this region in more-recent isolates, whereas the earliest strains (Quebec and Mebus) possessed three unique changes (at positions 59, 96, and 104). Only one substitution common for sambar deer, waterbuck, and bovine DB2 CoV sequences was located in the region adjacent to the −1 ribosomal shift site between ORF1a and ORF1b: a silent mutation 83 nucleotides downstream of the heptanucleotide slippery site. Comparison of the 3′-UTR for our isolates and for reference BCoV strains revealed only two mutations unique for WtDCoV (nucleotide positions given are for the Mebus strain): position 30778 of WtDCoV contained thymine; for other strains, cytosine; and for Mebus and Quebec BCoVs, guanine; in position 30855, adenine was replaced by guanine for the WtDCoV strain. There were no differences in length within the coding region of genomes for CoV sequences except for a single-nucleotide deletion in codon 39 of the 4.8-kDa nonstructural protein (nsp) of the SACoV. All CWR isolates contained the same deletion of AATT, 30 to 33 nucleotides upstream of the 12.7-kDa nsp start codon (28077 to 28080 in the Mebus genome) and had a single-nucleotide insertion after guanine 30956, a common feature for recent BCoV and CWR CoV strains that is in contrast to the earlier Mebus and Quebec BCoV strains.
Amino acid differences within the putative CoV proteins.
Next we compared the amino acid sequences of all proteins between the CWR CoV and reference BCoV strains: BCoV-Mebus, BCoV-Quebec, BCoV-DB2, BCoV-LUN, BCoV-ENT, and the GiCoV-OH3 strain (Fig. 3). The most-variable positions were detected when comparing recent bovine and bovine-like CoV isolates with the early BCoV-Mebus and Quebec strains. Among the 81 variable positions in the ORF1a amino acid sequence, only 14 positions were unique for one or more CWR bovine-like CoVs, including the GiCoV strain (27).
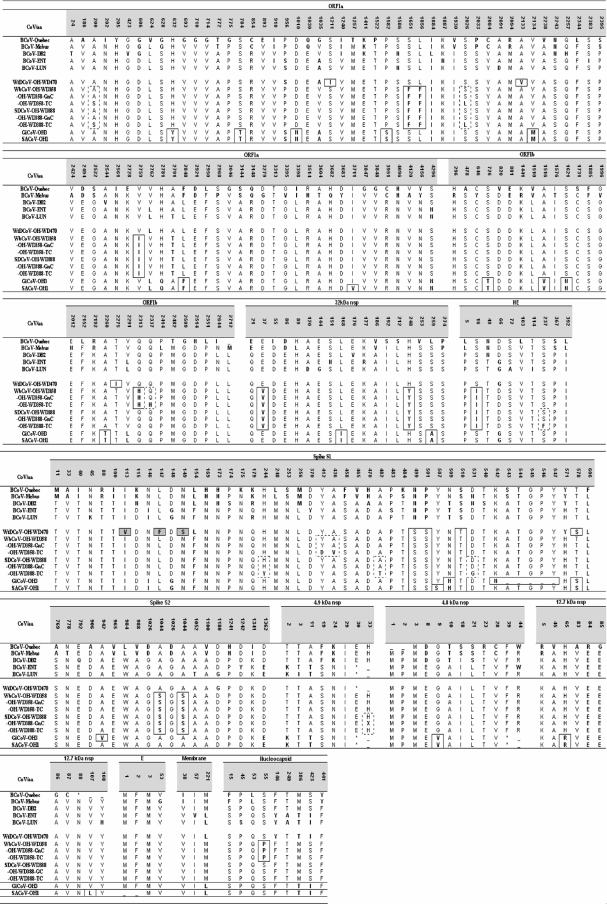
FIG. 3.
Amino acid variations observed in structural proteins and nsp's of CWR CoV and reference BCoV sequences. Differences from consensus sequence amino acids are marked in bold. Amino acid substitutions common for CWR bovine-like CoVs from the same outbreak are boxed with solid lines. Amino acid substitutions distinguishing TC- or GnC-adapted viruses from the original strain are boxed with dashed lines. Three amino acid substitutions in the S1 domain of WtDCoV spike protein that are potentially responsible for altering the virus-neutralizing antibody binding site are highlighted with gray and boxed with solid lines. Notable differences in sequence variations include the following. (i) The BCoV-Quebec strain has a 10-aa insertion (TILRQWLPAG) after the G2509 position in ORF1b. (ii) The BCoV-Quebec strain has several nucleotide deletions, starting from codon 2551 in ORF1b (nucleotide positions on the BCoV-Mebus sequence are 20995, 21004, 21007, 21014 to 21015, 21022, 21027, 21030, 21037 to 21038, 21042, and 21054). This leads to a shorter amino acid sequence length for the Quebec strain in the region 2551 to 2570, 16 aa instead of 20 as in all other strains, and difference in residues due to frameshifts (Quebec has the sequence LLLDIGVHVVRCSYIH, while other CoVs have PITKNIGEYNVSKDGFFTYI). (iii) The BCoV-Quebec strain has a single-nucleotide deletion (cytosine in position 21272 in the Mebus genome) in ORF1b, causing frameshift and early translation termination at codon 41 upstream compared to the Mebus sequence end (stop codon at aa position 2676). (iv) In the 32-kDa nsp, a single-nucleotide deletion in BCoV-Quebec (guanine 21666 in BCoV-Mebus genome) causes frameshift and mismatch between Quebec and other BCoV sequences in the region of aa 55 to 71. The guanine insertion after position 21714 (according to the Mebus sequence) restores the match between the Quebec sequence and other sequences. (v) A single-nucleotide deletion in the BCoV-Quebec strain (adenine 22259 in the BCoV-Mebus genome) causes frameshift and mismatch between the Quebec sequence and other BCoV sequences in the region of aa 253 to 261. A 2-nucleotide deletion (cytosine 22285 and adenine 22286 in the BCoV-Mebus genome) restores the match between the Quebec sequence and other sequences. (vi) The Y507S substitution in the S1 subunit of spike might have contributed to the failure of SACoV replication in cell culture and in Gn calves. (vii) The guanine-to-cytosine substitution in the start codon of the E protein of SACoV probably shifts the start of protein synthesis to codon 3, also methionine. *, stop codon.
Among the predicted amino acid sequences for all viral proteins, three substitutions in the spike glycoprotein were unique to all CWR bovine-like CoVs compared to this protein in the reference BCoV strains N499S, P501S, and S510T (Fig. 3). Amino acid differences among the CWR CoVs were represented by point mutations, excluding the nucleotide deletion in the 4.8-kDa nsp leading to protein synthesis termination at amino acid (aa) 39 for both SACoV and GiCoV.
The isolates that originated from the same location and outbreak were very similar. However, for the SACoV-OH1 strain from the sable antelope, the 5-aa deletion in the spike protein (as detected for the contemporary GiCoV) was not observed. Throughout all the protein sequences, these isolates differed in only five positions: aa 3711 in ORF1a; aa 507, 542, and 942 in spike; and aa 107 in the 12.7 kDa nsp. In addition, in the small membrane protein of SACoV, a single-nucleotide substitution of G for C was present in the start codon, probably leading to start of translation from the methionine codon 3. Five of these seven mutations were typical only for the SACoV (Fig. 3). In addition, a serine at position 507 of the spike protein was unique for the SACoV as compared to the other BCoV and bovine-like CoV sequences available from GenBank for this region of spike (91 total). Remarkably, only two amino acid differences were observed between the WbCoV and SDCoV 1994 isolates and these were located in the most-conserved proteins, ORF1b (aa 2313) and N (aa 55).
Compared to other CoVs, the WtDCoV possessed three unique amino acid changes (I113V, L147F, and N149S) localized in the spike protein (aa 100 to 150), which is part of the putative receptor binding domain (RBD). Interestingly, two of these changes (L147F and N149S) were present only in WtDCoV and in all recent (2002 to 2003 season) NCD (isolated from newborn calves) and WD (isolated from cattle 1 to 8 years old) BCoV isolates from South Korea (total of 29 sequences) (data not shown). Another substitution, I113V, was only present in half of the Korean BCoVs (data not shown) and additionally in strains R-AH65 (respiratory) and E-AH65 (enteric), BCoVs from young adult cattle recently characterized in our laboratory (71).
Among all CoV strains compared, ORF1b (except for BCoV-Quebec), membrane (M), the proteins of the replicative complex (ORF1a), nucleocapsid (N), and hemagglutinin-esterase (HE) were the most-conserved proteins, with 1.0 to 1.8 substitutions per 100 aa, whereas the nsp's (4.8 kDa, 4.9 kDa, 12.7 kDa, and 32 kDa) with unknown function and subunit S1 of the spike protein were the most variable: the substitution rate for subunit S1 of the spike protein was 6.1 per 100 aa according to the consensus sequence. The amino acid differences among the small nsp's were even higher than for S1, but these differences were caused by a single-nucleotide deletion (4.8 kDa nsp) or substitution (4.9 kDa nsp). For example, the amino acid substitution rate for the 4.9-kDa nsp of the WtDCoV was 32.6 per 100 aa, whereas the nucleotide substitution rate was only 3 substitutions per 100 nucleotides. Although small nsp's are highly variable among BCoV and bovine-like CoV strains, these differences are the result of minor changes at the nucleotide level.
Adaptational changes in putative CoV proteins.
To define the effect of cell culture adaptation in the cloned HRT-18 cell line and Gn-calf passage at the amino acid level, we compared SDCoV and WbCoV sequences from the original fecal sample to the Gn-calf-passaged and cell culture-adapted CoV genomes. Five amino acid differences were observed between the original and the cell culture-adapted WbCoV (aa positions 200 and 2026 in ORF1a, aa 2337 in ORF1b, and aa 412 and 436 in the spike protein), whereas no differences were observed between the original CoV and the calf-passaged strain. For the SDCoV, there were six amino acid differences (aa 200 and 2026 in ORF1a; aa 237 in HE; and aa 244, 482 and 531 in spike) between the original and cell culture-adapted strains and only one difference was observed in the Gn-calf-passaged genome, an ambiguity in aa 33 in the 4.9-kDa nsp (Fig. 3). Of interest, both changes in the ORF1a sequence of the cell culture-adapted strains, amino acid substitutions A200S and S2026L, were common for both WbCoV and SDCoV.
Also noteworthy, the phylogenetic tree based on the full-length spike protein sequences of all BCoV strains available in GenBank (total of 48 strains) (data not shown) resulted in the WtDCoV 1994 isolate grouping with the nine recent Korean WD strains (from 1- to 8-year-old cattle) that are distantly related to most U.S. and Canadian BCoV reference strains, including WD isolates. However, the rest of the CWR CoV isolates analyzed in the present study clustered together with North American reference strains.
DISCUSSION
CoVs (CoVs) cause clinically diverse diseases in humans and animals. Their zoonotic potential and ability to cause life-threatening disease are greatly more appreciated after the SARS epidemic in 2002 to 2003 (24). Previously, we confirmed the experimental interspecies transmission of BCoVs and bovine-like CoVs from wild ruminants to turkey poults and Gn calves, respectively (27, 30, 59). Additionally, a bovine-like CoV, HECV-4408, was reported to cause acute diarrhea in a child in 1994 (72), demonstrating the potential for transmission of bovine-like CoVs to humans. Also, BCoVs are closely related to respiratory HCoV isolate OC43, suggesting that this strain may have originated from BCoV (63). More recently, canine respiratory CoV was suggested to originate from the transmission of BCoV to dogs (20). Like the group 2b SARS-CoV, group 2a BCoVs also possess a dual pneumoenteric tropism (12, 40, 52). These findings not only make bovine and bovine-like CoVs a useful model to study the evolution of CoVs and their propensity for interspecies transmission but also stress the importance of understanding the ecology of bovine-like CoV strains previously associated with zoonotic CoV outbreak in humans (72).
In our study, we analyzed the full-length genome sequences of bovine-like CoVs isolated directly from feces of CWRs and the cell culture-adapted and the Gn-calf-passaged strains of two of the corresponding CWR CoVs (WbCoV and SDCoV) to determine their relatedness to BCoV reference strains and to identify sequence changes that may relate to their interspecies transmission. The results of phylogenetic analysis confirmed that the four CWR bovine-like CoVs belong to group 2a, as was previously shown by antigenic characterization (59). Although these four strains originated during different years (1994 and 2003) from the same location, it was considered that they are approximately contemporary (or equally distant on the time scale) with the most-recent reference strains (BCoV-ENT and BCoV-LUN from 1998), based on high genetic identity. Considering the similar percent relatedness between CWR bovine-like CoVs from different animal hosts and the recent reference BCoV strains (LUN and ENT) and the divergence of these isolates from the early BCoV strains (Mebus and Quebec), we suggest that during the observed time interval (from 1972 to 2003), the evolution of CWR CoVs in the United States was not independent or isolated from that of BCoVs. Alternatively, the CWR bovine-like CoVs may have recently split from BCoVs and at the time of isolation were still in the process of adaptation to the new hosts, as was observed for the earliest human SARS-CoV isolates with the closest relationship to SARS-like CoVs from civets (54).
The high amino acid substitution rates observed for the small nsp's of 4.9 and 4.8 kDA (13 and 26%, respectively) did not seem to correlate with the biological or adaptive characteristics of the CoVs from wild ruminants. The biological function of these nsp's is not yet known; however, they were suggested to be nonessential for CoV replication because they are lacking in HCoV-OC43 and porcine hemagglutinating encephalitis virus (46, 61). According to Abraham et al., the 4.9- and 4.8-kDa proteins appear to be vestiges of an 11-kDa protein for which a single-nucleotide deletion event in the central part of the gene gave rise to a stop codon (1, 65).
Of the three amino acid substitutions (N499S, P501S, and S510T, located in the highly variable subunit S1 region of the spike protein) that are common and unique for the CWR CoVs studied (Fig. 3) compared to the sequences of the reference BCoV strains (27), none were unique compared to sequences of the same region of 91 BCoV strains available in GenBank. Therefore, our results suggest that there are no consistent genetic markers for discrimination between CWR and bovine CoVs. It is also noteworthy that there were no amino acid substitutions between the original and Gn-calf-passaged strains of the WbCoV and SDCoV isolates, which was also shown for a GiCoV (27), suggesting that these strains may not require genetic modification for experimental transmission to and infection of cattle.
The sable antelope (Bovidae family) SACoV-OH1 that originated from the same outbreak as GiCoV-OH3 differed in only seven amino acids (in addition to the 5-aa deletion in the GiCoV spike) from the GiCoV (Fig. 3) that was highly pathogenic in Gn calves and replicated to high levels in HRT-18 cells (27). Two of these changes were potentially significant because of their genomic location: a hydrophobic tyrosine at position 507 of the spike protein was replaced by a polar serine, and this substitution was unique to the SACoV (Fig. 3). A single-nucleotide substitution in the start codon of the SACoV E protein (Fig. 3) could lead to protein synthesis starting from the third codon, also a methionine, and the absence of the first two amino acids (or even knockout of this protein), which may have dramatic consequences. This is because the small envelope protein of MHV was shown to play a pivotal role in the assembly of the virion (37). We could not clarify the role of these amino acid substitutions, because the SACoV failed to replicate in cell culture or to infect Gn calves, suggesting that the strain was nonviable after storage in a frozen state.
The two amino acid differences in the conserved proteins (ORF1b and N) between the waterbuck (Bovidae family) and sambar deer (Cervidae family) CoVs (Fig. 3) did not appear to be associated with species-specific adaptation. This suggests that the adaptation of bovine-like CoVs to a new host might be defined by a unique combination of amino acid substitutions scattered throughout the entire genome rather than by specific changes in known key residues or that no unique host adaptation changes are needed for CoV interspecies transmission among ruminant species.
The spike protein RBD of MHV is located within the first 330 aa of the N terminus of the S protein and plays a central role in MHV pathogenicity (36, 58). Considering the presence of unique mutations in this region of the WtDCoV spike protein and the results of the phylogenetic analysis that demonstrated the closer relationship of the WtDCoV spike to recent Korean WD strains (31) than to North American BCoV strains, we speculate that the bovine-like CoV from white-tailed deer had a longer history of host adaptation than other CWR bovine-like CoVs and had time to accumulate host-specific changes in its genome. Korean WD BCoV strains were also more homologous to each other and were sharply distinct from the other known BCoVs (American and Canadian), based on phylogenetic analyses of the spike and HE genes (but not for M and E genes). These results suggest that Korean WD strains had an evolutionarily distinct pathway (31, 34), as did the WtDCoV. Our speculation is further supported by the fact that the white-tailed deer is a wild ruminant that is endemic in Ohio, with sporadic diarrheic cases and with frequent contact with cattle herds in Ohio, whereas the other CoVs were from CWR species exotic to Ohio that were more recently transported to their present Ohio location. Possible divergence of WtDCoV from other CWR CoVs and from BCoV isolates was also suggested when CWR CoVs were characterized antigenically and biologically in our laboratory (59). Six and six-tenths percent of serum samples from wild white-tailed deer were seropositive to BCoV. The two positive white-tailed deer serum samples cross-reacted with BCoV in indirect IF staining but failed to neutralize BCoV Mebus and DBA strains, as well as SDCoV and WbCoV. Unfortunately, no serum from the deer which was the source of the WtDCoV isolate was available for neutralization tests. It is possible that one or more of the three unique substitutions within the putative RBD (I113V, L147F, and N149S) of the WtDCoV spike (Fig. 3) might have a dramatic impact on the induction of neutralizing antibodies, but no definitive conclusions are possible until CoVs from wild white-tailed deer are isolated and sequenced. These substitutions most likely reflect the adaptation of a CoV to a new host and therefore also suggest that WtDCoV had a longer prior history of adaptation in the deer population. However, the genomic changes identified in the WtDCoV strain did not affect its infectivity for Gn calves (59), demonstrating the high adaptability of the spike protein in virus-receptor interactions or an alternative role for the conserved HE in such interactions, highlighting the potential of bovine-like CoVs for interspecies transmission.
In the present study, we did not find specific genetic markers for the discrimination of CWR CoVs from reference BCoV strains, based on the complete genome sequencing. Therefore, we conclude that the CWR bovine-like CoVs are not only antigenically indistinguishable (59) but also are very similar genetically to reference BCoV strains. Interestingly, Korean scientists recently tried to identify certain genetic markers responsible for the absence of receptor-destroying enzyme activity against mouse erythrocytes of BCoV isolates obtained during summer months. However, sequence analyses failed to reveal any distinct divergences in the HE or in the S, M, and E genes among all of the field isolates (47).
Additionally, no genetic markers potentially responsible for cell culture adaptation were shown by genetic analysis of the WbCoV and SDCoV. Although both of them shared two common amino acid substitutions, no corresponding substitutions were found for BCoV strains isolated and adapted to cell culture in our laboratory earlier (71). Other individual substitutions for these two strains were noted, but considering their adaptation to the same HRT-18 cells, these mutations are likely not specific for cell culture adaptation. The substitution D531G in the SDCoV strain OH-WD388-TC (Fig. 3) was located only two codons downstream from aa 528 whose replacement resulted in a neutralizing-antibody-resistant mutant strain (70). But position 531 is highly variable among BCoV strains, and four different amino acids (including glycine) were found in this position for the 91 BCoV strains compared. Glycine in this position was observed in one of the Korean (KWD-13) isolates (31) that was sequenced without adaptation to cell culture. Also, most Brazilian BCoV isolates possess a 6-aa deletion in position 526 to 531 of the spike protein that does not seem to affect their ability to cause disease in calves (5).
Chouljenko et al. (14) reported five amino acid substitutions in the spike of BCoV-Mebus after adaptation to HRT-18G cells. However, none of these substitutions was observed in the cell culture-adapted WbCoV strains OH-WD358-TC and SDCoV OH-WD388-TC. As for host adaptation, no specific genetic markers were found for the cell culture-adapted strains and the amino acid changes accumulated during cell culture adaptation seem to be individual for different isolates. It is important to note that most reference BCoV strains are cell culture adapted and the early strains sequenced, Quebec and Mebus, were both attenuated strains after serial passage in cell culture.
Several researchers have attempted to identify genetic markers in the spike protein gene that discriminate between WD and NCD or between enteric and respiratory BCoV strains (14, 15, 22, 35). Several attempts to pinpoint genetic determinants for tissue tropism were also performed (14, 22). However, among all positions identified in these studies, we were only able to confirm D531G as possibly respiratory BCoV specific, because none of the enteric CWR CoV strains, including GiCoV (27) and the previously described enteric BCoV isolates AH65-E and AH187-E (71), had this substitution. Although we found this substitution in the SDCoV OH-WD388-TC sequence, this observation agrees with previous findings that the enteric strains can drift toward the respiratory counterpart sequences in the process of adaptation to cell culture (71). However, additional comparisons of the S1 subunit of spike proteins did not confirm this substitution to be respiratory BCoV specific, because one of the Korean BCoV strains (KWD-13) isolated from a fecal specimen of an adult animal had the same D531G substitution (47). The 4.9-kDa nsp was also suggested to contain respiratory genetic markers because known respiratory BCoV strains had four amino acid substitutions unique for respiratory BCoV and a point mutation resulting in a truncated protein (22). However, in contrast with this published data, we found the latter in the enteric SACoV and WtDCoV, and of the four amino acid substitutions, only P28L was confirmed to be unique for respiratory BCoV strains. In addition, four amino acid substitutions in the 32-kDa nsp were proposed to be respiratory BCoV specific (14), but all four were present in the enteric CWR BCoV-like strains. Finally, the small membrane protein was suggested as a marker for discrimination between virulent and attenuated strains (22), because a V53G substitution was unique to the virulent strains analyzed. Again, all of the CWR BCoV-like isolates from our laboratory were shown to be virulent for Gn calves but did not possess this substitution. Thus, we conclude that most of the amino acid changes are spontaneous and scattered among different viral proteins and appear to be strain specific, which is in agreement with previous findings for the S protein of Japanese BCoV strains (32). If any tropism-specific changes exist, they are more likely to be a combination of changes unique for individual strains.
Based on our data, ruminant host species exotic to North America may be more likely to acquire CoVs from cattle farms in the vicinity of the wild-animal habitat or from endemic wildlife species, such as white-tailed deer. The isolation and complete genomic sequencing of CoVs from free-ranging deer, including ones near the habitat, could help to clarify the origin of the captive-deer WtDCoV isolate and its relationship to other BCoVs and to the CWR bovine-like strains.
The results of our previous studies (27, 30, 59) demonstrated the interspecies transmission of bovine-like CoVs. The spike protein has been shown to be responsible for virus tropism and pathogenicity (6), and it is continuously under selective pressure in new host species. Overall, the adaptational changes occurring after CoV transmission to a new host or adaptation to cell culture are likely to be strain specific. Both BCoV and BCoV-like CWR strains may coevolve, with cross-infections culminating in genetically divergent (including recombinant) strains. The accumulated genetic diversity of BCoVs and bovine-like CoVs increases the possibility of transmission to other species, including humans, as demonstrated for a case of diarrhea in a child associated with a bovine-like CoV (71). The isolation and characterization of bovine-like CoVs from other heterologous hosts is important to better understand the interspecies transmission and zoonotic potential of such strains.
In conclusion, the comparative sequence analysis of WbCoV, SDCoV, WtDCoV, and SACoV failed to identify specific genomic markers in their genomes to allow discrimination between reference bovine strains and bovine-like CoVs from CWRs. In addition, no consistent changes occurred in the genome after Gn-calf passage of these CoVs. Our data suggest that BCoV and BCoV-like CoVs may represent a pool of common pathogens cross-infecting cattle and wild ruminants. Additionally, we found that only the WtDCoV possessed three unique amino acid substitutions in the putative RBD region of the spike protein that were likely responsible for the failure of deer serum to neutralize BCoV, suggesting a longer history and a higher level of adaptation of this CoV to the host than for the other isolates from CWRs.
Acknowledgments
We thank M. Azevedo, G. Myers, J. Hanson, J. McCormick, and T. Root for technical assistance.
This work was supported by grant R21 AI062763 from the NIAID, NIH. Salaries and research support were provided by state and federal funds provided to the Ohio Agricultural Research and Development Center, The Ohio State University.
Footnotes
Published ahead of print on 8 October 2006.
REFERENCES
- 1.Abraham, S., T. E. Kienzle, W. E. Lapps, and D. A. Brian. 1990. Sequence and expression analysis of potential nonstructural proteins of 4.9, 4.8, 12.7, and 9.5 kDa encoded between the spike and membrane protein genes of the bovine coronavirus. Virology 177488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida, J. D., D. M. Berry, C. H. Cunningham, D. Hamre, M. S. Hofstad, L. Mallucci, K. McIntosh, and D. A. J. Tyrrell. 1968. Coronaviruses. Nature 220650. [Google Scholar]
- 3.Baric, R. S., K. Fu, M. C. Schaad, and S. A. Stohlman. 1990. Establishing a genetic recombination map for murine coronavirus strain A59 complementation groups. Virology 177646-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benfield, D. A., and L. J. Saif. 1990. Cell culture propagation of a coronavirus isolated from cows with winter dysentery. J. Clin. Microbiol. 281454-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandao, P. E., F. Gregori, L. J. Richtzenhain, C. A. Rosales, L. Y. Villarreal, and J. A. Jerez. 2006. Molecular analysis of Brazilian strains of bovine coronavirus (BCoV) reveals a deletion within the hypervariable region of the S1 subunit of the spike glycoprotein also found in human coronavirus OC43. Arch. Virol. 1511735-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavanagh, D. 1995. The coronavirus surface glycoprotein, p. 73-114. In S. Siddell (ed.), The coronaviridae. Plenum Press, New York, NY.
- 7.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142629-633. [PubMed] [Google Scholar]
- 8.Cebra, C. K., D. E. Mattson, R. J. Baker, R. J. Sonn, and P. L. Dearing. 2003. Potential pathogens in feces from unweaned llamas and alpacas with diarrhea. J. Am. Vet. Med. Assoc. 2231806-1808. [DOI] [PubMed] [Google Scholar]
- 9.Chang, R. Y., R. Krishnan, and D. A. Brian. 1996. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J. Virol. 702720-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chasey, D., D. J. Reynolds, J. C. Bridger, T. G. Debney, and A. C. Scott. 1984. Identification of coronaviruses in exotic species of Bovidae. Vet. Rec. 115602-603. [DOI] [PubMed] [Google Scholar]
- 11.Chen, W., M. Yan, L. Yang, B. Ding, B. He, Y. Wang, X. Liu, C. Liu, H. Zhu, B. You, S. Huang, J. Zhang, F. Mu, Z. Xiang, X. Feng, J. Wen, J. Fang, J. Yu, H. Yang, and J. Wang. 2005. SARS-associated coronavirus transmitted from human to pig. Emerg. Infect. Dis. 11446-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu, Y. C., K. L. Wu, Y. P. Chou, T. V. Fong, T. L. Tsai, C. M. Kuo, C. H. Kuo, K. W. Chiu, J. W. Liu, H. L. Eng, B. Jawan, Y. F. Cheng, and C. L. Chen. 2004. Diarrhea in medical care workers with severe acute respiratory syndrome. J. Clin. Gastroenterol. 38880-882. [DOI] [PubMed] [Google Scholar]
- 13.Cho, K. O., A. E. Hoet, S. C. Loerch, T. E. Wittum, and L. J. Saif. 2001. Evaluation of concurrent shedding of bovine coronavirus via the respiratory tract and enteric route in feedlot cattle. Am. J. Vet. Res. 621436-1441. [DOI] [PubMed] [Google Scholar]
- 14.Chouljenko, V. N., K. G. Kousoulas, X. Lin, and J. Storz. 1998. Nucleotide and predicted amino acid sequences of all genes encoded by the 3′ genomic portion (9.5 kb) of respiratory bovine coronaviruses and comparisons among respiratory and enteric coronaviruses. Virus Genes 1733-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chouljenko, V. N., X. Q. Lin, J. Storz, K. G. Kousoulas, and A. E. Gorbalenya. 2001. Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J. Gen. Vir. 822927-2933. [DOI] [PubMed] [Google Scholar]
- 16.Clark, M. A. 1993. Bovine coronavirus. Brit. Vet. J. 14951-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domingo, E., E. Baranowski, C. M. Ruiz-Jarabo, A. M. Martin-Hernandez, J. C. Saiz, and C. Escarmis. 1998. Quasispecies structure and persistence of RNA viruses. Emerg. Infect. Dis. 4521-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 9613910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elazhary, M. A., J. L. Frechette, A. Silim, and R. S. Roy. 1981. Serological evidence of some bovine viruses in the caribou (Rangifer tarandus caribou) in Quebec. J. Wildlife Dis. 17609-612. [DOI] [PubMed] [Google Scholar]
- 20.Erles, K., K.-B. Shiu, and J. Brownlie. 2007. Isolation and sequence analysis of canine respiratory coronavirus. Virus Res. 12478-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erles, K., C. Toomey, H. W. Brooks, and J. Brownlie. 2003. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology 310216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gelinas, A. M., M. Boutin, A. M. Sasseville, and S. Dea. 2001. Bovine coronaviruses associated with enteric and respiratory diseases in Canadian dairy cattle display different reactivities to anti-HE monoclonal antibodies and distinct amino acid changes in their HE, S and ns4.9 protein. Virus Res. 7643-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorbalenya, A. E., E. J. Snijder, and W. J. Spaan. 2004. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 787863-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groneberg, D. A., L. Zhang, T. Welte, P. Zabel, and K. F. Chung. 2003. Severe acute respiratory syndrome: global initiatives for disease diagnosis. QJM 96845-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302276-278. [DOI] [PubMed] [Google Scholar]
- 26.Haring, J., and S. Perlman. 2001. Mouse hepatitis virus. Curr. Opin. Microbiol. 4462-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasoksuz, M., K. Alekseev, A. Vlasova, X. Zhang, D. Spiro, R. Halpin, S. Wang, E. Ghedin, and L. J. Saif. 2007. Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe. J. Virol. 814981-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasoksuz, M., S. L. Lathrop, K. L. Gadfield, and L. J. Saif. 1999. Isolation of bovine respiratory coronaviruses from feedlot cattle and comparison of their biological and antigenic properties with bovine enteric coronaviruses. Am. J. Vet. Res. 601227-1233. [PubMed] [Google Scholar]
- 29.Herrewegh, A. A., I. Smeenk, M. C. Horzinek, P. J. Rottier, and R. J. de Groot. 1998. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 724508-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ismail, M. M., K. O. Cho, L. A. Ward, L. J. Saif, and Y. M. Saif. 2001. Experimental bovine coronavirus in turkey poults and young chickens. Avian Dis. 45157-163. [PubMed] [Google Scholar]
- 31.Jeong, J. H., G. Y. Kim, S. S. Yoon, S. J. Park, Y. J. Kim, C. M. Sung, S. S. Shin, B. J. Lee, M. I. Kang, N. Y. Park, H. B. Koh, and K. O. Cho. 2005. Molecular analysis of S gene of spike glycoprotein of winter dysentery bovine coronavirus circulated in Korea during 2002-2003. Virus Res. 108207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanno, T., S. Hatama, R. Ishihara, and I. Uchida. 2007. Molecular analysis of the S glycoprotein gene of bovine coronaviruses isolated in Japan from 1999 to 2006. J. Gen. Virology. 881218-1224. [DOI] [PubMed] [Google Scholar]
- 33.Keck, J. G., G. K. Matsushima, S. Makino, J. O. Fleming, D. M. Vannier, S. A. Stohlman, and M. M. Lai. 1988. In vivo RNA-RNA recombination of coronavirus in mouse brain. J. Virol. 621810-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ko, C. K., M. I. Kang, G. K. Lim, G. Y. Kim, S. S. Yoon, J. T. Park, C. Jeong, S. H. Park, S. J. Park, Y. J. Kim, J. H. Jeong, S. K. Kim, S. I. Park, H. H. Kim, K. Y. Kim, and K. O. Cho. 2006. Molecular characterization of HE, M, and E genes of winter dysentery bovine coronavirus circulated in Korea during 2002-2003. Virus Genes 32129-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kourtesis, A. B., A. M. Gelinas, and S. Dea. 2001. Genomic and antigenic variations of the HE glycoprotein of bovine coronaviruses associated with neonatal calf diarrhea and winter dysentery. Arch. Virol. 1461219-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubo, H., Y. K. Yamada, and F. Taguchi. 1994. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 685403-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo, L., K. R. Hurst, and P. S. Masters. 2007. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J. Virol. 812249-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lathrop, S. L., T. E. Wittum, K. V. Brock, S. C. Loerch, L. J. Perino, H. R. Bingham, F. T. McCollum, and L. J. Saif. 2000. Association between infection of the respiratory tract attributable to bovine coronavirus and health and growth performance of cattle in feedlots. Am. J. Vet. Res. 611062-1066. [DOI] [PubMed] [Google Scholar]
- 39.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 10214040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung, W. K., K. F. To, P. K. Chan, H. L. Chan, A. K. Wu, N. Lee, K. Y. Yuen, and J. J. Sung. 2003. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology 1251011-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310676-679. [DOI] [PubMed] [Google Scholar]
- 42.Majhdi, F., H. C. Minocha, and S. Kapil. 1997. Isolation and characterization of a coronavirus from elk calves with diarrhea. J. Clin. Microbiol. 352937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McIntosh, K., W. B. Becker, and R. M. Chanock. 1967. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. USA 582268-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNulty, M. S., D. G. Bryson, G. M. Allan, and E. F. Logan. 1984. Coronavirus infection of the bovine respiratory tract. Vet. Microbiol. 9425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mebus, C. A., E. L. Stair, M. B. Rhodes, and M. J. Twiehaus. 1973. Neonatal calf diarrhea: propagation, attenuation, and characteristics of a coronavirus-like agent. Am. J. Vet. Res. 34145-150. [PubMed] [Google Scholar]
- 46.Mounir, S., and P. J. Talbot. 1993. Human coronavirus OC43 RNA 4 lacks two open reading frames located downstream of the S gene of bovine coronavirus. Virology 192355-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park, S. J., C. Jeong, S. S. Yoon, H. E. Choy, L. J. Saif, S. H. Park, Y. J. Kim, J. H. Jeong, S. I. Park, H. H. Kim, B. J. Lee, H. S. Cho, S. K. Kim, M. I. Kang, and K. O. Cho. 2006. Detection and characterization of bovine coronaviruses in fecal specimens of adult cattle with diarrhea during the warmer seasons. J. Clin. Microbiol. 443178-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saif, L. J. 2004. Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? Rev. Sci. Tech. 23643-660. [DOI] [PubMed] [Google Scholar]
- 49.Saif, L. J. 1990. A review of evidence implicating bovine coronavirus in the etiology of winter dysentery in cows: an enigma resolved? Cornell Vet. 80303-311. [PubMed] [Google Scholar]
- 50.Saif, L. J., K. V. Brock, D. R. Redman, and E. M. Kohler. 1991. Winter dysentery in dairy herds: electron microscopic and serological evidence for an association with coronavirus infection. Vet. Rec. 128447-449. [DOI] [PubMed] [Google Scholar]
- 51.Saif, L. J., D. R. Redman, P. D. Moorhead, and K. W. Theil. 1986. Experimentally induced coronavirus infections in calves: viral replication in the respiratory and intestinal tracts. Am. J. Vet. Res. 471426-1432. [PubMed] [Google Scholar]
- 52.Shi, X., E. Gong, D. Gao, B. Zhang, J. Zheng, Z. Gao, Y. Zhong, W. Zou, B. Wu, W. Fang, S. Liao, S. Wang, Z. Xie, M. Lu, L. Hou, H. Zhong, H. Shao, N. Li, C. Liu, F. Pei, J. Yang, Y. Wang, Z. Han, X. Shi, Q. Zhang, J. You, X. Zhu, and J. Gu. 2005. Severe acute respiratory syndrome associated coronavirus is detected in intestinal tissues of fatal cases. Am. J. Gastroenterol. 100169-176. [DOI] [PubMed] [Google Scholar]
- 53.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song, H. D., C. C. Tu, G. W. Zhang, S. Y. Wang, K. Zheng, L. C. Lei, Q. X. Chen, Y. W. Gao, H. Q. Zhou, H. Xiang, H. J. Zheng, S. W. Chern, F. Cheng, C. M. Pan, H. Xuan, S. J. Chen, H. M. Luo, D. H. Zhou, Y. F. Liu, J. F. He, P. Z. Qin, L. H. Li, Y. Q. Ren, W. J. Liang, Y. D. Yu, L. Anderson, M. Wang, R. H. Xu, X. W. Wu, H. Y. Zheng, J. D. Chen, G. Liang, Y. Gao, M. Liao, L. Fang, L. Y. Jiang, H. Li, F. Chen, B. Di, L. J. He, J. Y. Lin, S. Tong, X. Kong, L. Du, P. Hao, H. Tang, A. Bernini, X. J. Yu, O. Spiga, Z. M. Guo, H. Y. Pan, W. Z. He, J. C. Manuguerra, A. Fontanet, A. Danchin, N. Niccolai, Y. X. Li, C. I. Wu, and G. P. Zhao. 2005. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA 1022430-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spaan, W., D. Cavanagh, and M. C. Horzinek. 1988. Coronaviruses: structure and genome expression. J. Gen. Virol. 692939-2952. [DOI] [PubMed] [Google Scholar]
- 56.Storz, J., L. Stine, A. Liem, and G. A. Anderson. 1996. Coronavirus isolation from nasal swab samples in cattle with signs of respiratory tract disease after shipping. J. Am. Vet. Med. Assoc. 2081452-1455. [PubMed] [Google Scholar]
- 57.Sturman, L. S., and K. V. Holmes. 1983. The molecular biology of coronaviruses. Adv. Virus Res. 2835-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taguchi, F., H. Kubo, H. Takahashi, and H. Suzuki. 1995. Localization of neurovirulence determinant for rats on the S1 subunit of murine coronavirus JHMV. Virology 20867-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsunemitsu, H., Z. R. el-Kanawati, D. R. Smith, H. H. Reed, and L. J. Saif. 1995. Isolation of coronaviruses antigenically indistinguishable from bovine coronavirus from wild ruminants with diarrhea. J. Clin. Microbiol. 333264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tzipori, S., M. Smith, T. Makin, and C. McCaughan. 1978. Enteric coronavirus-like particles in sheep. Aust. Vet. J. 54320-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vieler, E., T. Schlapp, and W. Herbst. 1996. The region between the M and S genes of porcine haemagglutinating encephalomyelitis virus is highly similar to human coronavirus OC43. J. Gen Virol. 771443-1447. [DOI] [PubMed] [Google Scholar]
- 62.Vijgen, L., E. Keyaerts, P. Lemey, P. Maes, K. Van Reeth, H. Nauwynck, M. Pensaert, and M. Van Ranst. 2006. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 807270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vijgen, L., E. Keyaerts, E. Moes, I. Thoelen, E. Wollants, P. Lemey, A. M. Vandamme, and M. Van Ranst. 2005. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 791595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vlasova, A. N., X. Zhang, M. Hasoksuz, H. S. Nagesha, L. M. Haynes, Y. Fang, S. Lu, and L. J. Saif. 2007. Two-way antigenic cross-reactivity between severe acute respiratory syndrome coronavirus (SARS-CoV) and group 1 animal CoVs is mediated through an antigenic site in the N-terminal region of the SARS-CoV nucleoprotein. J. Virol. 8113365-13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiss, S. R., P. W. Zoltick, and J. L. Leibowitz. 1993. The ns 4 gene of mouse hepatitis virus (MHV), strain A 59 contains two ORFs and thus differs from ns 4 of the JHM and S strains. Arch. Virol. 129301-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woo, P. C., S. K. Lau, K. S. Li, R. W. Poon, B. H. Wong, H. W. Tsoi, B. C. Yip, Y. Huang, K. H. Chan, and K. Y. Yuen. 2006. Molecular diversity of coronaviruses in bats. Virology 351180-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wunschmann, A., R. Frank, K. Pomeroy, and S. Kapil. 2002. Enteric coronavirus infection in a juvenile dromedary (Camelus dromedarius). J. Vet. Diagn. Investig. 14441-444. [DOI] [PubMed] [Google Scholar]
- 69.Yeh, E. A., A. Collins, M. E. Cohen, P. K. Duffner, and H. Faden. 2004. Detection of coronavirus in the central nervous system of a child with acute disseminated encephalomyelitis. Pediatrics 113e73-e76. [DOI] [PubMed] [Google Scholar]
- 70.Yoo, D., and D. Deregt. 2001. A single amino acid change within antigenic domain II of the spike protein of bovine coronavirus confers resistance to virus neutralization. Clin. Diagn. Lab. Immunol. 8297-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, X., M. Hasoksuz, D. Spiro, R. Halpin, S. Wang, A. Vlasova, D. Janies, L. R. Jones, E. Ghedin, and L. J. Saif. 2007. Quasispecies of bovine enteric and respiratory coronaviruses based on complete genome sequences and genetic changes after tissue culture adaptation. Virology 3631-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, X. M., W. Herbst, K. G. Kousoulas, and J. Storz. 1994. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J. Med. Virol. 44152-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhong, N. S., and G. W. Wong. 2004. Epidemiology of severe acute respiratory syndrome (SARS): adults and children. Paediatr. Respir. Rev. 5270-274. [DOI] [PMC free article] [PubMed] [Google Scholar]