Abstract
The cell cycle requires cells to duplicate their chromatin, DNA, and histones, while retaining a subset of epigenetic marks, in a highly coordinated manner. The WEE1 kinase was identified as an important regulator during S phase, preventing entry into mitosis until DNA replication has been completed. Interestingly, WEE1 has also emerged as a key player in regulating histone synthesis. It phosphorylates histone H2B at tyrosine 37 in the nucleosomes found upstream of the histone gene cluster, which suppresses histone transcription in late S phase. These observations highlight a dual role for WEE1 as both a mitotic gatekeeper and a surveyor of chromatin synthesis, providing a direct link between epigenetics and cell cycle progression. Importantly, this link has implications for the design of novel epigenetic inhibitors targeting cancers that display elevated expression of this kinase.
Keywords: WEE1, Histones, Tyrosine phosphorylation, Epigenetics, Cancer, Cell cycle
Chromatin integrity and histone synthesis
Chromatin integrity - the synthesis and packaging of the nascent DNA with histones - is critical for proper chromosome condensation, segregation, epigenetic inheritance, and genome stability. Eukaryotic cells tightly regulate synthesis of core chromatin components, during each cell cycle. Although processes that interfere with DNA replication compromise genetic integrity, alterations in histone stoichiometry or mutations in histone genes are linked to chromosome loss, altered chromatin architecture, and cancer [1–6]. In addition, both histones and DNA are modified epigenetically, and inheritance of these epigenetic marks is essential to maintain cell lineage during development [7, 8]. Understanding how cells coordinate the synthesis of chromatin components and retain a subset of epigenetic marks during each cell cycle as well as identifying the key regulatory factors governing these processesa re active areas of research.
All eukaryotic cells maintain a precise ratio of core histones to the newly synthesized DNA; both higher and lower ratios of histones to the DNA have deleterious effects (Table 1). The importance of regulating this histone/DNA ratio comes from elegant genetic and biochemical studies [1, 2, 9]. In the model eukaryotic organism Saccharomyces cerevisiae, histone imbalance created synthetically by selectively over-expressing either the H2A-H2B or H3-H4 dimers, affected mitotic fidelity leading to the loss of chromosomes [1]. Moreover, histone levels are also critical for proper partitioning of chromosomes to the daughter cells [10]. Another undesirable outcome of overproduction of histones, observed in Schizosaccharomyces pombe, is a marked increase in chromosomal instability due to the inclusion of canonical H3 along with the centromere-specific H3 variant CENP-A at centromeres, which consequently increases nucleosome density and alters the chromatin structure [10]. Conversely, incorporation of CENP-A (CID in Drosophila) at non-centromeric sites leads to the formation of dicentric chromosomes, chromosome breakage during mitosis, and genome instability, which is associated with malignancy [11, 12]. Consequently, cells have developed specific mechanisms to prevent mis-incorporation of CENP-A and maintain chromatin integrity [13]. In addition, transcription of histones is tightly regulated and coordinated with the cell cycle. Genetic studies in yeast revealed presence of a surveillance machinery mediated by the checkpoint kinase, RAD53, which monitors histone dosage during replication [14].
Table 1.
Known phenotypes associated with histone alterations
| Histone | Species | Molecular Phenotypes | Refs |
|---|---|---|---|
| Alteration of histone stoichiometry | S. cerevisiae | Chromosome loss, DNA damage sensitivity, and slow growth | [9, 97] |
| Repression of H2B RNA synthesis is yeast | S. cerevisiae | Cell cycle arrest | [98] |
| Change in dosage of H2B | S. cerevisiae | Altered gene expression | [99] |
| Histone H1o variant overexpression | Mouse | Reduction in steady state transcription of all RNA pol II regulated genes and transient inhibition of G1 and S phase progression; increase in nucleosome spacing | [100, 101] |
| Histone H1c variant overexpression | Mouse | Marked increase in gene expression at some loci | [100] |
| Synthesis of histones in non-replicating cells | Drosophila | Developmental arrest and zygotic lethality | [102] |
| Embryos lacking the three H1 subtypes (H1c, H1d, and H1e), | Drosophila | Die by mid-gestation with a broad range of defects | [103] |
| Partial depletion of Histone H4 | S. cerevisiae | Accumulation of recombinogenic DNA lesions | [104] |
| Overproduction of histones | S. pombe | Incorporation of canonical H3 along with H3 variant CENP-A at centromeres | [10] |
| Somatic mutation in H3F3A gene, K27M and G34R/G34V, encoding histone H3.3 | Human | Mutations unique to pediatric and young adult glioblastomas, associated with lengthening of telomeres and specific gene expression profiles | [3–5] |
| Somatic mutation in HIST1H3B, K27M and G34R, encoding histone H3.1 | Human | Mutations unique to pediatric glioblastomas | [4] |
| D123N mutation in H3F3a | Zebrafish | Eliminates the cranial neural crest-derived head skeleton | [105] |
Termination of histone gene expression
Just as histone synthesis is exquisitely regulated during the cell cycle, its termination is also precisely synchronized with cell cycle progression [2, 15–17]. Higher eukaryotes have multiple copies of histone genes for the core histones, each encoding a fraction of the total histone protein to deal with the large-scale histone synthesis required for packaging newly synthesized DNA into chromatin during S-phase [18]. These genes are organized in three major clusters in humans, HIST1 (55 histone genes), HIST2 (6 histone genes), and HIST3 (3 histone genes), with copies being arranged in tandem. At the end of S-phase or upon replication inhibition, histone levels are rapidly lowered [2, 19, 20]. In addition, histones are regulated post-transcriptionally in higher eukaryotes through a reduction in mRNA half life at the end of S phase [21–23]. However, the rapid turnover of histone mRNA has to be coupled with cessation of histone mRNA synthesis to achieve the precipitous drop in histone transcript levels observed at the end of DNA synthesis. The process by which higher eukaryotic cells halt histone transcription has remained elusive for a number of years. Recent work demonstrated that transcription of the histone genes is itself regulated by histone modification [17]. Nucleosomes upstream of the mouse histone gene cluster I (HistI) were found to be decorated by histone H2B tyrosine phosphorylation at 37 residue (pY37-H2B) precisely at the end of S-phase when DNA synthesis is completed. This mechansism is conserved in S. cerevisiae, and mutation of the equivalent site, Y40 to alanine, (H2BY40A) leads to a significant increase in all the core histone mRNA levels [17].
A dual role for WEE1 in S phase regulation and histone synthesis
WEE1 is an evolutionarily conserved nuclear tyrosine kinase (Table 2) that is markedly active during the S/G2 phase of the cell cycle [24, 25]. It was first discovered 25 years ago as a cell division cycle (cdc) mutant-wee1- in the fission yeast, Schizosaccharomyces pombe [26]. Fission yeast lacking WEE1 are characterized by a smaller cell size, and this phenotype has been attributed to the ability of WEE1 to negatively regulate the activity of cyclin dependent kinase, Cdc2 (Cdc28 in budding yeast and CDK1 in human), in the Cdc2/CyclinB complex [27]. Recently, WEE1 was shown to directly phosphorylate the mammalian core histone H2B at tyrosine 37 in a cell cycle dependent manner. Inhibition of WEE1 kinase activity either by a specific inhibitor (MK-1775) or suppression of its expression by RNA interference abrogated H2B Y37-phosphorylation with a concurrent increase in histone transcription [17]. Interestingly, S. cerevisiae lacking the WEE1 homolog, Swe1, also exhibited loss of the equivalent H2B Y40-phosphorylation and histone gene dysregulation [17]. Collectively, these data suggest a role for WEE1 as a ‘chromatin synthesis sensor’ by two sequential phosphorylation events: (i) Y15-phosphorylation of CDK1 throughout S phase to prevent exit from S phase until DNA replication is completed [17, 28], (ii) Y37-phosphorylation of H2B at the end of S phase to terminate histone synthesis, thus maintaining the right histone-DNA stoichiometry prior to mitotic entry [17].
Table 2.
Histone Tyrosine Kinases
| Kinase | Histone | Tyrosine residue | Interactors | Function | Lethality in budding yeast | Human disease association | Refs |
|---|---|---|---|---|---|---|---|
| WSTF | H2AX | 142 | APBB1, PTB-domain containing adaptor protein Fe65 | Maintains DNA damage induced foci formation | Site is absent in yeast | Williams syndrome | [90] |
| WEE1 | H2B | 37 | Recruitment of HIRA | Transcriptional Suppression of histone genes | Not lethal in normal growth conditions | Glioblastoma, melanomas, TNBC | [17, 48, 67, 68, 70, 106] |
| JAK2 | H3 | 41 | Prevents recruitment of HP1 | Transcriptional activation | Lethal | Leukemia | [89] |
| * RAD53 (homolog of CHK2) | * H3 | *99 | Recruitment of ubiquitin conjugating enzymes Ubc4-Ubc5 | Degradation of excess histones | Not lethal in normal growth conditions | Not known | [6] |
identified only in S. cerevisiae
With more than 50 genes coding for core histones in the Hist1 cluster in mouse and humans, eukaryotic cells require a strict mechanism to suppress transcription of so many genes. WEE1 deposits pY37-H2B marks at nucleosomes located upstream of HistI cluster to disengage NPAT (Nuclear protein, ataxia-telangiectasia locus) [17], a transcriptional activator of mammalian histone genes and RNA polymerase II [29, 30]. In addition, this epigenetic modification acts as a beacon for the recruitment of a transcriptional repressor, HIRA (histone regulatory homolog A) [17]. HIRA is the mammalian homologue of the yeast HIR proteins and is a component of the histone chaperone complex that spreads across silenced domains to enforce transcriptional repression [31]. HIR proteins bind to the negative regulatory site NEG located at the promoters of seven of the eight yeast histone genes to repress histone transcription [32, 33]. H2B Y37 phosphorylation is enriched in the histone promoters containing the NEG site [17], consistent with a mechanism for HIR recruitment to suppress histone gene transcription.
Cell cycle analysis indicated that cells rapidly exit S-phase once the histone transcription is completed and the pY37-H2B marks are quickly erased [17]. The temporal and transient nature of pY37-H2B suggests that cells may actively recruit a phosphatase to dephosphorylate pY37-H2B before the cells enter mitosis. Furthermore, continuous repression of histone transcription is likely to cripple cells due to a lack of histones to package nascent DNA. Consistent with this, ectopic expression of HIRA which functionally approximates the continuously phosphorylated state of histone H2B, caused arrest in S phase [34]. Although a pY37-H2B-specific phosphatase has not yet been identified, the members of the CDC14 tyrosine phosphatase family are potential candidates. CDC14 is an evolutionarily conserved dual specificity phosphatase [35, 36] that was recently found to interact with Swe1 [37]. Based on the transient nature of H2B Y37-phosphorylation, WEE1 interaction with a tyrosine phosphatase at specific chromatin loci, wherein a kinase recruits the partner phosphatase, seems likely. Other potential candidates include the EYA family of tyrosine phosphatases, which have been shown to dephosphorylate the variant histone H2AX at tyrosine 142 (Box 1).
Box 1. Variant histone tyrosine phosphorylation.
Although it has long been known that tyrosine kinases regulate critical cellular processes, it was not discovered until 2009 that they also directly phosphorylate histones [6, 17, 89, 90] (see Table 2). Williams-Beuren syndrome transcription factor (WSTF), a component of the WICH complex (WSTF-ISWI ATP-dependent chromatin-remodeling complex) was the first kinase shown to phosphorylate histone variant H2A.X at tyrosine 142 [90]. This post-translational modification by an atypical tyrosine kinase was shown to play a decisive role when cells were confronted with DNA damage, determining if they initiate DNA repair or undergo apoptosis. The significance of WSTF/pY142-H2A.X signaling became quickly relevant when a protein tyrosine phosphatase, EYA, was found to dephosphorylate pY142-H2A.X in a DNA damage signal-dependent manner [91]. WSTF physically interacts with H2AX and deposits the pY142-H2AX marks specifically in undamaged cells, however, upon DNA damage, formation of a double-strand break (DSB) leads to the recruitment of the meiotic recombination protein-11 (MRE11)–RAD50–Nijmegen breakage syndrome protein-1 (NBS1) (MRN) complex and ataxia-telangiectasia mutated (ATM) to the DSBs. ATM kinase rapidly phosphorylates the C-terminal tail of H2AX at serine 139 (γH2AX) [92]. When cells were subjected to ionizing radiation, pY142-H2AX marks were removed by EYA1 and EYA3 phosphatase, promoting a damage repair response by recruitment of MDC1 and associated repair factors [91]. However, failure to remove pY142-H2AX marks in damaged cells leads to the recruitment of pro-apoptotic factors such as JNK1 to H2AX, while inhibiting the recruitment of the damage repair complex, promoting apoptotic response to genotoxic stress.
Epigenetic marks manifested during the cell cycle
In addition to pY37-H2B, a number of other histone marks are regulated in a cell-cycle dependent manner. For example, phosphorylation of serine 10 in the N-terminal tail of histone H3 was shown to be crucial for chromosome condensation and cell-cycle progression during mitosis and meiosis in Tetrahymena [38]. Other marks, such as phosphorylation of histone H3 at serine 28 and threonine 11, were also identified to be mitosis-specific histone modifications in mammalian cells [39, 40]. Methylation of histone H3 (K4me3, K9me1, K9me2) and H4 (K20me1) and acetylation of H4 (K5, K16 and K56) are also regulated in cell cycle dependent manner [41]. Although H4K5 acetylation marks are critical for deposition of nascent histones during chromatin assembly by histone chaperones during S phase [42]; they are likewise erased after nucleosome assembly to restore the chromatin structure [41]. The histone deacetylases, HDAC1, HDAC2, or HDAC3, remove H4K5 acetyl marks and their recruitment to newly synthesized DNA is a regulated process [43]. Chicken HIRA has been shown to interact with HDAC1 and 2 [44]. Whether recruitment of HIRA by pY37-H2B establishes a binding platform for the further recruitment of HDACs to deacetylate H4K5ac in the chromatin remains to be established.
In human embryonic stem cells and induced pluripotent stem (iPS) cells the transcriptionally active chromatin marks, H3K4me3 and H3K9ac, were found to be enriched upstream and downstream of the H3 and H4 gene loci but not at the transcription start sites (TSS) in cell cycle- and transcription-dependent manner [8]. Whether pY37-H2B marks occur in pluripotent stem cells and have an active role in cell cycle-dependent transcriptional suppression of histones genes remains to be examined. Because pluripotent stem cells can differentiate into many different cell types, these cells in particular need to tightly regulate chromatin status, which can significantly impact gene expression profiles. Thus, a regulatory mechanism that maintains balanced histone pools during the cell cycle will be critical in stem cells, and given the functional and evolutionary conservation of pY37-H2B marks during cell cycle, a role in regulating histone gene expression in stem cells seems likely.
WEE1 acts as a hub for regulating chromatin integrity
During S-phase, WEE1 appears to mark the completion of not only DNA replication and genetic integrity via Y15-phosphorylation of CDK1 but also histone synthesis by marking chromatin with H2B Y37-phosphorylation before cells enter mitosis [17, 45–47]. Because coupling of DNA synthesis with histone transcription is critical for proper chromatin formation, by regulating both of these processes WEE1 acts as a master regulator of chromatin integrity. Agents that compromise WEE1 function cause mitotic infidelity, chromosome loss, and apoptosis, effect commonly referred to as mitotic catastrophe [48]. Interestingly, in contrast to single-celled eukaryotes such as budding or fission yeast that can survive without Wee1, knockout of Wee1 in mice is embryonic lethal [49]. Wee1 null embryos do not proceed past the blastocyst stage due to defects in cell proliferation. Moreover, the abnormalities exhibited by Wee1-deficient cells are more severe than that noted for other checkpoint proteins such as ATM, which regulates G2/M entry, suggesting that the loss of proliferation may not be solely attributed to early entry into mitosis [49]. Consistent with this notion, Wee1-deficient cells exhibit increased DNA damage in S-phase as evidenced by increased H2AX S139-phosphorylation, a hallmark of DNA damage, and chromosome aneuploidy [49, 50].
When DNA replication is disrupted by endogenous or exogenous factors such as alkylating agents, oxidative free radicals, ultraviolet-radiation, or nucleotide depletion, cells rapidly mobilize checkpoint and DNA repair proteins to sites of damage. In response to stalled replication forks, the DNA replication checkpoint kinase, ATR (ATM and RAD3-related) is activated. Mammalian ATR regulates activation of the checkpoint kinase CHK1 [51, 52]. CHK1, a serine/threonine kinase not only negatively regulates the CDC25 phosphatase that dephosphorylates CDK1 but also phosphorylates and stabilizes WEE1 [53–55]. Consequently, WEE1 phosphorylates CDK1 in the CDK1/cyclin B complex at tyrosine 15 to prevent mitotic entry until the damaged DNA is repaired (G2/M checkpoint). In addition, WEE1 function appears to be important for the intra-S phase checkpoint as evident from the studies demonstrating that WEE1 inhibitors synergize with CHK1 inhibitors to induce cytotoxicity [56, 57]. During S-phase WEE1 also modulates the activity of CDK1 and CDK2 present at replication origins to control replication initiation (Figure 2). Untimely CDK activity in WEE1 inhibited cells led to a marked increase in the loading of the replication factor, CDC45, at chromatin [58]. As a consequence of augmented replication origin firing, the replication forks slow down and MUS81, a structure-specific endonuclease that directly interacts with WEE1, is recruited to cleave the DNA at the stalled forks generating double strand breaks (DSBs) [58, 59]. It is plausible that WEE1 negatively regulates MUS81 activity at replication forks to prevent untimely DSB formation during DNA replication. Further, the ability of activated WEE1 to phosphorylate Y37-H2B is likely to halt histone mRNA synthesis concomitant with inhibition of DNA synthesis. This is consistent with the earlier observation that showed co-ordinated repression of these two macromolecular processes in response to genotoxic insults [19].
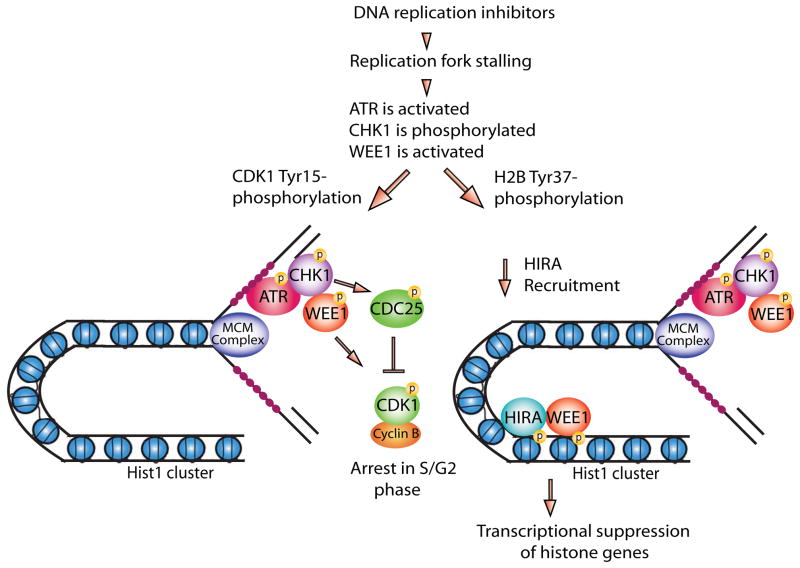
Figure 2. A model for WEE1 in chromatin integrity during replication stress.
Replication protein A (RPA) (small magenta circles) coats ssDNA formed during DNA replication and DNA repair which facilitates the localization of Ataxia-Telangiectasia mutated and RAD3-related (ATR) kinase to sites of DNA damage in both human and yeast systems. Activated ATR phosphorylates the downstream checkpoint kinase-1 (CHK1) which in turn phosphorylates and activates WEE1. In addition to WEE1 activation, CHK1 also phosphorylates and inactivates CDC25, a phosphatase that dephosphorylates CDK1. WEE1 plays a crucial role in maintaining chromatin integrity during DNA damage or replication stress by performing two distinct phosphorylations; first, it phosphorylates CDK1 at Y15 preventing entry into mitosis so as to allow repair of damaged DNA. Second, it actively downregulates histone gene transcription by marking H2B at Tyr37 in nucleosomes upstream of histone cluster 1 (Hist1). H2B Tyr37 phosphorylation prevents binding of the transcriptional coactivator NPAT and RNA polymerase II and recruits the histone chaperone HIRA upstream of the Hist1 cluster thereby avoiding overproduction of histones. The collective outcome of these events is initiation of intra-S phase and G2 checkpoints in response to DNA damage or replication stress and maintenance of chromatin integrity during repair of damaged DNA, prior to entry into mitosis.
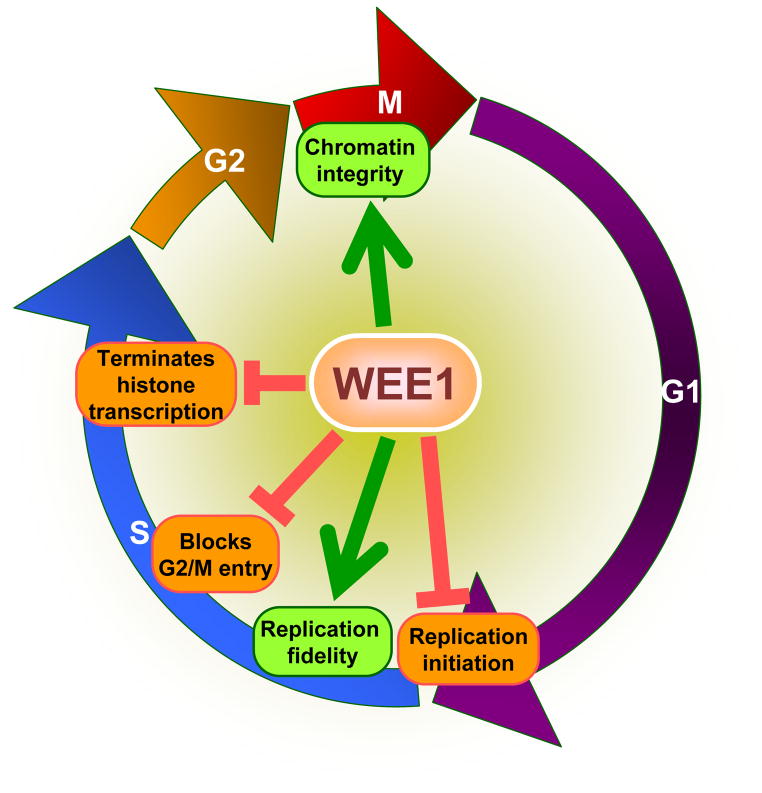
In addition to histone synthesis, chromosome condensation also appears to be regulated by WEE1 kinase [54]. Drosophila Wee1 and Grp (the Drosophila homolog of CHK1) were found to be crucial for the delay in chromosome condensation when DNA replication was perturbed by treatment with specific inhibitors that interfere with S phase progression or to delay metaphase in cells treated with topoisomerase inhibitors [54]. These observations imply that WEE1 co-ordinates completion of multiple chromosomal processes before cells divide by dynamically surveying events to ensure proper chromatin duplication and segregation (Figure 1). These studies suggest that WEE1 has a critical function in S-phase that is outside its well-established role in regulating mitotic entry. Thus, WEE1 is an important regulator in DNA replication, histone transcription, and chromosome condensation; perturbation of any of these processes profoundly affects chromatin integrity and faithful transmission of genetic and epigenetic information to the progeny (Figure 1).
Figure 1. Schematic outlining WEE1 functions.
WEE1 is emerging as a critical surveyor of chromatin integrity across the evolutionary landscape by regulating histone synthesis and the cell cycle. It controls genome stability by regulating the activity of factors such as CDKs involved in replication initiation during S phase. During DNA replication, it negatively regulates the activity of the Mus81 endonuclease at stalled replication forks to prevent formation of DNA double strand breaks and preserve genetic integrity. Importantly, it appears to couple DNA synthesis with histone synthesis due to its ability to prevent mitotic entry until DNA synthesis is completed and to switch off histone transcription, by modifying core histone H2B, at the end of S phase. These functions are likely to be critical for its ability to delay chromosome condensation and prevent aneuploidy.
One significant question is how the WEE1-mediated cytoplasmic signaling cascade (WEE1/CDK1) converges with WEE1-mediated nuclear events (WEE1/H2B) to coordinately regulate cell cycle progression. WEE1 phosphorylates H2B at the end of S phase when the peak phosphorylation of its other substrate, CDK1, is receding [17]. This opens up the possibility that when WEE1 senses completion of DNA synthesis, it switches its substrate preference from CDK1 to H2B. In eukaryotes the origin recognition complex (ORC), Cdc6 protein, and the minichromosome maintenance (MCM) protein complex assemble on chromatin before initiation of DNA replication. CDK1 has been shown to interact specifically with the ORCs in mammalian, Xenopus, and yeast cells [60–62]. CDKs regulate DNA replication positively by inducing the initiation of DNA replication at the G1/S transition and negatively by preventing further rounds of origin firing. WEE1 phosphorylates CDK1 in S phase thus preventing further rounds of origin firing within the same cell cycle [58]. Thus, it is likely that upon completion of DNA synthesis, when WEE1 is prevented from phosphorylating CDK1, it switches to a new substrate, H2B. Thus, by integrating these two seemingly distinct temporal events, WEE1 promotes progression into mitosis with accurately duplicated chromatin.
WEE1-independent modes of histone regulation
Despite its central role in coordinating DNA synthesis and histone transcription, WEE1 is not the only factor regulating these processes. Genetic studies using budding yeast revealed a critical role for the RAD53 checkpoint kinase in detecting excess histones that are not incorporated into the chromatin. Excess histones bind Rad53, which targets them for degradation by the ubiquitin-proteosome machinery [6, 14]. Furthermore, excess histones in budding yeast sensitized cells to DNA replication inhibitors and lead to loss of chromosomes; conversely, decreasing histone pools by deleting specific histone genes facilitates radioresistance by promoting access of the DNA repair factors to the damaged sites [14, 63, 64]. A recent study identified multiple novel ubiquitin ligases (Hel1, Hel2, Pep5 and Snt2) in S. cerevisiae that ubiquitylate ectopically overexpressed histone H3 and targets it for degradation [65]. Although viable, these E3 ligase mutants displayed differential sensitivity to the replication inhibitor, hydroxy urea (HU). Notably, the rad53 hel1 double mutants as well as some of the other double mutant combinations did not display increased HU sensitivity compared with the single rad53 mutant, suggesting that they are either functional redundant or epistatic. Interestingly, increased association of endogenous histones with the histone chaperone Asf1 was observed even in the absence of DNA damage in the hel1, hel2, pep5, and snt2 deletion strains. These results suggest that defects in histone protein turnover exacerbate cellular sensitivity to genotoxic agents- in part by interfering with the function of histone chaperone/chromatin remodeling enzymes by sequestering them with excess histones.
Other nuclear processes that alter histone stoichiometry can also hamper genome stability [14, 64, 66]. This is evident from several different yeast strains carrying mutations that affect histone levels. For example, excessive core histone mRNA expression has been reported in S. cerevisiae lacking Trf4, which encodes a poly(A) polymerase, and these mutants are characterized by acute sensitivity to DNA replication inhibitors and defects in chromosome condensation [66]. Although, trf4 mutants contain wild-type Rad53 that can detect excess histone proteins and target them for degradation by the ubiquitin proteosome machinery, histone mRNA overexpression is detrimental to trf4 mutants as evidenced by their sensitivity to genotoxic agents and a longer than average S phase. This finding is echoed by the observation that S. cerevisiae H2B Y40A mutants accumulate excess core histone mRNAs, again suggesting that both transcriptional and post-transcriptional mechanisms are required for maintaining balanced histone pools [17]. Collectively these studies illustrate that eukaryotic cells utilize both genetic and epigenetic controls to tightly regulate histone mRNA levels to ensure genome stability.
WEE1 and cancer epigenetics
The central role of WEE1 in integrating various aspects of cell cycle progression, histone synthesis, and genomic stability makes it an important target for cancer treatment. Gene expression profiling of various tumors revealed that the Wee1 kinase is overexpressed in Glioblastoma multiforme (GBM) [48, 67], luminal, and triple negative breast cancers (TNBC) [68, 69] as well as malignant melanomas [70]. WEE1 overexpression and the resultant decrease in histone levels could lead to inefficient chromatin packaging, making the DNA more accessible to the DNA damage repair machinery and promoting radioresistance [64]. The ability of WEE1 to downregulate histone levels could explain why cancer cells become dependent on its epigenetic activity. In addition to acquiring radioresistance, decreased nucleosomal packaging and consequently local alterations in chromatin architecture may activate transcription of pro-proliferative genes or even oncogenes that are otherwise kept in check in normal cells.
Overexpression of WEE1 in certain cancer types could be exploited by using WEE1-specific small molecule inhibitors such as MK-1775 that is in clinical trials [71]. Such inhibition of WEE1 in actively replicating cancer cells would increase histone dosage and, when combined with DNA damaging agents, would interfere with the DNA repair machinery and compromise genome integrity. Consistent with this hypothesis, the WEE1 inhibitor MK-1775 was able to radiosensitize human lung, breast, skin, brain, and prostate cancer cells to DNA damaging agents, and WEE1 inhibitors were shown synergize with CHK1 inhibitors to induce cytotoxicity [56, 72–76]. Moreover, breast cancer cells treated with hydroxyl urea (which inhibits DNA synthesis) and WEE1 inhibitors were found to contain a marked increase in disorganized mitotic spindles and abnormal mitoses [56].
Enzymes that promote or reverse histone modifications have emerged as major targets for the development of small molecule inhibitors, commonly referred as epigenetic inhibitors. Many of these are already in different stages of clinical trials with significant success in hematologic cancers [77]. The precision with which histone modifying enzymes modify a specific amino acid residue in a histone makes them ideal candidates for drug discovery efforts, however, because of a highly heterogeneous nature of disease, the efficacy of the epigenetic inhibitor is likely to be seen in only a subset of cancer patients that are addicted to the activity of the histone modifying enzyme. One well studied example is histone methyltransferase EZH2 that deposits the repressive mark H3K27me3. Two somatic heterozygous mutations in EZH2, Y641 and A677, have been identified in about 22% of diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma, which caused increased H3K27me3 levels [78–82]. Recently, two small-molecule inhibitors of EZH2 were identified, EPZ005687 and GSK126, which suppressed global H3K27me3 levels and reactivated silenced EZH2 target genes leading to inhibition of EZH2 mutant DLBCL cell line proliferation and the growth of xenograft tumors [83, 84]. This and other early successes, suggest that WEE1 inhibition may be a useful strategy for treatment of a subset of cancers.
Cancer is a complex disease, differences in biology and outcomes exist not only among various clinical states but also within each patient. Personalized medicine offers the potential to optimize treatment for a given patient, based on molecular biomarkers that drive individual variability or drug responses. Thus, elucidation of abundant H2B Y37-phosphorylation in a subset of GBM, TNBC or malignant melanoma tumor biopsies could be a ‘companion diagnostic’ test for administration of WEE1 inhibitor such as MK-1775. This ‘personalized therapy’ for a subset of WEE1-positive GBM, TNBC or malignant melanoma patients could be a significant development as limited therapeutic options and no targeted therapeutic modalities are currently available for these cancers with low survival rates.
Defects in chromatin architecture were recently discovered to play a critical role in pediatric and young adult GBM pathogenesis, a lethal brain tumor in both adults and children [3–5, 85]. Exon sequencing revealed two somatic mutations, K27M and G34R/G34V in the histone H3 variants, H3.3 and H3.1. Interestingly, mutations in ATRX (α-thalassaemia/mental retardation syndrome X-linked) and DAXX (death-domain associated protein) were identified in every tumor harboring a G34R/G34V mutation. ATRX and DAXX are subunits of a chromatin remodeling complex that is essential for H3.3 incorporation at pericentric heterochromatin and telomeres. It has been suggested that the alterations in heterochromatinization is the major consequence of the mutations in H3.1, H3.3, and the ATRX-DAXX chromatin remodeling pathway, leading to alternative lengthening of telomeres and expression of specific gene profiles [3–5]. Whether, GBM exhibits nucleosomal packaging defects selectively in children by acquiring histone mutations, and in adults by WEE1 overexpression would require more studies. However, the alteration in heterochromatic states appears to be the major mechanism of pathogenesis in GBM.
Concluding remarks
The discovery of H2B Y37-phosphorylation by WEE1 provides a direct link between the cell cycle kinase and an epigenetic mark that helps to maintain chromatin homeostasis. This finding raises an important question of whether WEE1 utilizes its ability to phosphorylate H2B to accomplish its other well known function as a mitotic gatekeeper. This expands our perspective of how we define the role of WEE1 in cell cycle regulation and its role in various malignancies, especially those which over express WEE1 kinase; these cancer cells could utilize alterations in histone levels to confer selective proliferative advantage and radioresistance.
Identification of WEE1 epigenetic activity raises several important questions in normal physiology as well as in disease pathology. Are there auxiliary loci epigenetically marked by pY37-H2B modification in cancers with aberrant WEE1 expression? If so, do cancer cells employ a different set of epigenetic readers to read H2B Y37-phosphorylation and are there common motifs that distinguish the promoters marked with this specific modification? It is still unclear at this point if pY37-H2B modulates chromatin architecture by recruiting chromatin-remodeling proteins and thereby impacting patterns of local gene expression. Another area to explore is whether this histone Tyr-modification potentially cross talks with other histone modifications such as the H2B ubiquitylation and H4K5 and H3K56 acetylation, which have known roles in chromatin replication or repair [8, 41, 86–88]. Further research aimed at answering these questions is critical for a clear understanding of the global regulatory role of histone Tyr-phosphorylation and the development of WEE1 inhibitors as potential cancer therapeutics.
Box 2. Core histone tyrosine phosphorylation.
The first core histone shown to be Tyr-phosphorylated in mammalian cells was histone H3. A non-receptor tyrosine kinase, Janus kinase 2 (JAK2) directly phosphorylates H3 at tyrosine 41, preventing binding of heterochromatin protein 1α (HP1α) [89]. HP1α represses the transcription of heterochromatic genes and preserves centromeric architecture [93, 94]. JAK2 is frequently hyperactivated in hematological malignancies, leading to the upregulation of LMO2, a gene important for tumor angiogenesis [89]. In addition, pY41-H3 marks other key lineage-specific hematopoietic genes [95]. Treatment with JAK2 inhibitors abrogates pY41-H3 marks, reduces levels of the pluripotency regulator Nanog, and increases recruitment of HP1α levels at the Nanog promoter [96]. In addition to Y41, H3 was also shown to be phosphorylated at Y99 in S. cerevisiae[6]. Interestingly, it was the non-chromatin bound histones, which were phosphorylated in a Rad53-dependent manner leading to their polyubiquitylation and degradation by proteosomal machinery [6]. The ubiquitin conjugating enzymes (E2) Ubc4 and Ubc5 and the ubiquitin ligase (E3) Tom1 (temperature dependent organization in mitotic nucleus 1) were shown to be involved in the polyubiquitylation of excess histones [6]. Taken together with histone transcriptional suppression role of H2B Tyr37-phosphorylation [17], these data indicate that cells not only shut down histone transcription upon completion of DNA synthesis or when exposed to DNA damaging agents, but they also degrade excess histones that are not incorporated into chromatin to prevent harmful effects of non -chromatin bound histones.
It is likely that there are other modes of histone Tyr-phosphorylation, especially in mammalian cells where many non-receptor tyrosine kinases could potentially target 15 distinct tyrosine residues in the core histone- (H2A- Y39, Y50, and Y57; H2B- Y37, Y40, Y42, Y83, and Y121; H3- Y41, Y54, and Y99; H4- Y51, Y72, Y88, and Y98), and 2 in the linker histone H1 (Y88 and Y119). The transient nature of histone Tyr-phosphorylation may be the reason why only a few examples these epigenetic changes have been identified to date. However, these tyrosine residues are highly conserved (Table 2), suggesting that new examples found in yeast or other tractable genetic systems are likely to be relevant in mammalian cells as well.
Highlights.
WEE1 phosphorylates histone H2B at tyrosine 37 in the late S phase
H2B Tyr37-phosphorylation downregulates expression of multiple histone genes
WEE1 kinase has emerged as a novel epigenetic writer
pY37-H2B epigenetic marks decorate several DNA and histone modifying genes
Acknowledgments
We apologize to those authors whose work could not be cited owing to the space constraint of reference citations. We thank Dr Akash Gunjan for critical reading of manuscript, Dr. Jozef Spychala, VisiScience for help with Figure 1. We declare competing financial interests; we are named as inventors on US patent application 61/583,864 and international application PCT/US2013/020395 titled “Antibodies specific for phosphorylated histones and uses thereof.” This work was supported in part by Department of Defense (W81XWH-12-1-0248) to K.M. and by the National Cancer Institute, NIH (1R01CA135328) and Moffitt Lung Cancer Spore to N.P.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meeks-Wagner D, Hartwell LH. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44:43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- 2.Osley MA. The regulation of histone synthesis in the cell cycle. Annual review of biochemistry. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 3.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 4.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khuong-Quang DA, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta neuropathologica. 2012;124:439–447. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh RK, et al. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nature cell biology. 2009;11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran V, et al. Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science (New York, N Y. 2012;338:679–682. doi: 10.1126/science.1226028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medina R, et al. Epigenetic control of cell cycle-dependent histone gene expression is a principal component of the abbreviated pluripotent cell cycle. Molecular and cellular biology. 2012;32:3860–3871. doi: 10.1128/MCB.00736-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meeks-Wagner D, et al. Isolation of two genes that affect mitotic chromosome transmission in S. cerevisiae. Cell. 1986;44:53–63. doi: 10.1016/0092-8674(86)90484-8. [DOI] [PubMed] [Google Scholar]
- 10.Takayama Y, et al. Hsk1- and SCF(Pof3)-dependent proteolysis of S. pombe Ams2 ensures histone homeostasis and centromere function. Developmental cell. 2010;18:385–396. doi: 10.1016/j.devcel.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackinnon RN, Campbell LJ. The role of dicentric chromosome formation and secondary centromere deletion in the evolution of myeloid malignancy. Genetics research international. 2011;2011:643628. doi: 10.4061/2011/643628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heun P, et al. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Developmental cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi ES, et al. Factors That Promote H3 Chromatin Integrity during Transcription Prevent Promiscuous Deposition of CENP-A(Cnp1) in Fission Yeast. PLoS Genet. 2012;8:e1002985. doi: 10.1371/journal.pgen.1002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 15.Heintz N, et al. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Molecular and cellular biology. 1983;3:539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma T, et al. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes & development. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahajan K, et al. H2B Tyr37 phosphorylation suppresses expression of replication-dependent core histone genes. Nature structural & molecular biology. 2012;19:930–937. doi: 10.1038/nsmb.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marzluff WF, et al. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- 19.Hereford LM, et al. Cell-cycle regulation of yeast histone mRNA. Cell. 1981;24:367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- 20.Hereford L, et al. Periodic transcription of yeast histone genes. Cell. 1982;30:305–310. doi: 10.1016/0092-8674(82)90036-8. [DOI] [PubMed] [Google Scholar]
- 21.Marzluff WF, et al. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nature reviews. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koseoglu MM, et al. Phosphorylation of threonine 61 by cyclin a/Cdk1 triggers degradation of stem-loop binding protein at the end of S phase. Molecular and cellular biology. 2008;28:4469–4479. doi: 10.1128/MCB.01416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnan N, et al. The Prolyl Isomerase Pin1 Targets Stem-Loop Binding Protein (SLBP) To Dissociate the SLBP-Histone mRNA Complex Linking Histone mRNA Decay with SLBP Ubiquitination. Molecular and cellular biology. 2012;32:4306–4322. doi: 10.1128/MCB.00382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Featherstone C, Russell P. Fission yeast p107wee1 mitotic inhibitor is a tyrosine/serine kinase. Nature. 1991;349:808–811. doi: 10.1038/349808a0. [DOI] [PubMed] [Google Scholar]
- 25.McGowan CH, Russell P. Cell cycle regulation of human WEE1. The EMBO journal. 1995;14:2166–2175. doi: 10.1002/j.1460-2075.1995.tb07210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- 27.Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe N, et al. Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the cell cycle. Embo J. 1995;14:1878–1891. doi: 10.1002/j.1460-2075.1995.tb07180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J, et al. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes & development. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, et al. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes & development. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamane K, et al. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol Cell. 2011;41:56–66. doi: 10.1016/j.molcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osley MA, et al. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell. 1986;45:537–544. doi: 10.1016/0092-8674(86)90285-0. [DOI] [PubMed] [Google Scholar]
- 33.Xu H, et al. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Molecular and cellular biology. 1992;12:5249–5259. doi: 10.1128/mcb.12.11.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall C, et al. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Molecular and cellular biology. 2001;21:1854–1865. doi: 10.1128/MCB.21.5.1854-1865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartwell LH, et al. Genetic control of the cell division cycle in yeast. Science (New York, N Y. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- 36.Gray CH, et al. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. Embo J. 2003;22:3524–3535. doi: 10.1093/emboj/cdg348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breitkreutz A, et al. A global protein kinase and phosphatase interaction network in yeast. Science (New York, N Y. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei Y, et al. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7480–7484. doi: 10.1073/pnas.95.13.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goto H, et al. Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J Biol Chem. 1999;274:25543–25549. doi: 10.1074/jbc.274.36.25543. [DOI] [PubMed] [Google Scholar]
- 40.Preuss U, et al. Novel mitosis-specific phosphorylation of histone H3 at Thr11 mediated by Dlk/ZIP kinase. Nucleic Acids Res. 2003;31:878–885. doi: 10.1093/nar/gkg176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Sullivan RJ, et al. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nature structural & molecular biology. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobel RE, et al. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sirbu BM, et al. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes & development. 2011;25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmad A, et al. WD dipeptide motifs and LXXLL motif of chicken HIRA are essential for interactions with the p48 subunit of chromatin assembly factor-1 and histone deacetylase-2 in vitro and in vivo. Gene. 2004;342:125–136. doi: 10.1016/j.gene.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 45.Chow JP, et al. Differential contribution of inhibitory phosphorylation of CDC2 and CDK2 for unperturbed cell cycle control and DNA integrity checkpoints. The Journal of biological chemistry. 2003;278:40815–40828. doi: 10.1074/jbc.M306683200. [DOI] [PubMed] [Google Scholar]
- 46.Aligue R, et al. Regulation of Schizosaccharomyces pombe Wee1 tyrosine kinase. The Journal of biological chemistry. 1997;272:13320–13325. doi: 10.1074/jbc.272.20.13320. [DOI] [PubMed] [Google Scholar]
- 47.Rhind N, et al. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes & development. 1997;11:504–511. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- 48.Mir SE, et al. In silico analysis of kinase expression identifies WEE1 as a gatekeeper against mitotic catastrophe in glioblastoma. Cancer Cell. 2010;18:244–257. doi: 10.1016/j.ccr.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tominaga Y, et al. Murine Wee1 plays a critical role in cell cycle regulation and pre-implantation stages of embryonic development. International journal of biological sciences. 2006;2:161–170. doi: 10.7150/ijbs.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beck H, et al. Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase. The Journal of cell biology. 2010;188:629–638. doi: 10.1083/jcb.200905059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sorensen CS, Syljuasen RG. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic acids research. 2012;40:477–486. doi: 10.1093/nar/gkr697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith J, et al. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Advances in cancer research. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 53.Sorensen CS, et al. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3:247–258. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 54.Fasulo B, et al. Chk1 and Wee1 kinases coordinate DNA replication, chromosome condensation, and anaphase entry. Molecular biology of the cell. 2012;23:1047–1057. doi: 10.1091/mbc.E11-10-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Connell MJ, et al. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. The EMBO journal. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aarts M, et al. Forced mitotic entry of S-phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer discovery. 2012;2:524–539. doi: 10.1158/2159-8290.CD-11-0320. [DOI] [PubMed] [Google Scholar]
- 57.Davies KD, et al. Chk1 inhibition and Wee1 inhibition combine synergistically to impede cellular proliferation. Cancer biology & therapy. 2011;12:788–796. doi: 10.4161/cbt.12.9.17673. [DOI] [PubMed] [Google Scholar]
- 58.Beck H, et al. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Molecular and cellular biology. 2012;32:4226–4236. doi: 10.1128/MCB.00412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dominguez-Kelly R, et al. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J Cell Biol. 2011;194:567–579. doi: 10.1083/jcb.201101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katsuno Y, et al. Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3184–3189. doi: 10.1073/pnas.0809350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Romanowski P, et al. Interaction of Xenopus Cdc2 x cyclin A1 with the origin recognition complex. J Biol Chem. 2000;275:4239–4243. doi: 10.1074/jbc.275.6.4239. [DOI] [PubMed] [Google Scholar]
- 62.Wuarin J, et al. Stable association of mitotic cyclin B/Cdc2 to replication origins prevents endoreduplication. Cell. 2002;111:419–431. doi: 10.1016/s0092-8674(02)01042-5. [DOI] [PubMed] [Google Scholar]
- 63.Singh RK, et al. Generation and management of excess histones during the cell cycle. Front Biosci. 2009;14:3145–3158. doi: 10.2741/3441. [DOI] [PubMed] [Google Scholar]
- 64.Liang D, et al. Histone dosage regulates DNA damage sensitivity in a checkpoint-independent manner by the homologous recombination pathway. Nucleic Acids Res. 2012;40:9604–9620. doi: 10.1093/nar/gks722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh RK, et al. Novel E3 ubiquitin ligases that regulate histone protein levels in the budding yeast Saccharomyces cerevisiae. PLoS One. 2012;7:e36295. doi: 10.1371/journal.pone.0036295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reis CC, Campbell JL. Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics. 2007;175:993–1010. doi: 10.1534/genetics.106.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wuchty S, et al. Prediction of Associations between microRNAs and Gene Expression in Glioma Biology. PLoS One. 2011;6:e14681. doi: 10.1371/journal.pone.0014681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iorns E, et al. Integrated functional, gene expression and genomic analysis for the identification of cancer targets. PLoS One. 2009;4:e5120. doi: 10.1371/journal.pone.0005120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murrow LM, et al. Identification of WEE1 as a potential molecular target in cancer cells by RNAi screening of the human tyrosine kinome. Breast cancer research and treatment. 2010;122:347–357. doi: 10.1007/s10549-009-0571-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Magnussen GI, et al. High expression of Wee1 is associated with poor disease-free survival in malignant melanoma: potential for targeted therapy. PLoS One. 2012;7:e38254. doi: 10.1371/journal.pone.0038254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hirai H, et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther. 2009;8:2992–3000. doi: 10.1158/1535-7163.MCT-09-0463. [DOI] [PubMed] [Google Scholar]
- 72.Sarcar B, et al. Targeting radiation-induced G(2) checkpoint activation with the Wee-1 inhibitor MK-1775 in glioblastoma cell lines. Mol Cancer Ther. 2011;10:2405–2414. doi: 10.1158/1535-7163.MCT-11-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hirai H, et al. MK-1775, a small molecule Wee1 inhibitor, enhances anti-tumor efficacy of various DNA-damaging agents, including 5-fluorouracil. Cancer Biol Ther. 2010;9:514–522. doi: 10.4161/cbt.9.7.11115. [DOI] [PubMed] [Google Scholar]
- 74.Stathis A, Oza A. Targeting Wee1-like protein kinase to treat cancer. Drug news & perspectives. 2010;23:425–429. doi: 10.1358/dnp.2010.23.7.1490760. [DOI] [PubMed] [Google Scholar]
- 75.Bridges KA, et al. MK-1775, a novel Wee1 kinase inhibitor, radiosensitizes p53-defective human tumor cells. Clinical cancer research. 2011;17:5638–5648. doi: 10.1158/1078-0432.CCR-11-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.De Witt Hamer PC, et al. WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:4200–4207. doi: 10.1158/1078-0432.CCR-10-2537. [DOI] [PubMed] [Google Scholar]
- 77.Mack GS. To selectivity and beyond. Nature biotechnology. 2010;28:1259–1266. doi: 10.1038/nbt.1724. [DOI] [PubMed] [Google Scholar]
- 78.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCabe MT, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wigle TJ, et al. The Y641C mutation of EZH2 alters substrate specificity for histone H3 lysine 27 methylation states. FEBS letters. 2011;585:3011–3014. doi: 10.1016/j.febslet.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 82.Yap DB, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Knutson SK, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nature chemical biology. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 84.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 85.Sturm D, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 86.Trujillo KM, Osley MA. A Role for H2B Ubiquitylation in DNA Replication. Mol Cell. 2012;48:734–746. doi: 10.1016/j.molcel.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee JS, et al. Codependency of H2B monoubiquitination and nucleosome reassembly on Chd1. Genes & development. 2012;26:914–919. doi: 10.1101/gad.186841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moyal L, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dawson MA, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao A, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cook PJ, et al. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nature reviews. Molecular cell biology. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 93.Yamagishi Y, et al. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature. 2008;455:251–255. doi: 10.1038/nature07217. [DOI] [PubMed] [Google Scholar]
- 94.Panteleeva I, et al. HP1alpha guides neuronal fate by timing E2F-targeted genes silencing during terminal differentiation. Embo J. 2007;26:3616–3628. doi: 10.1038/sj.emboj.7601789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dawson MA, et al. Three Distinct Patterns of Histone H3Y41 Phosphorylation Mark Active Genes. Cell reports. 2012;2:470–477. doi: 10.1016/j.celrep.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Griffiths DS, et al. LIF-independent JAK signalling to chromatin in embryonic stem cells uncovered from an adult stem cell disease. Nature cell biology. 2011;13:13–21. doi: 10.1038/ncb2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liang D, et al. Histone dosage regulates DNA damage sensitivity in a checkpoint-independent manner by the homologous recombination pathway. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Han M, et al. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell. 1987;48:589–597. doi: 10.1016/0092-8674(87)90237-6. [DOI] [PubMed] [Google Scholar]
- 99.Clark-Adams CD, et al. Changes in histone gene dosage alter transcription in yeast. Genes & development. 1988;2:150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- 100.Brown DT, et al. Differential effect of H1 variant overexpression on cell cycle progression and gene expression. Nucleic Acids Res. 1996;24:486–493. doi: 10.1093/nar/24.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gunjan A, et al. Effects of H1 histone variant overexpression on chromatin structure. J Biol Chem. 1999;274:37950–37956. doi: 10.1074/jbc.274.53.37950. [DOI] [PubMed] [Google Scholar]
- 102.Sullivan E, et al. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes & development. 2001;15:173–187. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fan Y, et al. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Molecular and cellular biology. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Prado F, Aguilera A. Partial depletion of histone H4 increases homologous recombination-mediated genetic instability. Molecular and cellular biology. 2005;25:1526–1536. doi: 10.1128/MCB.25.4.1526-1536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cox SG, et al. An essential role of variant histone h3.3 for ectomesenchyme potential of the cranial neural crest. PLoS Genet. 2012;8:e1002938. doi: 10.1371/journal.pgen.1002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Murrow LM, et al. Identification of WEE1 as a potential molecular target in cancer cells by RNAi screening of the human tyrosine kinome. Breast Cancer Res Treat. 2010;122:347–357. doi: 10.1007/s10549-009-0571-2. [DOI] [PMC free article] [PubMed] [Google Scholar]