Abstract
To understand the evolution, attenuation, and variable protective efficacy of bacillus Calmette–Guérin (BCG) vaccines, Mycobacterium bovis BCG Pasteur 1173P2 has been subjected to comparative genome and transcriptome analysis. The 4,374,522-bp genome contains 3,954 protein-coding genes, 58 of which are present in two copies as a result of two independent tandem duplications, DU1 and DU2. DU1 is restricted to BCG Pasteur, although four forms of DU2 exist; DU2-I is confined to early BCG vaccines, like BCG Japan, whereas DU2-III and DU2-IV occur in the late vaccines. The glycerol-3-phosphate dehydrogenase gene, glpD2, is one of only three genes common to all four DU2 variants, implying that BCG requires higher levels of this enzyme to grow on glycerol. Further amplification of the DU2 region is ongoing, even within vaccine preparations used to immunize humans. An evolutionary scheme for BCG vaccines was established by analyzing DU2 and other markers. Lesions in genes encoding σ-factors and pleiotropic transcriptional regulators, like PhoR and Crp, were also uncovered in various BCG strains; together with gene amplification, these affect gene expression levels, immunogenicity, and, possibly, protection against tuberculosis. Furthermore, the combined findings suggest that early BCG vaccines may even be superior to the later ones that are more widely used.
Keywords: glycerol metabolism, live vaccines, tandem duplications, tuberculosis
More than 3 billion individuals have been immunized with bacillus Calmette–Guérin (BCG), the “Bacille de Calmette et Guérin,” an attenuated derivative of Mycobacterium bovis (1). BCG is part of the WHO's Expanded Program on Immunization because of its proven efficacy at preventing extrapulmonary tuberculosis in children (2). However, in adults, its efficacy against pulmonary disease is variable (3, 4), possibly as a result of environmental, operational, demographic, and genetic factors (5). For instance, prior exposure to environmental mycobacteria severely compromises protection afforded by BCG (6), and this is influenced by the extent of cross-recognition of antigens shared with the vaccine (7).
Another possible explanation for variable efficacy lies in the use of different daughter strains, and a brief reminder of their history is required (8–10). For 13 years, Calmette and Guérin serially passaged their strain on potato slices imbibed with glycerol and monitored loss of virulence (1). Once safety had been confirmed, BCG was disseminated, and different laboratories maintained their own daughter strains by passaging, until the introduction of archival seed lots in the 1960s. Since then, it has been recommended that vaccine preparations undergo no more than 12 passages from each seed lot (2). Thus, M. bovis BCG Pasteur 1173P2 corresponds to the archive established after 1,173 passages.
Recently, the various daughter strains have been studied by comparative genomics (11–14), and this uncovered regions of difference (RD) such as deletions and insertions, plus some SNPs. BCG vaccines were thus divided into the early strains, represented by BCGs Japan, Birkhaug, Sweden, and Russia and the late strains, including BCGs Pasteur, Danish, Glaxo, and Prague (8).
The most obvious reason for the attenuation of BCG was the loss of the protein secretion system ESX-1, absent from all strains, due to deletion of RD1 (15–20). However, because reintroduction of ESX-1 to BCG Pasteur or Russia does not restore full virulence (17), there are likely to be other lesions. Here, in an attempt to refine the genealogy of BCG, elucidate the basis of attenuation, and understand variable vaccine efficacy, we present the complete genome sequence of M. bovis BCG Pasteur 1173P2, details of its bioinformatic and functional-genomic analysis, and evidence for tandem duplications, DU1 and DU2.
Results
The Genome Sequence.
By using gene prediction and genome comparison approaches (21, 22), a total of 3,954 genes coding for proteins (CDS) were identified in the 4,374,522-bp circular chromosome of BCG Pasteur, together with 34 pseudogenes (Fig. 1). Although the BCG genome has incurred several deletions since diverging from its parent M. bovis (11), it is nonetheless almost 30 kb larger than that of M. bovis AF2122/97, which contains 4,345,492 bp (22), as a result of two independent tandem duplications, DU1 and DU2 (23). Consequently, BCG Pasteur is diploid for 58 CDS and two tRNA genes. There are 48 repetitive elements corresponding to insertion sequences and 13E12 repeats but none of the known prophages associated with M. tuberculosis (21, 24).
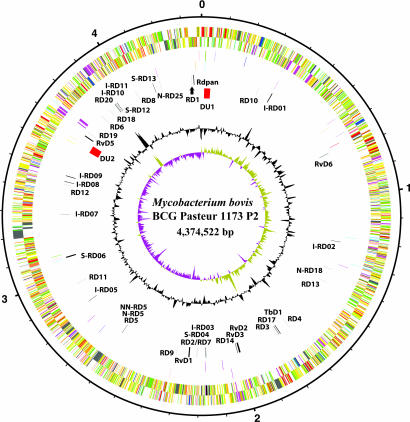
Fig. 1.
Circular representation of the M. bovis BCG Pasteur chromosome. The scale is shown in megabases in the outer black circle. Moving inward, the next two circles show forward and reverse strand CDS, respectively, with colors representing the functional classification (red, replication; light blue, regulation; dark blue, virulence; light green, hypothetical protein; dark green, cell wall and cell processes; orange, conserved hypothetical protein; cyan, IS elements; yellow, intermediate metabolism; gray, lipid metabolism; purple, PE/PPE). The following two circles show forward and reverse strand pseudogenes (colors represent the functional classification), the next circle shows RD (black) and DU (red), followed by the G+C content, and finally the GC skew (G-C)/(G+C) plotted by using a 10-kb window. For more details see SI Table 2.
Comparative Genomics.
Considerable insight into the evolution of tubercle bacilli has been obtained from studying polymorphisms like RD (25–28). On comparison of the genome sequences of M. tuberculosis strains H37Rv and CDC1551 (29) with those of M. bovis AF2122/97 and BCG Pasteur, 42 RD were uncovered, 28 of which had been detected previously [Fig. 1; and see supporting information (SI) Table 2]. These affect ≈170 genes, of which BCG Pasteur has lost 133. Of the 14 new RD, 1 is intergenic, 11 affect PE_PGRS or PPE genes, and another corresponds to amplification of a 57-bp tandem repeat in leuA. The finding that BCG Pasteur contains RD17 and RDpan, whereas M. bovis AF2122/97 does not, is consistent with the scheme in which the parental M. bovis strain preceded M. bovis AF2122/97, as borne out by spoligotyping (27).
On inspection of the complete SNP catalog (SI Table 3), it was found that there were only 736 SNP between the two M. bovis strains, indicating a close relationship, and ≈2,400 SNP between BCG and the M. tuberculosis strains, which also appear more divergent than the bovine strains. The majority of the SNP between M. bovis and BCG Pasteur occur within genes (83%), and 68% of these are nonsynonymous, consistent with the delayed effects of purifying selection on recent mutations (30).
Tandem Duplications in BCG Pasteur.
The most prominent genomic polymorphisms are the tandem duplications, DU1 and DU2. Both copies of DU1 are identical, whereas there is a single nonsynonymous base difference in DU2, within gene BCG3258. DU1 is 29,667 bp in length and spans the chromosomal origin of replication, oriC. As can be seen from Fig. 2, this duplication is restricted to BCG Pasteur, unlike DU2, which occurs in one of four different forms, DU2-I–DU2-IV, in all daughter BCG strains examined. In BCG Pasteur, DU2-IV, comprising 36,163 bp, arose as a result of tandem duplication of a 141-kb stretch and two subsequent internal deletions, one of which (Δint) is common to BCG groups DU2-II–DU2-IV. Several regulatory genes, including sigH and whiB1, occur in DU2-IV that might affect gene expression levels when diploid.
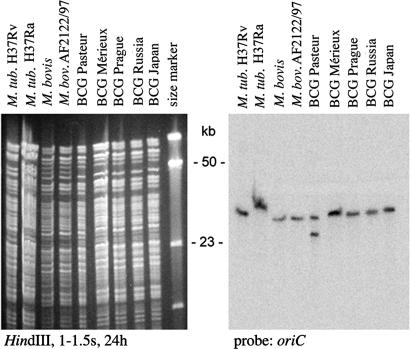
Fig. 2.
Mapping duplications in BCG strains. DU1 is confined to M. bovis BCG Pasteur as shown by Southern blotting of HindIII restriction digests of various BCG vaccines and hybridization with a probe for the oriC region. Note that the difference in size of the fragment hybridizing in M. tuberculosis (M. tub.) H37Ra is due to an IS6110 insertion in the fragment.
DU2 in Other BCG Strains.
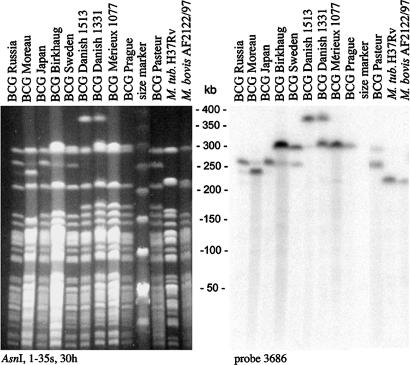
Detailed mapping of representative early and late BCG daughter strains was performed by pulsed-field gel electrophoresis and locus-specific Southern blotting. DU2-IV of BCG Pasteur is contained entirely within an AsnI fragment that spans region 3,529–3,753 kb (M. tuberculosis H37Rv coordinates); consequently, a change in its size reflects internal amplification events. As shown in Fig. 3, the corresponding AsnI fragments of the BCG strains are all larger than those of M. tuberculosis and M. bovis by ≈80 kb in BCGs Prague, Mérieux, and Danish, by ≈75 kb in BCGs Sweden and Birkhaug, and by ≈20 or 40 kb in BCG Moreau and BCGs Japan and Russia, respectively.
Fig. 3.
Genomic variations occur in vaccine preparations intended for human use. Variation in the DU2 region revealed by Southern blotting of AsnI restriction digests and hybridization with a probe for the 3,686-kb region. Note that the AsnI sites in the DU2 region are outside the duplicated region and that additional hybridizing bands are due to ongoing amplification events, such as triplications.
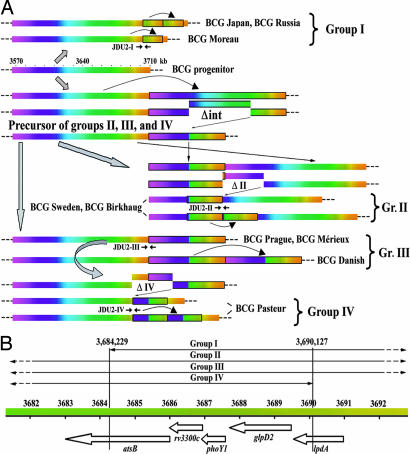
The endpoints of the DU2 duplications in the various BCG strains were pinpointed by mapping, cloning, and sequencing by using BAC clones. In the early strains BCGs Moreau, Russia, and Japan, the ≈20-kb duplicated segment corresponds to positions 3,684,229–3,704,932 (M. tuberculosis H37Rv coordinates), and this duplication was named DU2-I. There was extensive evidence of amplification of DU2-I because BCGs Russia and Japan have predominantly three copies, whereas BCG Moreau has two (Fig. 3 and SI Fig. 6), as summarized in Fig. 4A. PCR assays for the junction, JDU2-I, demonstrated the presence of DU2-I in all three strains but not in the remaining 11 BCG vaccines.
Fig. 4.
Scheme showing the appearance of DU2-I through DU2-IV. (A) Duplicated regions use a color scheme, and each duplication is boxed. Genomic coordinates based on M. tuberculosis H37Rv are indicated together with the positions of junctions (JDU2-I–JDU2-IV). (B). Identification of genes present in the region common to DU2-I through DU2-IV. Note the color scheme of A also applies to B.
BCGs Birkhaug and Sweden displayed identical hybridization patterns but their DU2 endpoints differed from those of other groups. Furthermore, the duplicated segment harboring Δint had switched position relative to groups III and IV. This configuration was termed DU2-II (Fig. 4A). In the case of BCGs Mérieux, Prague, and Danish, the duplicated fragment bore 78.5 kb of extra DNA, corresponding to regions 3,567,459–3,608,472 and 3,671,536–3,709,097 (M. tuberculosis H37Rv coordinates). This duplication, termed DU2-III, is a precursor to DU2-IV of BCG Pasteur, which has Δint but shows a different duplication junction because of the second deletion event (Fig. 4A and SI Fig. 6).
Systematic PCR screening with appropriate JDU2 primers (SI Table 4) classed each of the 14 BCG vaccines into one of four groups (Fig. 5). In this scheme, DU2-II–DU2-IV are closely related, whereas DU2-I is quite distinct. Early BCG strains with DU2-I have two or three copies of genes BCG3365–BCG3383, whereas late vaccines harboring DU2-III are diploid or triploid for genes BCG3221c–BCG3260c and BCG3356c–BCG3388c.
Fig. 5.
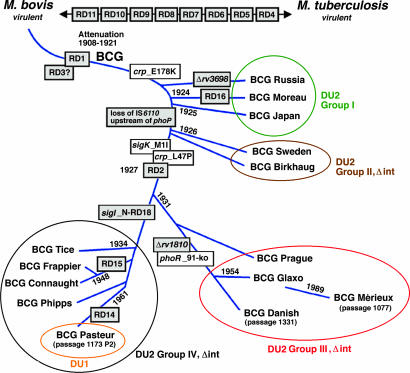
Refined genealogy of BCG vaccines. The scheme shows the position of genetic markers identified in this work, RD markers, some strain-specific deletions, and the distribution of vaccines into the four groups. Details of primers used for differentiation are listed in SI Table 4.
Amplification Is Ongoing.
Because gene amplification might alter immunogenicity and vaccine efficacy, we monitored current vaccine lots for triplications. A most striking example was found in the widely used vaccine BCG Danish 1331. A sample grown directly from a vial used in immunization programs was found to be positive for JDU2-III (Fig. 4B) and to contain AsnI fragments that were 80 or 160 kb larger than their counterparts in M. tuberculosis and M. bovis (Fig. 3), indicating that cells with either duplication or triplication of DU2-III coexisted in the population. In addition, we also investigated a BCG Danish strain that had been grown continuously for 1,513 passages and found only the triplicated form of DU2-III (Fig. 3 and SI Fig. 6). These results indicate that copy number may increase with additional passaging.
What Drives Amplification?
To find clues to the selective pressure, which led to tandem duplications, the region of overlap between DU2-I and DU2-IV was compared and found to comprise a mere 5,899 bp and contain only three intact genes: Rv3300c, encoding a member of the RNA pseudouridylate synthase family; phoY1, coding for a phosphate transport system regulator, and glpD2, encoding glycerol-3-phosphate dehydrogenase (Fig. 4B). Higher levels of the latter enzyme likely afforded an advantage to strains with duplications for growth in Calmette's glycerol-containing medium, and this is supported by the 2.7-fold increase in glpD2 expression levels in BCG strains compared with M. bovis (Table 1). Furthermore, because glycerol is still used in the medium for vaccine production, it is possible that amplification of the glpD2 region enhances the growth rate.
Table 1.
Selected gene expression data, including DU2
| Systematic name Mb | Systematic name BCG | Common name | BCG Pasteur* | BCG Japan | M. bovis 1121/01 | M. bovisAF2122/97 |
|---|---|---|---|---|---|---|
| Mb0674 | BCG0704 | mceG (mkl) | 2.2 (1.4–3.3) | 2.9 (1.8–4.3) | 23.8 (19.5–29.6) | 16.7 (9.3–31.3) |
| Mb0847c | BCG0877c | desA1 | 5.5 (1.9–13.1) | 6.1 (3.8–9.3) | 24.4 (15.9–39.3) | 14.67 (2.0–32.9) |
| Mb3245 | BCG3246/BCG3339 | whiB1 | 32.3 (16.7–58.1) | 22.9 (12.4–40.3) | 7.2 (3.5–11.1) | 8.8 (5.3–15.8) |
| Mb3250c | BCG3251/BCG3344 | sigH | 7.2 (2.7–11.6) | 3.0 (2.2–3.9) | 1.8 (1.2–6.1) | 1.8 (0.7–2.8) |
| Mb3251 | BCG3252/BCG3345 | Rv3224 | 7.9 (3.3–11.5) | 4.2 (2.7–6.7) | 4.0 (1.5–5.3) | 5.1 (81.8–7.9) |
| Mb3258c | BCG3259/BCG3352 | desA3 | 1.1 (0.7–1.9) | 1.4 (1.1–1.8) | 18.9 (3.9–35.1) | 18.2 (5.2–42.9) |
| Mb3259c | BCG3260/BCG3353 | Rv3230c | 0.7 (0.4–1.0) | 0.6 (0.4–0.8) | 2.9 (1.9–4.1) | 2.5 (1.2–5.3) |
| Mb3327c | BCG3328/BCG3364 | atsB | 1.4 (0.8–1.9) | 3.4 (2.4–4.4) | 0.8 (0.6–1.8) | 0.7 (0.3–1.1) |
| Mb3328c | BCG3329/BCG3365 | Rv3300c | 1.0 (0.5–2.8) | 1.6 (1.1–2.5) | 0.8 (0.4–1.1) | 0.8 (0.2–2.9) |
| Mb3329c | BCG3330/BCG3366 | phoY1 | 2.1 (1.3–2.6) | 2.0 (1.0–2.6) | 0.8 (0.5–1.3) | 1.0 (0.5–1.8) |
| Mb3330c | BCG3331/BCG3367 | glpD2 | 4.5 (2.8–5.8) | 2.7 (1.9–3.6) | 1.1 (0.8–1.5) | 1.2 (0.7–1.9) |
| Mb3331c | BCG3332/BCG3368 | lpdA | 1.8 (1.3–2.3) | 1.9 (1.3–2.6) | 0.3 (0.2–0.5) | 0.4 (0.2–0.6) |
| Mb3332 | BCG3333/BCG3369 | Rv3304 | 0.7 (0.4–1.2) | 2.4 (1.4–3.3) | 0.4 (0.2–0.5 | 0.35 (0.01–0.6) |
| Mb3336 | BCG3373 | pmmB | 0.4 (0.01–0.7) | 1.9 (1.5–2.5) | 0.4 (0.3–0.5) | 0.5 (0.2- 0.8) |
| Mb3345 | BCG3382 | sdhC | 1.1 (0.7–1.9) | 2.8 (1.9–3.8) | 1.5 (0.8–14.0) | 1.1 (0.6–2.3) |
| Mb3346 | BCG3383 | sdhD | 1.2 (0.5–2.1) | 4.4 (2.6–7.5) | 1.6 (1.1–2.0) | 1.2 (0.5–2.32) |
| Mb3450 | BCG3486 | whiB3 | 8.8 (3.6–14.7) | 1.0 (0.5–1.4) | 0.6 (0.4–1.3) | 0.6 0.2–3.2) |
*Values shown in boldface are more than twice those of the comparator group.
Gene Regulation and Phenotypic Differences.
Initial analysis of diversity in BCG revealed overrepresentation of regulatory genes in the RD (11). This trend was extended on analysis of the genome sequence because 3 of the 10 extracytoplasmic function (ECF) σ-factor genes (31) have been lost or inactivated. As a result of the N-RD18 deletion, the 5′ end of sigI (Rv1189) has been fused in-frame to the 3′ end of the Rv1191 ortholog. The resultant fusion protein is unlikely to function because the promoter recognition domain is located in the C-terminal part of ECF σ-factors. The sigI gene is intact in the other three BCG groups (Fig. 5).
The sigK gene of BCG Pasteur has incurred a missense mutation in its start codon that replaces the ATG by ATA. This mutation is present in all BCG strains except the early ones (Fig. 5). One of the consequences of this mutation is loss of expression of the major antigens MPB70 and MPB83 (32). Another interesting regulatory locus is that of the two-component system, PhoP-PhoR, as lesions in phoP in M. tuberculosis result in marked attenuation and a profoundly altered cell envelope (33, 34). Here, DU2-I strains differ from the others as they have an IS6110 element located upstream of phoP and this may influence expression levels as more phoP mRNA was detected in BCG Tokyo than BCG Pasteur (SI Table 5). This element was subsequently lost (Fig. 5).
Importantly, a 10-bp deletion was found in codon 91 of phoR from the DU2-III strains BCG Glaxo, Mérieux, and Danish, that would disrupt expression (Fig. 5). Inactivation of the PhoP-PhoR system leads to loss of diacyltrehaloses, polyacyltrehaloses, and sulfolipids (33, 35), all of which are missing from BCG, although the genes encoding the corresponding biosynthetic machinery are identical to those of M. bovis. Although the lesion in phoR might account for their absence from the DU2-III strains, it does not explain their loss from the other BCG vaccines, thus raising the possibility that other regulatory genes intervene. However, it is noteworthy that the DU2-III strains are considered to be the most attenuated.
In most bacteria, but possibly not M. tuberculosis (35), the PhoP-PhoR system generally regulates genes involved in phosphate metabolism, and genome analysis predicts that the high-affinity phosphate-uptake system may be inactive in BCG, because both the pstB and phoT genes, encoding key components, are frameshifted (36). BCG also lacks the alternative system for capturing phosphate, uptake of sn-glycerol-3-phosphate via the ugpABC system, because ugpB has a frameshift mutation. Intriguingly, although M. bovis AF2122/97 has an intact ugpB, it is mutated in ugpA, which is functional in BCG. Overall, BCG may be challenged for growth under conditions where phosphate concentrations are limiting, and, in vivo this would constitute a distinct growth disadvantage.
Another regulatory locus that has accrued down-regulating mutations is BCG3734, encoding the cAMP-receptor protein, Crp. Once again, there are variations between different BCG daughter strains (Fig. 5) because, although they all have an E178K substitution affecting the DNA-binding domain, the “late” strains also have the L47P replacement in the cAMP-binding site (37). Because Rv3676, the M. tuberculosis ortholog, mediates responses to low glucose concentrations and nutrient starvation, loss of BCG3734 function might perturb intermediary metabolism and growth under microaerophilic conditions.
Comparative Transcriptomics: Impact on Virulence and Metabolism.
To explore global gene expression differences, we performed comparative in vitro transcriptome analysis across the early and late BCGs, Japan and Pasteur, respectively, versus two M. bovis strains and highlighted a subset of genes showing significant differences in both comparisons (Table 1, SI Table 5, and SI Fig. 7).
A key selective pressure during in vitro adaptation of the M. bovis progenitor was the switch to a glycerol carbon source, evidenced by the presence of a functional pyruvate kinase enzyme in BCG (38) and significantly higher levels of transcription of glpD2 compared with M. bovis (Table 1). Further confirmation of metabolic remodeling is seen in the divergent regulation of genes associated with fatty acid degradation (SI Table 5). Hence, fadD2, fadE35, and the β-oxidation complex genes fadAB, are all down-regulated in BCG. Likewise, the desA1 and desA3 genes, encoding two desaturases involved in fatty acid modification (39), were down-regulated in BCG compared with M. bovis. Expression of desA3 is at least 12-fold higher in vitro in M. bovis compared with both BCG strains (Table 1). Inactivation of desA3 attenuates M. tuberculosis (40), and increased expression of desA1 and desA3 occurs in patients with active tuberculosis (41). These observations suggest a key role for these desaturases in virulence, so their decreased expression in BCG may be relevant to attenuation. Reduced expression of desA3 is surprising because it is located in DU2-IV (23); other DU2-IV genes showed ≈2-fold-increased relative expression, as expected for diploidy, and the same trend was observed with DU2-I genes (Table 1).
In line with the mutation of transcriptional activators, expression of multiple regulators was divergent between M. bovis and BCG (SI Table 5). Genes Mb0484, Mb0846c, Mb1433c, Mb2354, lexA, Mb3122, and Mb3614c were all at least 3-fold down-regulated in both BCG strains, whereas Mb3277 showed increased expression. Interestingly, three WhiB family transcription factors, which regulate virulence, cell division, and stress responses (41–44), were differentially expressed. In both BCGs, whiB4 was down-regulated, whereas whiB1 was overexpressed because of increased gene dosage; whiB3 showed 9-fold increased expression in BCG Pasteur over BCG Japan and M. bovis (Table 1).
One of the genes showing the greatest difference in expression between BCG and M. bovis is mceG, which is essential for the function of multiple mce loci involved in cell entry (45). Mutation of mceG had the same effect on in vivo growth as the simultaneous inactivation of both mce1 and mce4 loci, resulting in severe attenuation of M. tuberculosis (45). Our results show that mceG is highly repressed in BCG compared with M. bovis (Table 1), providing another possible explanation for attenuation of the vaccine.
Variable expression of genes encoding immunogenic surface and secreted proteins was revealed. Seven PE and PPE family members (pe-pgrs3, pe-pgrs5, pe-pgrs15, pe13, ppe18, ppe40, and pe-pgrs54) had reduced expression in BCG; ppe50 was up-regulated in both BCG strains, with ppe27 and pe19 up-regulated in BCG Pasteur versus BCG Japan. Expression of the serodominant Mpb83 and Mpb70 antigens (32) was repressed in BCG Pasteur, whereas five ESAT-6 genes (esxI, esxJ, esxK, esxM, and esxN) showed increased expression compared with BCG Japan (SI Table 5). Indeed, the cumulative effect of variation in secreted and cell wall antigens across BCG strains may contribute to variable vaccine efficacy.
Discussion
A large body of evidence is presented for diversity among BCG strains, and this stems from both genomic modifications and alterations to pleiotropic regulators of gene expression. We postulate that the resultant phenotypic differences contribute to variable vaccine efficacy. When Calmette and Guérin grew the progenitor M. bovis strain on glycerinated potato slices, they unknowingly imposed selective pressure for genetic alterations to this natural mutant for glycerol metabolism, so that glycerol could be used as a carbon and energy source. Some of these changes were simple point mutations, such as selection for an active pyruvate kinase (22, 38), whereas others, like tandem duplications, were more complex, with potentially far-reaching effects because of their size and instability. As part of the metabolic improvement, one direct consequence of DU2 amplifications was higher levels of glycerol-3-phosphate dehydrogenase, another key enzyme required for growth on glycerol, as confirmed by transcriptomics.
A study of the transcriptome also revealed extensive variation in gene expression both between early and late BCG daughter strains and with respect to virulent tubercle bacilli. This results from increased gene dosage or from altered activity of pleiotropic regulators leading to over- or underproduction of certain proteins, including virulence factors and enzymes. Consequently, the corresponding vaccine strain may have an imbalanced antigenic repertoire that does not accurately reflect that of bovine or human tubercle bacilli. As can be seen from SI Table 5, there are extensive differences in the level of expression of known surface proteins and immunodominant protein antigens between BCGs Pasteur and Japan that may induce protective responses.
Tandem gene duplications provide increased activity in response to strong selective pressure. From the known BCG chronology (9), it appears that DU2-I arose in prototypical BCG and was then lost before the emergence of the DU2-II precursor, which later incurred Δint. This, in turn, served as a substrate for further duplication events in vaccines of groups II, III, and IV (Fig. 5). If our model is correct, DU2 duplication arises as a function of growth rate on glycerol. Gene amplification, and the subsequent diversity in cellular populations and immunogenicity, could be avoided by producing the vaccine in a more controlled way, by using tools developed here for quality control and assurance, because the manufacturing process has scarcely changed since 1921. These innovations will also be useful for monitoring recombinant BCG vaccines.
From Fig. 5 it seems that, after 1925, Calmette's team may have replaced the strain initially distributed (now represented by BCG Japan) by another derivative, possibly less virulent or reactogenic. The later strains fall into three groups of which DU2-II is intermediate and followed by the major vaccine-production strains BCG Danish, Glaxo, and Pasteur. These three accounted for 66% of the 335 million doses administered in 1996 (2) and all of the metaanalyses of BCG vaccine efficacy use data sets from trials performed with them (3). We note that, in a comparative study of immune responses in babies, BCG Japan induced significantly higher levels of Th1-cytokines (IFN-γ, TNF-α, IL-2) and lower secretion of the Th2-cytokine, IL-4, accompanied by greater CD4 and CD8 T cell proliferation, than did BCG Danish (46). Taken together with our findings, this suggests that early BCG vaccines may confer better protection against tuberculosis, a possibility that would benefit from formal evaluation in clinical trials.
Materials and Methods
Bacteria, Genome Mapping, and Sequencing.
BCG vaccines were from the collections at the Institut Pasteur or McGill University, and grown in Middlebrook 7H9 medium (Difco) with 0.05% Tween 80 and Albumin Dextrose Catalase (ADC) supplement. An initial shotgun of BCG Pasteur was generated from ≈21,700 paired-end sequences from three pUC19 libraries with insert sizes ranging from 2.8 to 5.5 kb, 7,500 paired-end sequences from two pMAQ1b libraries with insert sizes of 5.5–10 kb, and 3,500 single-end sequences from an M13 library. This produced an initial 4-fold coverage of the genome. This was supplemented with 30,000 reads obtained from a whole-genome shotgun library prepared in the vector pCDNA2.1 in Paris. Plasmids were sequenced by using ABI 3700 DNA sequencers (PerkinElmer, Foster City, CA), and sequences assembled by using PHRAP and GAP4 as described (21). Annotation and database presentation were by means of Artemis (47) and BCGList (http://genolist.pasteur.fr/BCGList/). Genomic DNA analysis by pulsed-field gel electrophoresis and BAC library construction were as described (23, 48, 49).
Transcriptome Analysis.
M. bovis AF2122/97 (GB spoligotype 9, international code SB0140 as defined at www.mbovis.org), M. bovis 1121/01 (GB spoligotype 17, the second most frequent isolate in the U.K.; intl. code SB0263), M. bovis BCG Pasteur and M. bovis BCG Japan were grown as above. Total RNA from each strain was extracted, purified, reverse-transcribed, and labeled with Cy5-dCTP as described [(50) http://www.bugs.sgul.ac.uk/bugsbase/]. Cy3-labeled DNA (AF2122/97) was used for control purposes. Probes were hybridized to whole-genome M. bovis/M. tuberculosis composite microarrays (TbV2; St. Georges Hospital, U.K.), the arrays were scanned with an Affymetrix 428 scanner, images were analyzed with Imagene 5.0, and median spot intensities were calculated by using Genespring 7.0 software (Silicon Genetics, Redwood City, CA). Significance values were calculated by using oneway ANOVA, followed by a Student–Newman–Keuls post hoc test. Final P values were obtained by Benjamini and Hochberg correction for multiple testing with a false discovery rate of 5%. Results for selected genes were confirmed by real-time RT-PCR analysis, and expression levels were normalized by using the sigA gene as an internal reference.
Accession Numbers.
The genome sequence has been deposited at EMBL under accession no. AM408590; fully annotated microarray data have been deposited in BμG@Sbase (accession no. E-BUGS-43; http://bugs.sgul.ac.uk/E-BUGS-43) and ArrayExpress (accession no. E-BUGS-43).
Supplementary Material
Acknowledgments
We thank Drs. K. Haslov, I. Kromann, (both at Statens Serum Institutet, Copenhagen, Denmark), G. Marchal (Institut Pasteur), and S. Yamamoto [National Institute of Health (NIH), Tokyo, Japan] for providing strains and the shotgun sequencing team from the Sanger Institute for generating reads. This work was supported by the Department for Environment, Food, and Rural Affairs (DEFRA, GB), The Wellcome Trust, the Association Française Raoul Follereau, and the Institut Pasteur. We acknowledge BμG@S (the Bacterial Microarray Group at St. George's, University of London) for supply of the microarray and advice and The Wellcome Trust for funding the multicollaborative microbial pathogen microarray facility under its Functional Genomics Resources Initiative.
Abbreviations
- BCG
bacillus Calmette–Guérin
- RD
regions of difference.
Footnotes
The authors declare no conflict of interest.
Data deposition: The genome sequence reported in this paper has been deposited at EMBL (accession no. AM408590). Fully annotated microarray data have been deposited in BμG@Sbase, http://bugs.sgul.ac.uk/E-BUGS-43 (accession no. E-BUGS-43) and ArrayExpress (accession no. E-BUGS-43).
This article contains supporting information online at www.pnas.org/cgi/content/full/0700869104/DC1.
References
- 1.Calmette A. La Vaccination Préventive Contre la Tuberculose. Paris: Masson; 1927. [Google Scholar]
- 2.Fine PEM, Carneiro IAM, Milstien JB, Clemens CJ. Issues Relating to the Use of BCG in Immunization Programmes. A Discussion Document. Geneva: World Health Organization; 1999. [Google Scholar]
- 3.Colditz GA, Berkey CS, Mosteller F, Brewer TF, Wilson ME, Burdick E, Fineberg HV. Pediatrics. 1995;96:29–35. [PubMed] [Google Scholar]
- 4.Fine PEM. Lancet. 1995;346:1339–1345. doi: 10.1016/s0140-6736(95)92348-9. [DOI] [PubMed] [Google Scholar]
- 5.Bloom BR, Fine PEM. In: Tuberculosis: Pathogenesis, Protection, and Control. Bloom BR, editor. Washington, DC: Am Soc Microbiol; 1994. pp. 531–557. [Google Scholar]
- 6.Brandt L, Cunha JF, Olsen AW, Chilima B, Hirsch P, Appelberg R, Andersen P. Infect Immun. 2002;70:672–678. doi: 10.1128/iai.70.2.672-678.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demangel C, Garnier T, Rosenkrands I, Cole ST. Infect Immun. 2005;73:2190–2196. doi: 10.1128/IAI.73.4.2190-2196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosch R, Behr MA. In: Tuberculosis and the Tubercle Bacillus. Cole ST, Eisenach KD, McMurray DN, Jacobs WR Jr, editors. Washington, DC: Am Soc Microbiol; 2005. pp. 155–164. [Google Scholar]
- 9.Behr MA, Small PM. Vaccine. 1999;17:915–922. doi: 10.1016/s0264-410x(98)00277-1. [DOI] [PubMed] [Google Scholar]
- 10.Oettinger T, Joergensen M, Ladefoged A, Hasloev K, Andersen P. Tubercle Lung Dis. 1999;79:243–250. doi: 10.1054/tuld.1999.0206. [DOI] [PubMed] [Google Scholar]
- 11.Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, Small PM. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 12.Gordon SV, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole ST. Mol Microbiol. 1999;32:643–656. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 13.Belley A, Alexander D, Di Pietrantonio T, Girard M, Jones J, Schurr E, Liu J, Sherman DR, Behr MA. Infect Immun. 2004;72:2803–2809. doi: 10.1128/IAI.72.5.2803-2809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mostowy S, Tsolaki AG, Small PM, Behr MA. Vaccine. 2003;21:4270–4274. doi: 10.1016/s0264-410x(03)00484-5. [DOI] [PubMed] [Google Scholar]
- 15.Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. J Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR. Mol Microbiol. 2004;51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pym AS, Brodin P, Brosch R, Huerre M, Cole ST. Mol Microbiol. 2002;46:709–717. doi: 10.1046/j.1365-2958.2002.03237.x. [DOI] [PubMed] [Google Scholar]
- 18.Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, et al. Proc Natl Acad Sci USA. 2003;100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, Behr MA, Sherman DR. J Infect Dis. 2003;187:117–123. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanley SA, Raghavan S, Hwang WW, Cox JS. Proc Natl Acad Sci USA. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, et al. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 22.Garnier T, Eiglmeier K, Camus J-C, Medina M, Mansoor H, Pryor M, Duthoy S, Grondin S, Lacroix C, Monsempe C, et al. Proc Natl Acad Sci USA. 2003;100:7877–7882. doi: 10.1073/pnas.1130426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brosch R, Gordon SV, Buchrieser C, Pym A, Garnier T, Cole ST. Comp Funct Genomics (Yeast) 2000;17:111–123. doi: 10.1002/1097-0061(20000630)17:2<111::AID-YEA17>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon SV, Heym B, Parkhill J, Barrell B, Cole ST. Microbiology. 1999;145:881–892. doi: 10.1099/13500872-145-4-881. [DOI] [PubMed] [Google Scholar]
- 25.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, et al. Proc Natl Acad Sci USA. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mostowy S, Inwald J, Gordon S, Martin C, Warren R, Kremer K, Cousins D, Behr MA. J Bacteriol. 2005;187:6386–6395. doi: 10.1128/JB.187.18.6386-6395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith NH, Dale J, Inwald J, Palmer S, Gordon SV, Hewinson RG, Smith JM. Proc Natl Acad Sci USA. 2003;100:15271–15275. doi: 10.1073/pnas.2036554100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith NH, Kremer K, Inwald J, Dale J, Driscoll JR, Gordon SV, van Soolingen D, Hewinson RG, Smith JM. J Theor Biol. 2006;239:220–225. doi: 10.1016/j.jtbi.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 29.Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, DeBoy R, Dodson R, Gwinn M, Haft D, et al. J Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocha EP, Smith JM, Hurst LD, Holden MT, Cooper JE, Smith NH, Feil EJ. J Theor Biol. 2006;239:226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 31.Helmann JD. Adv Microb Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- 32.Charlet D, Mostowy S, Alexander D, Sit L, Wiker HG, Behr MA. Mol Microbiol. 2005;56:1302–1313. doi: 10.1111/j.1365-2958.2005.04618.x. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalo Asensio J, Maia C, Ferrer NL, Barilone N, Laval F, Soto CY, Winter N, Daffe M, Gicquel B, Martin C, Jackson M. J Biol Chem. 2006;281:1313–1316. doi: 10.1074/jbc.C500388200. [DOI] [PubMed] [Google Scholar]
- 34.Perez E, Samper S, Bordas Y, Guilhot C, Gicquel B, Martin C. Mol Microbiol. 2001;41:179–187. doi: 10.1046/j.1365-2958.2001.02500.x. [DOI] [PubMed] [Google Scholar]
- 35.Walters SB, Dubnau E, Kolesnikova I, Laval F, Daffe M, Smith I. Mol Microbiol. 2006;60:312–330. doi: 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 36.Collins DM, Kawakami RP, Buddle BM, Wards BJ, de Lisle GW. Microbiology. 2003;149:3203–3212. doi: 10.1099/mic.0.26469-0. [DOI] [PubMed] [Google Scholar]
- 37.Spreadbury CL, Pallen MJ, Overton T, Behr MA, Mostowy S, Spiro S, Busby SJ, Cole JA. Microbiology. 2005;151:547–556. doi: 10.1099/mic.0.27444-0. [DOI] [PubMed] [Google Scholar]
- 38.Keating LA, Wheeler PR, Mansoor H, Inwald JK, Dale J, Hewinson RG, Gordon SV. Mol Microbiol. 2005;56:163–174. doi: 10.1111/j.1365-2958.2005.04524.x. [DOI] [PubMed] [Google Scholar]
- 39.Makinoshima H, Glickman MS. Nature. 2005;436:406–409. doi: 10.1038/nature03713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sassetti CM, Rubin EJ. Proc Natl Acad Sci USA. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rachman H, Strong M, Ulrichs T, Grode L, Schuchhardt J, Mollenkopf H, Kosmiadi GA, Eisenberg D, Kaufmann SH. Infect Immun. 2006;74:1233–1242. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomez JE, Bishai WR. Proc Natl Acad Sci USA. 2000;97:8554–8559. doi: 10.1073/pnas.140225297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris RP, Nguyen L, Gatfield J, Visconti K, Nguyen K, Schnappinger D, Ehrt S, Liu Y, Heifets L, Pieters J, et al. Proc Natl Acad Sci USA. 2005;102:12200–12205. doi: 10.1073/pnas.0505446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steyn AJ, Collins DM, Hondalus MK, Jacobs WR, Jr, Kawakami RP, Bloom BR. Proc Natl Acad Sci USA. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joshi SM, Pandey AK, Capite N, Fortune SM, Rubin EJ, Sassetti CM. Proc Natl Acad Sci USA. 2006;103:11760–11765. doi: 10.1073/pnas.0603179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davids V, Hanekom WA, Mansoor N, Gamieldien H, Gelderbloem SJ, Hawkridge A, Hussey GD, Hughes EJ, Soler J, Murray RA, et al. J Infect Dis. 2006;193:531–536. doi: 10.1086/499825. [DOI] [PubMed] [Google Scholar]
- 47.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A, Barrell B. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 48.Brosch R, Gordon SV, Billault A, Garnier T, Eiglmeier K, Soravito C, Barrell BG, Cole ST. Infect Immun. 1998;66:2221–2229. doi: 10.1128/iai.66.5.2221-2229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Philipp WJ, Nair S, Guglielmi G, Lagranderie M, Gicquel B, Cole ST. Microbiology. 1996;142:3135–3145. doi: 10.1099/13500872-142-11-3135. [DOI] [PubMed] [Google Scholar]
- 50.Stewart GR, Wernisch L, Stabler R, Mangan JA, Hinds J, Laing KG, Young DB, Butcher PD. Microbiology. 2002;148:3129–3138. doi: 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.