Heparan sulfate (HS) appeared early in metazoan evolution. As such, many of the structural motifs (variably sulfated disaccharide subunits) that characterize HS (and heparin) were established early on and have been preserved in modern organisms. Thus, many of the biological functions associated with HS either occurred early in evolution or have depended on the subsequent evolution of the protein ligands that bind to the polysaccharide. Today, we know of literally hundreds of heparin-binding proteins, and many interactions have profound consequences in vertebrate and invertebrate physiology. This Perspective aims to provide an overview of HS structure, function, and biosynthesis and to set the stage for discussing the relationship between structure and function of these fascinating molecules and how altered HS biosynthesis and catabolism can lead to human disorders.
HS proteoglycans
Virtually all cells, from simple invertebrates to humans, have the capacity to produce HS. These polysaccharides represent a type of glycosaminoglycan characterized by alternating uronic acid (D-glucuronic acid [GlcA] or L-iduronic acid [IdoA]) and D-glucosamine (GlcN) units (Figure 1) (1). The chains rarely occur as free entities but rather form covalent complexes with specific “core” proteins. The composite glycoproteins constitute the superfamily of HS proteoglycans. Two major subfamilies of plasma membrane–bound core proteins can be distinguished by primary amino acid sequence homologies, the syndecans and glypicans (2). The four syndecans (designated syndecan-1, -2, -3, and -4) have protein cores with characteristic structural domains (3). The variable ectodomain, exposed to the extracellular milieu, contains 3–5 HS chains in conserved motifs, although hybrid forms containing both HS and another glycosaminoglycan (chondroitin sulfate) are expressed in some tissues. A short hydrophobic transmembrane segment tethers the proteoglycan to the plasma membrane. A protease cleavage site exists proximal to the ectodomain, and proteolysis results in shedding of the ectodomains from the cell surface. Membrane-bound syndecans can also be internalized by endocytosis and degraded in lysosomes, suggesting that the membrane domain or the cytoplasmic tails contain sequences that can interact with proteins involved in endocytosis (4). The cytoplasmic domain also contains peptide sequences that bind to cytoskeletal proteins and that serve as substrates for cellular kinases. Thus, the syndecans can act as signaling molecules (5, 6).
Figure 1.
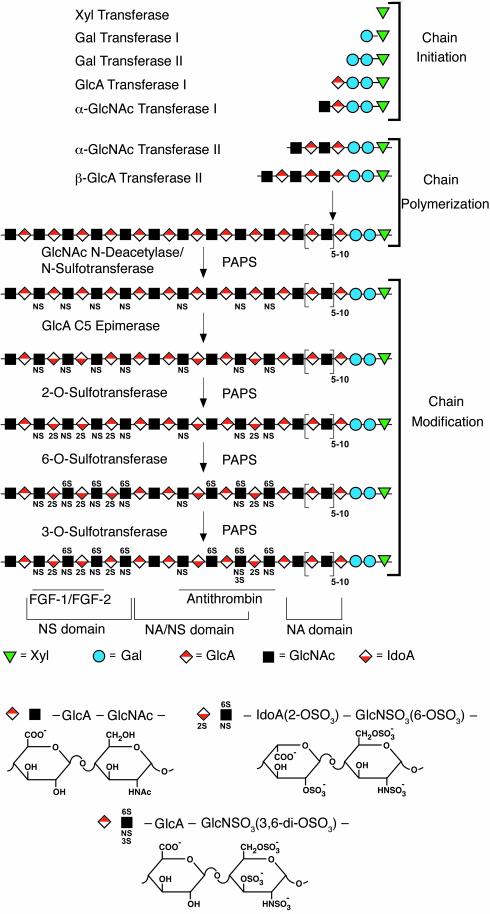
Scheme of HS chain biosynthesis. The symbols used are defined by the structures shown below the scheme. Structural domains (NA, NA/NS, NS) are defined with regard to the distribution of GlcN N-substituents as indicated. Also shown are regions that have been implicated in binding of specific ligands, such as FGF-1/FGF-2 and antithrombin.
The glypicans (six members) are distinguished by their cysteine-rich globular ectodomains, and the presence of 2–3 HS chains attached just between the globular domain and the glycosylphosphatidylinositol moiety that anchors it to the outer leaflet of the plasma membrane (7). No release of glypicans by a phospholipase C–type mechanism has been documented as yet, but like syndecans, the glypicans can be degraded by endocytosis and lysosomal degradation (8).
Several minor membrane proteoglycans containing HS chains have been described as well (epican, betaglycan, and others) (1). In addition, cells that produce basement membranes secrete the HS proteoglycans, perlecan, agrin, and collagen XVIII (9, 10). The individual glypicans, syndecans, and basement membrane HS proteoglycans are expressed in a tissue-specific manner.
Lower organisms, such as Drosophila melanogaster and Caenorhabditis elegans, express homologs of glypican, syndecan, and perlecan (11). In contrast to the multiple isoforms found in vertebrates, these organisms generally contain only one copy of syndecan and perlecan, and two copies of glypican are present in flies. Proteoglycans are also present in very ancient multicellular organisms such as Hydra, but the core proteins in these species have not yet been characterized (12). Thus, throughout evolution the HS proteoglycans have played fundamental roles in development and physiology common to a multitude of living creatures.
Structural diversity of HS
The tissue-specific expression of individual proteoglycan core proteins will obviously determine when and where HS chains are expressed. However, ligand binding by proteoglycans depends on the structure of the HS chains. As shown in Figure 1, HS can be analyzed in terms of disaccharide composition; the disaccharides are distinguished by the presence of variably sulfated or nonsulfated GlcA/IdoA and GlcN residues. Generally, the same set of disaccharides exists in most tissues, but their relative content varies quantitatively. For example, the disaccharide GlcA-GlcNS3S occurs predominantly in endothelial cells and connective tissue mast cells, as this unit is a critical substructure in the pentasaccharide sequence that binds antithrombin (13, 14). In contrast, kidney HS contains a large amount of IdoA2S-GlcNS3S, but the precise function of this unit in ligand binding is not known (15). Interestingly, the basic structural disaccharides of HS appear to be quite ancient.
Another way to characterize structure is in terms of the relative distribution of the major N-substituents of the GlcN residues: tracts of contiguous N-acetylated disaccharide units (NA domains), contiguous N-sulfated sequences of variable length (NS domains), and alternating N-acetylated and N-sulfated units (NA/NS domains) (Figure 1). Such N-substitution patterns appear to be characteristic of the cells/tissues from which the HS was obtained. Notably, heparin, the mast cell polysaccharide, may be considered essentially a single, unusually extended NS domain. Since other modifications, such as O-sulfation and epimerization of GlcA to L-IdoA, depend on prior N-sulfation of GlcN units, the modified disaccharide units tend to cluster in the NS or NA/NS domains (16).
The disaccharide composition and the arrangement of NA and NS domains do not by themselves define binding sites for specific ligands. Instead, binding occurs to specific sets of variably modified disaccharides usually within the NS or NA/NS domains (17, 18). The best-studied example to date is the “lock-and-key” interaction between HS/heparin and antithrombin, which leads to inactivation of thrombin, factor Xa, and other serine proteinases of the coagulation cascade. This interaction depends on a very specific structure of a pentasaccharide that contains a central 3-O-sulfated GlcN residue (GlcNAc6SGlcAGlcNS3S6SIdoA2SGlcNS6S) (see Figure 1, bottom) (13). Other examples are the interactions of glycoprotein gD from Herpes simplex virus (see Shukla and Spear, this Perspective series, ref. 19) with an oligosaccharide containing IdoA2S-GlcN3S and of FGF-1 and FGF-2 with N-sulfated pentasaccharide sequences containing IdoA2S and GlcN6S units in distinct combinations (see Gallagher, this series, ref. 20; and ref. 21). The distribution of binding sites for other ligands and their corresponding oligosaccharide sequences are less clear-cut. Recent studies have focused on sequences that mediate binding and/or activation of PDGF, platelet factor 4, HGF (scatter factor), lipoprotein lipase, Herpes simplex glycoprotein gC, laminin, and chemokines. Some of the binding sites involve discontinuous domains of the chains (e.g., IFN-γ, platelet factor 4, and IL-8) (see ref. 16 for relevant references). In other cases, the chain may act as a template, approximating a ligand with its binding partner (13, 22, 23). The expression of binding sites occurs in a tissue-specific manner and can change during development, aging, and disease. An unexplored question concerns the potential variation of structure in a given tissue in different individuals of the same species that might arise from differences in nutrition or genetic background.
HS biosynthesis depends on many enzymes
The fine structure of the chains ultimately depends on the regulated expression and action of multiple glycosyltransferases, sulfotransferases, and an epimerase, which are arrayed in the lumen of the Golgi apparatus (Figure 1). In addition, a series of cytoplasmic enzymes are needed to catalyze nucleotide sugar (UDP-Xyl, UDP-Gal, UDP-GlcA, UDP-GlcNAc) and nucleotide sulfate (PAPS) formation, and multiple membrane transporters to import the nucleotides from the cytosol to the lumen of Golgi apparatus (24, 25). The assembly process may also depend on the availability of GlcN or other sugars in the diet. Synthesis initiates through the assembly of a linkage tetrasaccharide, GlcAβ1,3Galβ1,3Galβ1,4Xylβ1-, on serine residues in the proteoglycan core polypeptide (26). This process is catalyzed by four enzymes that add individual sugar residues sequentially to the nonreducing end of the growing chain. The initiating reaction, catalyzed by xylosyltransferase, occurs at specific sites, defined by Ser-Gly residues flanked by one or more acidic residues (27). The linkage region also undergoes phosphorylation at C2 of xylose and sulfation at C4 or C6 of the galactose residues, but the functions of these modifications remain unclear (26). These linkage region modifications tend to be substoichiometric and are absent in some proteoglycans. Phosphorylation may be transient, suggesting a role in secretion or in regulation of the assembly process.
After assembly of the linkage region, one or more α-GlcNAc transferases add a single α1,4-linked GlcNAc unit to the chain, which commits the intermediate to the assembly of HS. Competition exists between this reaction and the addition of a β1,4-linked GalNAc residue catalyzed by a separate enzyme. When the latter reaction occurs, the intermediate serves as a primer for chondroitin sulfate formation. Evidence suggests that amino acid determinants lying close to the glycosaminoglycan attachment site or structural domains at some distance away regulate this process, with chondroitin sulfate representing the default pathway (27). Polymerization of HS then takes place by the alternating addition of GlcAβ1,4 and GlcNAcα1,4 residues, catalyzed by proteins now recognized as members of the exostosin family of tumor suppressors (see Duncan et al., this Perspective series, ref. 28). As the chain polymerizes, it undergoes a series of modifications that include GlcNAc N-deacetylation and N-sulfation, C5 epimerization of GlcA to IdoA, and variable O-sulfation at C2 of IdoA and GlcA, at C6 of GlcNAc and GlcNS units, and occasionally at C3 of GlcN residues (Figure 1). The concerted action of the enzymes catalyzing these reactions results in the formation and organization of NA, NS, and NA/NS domains (16).
Most of the enzymes involved in modifying the chain have now been purified and molecularly cloned. The GlcNAc N-deacetylase/N-sulfotransferase (NDST), the glucosaminyl 6-O-sulfotransferases (6OST), and the glucosaminyl 3-O-sulfotransferases (3OST) each represent a gene family whose members appear to be expressed in a tissue-specific and developmentally regulated pattern. Four NDSTs, five 3OSTs, and three 6OSTs are known. Substrate specificity studies performed in vitro indicate that the individual members of each enzyme subfamily catalyze the same reaction, but in different chemical contexts. For example, 3OST-1 is the only 3OST isozyme that can form the antithrombin binding sequence (i.e., domains containing GlcA-GlcNS3S). In contrast, 3OST-2 transfers sulfate to GlcA2S-GlcNS and IdoA2S-GlcNS, whereas 3OST-3A transfers sulfate to IdoA2S-GlcN, where the GlcN has an unsubstituted amino group, thus generating the binding site for the viral gD glycoprotein (29, 30). The three 6-O-sulfotransferases add sulfate to the C6 of GlcN units, but the preferred location of the target relative to GlcA and IdoA varies (31). The four NDST isozymes show variation in relative ratios of N-deacetylase and N-sulfotransferase activity. Modeling studies of the NDSTs against the crystal structure of the sulfotransferase domain of NDST1 (32) suggest that modulations of the binding cleft for the sugar chain may confer different substrate specificities for the enzymes (33).
In contrast to these sulfotransferases, only one 2-O-sulfotransferase (2OST) (34, 35) and one epimerase (36, 37) appear to exist in vertebrates. A survey of lower organisms has shown only single isozymes for the other sulfotransferases, suggesting that the ancestral forms of these enzymes perform all the basic reactions of HS biosynthesis required to generate the diversity of structure necessary for the various biological activities essential to the organisms.
In spite of the detailed information available about primary sequence of the isozymes and the evolving information about their substrate preferences, the formation of binding sites for antithrombin and glycoprotein gD remain the only known examples in which formation of a biologically significant structure can be correlated with expression of a specific isozyme (3OST-1 and 3OST-3A, respectively) (14, 30). Notably, these examples both involve a “rare” component, i.e. the 3-O-sulfated GlcN residue, and are therefore, in a sense, conceptually simple compared with ligands that depend on the differential topology of major building blocks that are present in most HS species. Examples of such ligands include members of the FGF family, FGF1 and FGF2, with different binding requirements for GlcN 6-O-sulfate and IdoA 2-O-sulfate groups that are expressed in a selective fashion on distinct HS species (see Gallagher, this Perspective series, ref. 20). The precise structures of the corresponding binding sites and their distribution in HS chains are unknown. Clearly, a major effort in the field should aim at understanding the functional properties of the biosynthetic enzymes, as required to generate specific (or sometimes overlapping) binding sites in HS chains for a variety of protein ligands.
New insights into this problem should emerge from multiple directions. First, it should be possible to refine our understanding of the catalytic specificities of the various isozymes using recombinant enzymes with chemically defined substrates. Recent advances in carbohydrate synthesis (38, 39) represent a step in the right direction, but ultimately large scale synthesis of oligosaccharide libraries will be needed to elucidate substrate specificities in depth. Secondly, we need to understand the topographic organization of the biosynthetic apparatus, including the localization of enzymes in the Golgi membrane, their interaction with each other and with any auxiliary proteins, and their mode of processing the polysaccharide substrate. This objective can be approached in a variety of ways, ranging from immunochemical localization of native and mutated proteins in the cell (M.A.S. Pinhal and J.D. Esko, unpublished work) to the not yet realized assembly of model biosynthetic systems, using artificial membranes and recombinant proteins.
Insight from the derangement of HS function
Further insights into the relationship of enzyme structure and function should emerge from genetic studies in both invertebrates and vertebrates. Several groups simultaneously discovered that mutations in D. melanogaster affecting HS biosynthetic enzymes homologous to those found in mammals had deleterious effects on developmental patterning dependent on the wingless signaling pathway (see Filmus and Selleck, this Perspective series, ref. 40) and Hedgehog diffusion (see Duncan et al., this series, ref. 28), consistent with the idea that matrix HS may play a role in forming gradients of morphogens and growth factors. Recently, C. elegans mutants showing deranged vulva development were found to harbor mutations in three genes involved in glycosaminoglycan biosynthesis (41, 42). Collectively, these studies provided the first evidence that glycosaminoglycan biosynthesis and the HS binding properties of various ligands are of physiological importance in vivo. However, due to the relative simplicity of the isozymes and core proteins expressed in flies and worms, these findings will not necessarily provide insight into the function of the multiple vertebrate homologs.
The development of homologous recombination techniques in murine ES cells provides a way to induce mutations in HS synthesis in mice (see Forsberg and Kjellén, this Perspective series, ref. 43). A mutation in the gene for the IdoA 2-O-sulfotransferase by insertional mutagenesis causes renal agenesis and neonatal lethality in the mutant mice (44). Analysis of tissues from these animals shows a complete loss of 2-O-sulfated IdoA containing disaccharides (J. Gallagher and C. Merry, unpublished work), consistent with the idea that only one 2OST exists. However, the surprising finding is that most tissues and organs develop normally, and many processes thought to depend on growth factor signaling occur apparently unhampered. Thus, signaling by way of FGFs does not obligatorily depend on the formation of HS sequences with the highest ligand affinity (45).
Two of the NDST isozymes have been inactivated by homologous recombination. NDST1 mutants exhibit neonatal lethality due to defective lung development (46, 47), whereas NDST2 defects have a restricted phenotype in which only heparin formation in connective tissue mast cells is affected (48, 49). These findings indicate that one cannot predict whether a particular isozyme will be essential for embryogenesis. In those cases where the mutations result in lethality, conditional mutations are needed in which expression of genes can be ablated in a tissue-specific manner or in adult animals. New methods for making targeted mutations in mice have recently become available using the Cre-loxP recombination system. Several key steps in the biosynthetic pathway are now in the process of being targeted in this way in order to examine the effect of ablating specific isozymes on structure and function of HS in specific tissues (J.D. Esko, unpublished results).
A priori, one can predict that mutations in every gene should be present in the human population. However, finding these individuals depends on whether the mutation led to death in utero or resulted in a clinical phenotype that an informed clinician would associate with altered HS formation. The defect in Simpson-Golabi-Behmel overgrowth syndrome has been traced to a chromosomal mutation in glypican-3 (see Filmus and Selleck, this Perspective series, ref. 40). Moreover, hereditary multiple exostosis, a bone disorder characterized by cartilage-capped bony outgrowths from the growth plates, has been associated with alterations in the copolymerase required for HS biosynthesis (see Duncan et al., this series, ref. 28). Although the actual molecular events underlying these diseases are unknown, one can already begin to glimpse the importance of HS in human growth and development. Harvesting the wealth of information that can be obtained from these unfortunate acts of heredity and from induced mutations in lower organisms remains an ongoing challenge.
Acknowledgments
We apologize to the many investigators whose articles we could not cite due to the journal’s space constraints. Interested readers should see the suggested reading list appended to the electronic version of this article (http://www.jci.org/cgi/content/full/108/2/169/DC1). This work was supported by grants from the NIH (R37GM33063 and PO1HL57345 to J.D. Esko).
References
- 1.Kjellén L, Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 2.Bernfield M, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 3.Bernfield M, et al. Biology of the syndecans: a family of transmembrane heparan sulfate proteoglycans. Annu Rev Cell Biol. 1992;8:365–393. doi: 10.1146/annurev.cb.08.110192.002053. [DOI] [PubMed] [Google Scholar]
- 4.Williams KJ, Fuki IV. Cell-surface heparan sulfate proteoglycans: dynamic molecules mediating ligand catabolism. Curr Opin Lipidol. 1997;8:253–262. doi: 10.1097/00041433-199710000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Tumova S, Woods A, Couchman JR. Heparan sulfate proteoglycans on the cell surface: versatile coordinators of cellular functions. Int J Biochem Cell Biol. 2000;32:269–288. doi: 10.1016/s1357-2725(99)00116-8. [DOI] [PubMed] [Google Scholar]
- 6.Rapraeger AC. Syndecan-regulated receptor signaling. J Cell Biol. 2000;149:995–998. doi: 10.1083/jcb.149.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filmus J. Glypicans in growth control and cancer. Glycobiology. 2001;11:19R–23R. doi: 10.1093/glycob/11.3.19r. [DOI] [PubMed] [Google Scholar]
- 8.Yanagishita M, Hascall V. Cell surface heparan sulfate proteoglycans. J Biol Chem. 1992;267:9451–9454. [PubMed] [Google Scholar]
- 9.Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annu Rev Biochem. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- 10.Halfter W, Dong SC, Schurer B, Cole GJ. Collagen XVIII is a basement membrane heparan sulfate proteoglycan. J Biol Chem. 1998;273:25404–25412. doi: 10.1074/jbc.273.39.25404. [DOI] [PubMed] [Google Scholar]
- 11.Selleck SB. Genetic dissection of proteoglycan function in Drosophila and C. elegans. Semin Cell Dev Biol. 2001;12:127–134. doi: 10.1006/scdb.2000.0242. [DOI] [PubMed] [Google Scholar]
- 12.Sarras MP, Jr, et al. Extracellular matrix (mesoglea) of Hydra vulgaris. I. Isolation and characterization. Dev Biol. 1991;148:481–494. doi: 10.1016/0012-1606(91)90266-6. [DOI] [PubMed] [Google Scholar]
- 13.Bourin MC, Lindahl U. Glycosaminoglycans and the regulation of blood coagulation. Biochem J. 1993;289:313–330. doi: 10.1042/bj2890313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg RD, Shworak NW, Liu J, Schwartz JJ, Zhang LJ. Heparan sulfate proteoglycans of the cardiovascular system. Specific structures emerge but how is synthesis regulated? J Clin Invest. 1997;99:2062–2070. doi: 10.1172/JCI119377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edge AS, Spiro RG. Characterization of novel sequences containing 3-O-sulfated glucosamine in glomerular basement membrane heparan sulfate and localization of sulfated disaccharides to a peripheral domain. J Biol Chem. 1990;265:15874–15881. [PubMed] [Google Scholar]
- 16.Lindahl U, Kusche-Gullberg M, Kjellén L. Regulated diversity of heparan sulfate. J Biol Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 17.Jackson RL, Busch SJ, Cardin AD. Glycosaminoglycans: molecular properties, protein interactions, and role in physiological processes. Physiol Rev. 1991;71:481–539. doi: 10.1152/physrev.1991.71.2.481. [DOI] [PubMed] [Google Scholar]
- 18.Spillmann D, Lindahl U. Glycosaminoglycan-protein interactions: a question of specificity. Curr Opin Struct Biol. 1994;4:677–682. doi: 10.1016/j.sbi.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Shukla, D., and Spear, P.G. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]
- 20.Gallagher, J.T. 2001. Heparan sulfate: growth control with a restricted sequence menu. J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]
- 21.Kreuger, J., Salmivirta, M., Sturiale, L., Gimenez-Gallego, G., and Lindahl, U. 2001. Sequence analysis of heparan sulfate epitopes with graded affinities for FGF-1 and FGF-2. J. Biol. Chem. In press. [DOI] [PubMed]
- 22.Stauber DJ, DiGabriele AD, Hendrickson WA. Structural interactions of fibroblast growth factor receptor with its ligands. Proc Natl Acad Sci USA. 2000;97:49–54. doi: 10.1073/pnas.97.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pellegrini L, Burke DF, Von Delft F, Mulloy B, Blundell TL. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature. 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- 24.Hirschberg CB, Robbins PW, Abeijon C. Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu Rev Biochem. 1998;67:49–69. doi: 10.1146/annurev.biochem.67.1.49. [DOI] [PubMed] [Google Scholar]
- 25.Berninsone PM, Hirschberg CB. Nucleotide sugar transporters of the Golgi apparatus. Curr Opin Struct Biol. 2000;10:542–547. doi: 10.1016/s0959-440x(00)00128-7. [DOI] [PubMed] [Google Scholar]
- 26.Sugahara K, Kitagawa H. Recent advances in the study of the biosynthesis and functions of sulfated glycosaminoglycans. Curr Opin Struct Biol. 2000;10:518–527. doi: 10.1016/s0959-440x(00)00125-1. [DOI] [PubMed] [Google Scholar]
- 27.Esko JD, Zhang L. Influence of core protein sequence on glycosaminoglycan assembly. Curr Opin Struct Biol. 1996;6:663–670. doi: 10.1016/s0959-440x(96)80034-0. [DOI] [PubMed] [Google Scholar]
- 28.Duncan, G., McCormick, C., and Tufaro, F. 2001. The link between heparan sulfate and hereditary bone disease: finding a function for the EXT family of putative tumor suppressor proteins. J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]
- 29.Liu JA, et al. Heparan sulfate D-glucosaminyl 3-O-sulfotransferase-3A sulfates N-unsubstituted glucosamine residues. J Biol Chem. 1999;274:38155–38162. doi: 10.1074/jbc.274.53.38155. [DOI] [PubMed] [Google Scholar]
- 30.Shukla D, et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 31.Habuchi H, et al. The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for hexuronic acid adjacent to the targeted N-sulfoglucosamine. J Biol Chem. 2000;275:2859–2868. doi: 10.1074/jbc.275.4.2859. [DOI] [PubMed] [Google Scholar]
- 32.Kakuta Y, Sueyoshi T, Negishi M, Pedersen LC. Crystal structure of the sulfotransferase domain of human heparan sulfate N-deacetylase/N-sulfotransferase 1. J Biol Chem. 1999;274:10673–10676. doi: 10.1074/jbc.274.16.10673. [DOI] [PubMed] [Google Scholar]
- 33.Aikawa J, Grobe K, Tsujimoto M, Esko JD. Multiple isozymes of heparan sulfate/heparin GlcNAc N-deacetylase/N-sulfotransferase: structure and activity of the fourth member, NDST4. J Biol Chem. 2001;276:5876–5882. doi: 10.1074/jbc.M009606200. [DOI] [PubMed] [Google Scholar]
- 34.Bai XM, Esko JD. An animal cell mutant defective in heparan sulfate hexuronic acid 2-O-sulfation. J Biol Chem. 1996;271:17711–17717. doi: 10.1074/jbc.271.30.17711. [DOI] [PubMed] [Google Scholar]
- 35.Rong J, Habuchi H, Kimata K, Lindahl U, Kusche-Gullberg M. Substrate specificity of the heparan sulfate hexuronic acid 2-O-sulfotransferase. Biochemistry. 2001;40:5548–5555. doi: 10.1021/bi002926p. [DOI] [PubMed] [Google Scholar]
- 36.Crawford BE, Olson SK, Esko JD, Pinhal MA. Cloning, Golgi localization, and enzyme activity of the full length heparin/heparan sulfate: glucuronic acid C5 epimerase. J Biol Chem. 2001;276:21538–21543. doi: 10.1074/jbc.M100880200. [DOI] [PubMed] [Google Scholar]
- 37.Li JP, Gong F, El Darwish K, Jalkanen M, Lindahl U. Characterization of the D-glucuronyl C5-epimerase involved in the biosynthesis of heparin and heparan sulfate. J Biol Chem. 2001;276:20069–20077. doi: 10.1074/jbc.M011783200. [DOI] [PubMed] [Google Scholar]
- 38.Seeberger PH, Danishefsky SJ. Solid-phase synthesis of oligosaccharides and glycoconjugates by the glycal assembly method: a five year retrospective. Acc Chem Res. 1998;31:685–695. [Google Scholar]
- 39.Koeller KM, Wong CH. Complex carbohydrate synthesis tools for glycobiologists: enzyme-based approach and programmable one-pot strategies. Glycobiology. 2000;10:1157–1169. doi: 10.1093/glycob/10.11.1157. [DOI] [PubMed] [Google Scholar]
- 40.Filmus, J., and Selleck, S.B. 2001. Glypicans: proteoglycans with a surprise. J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]
- 41.Herman T, Horvitz HR. Three proteins involved in Caenorhabditis elegans vulval invagination are similar to components of a glycosylation pathway. Proc Natl Acad Sci USA. 1999;96:974–979. doi: 10.1073/pnas.96.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bulik DA, et al. sqv-3 -7, and -8, a set of genes affecting morphogenesis in Caenorhabditis elegans, encode enzymes required for glycosaminoglycan biosynthesis. Proc Natl Acad Sci USA. 2000;97:10838–10843. doi: 10.1073/pnas.97.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forsberg E, Kjellén L. Heparan sulfate: lessons from knockout mice. J Clin Invest. 2001;108:175–180. doi: 10.1172/JCI13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bullock SL, Fletcher JM, Beddington RSP, Wilson VA. Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev. 1998;12:1894–1906. doi: 10.1101/gad.12.12.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang Z, Meyer K, Rapraeger AC, Friedl A. Differential ability of heparan sulfate proteoglycans to assemble the fibroblast growth factor receptor complex in situ. FASEB J. 2000;14:137–144. doi: 10.1096/fasebj.14.1.137. [DOI] [PubMed] [Google Scholar]
- 46.Fan G, et al. Targeted disruption of NDST-1 gene leads to pulmonary hypoplasia and neonatal respiratory distress in mice. FEBS Lett. 2000;467:7–11. doi: 10.1016/s0014-5793(00)01111-x. [DOI] [PubMed] [Google Scholar]
- 47.Ringvall M, et al. Defective heparan sulfate biosynthesis and neonatal lethality in mice lacking N-deacetylase/N-sulfotransferase-1. J Biol Chem. 2000;275:25926–25930. doi: 10.1074/jbc.C000359200. [DOI] [PubMed] [Google Scholar]
- 48.Forsberg E, et al. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature. 1999;400:773–776. doi: 10.1038/23488. [DOI] [PubMed] [Google Scholar]
- 49.Humphries DE, et al. Heparin is essential for the storage of specific granule proteases in mast cells. Nature. 1999;400:769–772. doi: 10.1038/23481. [DOI] [PubMed] [Google Scholar]