Abstract
Three-dimensional structures are now known for roughly half of all protein families. It is thus quite likely, in searching sequence databases, that one will encounter a homolog with known structure and be able to use this information to infer structure–function properties. The goal of Entrez’s 3D structure database is to make this information accessible and useful to molecular biologists. To this end, Entrez’s search engine provides three powerful features: (i) Links between databases; one may search by term matching in Medline®, for example, and link to 3D structures reported in these articles. (ii) Sequence and structure neighbors; one may select all sequences similar to one of interest, for example, and link to any known 3D structures. (iii) Sequence and structure visualization; identifying a homolog with known structure, one may view a combined molecular-graphic and alignment display, to infer approximate 3D structure. Entrez’s MMDB (Molecular Modeling DataBase) may be accessed at: http://www.ncbi.nlm.nih.gov/Entrez/structure.html
MMDB CONTENTS
Data sources
Experimental structure data for Entrez (1) are retrieved from the RCSB Protein Data Bank (PDB) (2). Agreement of 3D coordinate and chemical-sequence data is checked and, if necessary, sequence data are automatically modified to achieve exact agreement with coordinates. This validation allows Entrez to support communication between sequence and structure displays. Author-annotated features are recorded and mapped from PDB to MMDB, and uniformly defined secondary structure and domain features are added, to support structure neighbor calculations (3). MMDB currently contains about 10 000 structure records, corresponding to about 20 000 chains and 35 000 domains.
Links
Sequences derived from MMDB entries are merged into Entrez’s protein and nucleic acid sequence databases, preserving links to the corresponding 3D structure. Links to the Medline® scientific literature database are generated by processing citation data in MMDB. These links allow Entrez to provide instant access to publications describing the original structure determination, including links to publisher sites with full text. Links to NCBI’s taxonomy database are generated by semi-automatic processing of ‘source’ text provided by PDB. Taxonomy is assigned at the level of individual chains (and sequences), with new organisms added to the Taxon database as needed. Taxonomy links support queries based on phylogenetic relationships.
Neighbors
Neighbors of MMDB-derived sequences are identified automatically using the BLAST algorithm (4). Sequence–neighbor relationships are reciprocal, and MMDB-derived sequences thus also appear as neighbors of other sequences in Entrez. BLAST detects highly significant sequence similarities that are indicative of homology. Structure neighbors are identified using the VAST algorithm, a structure–structure alignment method (5). While VAST uses a conservative significance threshold, the structure similarities it detects very often represent remote relationships not detectable by sequence comparison. Structure similarities may also represent evolutionary convergence, particularly when they involve repetitive structural elements such as β-α units. Entrez supports molecular- graphics visualization of all structure neighbor relationships, so users may examine and interpret structural similarities for themselves. Links and neighbors in Entrez are summarized in Figure 1.
Figure 1.
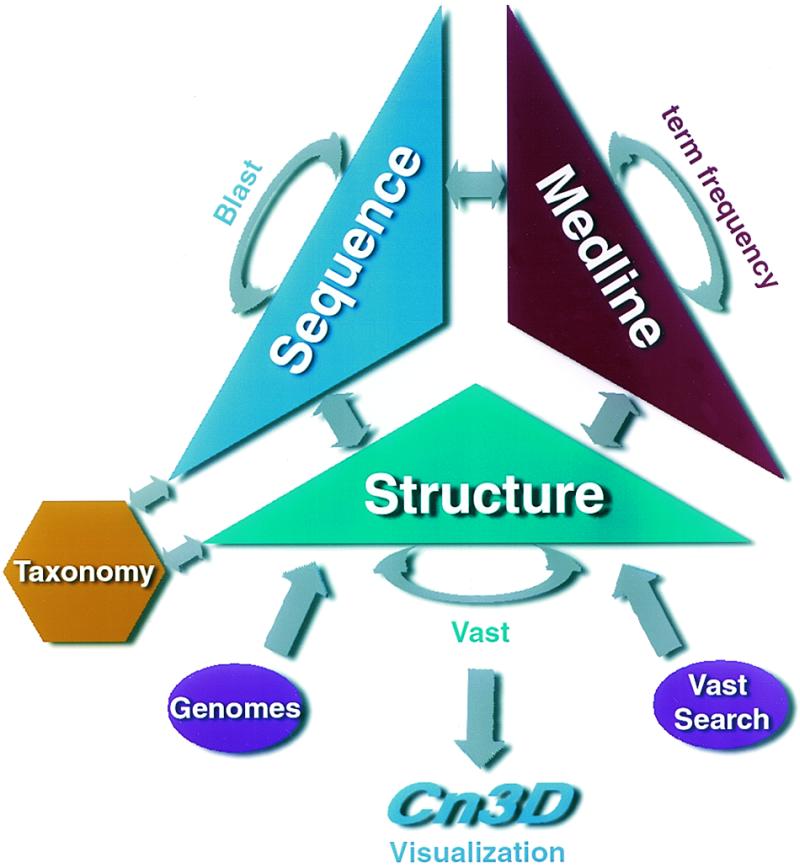
Connections between MMDB and other databases, including the neighboring mechanisms used to relate entries within the same database.
USING MMDB
Simple queries
MMDB is an integrated part of Entrez and can be accessed by querying Entrez’s ‘structure’ database for particular terms or keywords. This allows one to identify structures based on protein names, for example, or author names, publication dates or species names. A simple boolean query will produce a list of MMDB entries. One may browse this list, following links to other databases, for example those to Medline® abstracts.
Advanced queries
The latest version of Entrez supports a powerful query refinement feature, allowing one to logically combine the results of simple queries involving term-match hits, links or neighbors. Suppose, for example, one wishes to select from among the structure neighbors of a protein those that are also sequence neighbors. One query may select structure neighbors of the protein. Another may in turn select the protein’s sequence link, its sequence neighbors and those neighbors’ structure links. Since Entrez stores query results in its ‘history’, one may now combine the simple queries by asking for ‘<q1> AND <q2>’, where <q1> and <q2> represent selections from Entrez’s ‘history’.
3D visualization
Selecting the ‘Structure Summary’ for an MMDB entry, one finds a detailed choice of links and visualization options. The default ‘View’ will launch NCBI’s Cn3D molecular graphics viewer. One may also choose links to ‘Structure Neighbors’ of individual chains and/or domains. This leads to a detailed listing of VAST neighbor results, with ‘View’ options to display structure–structure superpositions, and numerical scores describing the extent of structural similarity. Using Cn3D one may view the corresponding sequence alignments in intercommunicating windows. For more information and downloading instructions for Cn3D, see: http://www.ncbi. nlm.nih.gov/Structure/CN3D
Special services
As a service to structural biologists, NCBI supports ‘on the fly’ structure neighbor calculation. Users may submit coordinates for a new structure or domain, and when the search is complete, browse a neighbor list of the same form as available in Entrez. VAST-Search results are accessible only to the user requesting the search. For further information see: http://www.ncbi.nlm.nih.gov/Structure/VAST/vastsearch.html
A large portion of the protein sequences in complete genomes share significant sequence similarities with proteins of known 3D structure. As a service to genome annotators, we have linked these protein sequences to BLAST sequence neighbors in MMDB. The new service lists sequence neighbors with links to structure, and allows users to view the corresponding sequence–structure alignments. See the ‘PDB neighbors’ for genomes listed in: http://www.ncbi.nlm.nih.gov/Entrez/Genome/ org.html
A BIOLOGICAL EXAMPLE
Suppose one wishes to ask whether structure data can help to characterize the mutations responsible for Nail–Patella Syndrome (NPS). One may begin by querying the OMIM database (6), http://www.ncbi.nlm.nih.gov/Omim , for information on NPS. There, one finds links to the relevant proteins in Entrez’s sequence database, for example the SWISS-PROT (7) entry LM12_HUMAN, human homeobox protein LMX-1.2, a transcription factor belonging to the LIM subfamily. Annotations in the sequence entry describe, for example, that mutations at positions 95 (C→F) and 246 (N→K) have been associated with NPS (8). No 3D structure is available for LMX-1.2, but one may query Entrez’s structure database to find out whether a homologous structure is known. One asks Entrez to display the more than 800 ‘protein neighbors’ of LMX-1.2. One may then ask for the ‘structure links’ for all of these neighbors, immediately identifying two LMX-1.2 homologs with known 3D structure. 1FJL, for example, is a homeodomain from the Drosophila paired protein, cocrystallized with a bound DNA oligonucleotide (9). Clicking the ‘View’ button on its structure summary page pops up a combined sequence/structure display. One may then use Cn3D’s sequence import features to align LMX-1.2 to the sequence of 1FJL chain A, as shown in Figure 2. The region including Asn246 of LMX-1.2 is part of the alignment, and one may see that the asparagine at this position is conserved between the two aligned proteins. In fact, examination reveals that in the paired homeodomain–DNA complex, this asparagine residue is involved in specific interactions with an A:T base pair in the transcription factor recognition site. This model thus suggests that mutations at this position may alter or even abolish specific protein–DNA interactions for LMX-1.2, interfering with its cellular and developmental functions. There are also numerous sequence and structure neighbors of 1FJL, and by browsing them, using Cn3D in the same fashion, one may examine conservation of these and other sequence features in this broader family of DNA-binding proteins.
Figure 2.
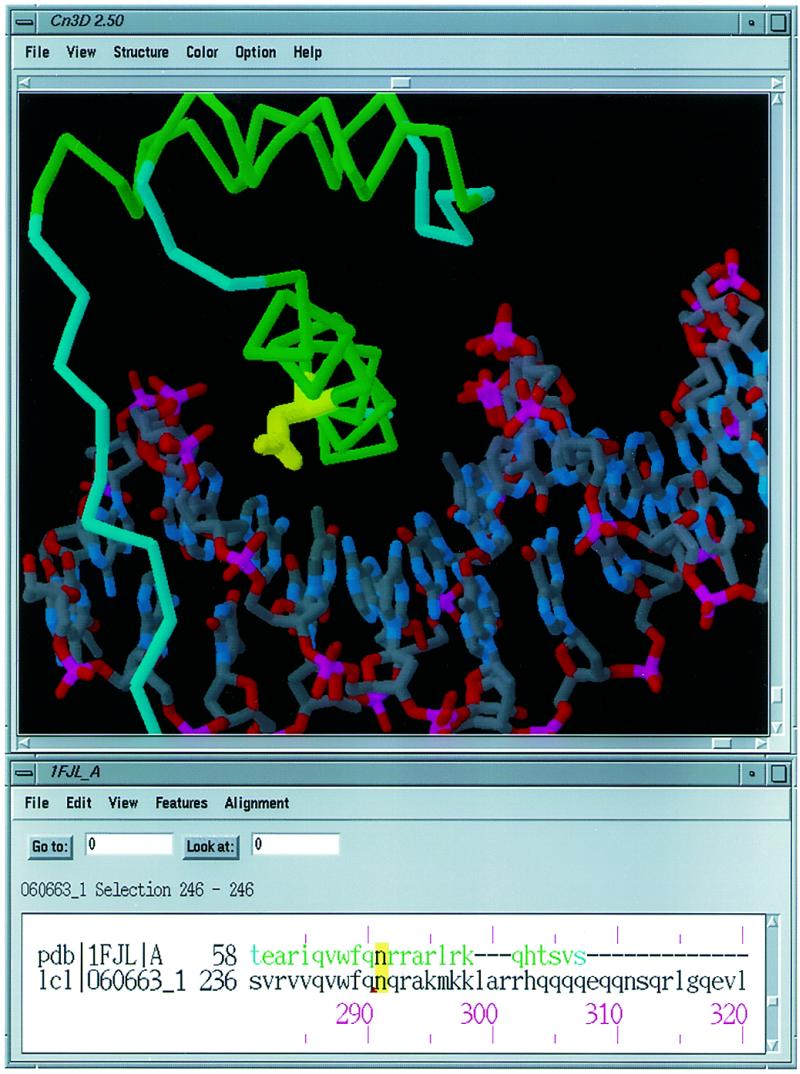
Cn3D image of the Drosophila paired protein and its alignment with human homeobox protein LMX-1.2. The site of the N→K mutation associated with NPS has been highlighted.
UPDATE AND AVAILABILITY
Entrez’s 3D structure database is currently updated once per month. Entrez’s VAST structure neighbor database is also updated monthly, but with a lag of about 2 weeks, due to the computer time necessary for structure–structure comparison and alignment. Researchers who wish to use the MMDB and VAST neighbor data in their own calculations may download them directly via FTP at: ftp://ncbi.nlm.nih.gov/mmdb/ . Source code for the Cn3D viewing program is also available as part of the NCBI toolkit distribution.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Colombe Chappey, Jonathan Kans, Tatiana Tatusova and the other members of the NCBI software team for their help in adding structure visualization to Entrez. We thank the NIH Intramural Research Program for support. Comments, suggestions and questions are welcome and should be addressed to: info@ncbi.nlm.nih.gov.
REFERENCES
- 1.Wheeler L., Chappey,C., Lash,A., Leipe,D.C., Madden,T.L., Schuler,G.D., Tatusova,T.,D. and Rapp,B.A. (2000) Nucleic Acids Res., 28. 10–14 (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) Nucleic Acids Res., 28, 235–242 (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchler-Bauer A., Addess,K.J., Chappey,C., Geer,L., Madej,T., Matsuo,Y., Wang,Y. and Bryant,S.H. (1999) Nucleic Acids Res., 27, 240–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 5.Gibrat J.F., Madej,T. and Bryant,S.H. (1996) Curr. Opin. Struct. Biol., 6, 377–385. [DOI] [PubMed] [Google Scholar]
- 6.McKusick V.A. (1998) Mendelian Inheritance in Man. Catalogs of Human Genes and Genetic Disorders. Johns Hopkins University Press, Baltimore, MD.
- 7.Bairoch A. and Apweiler,R. Nucleic Acids Res. (2000), 28, 45–48 (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dreyer S.D., Zhou,G., Baldini,A., Winterpacht,A., Zabel,B., Cole,W., Johnson,R.L. and Lee,B. (1998) Nature Genet., 19, 47–50. [DOI] [PubMed] [Google Scholar]
- 9.Wilson D.S., Guenther,B., Desplan,C. and Kuriyan,J. (1995) Cell, 82, 709–719. [DOI] [PubMed] [Google Scholar]


