Abstract
Eradication of poliomyelitis from large metropolis cities in India has been difficult due to high population density and the presence of large urban slums. Three paralytic poliomyelitis cases were reported in Mumbai, India, in 1999 and 2000 in spite of high immunization coverage and good-quality supplementary immunization activities. We therefore established a systematic environmental surveillance study by weekly screening of sewage samples from three high-risk slum areas to detect the silent transmission of wild poliovirus. In 2001, from among the 137 sewage samples tested, wild poliovirus type 1 was isolated from 35 and wild poliovirus type 3 was isolated from 1. Acute flaccid paralysis (AFP) surveillance indicated one case of paralytic poliomyelitis from the city. Phylogenetic analysis with complete VP1 sequences revealed that the isolates from environmental samples belonged to four lineages of wild polioviruses recently isolated from poliomyelitis cases in Uttar Pradesh and not to those previously isolated from AFP cases in Mumbai. Wild poliovirus thus introduced caused one case of paralytic poliomyelitis. The virus was detected in environmental samples 3 months before. It was found that wild polioviruses introduced several times during the year circulated in Mumbai for a limited period before being eliminated. Environmental surveillance was found to be sensitive for the detection of wild poliovirus silent transmission. Nucleotide sequence analysis helped identify wild poliovirus reservoir areas.
In 1988, the World Health Assembly committed the World Health Organization (WHO) to the global eradication of poliomyelitis by the year 2000 (31). Implementation of WHO-recommended strategies for poliomyelitis eradication resulted in a decrease in the number of globally reported poliomyelitis cases from 35,251 in 1988 to 2,971 by the year 2000 (11). The American, Western Pacific, and European regions of the WHO have been certified free from wild poliovirus (1, 3, 18). By the end of 2001, endemic wild poliovirus transmission was confined to 10 countries (4). In the last 2 years, the poliomyelitis eradication program has made significant progress in India. A total of 265 virologically confirmed paralytic poliomyelitis cases were recorded during the year 2000. Uttar Pradesh and Bihar are the two remaining major reservoirs of wild poliovirus transmission in India (5).
Less than 1% of all wild poliovirus infections result in paralytic disease, and the ratio may be greatly changed in highly immunized populations (15). An efficient acute flaccid paralysis (AFP) surveillance system supported by a WHO-accredited laboratory network for the virological diagnosis of wild poliovirus infections is essential not only for the detection of cases of poliomyelitis but also for documentation of the success of poliovirus eradication efforts (10). Supplementary surveillance methods involving healthy children stool surveys and screening of wastewater were successfully used to demonstrate wild poliovirus transmission as well as to judge the effectiveness of poliomyelitis immunization campaigns, especially during the last stages of poliovirus eradication (7, 13, 20, 26, 27). It is estimated that systematic environmental sampling would detect 1 individual excreting wild poliovirus among 10,000 inhabitants (9). However, the requirement of supplementary surveillance for the purpose of certification of poliovirus eradication is not yet clearly defined (31a).
Various combinations of methods, such as virus concentration by two-phase separation or precipitation, virus culturing at a supraoptimal temperature, the use of poliovirus-selective cell lines, and PCR, have been used to increase the sensitivity of wild poliovirus detection in sewage samples (2, 14, 22, 26, 30). Molecular analysis of wild poliovirus isolates either by restriction fragment length polymorphism analysis or by sequencing of the VP1-VP2A junction region and/or the complete VP1 gene has been used to define epidemiological links (21, 23, 29).
More than 1,000 cases of AFP were reported annually in the city of Mumbai, India, when the poliomyelitis eradication program began (12). Improved oral poliovirus vaccine (OPV) immunization coverage since 1988 and supplementary immunizations provided during National Immunization Days conducted since 1995-1996 resulted in a reduction in the number of confirmed paralytic poliomyelitis cases to less than 30 in 1996. However, intensive efforts could not eradicate poliomyelitis from Mumbai as quickly as anticipated. Three paralytic poliomyelitis cases due to wild poliovirus infection were detected by AFP surveillance in Mumbai in 1999 and 2000 (J. M. Deshpande, unpublished data).
We initiated sampling of sewage in selected slum areas in Mumbai to study the feasibility of long-term environmental surveillance, to detect the silent transmission of wild poliovirus, and to understand factors affecting polio eradication efforts in Mumbai. The period covered during the present study was considered a transition from the stage of elimination of poliovirus circulation to its eradication. During this phase, we expected to be able to compare the sensitivity of environmental surveillance with that of AFP surveillance. In this communication, we report the results of systematic monitoring of sewage samples for wild poliovirus in 2001.
MATERIALS AND METHODS
Mumbai.
Mumbai, an island city on the west coast of India, is the commercial capital of the country. Spread longitudinally over an area of ∼440 km2, it has more than 13 million inhabitants. The city is divided into administrative units called municipality wards identified by single-letter codes.
Sewage sampling sites.
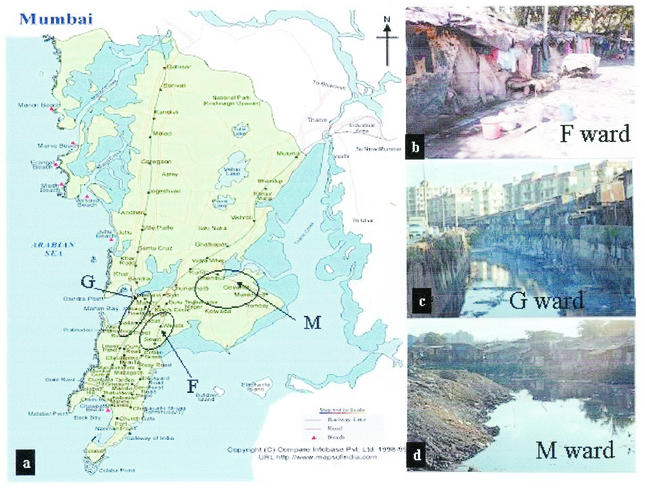
Three wards, F, G, and M, with large urban slums were selected on the basis of past epidemiological data on poliomyelitis. These slum areas with poor sanitation are densely populated, are inhabited by people of low socioeconomic status, and serve as settlements for laborers entering Mumbai from other provinces. Samples of untreated sewage were collected from three sites, Wadala sewage pumping station (F ward; population, 400,000), Dharavi slum (G ward; population, 590,000), and Shivajinagar slum (M ward; population, 762,000). F ward has a piped sewage system, and samples were collected from the wet well of the pumping station after large floating objects were removed by screening. G and M ward samples were collected from large trenches (more than 2 to 3 km long) in which domestic wastewater is drained. The water in these trenches finally drains into the sea. Sewage sampling sites and their locations in the city are shown in Fig. 1. Site selection was based on knowledge of the incidence of paralytic poliomyelitis in various municipality wards in Mumbai for several years.
FIG. 1.
Sewage sampling sites chosen for environmental surveillance. (a) Map of the city of Mumbai showing sewage sampling areas. (Copyright © Compare Infobase Pvt. Ltd., New Delhi, India.) (b) F ward roadside hutments. (c and d) G ward Dharavi and M ward Shivajinagar sewage sample collection sites, respectively.
Sampling frequency and method.
Sewage samples were collected once a week from each site. Sample collection from F and M wards began in January 2001. G ward sampling started in May 2001. Samples were collected in the morning after 10:00 a.m. A stainless steel bucket was lowered into the flowing water to collect approximately 2 liters of sewage sample. The sample was transferred to a clean sterile glass bottle at the collection site and transported to the laboratory within 1 h after collection. Adequate personal safety precautions were taken, and all materials used for sample collection were decontaminated by autoclaving. Samples were processed for virus isolation immediately as described in detail below.
Sewage sample processing.
The sewage sample was adjusted to pH 7.2, and 1 liter was centrifuged at 5,000 rpm (4,600 × g) for 30 min at 4°C with a JS-7.5 swinging-bucket rotor in a Beckman J2-21 M/E refrigerated centrifuge. The supernatant was collected and saved. The sediment was resuspended in a small volume of 3% beef extract (pH 7.5) and treated with 20% (vol/vol) chloroform (containing 0.001% Dithizone) to release virus particles bound to solids. After centrifugation, the supernatant was collected and added to the initial supernatant. Chloroform extraction of sewage sediment was repeated once. Sodium chloride (17.5 g per liter) and polyethylene glycol 8000 (PEG; 80 g per liter) were added to the sewage supernatant. The mixture was stirred with a magnetic stirrer for 30 min and then allowed to stand at 4°C for 16 h (overnight) for precipitation of virus. The precipitate was collected after centrifugation as described above and dissolved in 3 to 4 ml of phosphate-buffered saline (PBS) (the supernatant was discarded). The dissolved precipitate was extracted with chloroform (20% [vol/vol]) and centrifuged, and the supernatant aqueous layer containing the virus was collected. The nondissolved precipitate was extracted once with 3% beef extract, and the supernatant was added to the virus suspension. Finally, fetal bovine serum (final concentration, 2%) and antibiotics were added to the concentrated virus suspension (final resuspension volume, ∼5 ml). The concentrated virus suspension was stored at −20°C until used for poliovirus isolation. Normally, cell culture inoculation was done on the same day.
Determining the efficiency of the virus concentration procedure.
Raw sewage samples (1,000 ml) were spiked with 103 50% tissue culture infective doses (TCID50s) per ml of poliovirus type 1 (Sabin, attenuated vaccine strain). The virus was concentrated by PEG precipitation as described above. Virus titers in raw sewage, spiked sewage, and virus concentrates were determined by microtitration with 0.5-log10-dilution intervals and RD and L20B cells. Virus recovery in the concentrates was used to calculate the efficiency of the PEG precipitation method. Several raw sewage samples and virus concentrates were also titrated to determine virus loads in environmental samples.
Virus isolation.
Each virus concentrate was used to inoculate four 25-cm2 culture flasks of freshly confluent monolayers of L20B cells. The inoculum volume used were 0.1, 0.2, 0.3, and 0.5 ml (one flask each). A cell control flask was seeded with 0.3 ml of PBS. After 45 min of incubation to facilitate virus adsorption, the inoculum was removed, the cells were rinsed with 5 ml of PBS, and 7 ml of culture medium (minimal essential medium containing 2% fetal bovine serum) was added to each flask. The cultures were incubated at 36°C for at least 7 days and examined daily under a microscope for virus-induced cytopathic effects.
Simultaneously, 0.1 ml of a 1:10 dilution of virus concentrate was used to inoculate each of 10 RD cell monolayer cultures grown in test tubes. The tubes were incubated at 36°C for up to 7 days and examined under a microscope to record cytopathic effects.
Poliovirus typing and intratypic differentiation.
Virus-containing culture medium harvested from cytopathic effect-positive L20B cell cultures was used for poliovirus identification by the neutralization test with type-specific equine antipoliovirus sera (8). Homotypic mixtures of poliovirus type 1 were resolved by neutralization with high-titer calf anti-poliovirus type 1 (Sabin) serum.
Poliovirus isolates were further differentiated as vaccine-like or wild by using an enzyme-linked immunosorbent assay (ELISA) and RNA probe hybridization. A microwell ELISA (Nunc Immunostrips) was carried out by using sheep antipoliovirus antibodies (capture antibodies) and cross-absorbed strain-specific rabbit anti-Sabin vaccine poliovirus and anti-wild poliovirus antisera (28). For probe hybridization, viral RNA was spotted onto positively charged nylon membranes by using a dot blot manifold. The viral RNA was probed with digoxigenin-labeled RNA probes specific for Sabin vaccine poliovirus and wild virus (6). Hybridization was detected by reacting the probed membranes with anti-digoxigenin-Fab-alkaline phosphatase conjugate and nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate as the chromogen. The reagents for the ELISA and probe hybridization were obtained from H. van der Avoort, National Institute of Public Health and Environment Protection, Bilthoven, The Netherlands, and O. M. Kew, Centers for Disease Control and Prevention, Atlanta, Ga., respectively.
RNA sequence analysis.
The nucleotide sequence of the complete VP1 protein coding region of wild poliovirus isolates was determined by using a fully automated DNA sequencer (ABI Prism model 3100) and the cycle sequencing protocols supplied by the manufacturer (ABI Prism Big-Dye Terminator Cycle Sequencing Ready Reaction kit, version 2.0; part no. 4303237E; Applied Biosystems). An RNA segment encompassing the VP1 region was amplified by reverse transcription-PCR in two parts with primers Y7 (nucleotides [nt] 2399 to 2421) and S1 (nt 2987 to 3006) and primers S2/S2E (nt 2852 to 2871) and Q8 (nt 3485 to 3504). Primers were kindly provided by O. M. Kew. Primer S2E (5′-TTCACCTACTCCAGGTTT3-′) was a modified version of primer S2 designed by us. Phylogenetic analysis was done by using the computer program Tree-Puzzle, version 4.0.2 (25).
Nucleotide sequence accession numbers.
The sequences determined here were submitted to GenBank under accession numbers AY189834 to AY189894.
RESULTS
Efficiency of the concentration procedure.
The efficiency of the PEG precipitation method in concentrating enteroviruses from sewage was studied by using poliovirus-spiked sewage samples. One liter of raw sewage spiked with 103 TCID50s of poliovirus type 1 (Sabin vaccine strain) per ml was processed. Virus recovery was calculated from the total virus added to the sample and the total virus obtained in the resuspended PEG precipitate. At least 105.9 TCID50s (79%) of the spiked virus was recovered in the concentrates.
The exact virus concentration in raw sewage samples could not be determined by using a microtitration technique with a lowest dilution of 1:10 and RD cells (enteroviruses) or L20B cells (polioviruses) due to low virus titers. The enterovirus concentration in sewage samples was extrapolated from the virus recovered after PEG precipitation. Totals of 24, 10, and 24 concentrates from F, G, and M wards were titrated. The enterovirus concentration varied from 103.35 to 104.26 TCID50s per liter in the sewage specimens collected from these wards over the entire 1-year period. No substantial difference in enterovirus loads at the three sites was found. The results not only confirmed the efficiency of the virus concentration procedure but also provided some insights of enterovirus loads in environmental samples. However, these virus concentrations should be considered low estimates.
L20B cells were used to determine poliovirus titers in the virus concentrates because of their selective susceptibility to poliovirus infection. The poliovirus concentration was estimated to be less than 100 TCID50s per ml in the concentrates. This titer is below the levels that can be determined accurately by the microtitration method. Plaque assays were not performed. Although enterovirus concentrations in sewage were nearly constant at different sites, poliovirus (especially wild virus) loads could vary considerably from site to site.
Virus isolation.
During the period from January to December 2001, a total of 137 sewage samples (51, 51, and 35 from F, M, and G wards, respectively) were studied for poliovirus detection by the cell culture method. Sample collection from G ward was initiated in May 2001; hence, the number of samples for this site was smaller than those for the other two sites.
All virus concentrates were used to inoculate RD cells for quality control of the virus concentration procedure. Any sample not producing cytopathic effects in RD cells would have indicated deficiency in the procedure, since all sewage samples are expected to contain nonpoliovirus enteroviruses. All samples used to inoculate RD cells produced cytopathic effects in all tubes.
Four different inoculum volumes were used for poliovirus isolation from the concentrated samples to reduce the risk of cytotoxicity. None of the concentrated samples was found to be cytotoxic to L20B and RD cell cultures, even at an inoculum of 0.5 ml.
A total of 137 sewage samples were studied. As shown in Table 1, the 137 samples (100%) caused cytopathic effects in L20B cells, indicating the presence of poliovirus. Four different volumes of each concentrated sewage sample were used to inoculate L20B cell culture flasks. Samples positive for poliovirus cytopathic effects at small inocula were also positive at all larger inocula. No discrepancy, such as cytopathic effects with a smaller inoculum but no cytopathic effects with a larger inoculum, was observed for any specimen.
TABLE 1.
Poliovirus isolation from sewage sample concentrates at different inoculuma volumes
| Sampling site (ward) | No. of samples tested | No. of poliovirus-positive samples
|
||||
|---|---|---|---|---|---|---|
| Total | At an inoculum (ml) of:
|
|||||
| 0.1 | 0.2 | 0.3 | 0.5 | |||
| F | 51 | 51 | 29 | 12 | 6 | 4 |
| M | 51 | 51 | 38 | 8 | 4 | 1 |
| G | 35 | 35 | 23 | 6 | 3 | 3 |
| All | 137 | 137 | 90 | 26 | 13 | 8 |
Samples positive at small inoculum volumes were positive at all larger inoculum volumes.
Eight of the 137 virus concentrates (5.8%) required an inoculum of 0.5 ml to cause poliovirus cytopathic effects, i.e., poliovirus detection. A total of 13 (9.5%), 26 (19%), and 90 (65.7%) of the virus concentrates showed the presence of polioviruses with inocula of 0.3, 0.2, and 0.1 ml, respectively. These observations suggest differences in initial poliovirus concentrations in samples collected from the same sites at different times. From these data, the minimum and maximum concentrations of polioviruses in raw sewage were estimated to be 10 to 500 TCID50s per liter.
Of the eight samples that required a large inoculum (0.5 ml) to show the presence of poliovirus, two were collected during the month of May (summer high temperature) and five were collected during July and August (peak monsoon) from the open trenches. These results may reflect virus inactivation during summer and virus dilution during peak monsoon.
Table 2 shows that all three poliovirus serotypes were isolated from sewage samples at each of the three sites. A single serotype and bivalent and trivalent mixtures of poliovirus serotypes were isolated from 61 (44.5%), 41 (30%), and 35 (25.5%) of the samples, respectively. A temporal association between the frequency of detection of trivalent mixtures of poliovirus serotypes in sewage and OPV campaigns in the form of National Immunization Days was observed.
TABLE 2.
Polioviruses isolated from sewage samples with L20B cells
| Sampling site (ward) | No. of samples
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Tested | With the following poliovirus serotype(s)
|
|||||||
| Single type
|
Bivalent mixture
|
Trivalent mixture
|
||||||
| 1 | 2 | 3 | 1 + 2 | 1 + 3 | 2 + 3 | 1 + 2 + 3 | ||
| F | 51 | 14 | 7 | 8 | 2 | 4 | 6 | 10 |
| M | 51 | 2 | 6 | 6 | 6 | 2 | 10 | 19 |
| G | 35 | 0 | 11 | 7 | 2 | 2 | 7 | 6 |
| All | 137 | 16 | 24 | 21 | 10 | 8 | 23 | 35 |
Vaccine versus wild poliovirus.
Sabin vaccine polioviruses were isolated at all three sampling sites. As shown in Table 3, Sabin strains of poliovirus types 1, 2, and 3 were isolated from 37, 92, and 86 sewage samples, respectively. Wild poliovirus type 1 was isolated from a total of 35 sewage samples (16, 17, and 2 from F, M, and G wards, respectively). Homotypic mixtures (wild and vaccine strains) of poliovirus type 1 were recovered from three samples. No wild poliovirus type 2 was isolated. Wild poliovirus type 3 was detected in one sample collected from G ward in May 2001. The enterovirus concentration in G ward samples was comparable to those in samples from the other two wards. Moreover, the only wild poliovirus type 3 was recovered from a G ward sample. Therefore, technical deficits were not the reason for the fewer wild poliovirus-positive samples from G ward.
TABLE 3.
Wild and vaccine-like polioviruses isolated from sewage samples obtained in Mumbai in 2001
| Sampling site (ward) | No. of samples with the following poliovirus typea
|
|||||
|---|---|---|---|---|---|---|
| PV1
|
PV2
|
PV3
|
||||
| Wild | SL | Wild | SL | Wild | SL | |
| F | 16 | 14 | 0 | 25 | 0 | 28 |
| M | 17 | 15 | 0 | 41 | 0 | 37 |
| G | 2 | 8 | 0 | 26 | 1 | 21 |
| All | 35 | 37 | 0 | 92 | 1 | 86 |
SL, Sabin vaccine-like. Vaccine-like and wild poliovirus type 1 mixtures were recovered from three sewage samples collected from M ward.
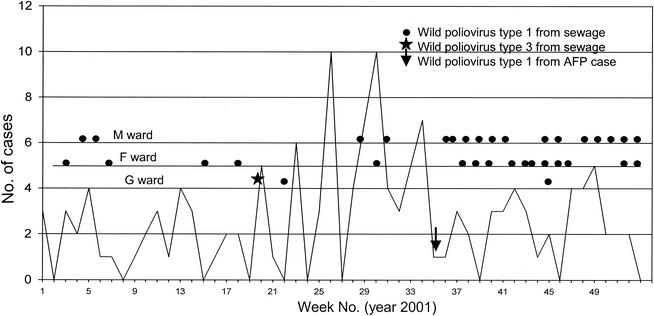
Figure 2 shows AFP cases reported by week in Mumbai through the AFP surveillance system, the single wild poliovirus-positive AFP case reported in week 36, and wild polioviruses isolated from the sewage samples obtained weekly from the three sites. A total of 90 AFP cases were reported by AFP surveillance in 2001. Stool specimens collected from 84 cases (93%) were virologically investigated for poliovirus detection. AFP surveillance identified one case of paralytic poliomyelitis due to wild poliovirus infection in Mumbai in 2001. Environmental surveillance isolated wild poliovirus of seven different clusters from 36 samples over the 12-month period. During the first 25 weeks (January to June), the environmental surveillance system detected wild poliovirus in four sewage samples from F ward and two samples each from G and M wards. From week 36 on, wild poliovirus was isolated from sewage in at least one of the three wards every week. Thus, environmental surveillance also detected an increased burden of wild poliovirus in Mumbai during and after the monsoon months.
FIG. 2.
AFP surveillance and environmental surveillance in Mumbai. The graph shows the weekly distribution of AFP cases reported in Mumbai in 2001.
Genomic analysis of wild poliovirus isolates.
In the years 2000 and 2001, wild poliovirus type 1 was isolated from 138 and 209 cases of AFP in India, respectively. The majority of the paralytic poliomyelitis cases were reported in Uttar Pradesh and Bihar. In Mumbai, wild poliovirus type 1 was isolated from two cases in the year 2000 and one case in the year 2001. The virus isolates were genetically distinct from those isolated in Mumbai in previous years (data not shown).
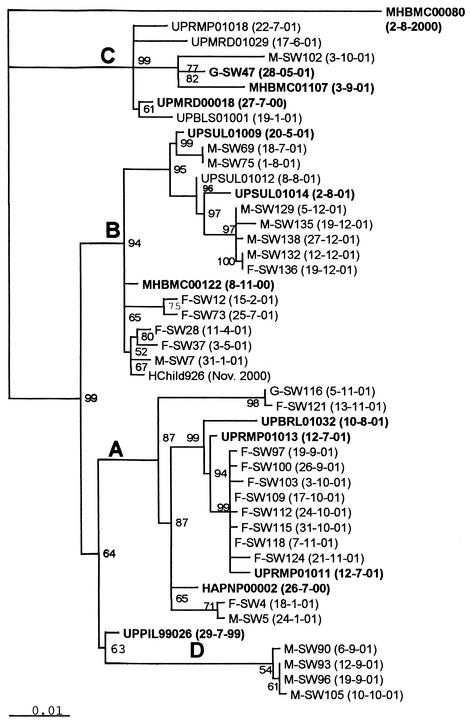
Complete VP1 nucleotide sequences of 30 of the 35 wild poliovirus type 1 isolates from sewage samples and those of isolates from paralytic poliomyelitis cases in India were compared to determine the sources of these viruses. Only isolates from AFP cases that had close sequence similarity with isolates from sewage samples are reported in this study. As shown in Fig. 3, four different genetic lineages of wild poliovirus type 1 were detected in Mumbai in 2001 by environmental sewage sampling.
FIG. 3.
Dendrogram showing the genetic relationship of wild poliovirus type 1 isolates from sewage samples collected from slums in Mumbai and closely related isolates from AFP cases. Wild poliovirus isolates from sewage samples are coded as follows: single letter identifying the sampling site (F, G, or M), serial number of the sewage sample, and date (day-month-year) of sample collection. Wild poliovirus isolates from AFP cases are coded according to the unique epidemiological case identification number used in the AFP surveillance system; the number includes state and district codes, year and serial number of the case, and date of isolation. The neighbor-joining tree was constructed after 1,000 puzzling steps with the Tree-Puzzle program. The scale indicates the fractional genetic distance (0.01 = 1%), and the numbers at the nodes indicate the frequency (as a percentage) at which the particular tree topology was found in the 1,000 puzzling steps. A to D indicate lineages.
None of the sewage isolates was genetically similar to the wild poliovirus isolated from an AFP case (MHBMC00080) reported in M ward in August 2000. The second wild poliovirus-positive case (MHBMC00122) was reported in G ward in November 2000. Genetically closely related viruses (99.45% sequence similarity) were isolated from stools of four healthy children in F ward (Hchild926) in a stool survey carried out in the same month. Viruses of the same lineage (lineage B) were isolated from sewage samples in M ward in January 2001 (M-SW7) and F ward in February, April, May, and July 2001. Circulation of the wild poliovirus was thus found for 5 to 6 months. Viruses of the same genetic lineage (lineage B) but of a different cluster in eastern Uttar Pradesh were recovered from a total of six sewage samples in M ward and one sample in F ward in July-August and December 2001. It can be seen from the dendrogram that these sewage isolates were more closely related to viruses isolated from AFP cases (UPSUL01009 and UPSUL01014) in Sultanpur district, Uttar Pradesh. The high support values (94%) for the tree topology indicate that wild poliovirus type 1 isolated during July-August and December from M ward represents a separate importation from Uttar Pradesh and not the continued undetected transmission of MHBMC00122 and related isolates. Wild poliovirus type 1 isolates of a different genetic lineage (lineage A) were recovered from both F and M wards in January 2001 (F-SW4 and M-SW5). Genetically most similar to these viruses was a virus isolated from a case of AFP in Haryana state (HAPNP00002). Viruses belonging to this lineage were frequently isolated from sewage during September-October 2001; however, their origin was found to be western Uttar Pradesh (UPRMP01011, UPRMP01013, and UPBRL01032). The sequence data thus documented two importations of wild poliovirus type 1 of lineage A from north India to Mumbai.
The third wild poliovirus lineage (lineage C) detected in May 2001 in G ward (G-SW47) was also related to a western Uttar Pradesh isolate (UPMRD00018). Wild poliovirus type 1 lineage C was predominant in western Uttar Pradesh during 2001. One paralytic case due to wild poliovirus type 1 of this lineage was reported in Mumbai in September 2001 (MHBMC01107). It was inferred that environmental surveillance detected silent circulation of the virus at least 3 months before a case of paralytic poliomyelitis was reported through AFP surveillance. A virus of this lineage was isolated from sewage in October 2001(M-SW102), indicating continued local circulation for about 6 months.
A fourth lineage (lineage D) detected in sewage samples in M ward in September-October 2001 had its origin in Uttar Pradesh (UPPIL99026). We did not find a virus that was genetically closer to the lineage D sewage viruses among the year 2001 isolates in Uttar Pradesh. The reservoir for the lineage between the years1999 and 2001 remains unknown, suggesting a surveillance gap. The lineage was eliminated from circulation within a very short time.
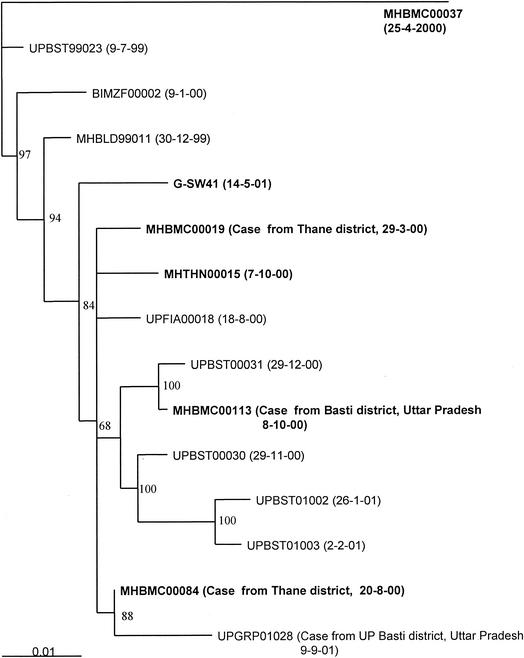
In the year 2000, four case of poliomyelitis due to wild poliovirus type 3 were reported in Mumbai. Of these, only one case occurred in a resident of the city and the remaining three cases were brought to Mumbai after onset for medical treatment. Figure 4 shows the genetic relationship of wild poliovirus type 3 isolated from poliomyelitis cases and sewage. MHBMC00037, isolated from the resident case (H ward), was genetically distinct from viruses isolated from cases known to have acquired infection outside Mumbai. Wild poliovirus type 3 was isolated from G ward sewage (G-SW41) in May 2001. The virus lineage was also detected in the year 2000 among poliovirus cases isolates from Thane district, close to Mumbai. This lineage was circulating in Basti district in Uttar Pradesh in the years 2000 and 2001. Epidemiological linkage of cases in Thane and Basti districts had been established. This virus was not isolated again, indicating its elimination from circulation. However, this lineage continued to circulate in Uttar Pradesh, as evident from viruses isolated from poliomyelitis cases in Basti district in Uttar Pradesh in the year 2001.
FIG. 4.
Dendrogram showing the genetic relationship of wild poliovirus type 3 isolates from one sewage sample and AFP cases. Isolates are coded as described in the legend to Fig. 3. The neighbor-joining tree was constructed after 1,000 puzzling steps with the Tree-Puzzle program. The scale indicates the fractional genetic distance (0.01 = 1%), and the numbers at the nodes indicate the frequency (as a percentage) at which the particular tree topology was found in the 1,000 puzzling steps.
Environmental surveillance detected wild poliovirus type 1 introduction from areas of high endemicity in Uttar Pradesh to Mumbai at least seven times and wild poliovirus type 3 once during the year 2001. It can be seen from Fig. 3 and 4 that wild virus transmission from Uttar Pradesh to Mumbai occurred within days after the occurrence of a paralytic case in Uttar Pradesh. Each of the introduced wild virus lineages was eliminated from circulation within 2 to 3 months. Recent cases of paralytic poliomyelitis in Mumbai were due to wild poliovirus from Uttar Pradesh. The introduced viruses were detected by environmental surveillance before a paralytic case occurred.
DISCUSSION
We initiated environmental surveillance for wild poliovirus in Mumbai because, despite high routine OPV immunization coverage and supplementary immunization activities, a very small number of paralytic poliomyelitis cases continue to be reported in the city. Molecular sequence analysis of wild viruses isolated from AFP cases in Mumbai since 1996 showed that indigenously circulating virus lineages had been eliminated by 1999 (unpublished data). Wild poliovirus-positive persons from other districts provided medical treatment in Mumbai hospitals and population influx from wild poliovirus reservoir communities pose major unavoidable risks to the polio eradication program in Mumbai (12). Viruses thus introduced into the city may initiate and establish wild poliovirus transmission in this city of 13 million inhabitants, about 50% of whom live in urban slums.
In many developing countries, well-organized sewage systems may not be present for carrying out environmental surveillance. We therefore selected one sewage pumping station and two large trenches draining domestic wastewater as sampling sites. Our studies showed that continuous monitoring of sewage samples for wild poliovirus is feasible under varied conditions of wastewater disposal and may be established for long-term applications.
Human cell surface molecule CD155, a member of the immunoglobulin superfamily, is used as the cell receptor by polioviruses but not by other enteroviruses (16). A genetically transformed murine cell line, L20B, expressing the human poliovirus receptor on the surface, is selectively susceptible to poliovirus infection while remaining refractory to infection by most other enteroviruses (19). With this advantage in mind, we used the L20B cell line for isolating polioviruses from sewage samples to eliminate the difficulties of separating polioviruses from other enteroviruses present in sewage, which were experienced by earlier investigators (14). Our data showed that L20B cells could be used for the isolation of poliovirus in the presence of more than a 1,000-fold excess of nonpoliovirus enteroviruses. Vaccine strains (Sabin attenuated poliovirus) were readily recovered from sewage, as observed by others, because OPV is used for child immunization in India (20). Moreover, wild polioviruses could be detected in mixtures of vaccine and wild viruses without serious technical problems.
As mentioned earlier, wild poliovirus type 1 isolated from the two AFP cases in the year 2000 were not indigenously circulating. Both of the affected persons had close interactions with people from Uttar Pradesh. One child (MHBMC00080) had a travel history. The other had come from Uttar Pradesh to Mumbai very recently and had not received the recommended doses of OPV. Nucleotide sequence analysis of the virus isolates provided further confirmatory evidence that the virus isolates were from two different lineages in north India. Our objective was to determine whether the wild poliovirus thus introduced established focal transmission.
In this study, evidence is presented to show that one of the two wild poliovirus lineages established transmission (MHBMC00122) for a short period but did not cause secondary poliomyelitis cases. We isolated wild poliovirus type 1 from 35 samples and poliovirus type 3 from 1 sample during the year 2001. Poliovirus transmission is facilitated during late summer and monsoon months (June to September), as observed from the seasonal variations of poliomyelitis cases in India (24). Wild poliovirus isolation from sewage increased in frequency after poliomyelitis cases reached a seasonal peak in north India, indicating a rapid exchange of circulating wild polioviruses between certain regions.
Four different poliovirus type 1 lineages, A, B, C, and D, were identified in sewage samples. Viruses grouped in lineages A, B, and C were isolated from AFP cases in Uttar Pradesh in the years 2000 and 2001. Lineage C was predominant during the year 2001. The poliovirus genome evolves at a rate of 1 to 2% nucleotide substitutions per year (17). The high mutation rate has been used to determine the transmission pattern for wild poliovirus. At least seven separate instances of wild poliovirus type 1 importations into Mumbai were identified by sequence analysis. In the phylogenetic analysis, branches showing a quartet puzzling reliability of 90 to 100% can be considered very strongly supported. Within lineage B, the virus of case MHBMC00122 circulated mainly in F ward from November 2000 to July 2001. Lineage B was reintroduced from Sultanpur (UPSUL01009) in July 2001 (M-SW69) and again from the same district (UPSUL01014) in December 2001 (M-SW129). Similarly, lineage A was introduced repeatedly in January (F-SW4 and M-SW5), September, and November 2001. It is noteworthy that the wild poliovirus lineages were eliminated from circulation within 2 to 3 months. The absence of divergent isolates of the same lineages also suggested limited circulation of the wild polioviruses and rapid elimination. For two virus introductions, however, transmission was detected for 5 to 7 months (MHBMC00122 and G-SW47). Importantly, environmental surveillance detected wild poliovirus type 1 3 months before AFP surveillance did (G-SW47 versus MHBMC01107). The supplementary surveillance system is a very useful tool for planning poliovirus eradication activities, especially during the last phase of the program.
Environmental surveillance is resource and labor intensive, but careful selection of sampling sites based on previous epidemiological data can logically reduce their numbers, as was accomplished in the present study. A single 1-liter grab sample was found to be sufficient for the detection of wild poliovirus, when present, but this quantity can be reduced further, according to recent reports (9). In our studies, wild viruses of the same genetic lineage were isolated from consecutive weekly sewage samples. It is therefore possible to adjust sampling frequency without compromising the sensitivity of poliovirus detection.
In environmental surveillance, a negative (wild poliovirus) finding may not be meaningful unless long-term follow-up results become available, but a positive result could prompt programmatic action. Guidelines are needed for incorporating wild poliovirus isolation data accrued through supplementary surveillance methods and programmatic action required in countries conducting routine OPV immunization and annual National Immunization Day campaigns.
Wild polioviruses detected during this study were promptly reported to the local health authorities and the poliovirus surveillance program. Wild polioviruses were isolated from sewage even after immunization rounds. High-quality “mop-up” immunization is a prerequisite for stopping wild poliovirus transmission. Moreover, the different lineages of wild polioviruses isolated also suggested a high population mobility between Uttar Pradesh and Mumbai bringing in different viruses each time. It is not unexpected that similar exchanges of wild polioviruses occur in other parts of the country. In addition to being effectively used to detect wild poliovirus transmission, environmental surveillance may become a powerful tool in the early detection of circulating vaccine-derived poliovirus strains (32).
Our results highlight the vulnerability of the poliovirus eradication program in India until wild poliovirus is no longer endemic in any part of the country. The findings from this study may lead to a better definition of the role of environmental sewage sampling in the evaluation of the status of poliomyelitis eradication activities and in the process of certification.
Acknowledgments
We are grateful to Walter Dowdle and Ray Sanders for critical evaluation of the project and constructive suggestions. V. B. Mandke provided AFP surveillance data and isolates from cases in Mumbai, and T. N. Dhole provided isolates from cases in Uttar Pradesh; we gratefully acknowledge their contributions. Thanks are due to R. L. More and N. A. Raikar for intratypic differentiation tests and D. S. Jagtap for sewage sample collection.
Financial support received from the World Health Organization, Geneva, Switzerland (TSA I8/181/307 and 395), is gratefully acknowledged.
REFERENCES
- 1.Adams, T. 2000. Farewell to polio in the Western Pacific. Bull. W. H. O. 78:1375. [PMC free article] [PubMed]
- 2.Albertsson, P. 1967. Two phase separation of viruses, p. 303-321. In K. Maramorosch and H. Koprowski (ed.), Methods in Virology, vol. 2. Academic Press, New York, N.Y.
- 3.Anonymous. 2002. Certification of poliomyelitis eradication, European region, June 2002. Wkly. Epidemiol. Rep. 77:221-228. [Google Scholar]
- 4.Anonymous. 2002. Progress towards the global eradication of poliomyelitis, 2001. Wkly. Epidemiol. Rep. 77:98-107. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2001. Progress towards poliomyelitis eradication—South-East Asia, January 2000-June 2001. Morb. Mortal. Wkly. Rep. 50:738-751. [PubMed] [Google Scholar]
- 6.DeL., B. Nottay, L., C. F. Yang, B. P. Holloway, M. Pallansch, and O. M. Kew. 1995. Identification of vaccine-related polioviruses by hybridization with specific RNA probes. J. Clin. Microbiol. 33:562-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshpande, J. M., V. K. Rao, M. V. Karnataki, S. S. Nadkarni, V. K. Saxena, R. R. Karambelkar, S. G. Ramdasi, K. S. Patil, and J. J. Rodrigues. 1996. Absence of wild poliovirus circulation among healthy children in a rural area with high oral poliovirus vaccination coverage. Indian J. Med. Res. 103:289-293. [PubMed] [Google Scholar]
- 8.Expanded Programme on Immunization. 1997. Manual for virological investigation of polio. WHO/EPI/GEN/97.01. World Health Organization, Geneva, Switzerland.
- 9.Hovi, T., M. Stenvik, H. Partanen, and A. Kangas. 2001. Poliovirus surveillance by examining sewage specimens. Quantitative recovery of virus after introduction into sewerage at remote upstream location. Epidemiol. Infect. 127:101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hull, B. P., and W. R. Dowdle. 1997. Poliovirus surveillance: building the global polio laboratory network. J. Infect. Dis. 175(Suppl. 1):S113-S116. [DOI] [PubMed]
- 11.Hull, H. F., N. A. Ward, B. P. Hull, J. B. Milstein, and C. de Quadros. 1994. Paralytic poliomyelitis: seasoned strategies, disappearing disease. Lancet 343:1331-1333. [DOI] [PubMed] [Google Scholar]
- 12.Kim-Farley, R. J., K. H. Dave, J. Sokhey, and V. B. Mandke. 1989. Poliomyelitis surveillance and vaccine efficacy in Bombay. Bull. W. H. O. 67:663-667. [PMC free article] [PubMed] [Google Scholar]
- 13.Manor, Y., R. Handsher, T. Halmut, M. Neuman, A. Bobrov, H. Rudich, A. Vansover, L. Shulman, O. Kew, and E. Mendelson. 1999. Detection of poliovirus circulation by environmental surveillance in the absence of clinical cases in Israel and the Palestinian authority. J. Clin. Microbiol. 37:1670-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manor, Y., R. Handsher, T. Halmut, M. Neuman, B. Abramovitz, A. Mates, and E. Mendelson. 1999. A double-selective tissue culture system for isolation of wild-type poliovirus from sewage applied in a long-term environmental surveillance. Appl. Environ. Microbiol. 65:94-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melnick, J. L. 1985. Enteroviruses: polioviruses, coxsackieviruses, echoviruses and newer enteroviruses, p. 739-794. In B. N. Fields, D. M. Knipe, R. M. Chanock, J. L. Melnick, B. Roizman, and R. E. Shope (ed.), Virology. Raven Press, New York, N.Y.
- 16.Mendelsohn, C. L., E. Wimmer, and V. R. Racaniello. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence and expression of a new member of the immunoglobulin superfamily. Cell 56:855-865. [DOI] [PubMed] [Google Scholar]
- 17.Nottay, B. K., O. M. Kew, M. H. Hatch, J. T. Heyward, and J. F. Obijeski. 1981. Molecular variation of type 1 vaccine-related and wild polioviruses during replication in humans. Virology 108:405-423. [DOI] [PubMed] [Google Scholar]
- 18.Pan American Health Organization. 1994. Final report of the 3rd Meeting of the International Commission for the Certification of Eradication of Poliomyelitis in the Americas. Pan American Health Organization, Washington, D.C.
- 19.Pipkin, P. A., D. J. Wood, V. R. Racaniello, and P. D. Minor. 1993. Characterization of L cells expressing the human poliovirus receptor for the specific detection of poliovirus. J. Virol. Methods 41:333-340. [DOI] [PubMed] [Google Scholar]
- 20.Pöyry, T., M. Stenvik, and T. Hovi. 1988. Viruses in sewage waters during and after a poliomyelitis outbreak and subsequent nationwide oral poliovirus vaccination campaign in Finland. Appl. Environ. Microbiol. 30:212-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rico-Hesse, R., M. A. Pallansch, B. K. Nottay, and O. M. Kew. 1987. Geographic distribution of wild poliovirus type 1 genotypes. Virology 160:311-322. [DOI] [PubMed] [Google Scholar]
- 22.Shieh, Y.-S., D. Wait, L. Tai, and M. D. Sobsey. 1995. Methods to remove inhibitors and other fecal wastes for enterovirus detection by polymerase chain reaction. J. Virol. Methods 54:51-66. [DOI] [PubMed] [Google Scholar]
- 23.Shulman, L. M., R. Handsher, C. F. Yang, S. J. Yang, J. Manor, A. Vansover, Z. Grossman, M. Pallansch, E. Mendelson, and O. M. Kew. 2000. Resolution of the pathways of poliovirus type 1 transmission during an outbreak. J. Clin. Microbiol. 38:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sokhey, J. 1992. Poliomyelitis surveillance in India. Indian Pediatr. 29:677-682. [PubMed] [Google Scholar]
- 25.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 26.Tambini, G., J. K. Andrus, E. Marques, J. Boshell, M. Pallansch, C. A. de Quadros, and O. Kew. 1993. Direct detection of wild poliovirus circulation by stool survey of healthy children and analysis of community wastewater. J. Infect. Dis. 168:1510-1514. [DOI] [PubMed] [Google Scholar]
- 27.van der Avoort, H. G., J. H. Reimerink, A. Ras, M. N. Mulders, and A. M. van Loon. 1995. Isolation of epidemic poliovirus from sewage during the 1992-3 type 3 outbreak in The Netherlands. Epidemiol. Infect. 114:481-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Wezel, A. L., and A. G. Hazendonk. 1979. Intratypic serodifferentiation of poliomyelitis virus strains by strain-specific antisera. Intervirology 11:2-8. [DOI] [PubMed] [Google Scholar]
- 29.Vonsover, A., R. Handsher, M. Neuman, S. Guillot, J. Balanant, H. Rudich, E. Mendelson, T. Swartz, and R. Crainic. 1993. Molecular epidemiology of polioviruses isolated in Israel and defined by restriction fragment length polymorphism assay. J. Infect. Dis. 176:199-203. [DOI] [PubMed] [Google Scholar]
- 30.Wood, D. J., and B. P. Hull. 1998. L20B cells simplify culture of polioviruses from clinical samples. J. Med. Virol. 58:188-192. [PubMed] [Google Scholar]
- 31.World Health Assembly. 1988. Global eradication of poliomyelitis by the year 2000. Resolution WHA 41.28. World Health Organization, Geneva, Switzerland.
- 31a.World Health Organization. 1997. Global eradication of poliomyelitis. Report of the technical consultation, 29-30 April 1996. World Health Organization, Geneva, Switzerland.
- 32.Yoshida, H., H. Horie, K. Matsuura, and T. Miyamura. 2000. Characterization of vaccine derived polioviruses isolated from sewage and river water in Japan. Lancet 356:1461-1463. [DOI] [PubMed] [Google Scholar]