Abstract
Ectopic expression of the transcription factor Fra-1 in transgenic mice leads to osteosclerosis, a bone disorder characterized by increased bone mass. The molecular basis for this phenotype is unknown and Fra-1 functions cannot be studied by a conventional loss-of-function approach, since fra-1-knockout mice die in utero likely due to placental defects. Here we show that the lethality of fra-1-knockout mice can be rescued by specific deletion of Fra-1 only in the mouse embryo and not in the placenta. Mice lacking Fra-1 (fra-1Δ/Δ) are viable and develop osteopenia, a low bone mass disease. Long bones of fra-1Δ/Δ mice appear to have normal osteoclasts but express reduced amounts of bone matrix components produced by osteoblasts and chondrocytes such as osteocalcin, collagen1a2 and matrix Gla protein. The gene for matrix Gla protein seems to be a specific target of Fra-1 since its expression was markedly increased in the long bones of fra-1-transgenic mice. These results uncover a novel function of Fra-1 in regulating bone mass through bone matrix production by osteoblasts and chondrocytes.
Keywords: AP-1, bone extracellular matrix, conditional Fra-1 knockout, osteoblasts, osteopenia
Introduction
The functions of bones include regulation of blood calcium levels, providing mechanical support to the body and supporting hematopoiesis in the bone marrow. All these functions require the maintenance of an adequate bone shape and bone density, which are mainly determined by the activities of bone-forming osteoblasts and bone-resorbing osteoclasts (Karsenty and Wagner, 2002). The presence of two cell types with opposing activities makes the bone a highly dynamic structure with a continuous tissue renewal called remodeling, but also requires a tight regulation of osteoblast and osteoclast activities in order to prevent bone diseases.
Bone resorption is carried out by hematopoietically derived osteoclasts. The bone-resorbing activity of osteoclasts is determined by proliferation and differentiation of osteoclast precursors and by the resorptive efficiency of mature osteoclasts. Several soluble factors like macrophage colony-stimulating factor (M-CSF) and receptor activator of NF-κB ligand (RANKL) as well as transcription factors such as c-Fos, NFATc1 and NF-κB have been identified to regulate osteoclast proliferation and differentiation (Teitelbaum and Ross, 2003). In addition, genetic studies in mice have revealed genes that regulate the resorptive activity of mature osteoclasts, which depends on their characteristic shape and on the ability to acidify and dissolve the bone matrix (Boyle et al, 2003).
Mesenchyme-derived osteoblasts rebuild the resorbed bone by depositing bone matrix that then becomes mineralized. Osteoblast activity is determined by osteoblast proliferation and differentiation and by the bone-forming activity of mature osteoblasts. A number of soluble molecules and transcription factors that include Ihh, FGF18, Runx2 (Cbfa-1) and Osterix regulate osteoblast differentiation (Karsenty and Wagner, 2002). The bone-forming activity of mature osteoblasts is stimulated by insulin-like growth factor (IGF)-II and transforming growth factor (TGF)-β, which are released from the extracellular matrix during bone resorption thereby affecting both osteoclasts and osteoblasts. In addition, several factors, such as parathyroid hormone, prostaglandin E, fibroblast growth factor and TGF-β, have been shown to stimulate both bone resorption and formation (Harada and Rodan, 2003). However, the transcription factors that act downstream of these anabolic signals in osteoblasts are largely unknown.
Activator protein-1 (AP-1) is a dimeric transcription factor formed by Jun proteins (c-Jun, JunB, JunD) and Fos proteins (c-Fos, Fra-1, Fra-2, FosB, ΔFosB) (Eferl and Wagner, 2003). Both, Jun and Fos protein family members are required for bone formation and remodeling. Ubiquitous balanced deletion of a conditional c-jun allele leads to malformations of the axial skeleton (Behrens et al, 2003), and JunB has recently been shown to be essential for osteoblast proliferation and differentiation (Kenner et al, 2004). Most Fos proteins are also implicated in proliferation and differentiation of osteoblasts and osteoclasts. Transgenic mice expressing c-Fos develop osteosarcomas due to increased osteoblast proliferation (Grigoriadis et al, 1993). In contrast, mice lacking c-Fos develop osteopetrosis caused by a differentiation defect in the osteoclast lineage (Wang et al, 1992; Grigoriadis et al, 1994). This differentiation defect can be rescued by expression of the Fos-related protein Fra-1, suggesting that Fos and Fra-1 have overlapping functions in osteoclast differentiation (Fleischmann et al, 2000). Ectopic Fra-1 expression leads to osteosclerosis likely due to accelerated differentiation of osteoprogenitors into mature osteoblasts (Jochum et al, 2000). A similar skeletal phenotype was described in transgenic mice expressing ΔFosB, a splice variant of FosB, suggesting that Fra-1 and ΔFosB promote osteoblast differentiation by regulating common transcriptional target genes in the osteoblast lineage (Sabatakos et al, 2000). However, the requirement of Fra-1 in bone formation was unclear, since fra-1-knockout mice die during embryogenesis likely due to placental defects (Schreiber et al, 2000).
We generated mice with conditional alleles of fra-1 simultaneously harboring a MORE-cre knock-in allele (Tallquist and Soriano, 2000), in order to study the importance of Fra-1 in postnatal development and in bone remodeling. Conditional deletion of fra-1 by the Cre-recombinase was efficient throughout the mouse embryo, but did not occur in extraembryonic tissues such as the placenta. The resulting fra-1Δ/Δ mice were viable but developed osteopenia, a bone disorder characterized by reduced bone mass. Dynamic histomorphometry of bone tissues demonstrated that the osteopenic phenotype was due to reduced bone formation. The numbers of the major bone cells, the osteoblasts and osteoclasts, were unchanged although in vitro and in vivo analysis revealed a severely reduced bone-forming activity of osteoblasts. This defect was most likely due to reduced expression of the bone matrix genes osteocalcin, collagen1a2 and matrix Gla protein.
Results
Embryo-specific deletion of Fra-1 rescues the embryonic lethality of fra-1-deficient fetuses
To study the role of Fra-1 in organ functions and in particular in bone remodeling, mice with a conditional floxed allele of fra-1 (fra-1f) were generated (Figure 1A). The correct gene targeting was confirmed by PCR (data not shown) and Southern blot analysis (Figure 1B). Homozygous fra-1f/f mice showed no overt phenotype, suggesting that the genetic manipulation of the fra-1 alleles did not interfere with gene functions. Heterozygous fra-1+/− mice with a germline deletion of the floxed fra-1 allele were generated using the SycP1-cre deleter mouse (Vidal et al, 1998). Intercrossing these mice gave no viable fra-1−/− offsprings at birth (data not shown), indicating that germline deletion of both fra-1 alleles causes embryonic lethality as recently described (Schreiber et al, 2000).
Figure 1.
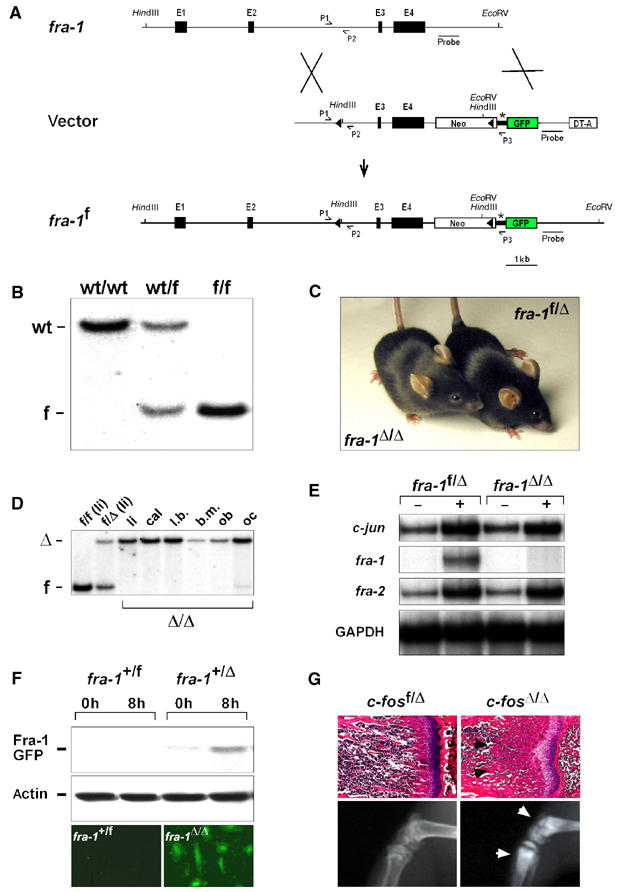
Strategy to generate mice with conditional alleles of fra-1. (A) Scheme of the gene targeting. Exons 3 and 4 of fra-1 were flanked by loxP sites (triangles). These exons contain the dimerization and the DNA-binding domains of Fra-1, and deletion of these exons renders Fra-1 inactive. A GFP reporter gene (GFP) is spliced to the residual N-terminal part of fra-1 upon deletion and is thereby activated. The relevant restriction sites used for Southern blot analysis and the location of the probe are indicated. fra-1: genomic locus of fra-1; vector: targeting vector for fra-1; fra-1f: floxed allele of fra-1; E1–4: exons 1–4 of fra-1; Neo: neomycin phosphotransferase gene; asterisk: splice acceptor of exon 2 of fra-1; DT-A: gene for diphtheria toxin; P1–3: PCR primers used for genotyping. (B) Southern blot analysis of offsprings demonstrating successful targeting of fra-1. For the Southern blot analysis, DNA was digested with HindIII/EcoRV and the bands were detected with the probe indicated in (A). wt: 12.5 kb wild-type allele; f: 4.5 kb floxed allele. (C) The embryonic lethality of fra-1-knockout mice is rescued by conditional deletion with the MORE-cre knock-in allele. The MORE-cre deletes the floxed fra-1 alleles in the whole mouse embryo but not in the placenta. MORE-cre fra-1f/f mice are therefore fra-1Δ/Δ mice that were viable and born at Mendelian frequency. (D) Deletion of fra-1 was confirmed in different tissues of 8-week-old mice by Southern blot analysis and in primary osteoblasts from newborn mice. li: liver; cal: calvaria; l.b.: long bones; b.m.: bone marrow; ob: osteoblasts differentiated in vitro; oc: osteoclasts differentiated in vitro; f: 4.5 kb floxed allele; Δ: 11.5 kb deleted allele. (E) RPA of c-jun, fra-1 and fra-2 in primary E12.5 MEFs. MEFs of the indicated genotypes were starved in 0.5% FCS (−) and then stimulated for 2 h with 10% FCS (+). Note that fra-1 mRNA is not detectable in stimulated fra-1Δ/Δ MEFs. The amount of GAPDH mRNA was used as a loading control. (F) A fusion protein between the residual N-terminal part of Fra-1 and GFP (43 kDa) is generated after deletion and was detected by Western blot analysis using an anti-GFP antibody (upper panel) and by direct immunofluorescence (lower panel). Note that only cells with at least one Δ allele contain the fusion protein. The amount of actin was used as a loading control in the Western blot analysis. (G) c-fosΔ/Δ mice (MORE-cre c-fosf/f) develop osteopetrosis as judged by H&E staining (upper panel, arrows) and X-ray analysis (lower panel, arrows) of long bones (6-week-old mice).
We next intercrossed fra-1f/f mice with MORE-cre knock-in mice in an attempt to rescue the embryonic lethality of fra-1-knockout mice, likely caused by placental defects (Schreiber et al, 2000). The MORE-cre knock-in allele is expressed at gastrulation throughout the epiblast of the early mouse embryo, but not in extraembryonic tissues such as the yolk sac and the placenta (Tallquist and Soriano, 2000). Therefore, Cre-mediated deletion of the floxed fra-1 alleles in MORE-cre fra-1f/f (fra-1Δ/Δ) mice was restricted to the embryo proper and did not occur in the placenta (data not shown). MORE-cre fra-1f/f (fra-1Δ/Δ) mice were born at Mendelian frequencies and did not show any overt phenotype at weaning age (Figure 1C). Efficient deletion of fra-1 was confirmed in internal organs (Figure 1D and data not shown), bone and isolated bone cells of adult fra-1Δ/Δ mice by Southern blot analysis (Figure 1D). RNase protection assay (RPA) demonstrated that fra-1 mRNA was absent in primary fra-1Δ/Δ mouse embryonic fibroblasts (MEFs; Figure 1E). Expression of fra-1 was hardly detectable by RPA in control fra-1f/Δ MEFs, but was inducible by serum stimulation. In contrast, no fra-1 mRNA was detectable in fra-1Δ/Δ MEFs even after serum stimulation (Figure 1E). The expression of the other AP-1 members and their serum induction was unchanged in fra-1Δ/Δ MEFs (Figure 1E and data not shown). In addition to isolated MEFs, RNAs from adult tissues of fra-1Δ/Δ mice including long bones and calvariae were tested for fra-1 expression by RT–PCR analysis. No fra-1 mRNA was detectable, whereas expression was readily detectable in corresponding control samples from fra-1f/Δ mice (data not shown). The deletion of the floxed fra-1 allele results in the formation of a Fra-1-GFP fusion protein (Figure 1A), which was detectable by Western blot analysis using an anti-GFP antibody and by direct GFP fluorescence in fra-1+/Δ, fra-1Δ/Δ, but not fra-1+/f MEFs (Figure 1F and data not shown). These data demonstrate the correct targeting and efficient embryo-specific deletion of the floxed fra-1 alleles and further show that Fra-1 is essential for placentation, but apparently dispensable for the development and function of most other organs after birth.
We have used conditional alleles of the JunB gene (Kenner et al, 2004) as well as the c-fos gene to test the applicability of the MORE-cre approach for the analysis of bone remodeling in vivo. Conditional deletion of c-fos in MORE-cre c-fosf/f (c-fosΔ/Δ) mice led to the identical osteopetrotic bone phenotype as described for the c-fos-knockout mice (Wang et al, 1992) (Figure 1G).
Decreased bone mass in fra-1Δ/Δ mice
Ectopic expression of Fra-1 in transgenic mice leads to increased bone mass with accelerated differentiation of osteoblasts (Jochum et al, 2000). However, the high level of Fra-1 expression in the transgenic mice can cause unphysiological effects thereby preventing a definitive conclusion about the requirement of Fra-1 in bone formation. Therefore, we have analyzed bone formation in 6-week-old and 3-month-old fra-1Δ/Δ mice. Radiography, von Kossa staining and microcomputed tomography (μCT) analysis revealed a significantly decreased bone mass in the lumbar vertebral bodies and long bones (Figure 2A–D and data not shown), whereas the structure of the epiphyseal growth plates in long bones was normal (data not shown). Static and dynamic histomorphometric analyses on lumbar vertebral bodies and tibial metaphyses of 1.5- and 3-month-old fra-1Δ/Δ and control mice demonstrated that bone mass is progressively lost in fra-1Δ/Δ mice leading to the osteopenic phenotype (Figure 2E and data not shown). In addition, trabecular thickness was reduced (Figure 2G), whereas trabecular numbers were maintained (Figure 2H). This phenotype is similar to low-turnover osteoporosis in humans, which results from a defect in osteoblast function. Given the reduced bone mass, the absolute numbers of osteoblasts (Figure 2F) and osteoclasts (data not shown) were reduced in fra-1Δ/Δ mice, but osteoblast- and osteoclast-covered bone surfaces were comparable to control fra-1f/Δ mice (Figure 2J–M). The number and morphology of osteocytes within the tibial cortex of fra-1Δ/Δ bones were unchanged and expressed the mineralization marker Phex (PHosphate-regulating gene with homology to Endopeptidases on the X chromosome) at similar levels to control fra-1f/Δ osteocytes (data not shown). Dynamic histomorphometry after calcein labeling revealed a significant decrease in bone formation rates (Figure 2I). These results demonstrate that fra-1Δ/Δ mice develop osteopenia, a bone loss phenotype, due to decreased bone formation.
Figure 2.
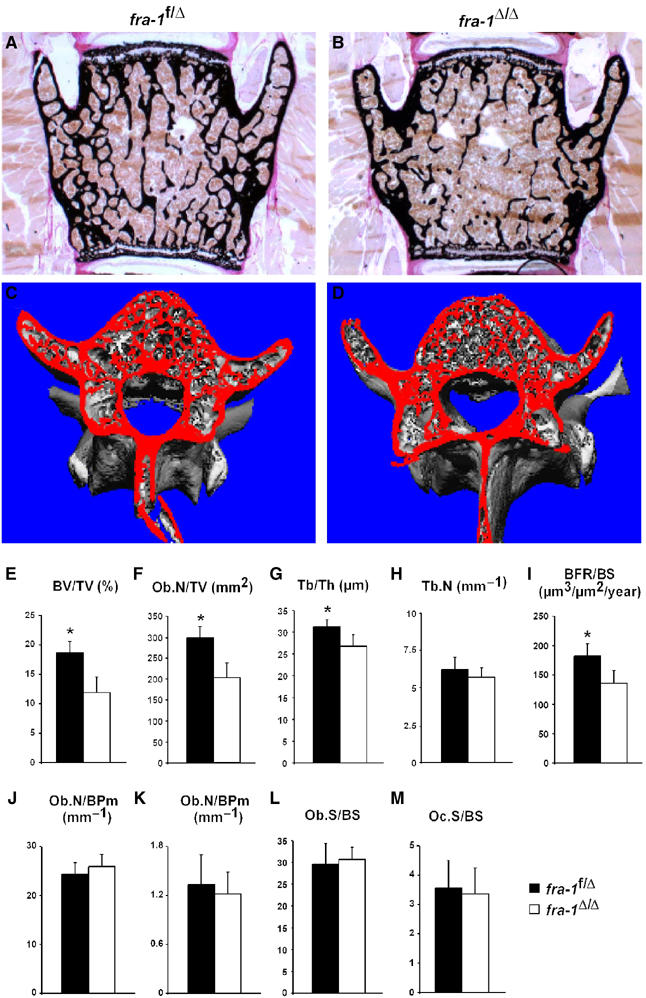
fra-1Δ/Δ mice develop osteopenia. (A–D) Decreased bone mass was detected in the vertebrae of fra-1Δ/Δ mice (3-month-old) by von Kossa staining (A, B) and μCT analysis (C, D). (E–M) Bone parameters measured by dynamic bone histomorphometry after calcein labeling demonstrated decreased bone mass (E), a decrease in absolute numbers of osteoblasts (F) and reduced trabecular thickness (G) in fra-1Δ/Δ mice, which is due to decreased bone formation (I). Numbers of trabeculae (H), osteoblasts per bone perimeter (J) and osteoclasts per bone perimeter (K) were not affected. The surface of osteoblasts (L) and osteoclasts (M) per bone surface was unchanged. BV/TV: trabecular bone volume per tissue volume; Ob.N/TV: osteoblast number per tissue volume; Tb/Th: trabecular thickness; Tb.N: trabecular number; BFR/BS: bone formation rate per bone surface; Ob.N/BPm: osteoblast number per bone perimeter; Oc.N/BPm: osteoclast number per bone perimeter; Ob.S/BS: osteoblast surface per bone surface; Oc.S/BS: osteoclast surface per bone surface. n=18 male mice; P<0.001.
No apparent osteoclast defect in fra-1Δ/Δ mice
To study the cellular mechanisms that account for the osteopenic phenotype of fra-1Δ/Δ mice, the differentiation and activity of osteoblasts and osteoclasts were analyzed in vivo and in vitro. Histochemical staining of fra-1Δ/Δ bones for the osteoclast marker protein tartrate-resistant acidic phosphatase (TRAP) showed mature osteoclasts in the bones of fra-1Δ/Δ mice (Figure 3A and B). Analysis of osteoclast marker gene expression revealed a slight reduction of all mRNAs in fra-1Δ/Δ bones (Figure 3C), which is most likely due to the reduced bone mass rather than due to an osteoclast differentiation defect.
Figure 3.
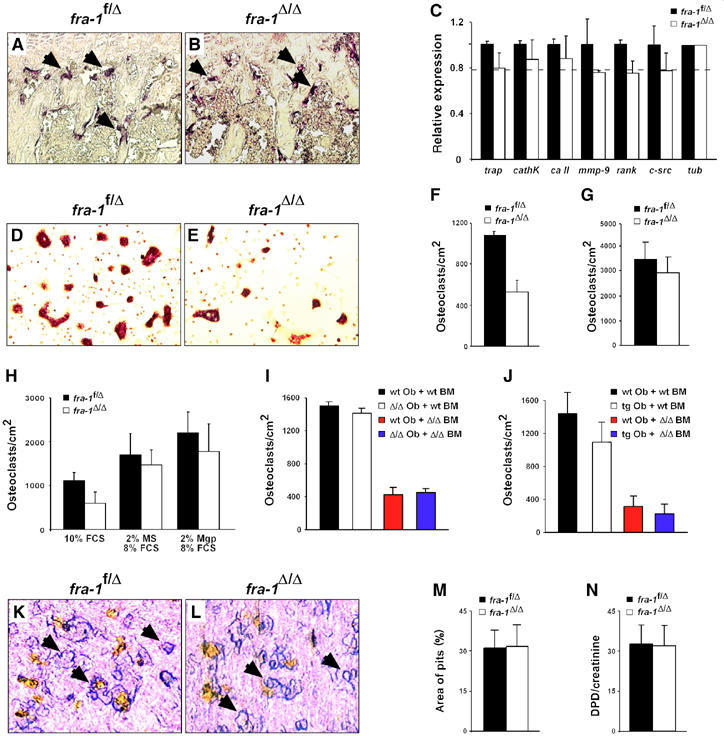
No overt osteoclast phenotype in fra-1Δ/Δ mice. (A, B) TRAP staining of osteoclasts (arrows) on the long bones of fra-1f/Δ and fra-1Δ/Δ mice (6-week-old) showed no major difference. (C) Real-time PCR analysis of mRNAs that are expressed in osteoclasts. Long bones of 6-week-old mice were used for RNA isolation (n=3). Relative expression levels normalized for tubulin with standard deviations indicated are shown. The broken line indicates the amount of bone mass reduction in the long bones of 6-week-old fra-1Δ/Δ mice as judged by bone histomorphometry. trap: tartrate-resistant acidic phosphatase; cathK: cathepsin K; ca II: carbonic anhydrase II; mmp-9: matrix metalloproteinase 9; rank: receptor and activator of NF-κB; tub: tubulin. (D, E) Osteoclast precursor cells derived from the bone marrow of adult mice were differentiated into mature multinucleated osteoclasts on plastic dishes and stained for the osteoclast marker protein TRAP (n=5). Differentiation of fra-1Δ/Δ precursors into osteoclasts was decreased on plastic dishes (E, F). (G) Plating of the precursors on bone slices rescued the osteoclast differentiation defect (n=5). (H) Addition of 2% mouse serum (MS) or 2% ApoE-Mgp serum (Mgp) rescued the osteoclast differentiation defect (n=4). A reciprocal co-culture assay on fra-1Δ/Δ osteoblasts (I) or fra-1-transgenic osteoblasts (J) could not rescue the osteoclast differentiation defect (n=4). Ob: osteoblast; BM: bone marrow; wt: wild type; Δ/Δ: fra-1Δ/Δ; tg: fra-1-transgenic. (K, L) Osteoclast precursor cells derived from the bone marrow of adult mice were differentiated into mature multinucleated osteoclasts on bone slices and the resorption pits (arrows) were stained with toluidine blue. (M) The resorption activity of fra-1Δ/Δ osteoclasts in vitro was analyzed by quantification of resorption pit areas on bone slices (n=5). (N) Deoxypyridinoline (DPD) crosslinks in the urine were measured as an osteoclast activity parameter in vivo. The values were normalized for muscle creatinine that is also excreted in the urine to account for urine concentration (n=9, 6-week-old mice).
When osteoclast precursors from the bone marrow were cultured under osteoclastogenic conditions in the presence of M-CSF and RANKL, the differentiation of fra-1Δ/Δ precursors into multinucleated osteoclasts was decreased to 50% of control cells (Figure 3D–F). This differentiation defect could not be rescued by reciprocal co-culture of osteoclast precursors on osteoblast feeder layers of different genotypes (Figure 3I and J), but was rescued on bovine cortical bone slices (Figure 3G). Surprisingly, addition of 2% of mouse serum as well as 2% serum from ApoE-Mgp-transgenic mice also rescued the differentiation defect (Figure 3H; see below). Functional analysis of osteoclasts in vitro by quantification of resorption pits on bone slices demonstrated that the resorption activity of control fra-1f/Δ and fra-1Δ/Δ osteoclasts was similar (Figure 3K–M). In addition, analysis of deoxypyridinoline crosslinks in urine samples, which reflects osteoclast activity in vivo, was not significantly changed in fra-1Δ/Δ mice (Figure 3N). These data demonstrate that Fra-1 is apparently not necessary for osteoclast differentiation in vivo, and that a molecular signal from the bone surface or a soluble component from the serum is able to rescue the osteoclast differentiation defect in vitro.
Fra-1 is required for osteoblast differentiation and function
To test whether the osteopenic phenotype is intrinsic to the osteoblast lineage, primary osteoblasts were prepared from calvariae of neonatal mice and differentiated in vitro. Staining for osteoblast differentiation marker protein alkaline phosphatase (ALP) and analysis of ALP activity in cell extracts revealed no major difference between fra-1Δ/Δ and control osteoblasts (Figure 4A and B and data not shown). The activity of osteoblasts was next analyzed by the deposition of mineralized extracellular matrix (Figure 4C–E). Despite the presence of ALP-positive cells in fra-1Δ/Δ cultures, the deposition of mineralized extracellular matrix as monitored by alizarin red staining of bone nodules was strongly reduced (Figure 4D and E). However, cell density and proliferation capacity were not significantly different between control and fra-1Δ/Δ osteoblasts and therefore cannot account for the reduced bone nodule formation in fra-1Δ/Δ cultures (Figure 4F and data not shown). These data indicate that Fra-1 positively regulates the bone-forming activity of osteoblasts.
Figure 4.
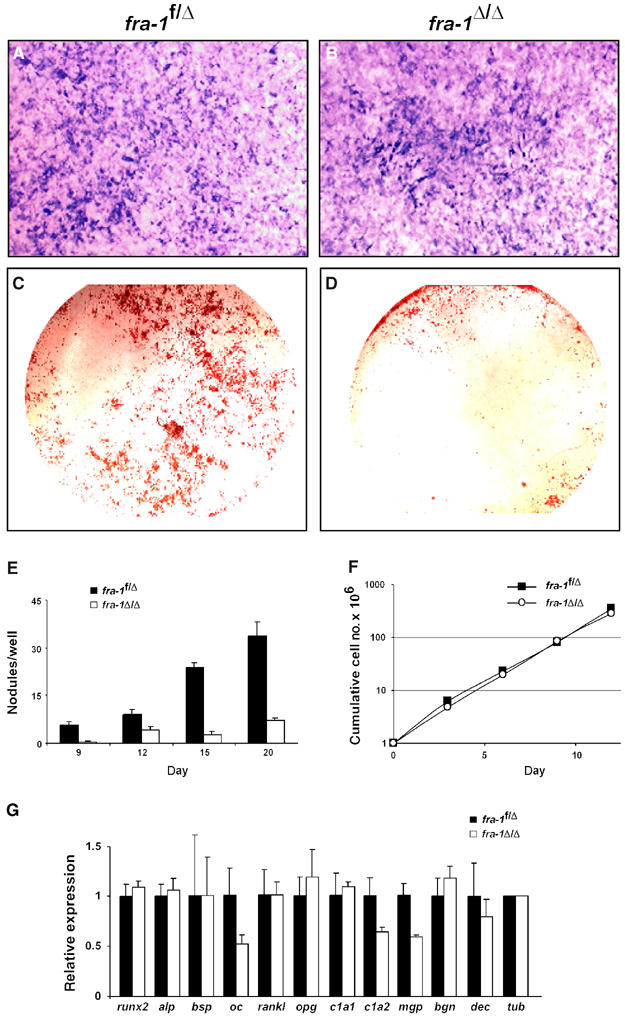
Formation of bone nodules is impaired in cultures of fra-1Δ/Δ osteoblasts. (A, B) Osteoblast precursor cells from the calvariae of newborn mice were differentiated for 21 days in vitro and stained for ALP activity (ALP-positive cells in blue). (C, D) Deposition of extracellular matrix material and bone nodule formation was analyzed by staining of osteoblast cultures (21 days) with alizarin red (bone nodules in red). (E) Time course of bone nodule formation by fra-1f/Δ and fra-1Δ/Δ osteoblasts (n=5). (F) Cumulative cell number (proliferation) of fra-1f/Δ and fra-1Δ/Δ osteoblasts. (G) Real-time PCR analysis of mRNAs for osteoblast differentiation factors and extracellular matrix components in osteoblast cultures (day 9 of differentiation). One representative example out of three independent primary culture experiments is shown. The relative expression levels were normalized for tubulin expression. alp: ALP, bsp: bone sialoprotein; oc: osteocalcin; rankl: receptor and activator of NF-κB; opg: osteoprotegerin; c1a1: collagen1a1; c1a2: collagen1a2; mgp: matrix Gla protein; bgn: biglycan; dec: decorin; tub: tubulin.
Bone matrix components are targets of Fra-1 in osteoblasts
We next analyzed the expression levels of osteoblast marker genes by real-time PCR during osteoblast differentiation in vitro, in order to identify candidate genes that might be responsible for the decreased bone-forming activity of fra-1Δ/Δ osteoblasts. The expression of bone matrix components osteocalcin (oc), collagen1a2 (col1a2) and matrix Gla protein (mgp) was consistently reduced in osteoblast cultures, whereas the expression of the differentiation markers runx2, ALP (alp), bone sialoprotein (bsp), rankl and osteoprotegerin (opg) was unchanged (Figure 4G). The mRNA levels for other bone matrix components such as collagen1a1, biglycan and decorin were unchanged in fra-1Δ/Δ osteoblasts (Figure 4G). It is worth noting that some variability of gene expression was observed, which might be due to the heterogeneous composition of primary calvarial cultures (Garcia et al, 2002).
Expression analysis was next performed in vivo using RNA from long bones. Consistent with the in vitro results, no major changes in the expression of differentiation markers runx2, alp, bsp, rankl and opg were observed (Figure 5A). In addition, the expressions of membrane-bound matrix metalloproteinase mt-1-mmp and genes implicated in signaling pathways important for osteoblast differentiation and activity were unchanged (Figure 5A). However, mRNA levels for oc, col1a2 and mgp were reduced to a similar extent to that in calvarial osteoblast cultures (Figure 5A and B). The unchanged expression of runx2 and the reduced expression of oc and col1a2 in osteoblasts were confirmed by in situ hybridization (ISH) experiments on sections of long bones (Figure 5C–J). In addition, the expression of mgp was reduced in hypertrophic chondrocytes of fra-1Δ/Δ bones (Figure 5I and J). These data suggest that the osteopenia in fra-1Δ/Δ mice could be due to reduced synthesis of bone matrix proteins by osteoblasts and chondrocytes identifying Fra-1 as a positive regulator of bone matrix production.
Figure 5.
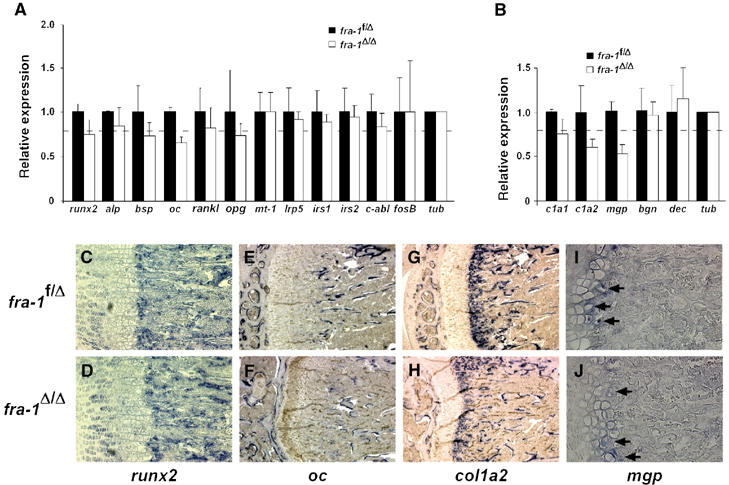
Reduced expression of bone matrix components in the long bones of fra-1Δ/Δ mice. (A, B) Real-time PCR analysis of mRNAs for osteoblast differentiation and activity factors (A) and bone matrix genes (B) in long bones (n=3, 6-week-old mice). The relative expression levels were normalized for tubulin expression with the standard deviations indicated. The broken line indicates the amount of bone mass reduction (and reduction in osteoblast numbers) in the long bones of 6-week-old fra-1Δ/Δ mice. Therefore, only the expression levels of osteocalcin and collagen1a2 are reduced in fra-1Δ/Δ osteoblasts. The reduced expression of matrix Gla protein needs no correction for reduced osteoblast numbers since it is mainly expressed in chondrocytes. alp: ALP, bsp: bone sialoprotein; oc: osteocalcin; rankl: receptor and activator of NF-κB; opg: osteoprotegerin; mt-1: membrane-bound matrix metalloproteinase mt-1-mmp; irs1,2: insulin receptor substrate 1,2; tub: tubulin; c1a1: collagen1a1; c1a2: collagen1a2; mgp: matrix Gla protein; bgn: biglycan; dec: decorin; tub: tubulin. (C–J) In situ hybridization experiments for expression of runx2 (C, D), osteocalcin (E, F), collagen1a2 (G, H), and matrix Gla protein (I, J, arrows show expression in hypertrophic chondrocytes) on long bones (n=3, 6-week-old mice).
We reasoned that increased production of extracellular bone matrix components might also account for the osteosclerosis previously described in fra-1-transgenic mice (Jochum et al, 2000). To test this hypothesis, the expression of bone matrix genes was analyzed in fra-1-transgenic osteoblasts and long bones. Intriguingly, RNA expression of mgp was several-fold increased in fra-1-transgenic long bones (Figure 6A–C), calvariae and primary osteoblast cultures (data not shown). Increased expression of mgp was specific for bone tissues and could not be observed in hearts and lungs of fra-1-transgenic mice (data not shown). In addition to mgp, expression levels for oc and col1a2 were also significantly increased in fra-1-transgenic long bones (Figure 6C) and primary calvarial cultures (data not shown) albeit not as prominent as mgp. Consistently, the expression of oc was also increased in the calvariae of fra-1-transgenic mice (Jochum et al, 2000). These data suggest that the osteosclerotic phenotype of fra-1-transgenic mice is likely caused by increased bone matrix production.
Figure 6.
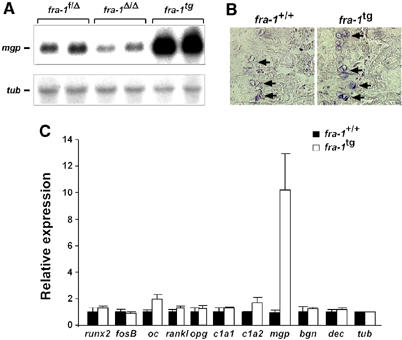
Increased expression of bone matrix components in the long bones of fra-1-transgenic (fra-1tg) mice. (A) Northern blot analysis of mgp expression in the long bones of fra-1f/Δ, fra-1Δ/Δ and fra-1tg mice. The corresponding littermate controls for the fra-1tg mice expressed mgp at similar levels compared with fra-1f/Δ mice (not shown). Expression of tubulin was used as loading control. (B) In situ hybridization for mgp in the long bones of fra-1+/+ (wild type) and fra-1tg mice. Note the increased blue staining indicative for increased mgp expression in hypertrophic chondrocytes (arrows) of fra-1tg mice. (C) Real-time PCR analysis of mRNAs for osteoblast differentiation markers and genes for bone matrix components in fra-1+/+ and fra-1tg mice (n=3, 3-week-old fra-1tg mice were used before first histological signs of increased bone mass became apparent). The relative expression levels were normalized for tubulin expression and standard deviations are indicated. oc: osteocalcin; rankl: receptor and activator of NF-κB; opg: osteoprotegerin; c1a1: collagen1a1; c1a2: collagen1a2; mgp: matrix Gla protein; bgn: biglycan; dec: decorin; tub: tubulin.
Discussion
Ectopic expression of Fra-1 and ΔFosB in transgenic mice causes osteosclerosis with increased bone mass (Jochum et al, 2000; Sabatakos et al, 2000). However, the molecular mechanism leading to this phenotype has not been analyzed. In this study, a conditional loss-of-function approach was employed to define functions of Fra-1 postnatally and in osteoclasts and osteoblasts. Conventional fra-1-knockout embryos have extraembryonic defects leading to embryonic lethality (Schreiber et al, 2000). Therefore, we have generated a floxed fra-1 allele and used the MORE-cre knock-in allele (Tallquist and Soriano, 2000) to specifically delete fra-1 in the embryo (fra-1Δ/Δ), but not in extraembryonic tissues. Conditional fra-1Δ/Δ mice were viable and showed no major defects in adulthood, demonstrating that Fra-1 is dispensable for most organs. The mutant mice, however, developed osteopenia with progressive loss of bone mass, which is the opposite phenotype as described for fra-1-transgenic mice.
In osteoblasts, the expression pattern of Fos proteins mainly suggested a role for Fra-1 in osteoblast proliferation (McCabe et al, 1995; 1996), but the absence of Fra-1 had no effect on osteoblast proliferation in vitro and on osteoblast numbers in vivo. Similarly, ectopic expression of Fra-1 in transgenic mice did not lead to accelerated proliferation of osteoblasts, demonstrating that Fra-1 is not important for osteoblast proliferation (Jochum et al, 2000). The latter study proposed Fra-1 to function as an osteoblast differentiation factor based on increased osteoblast surface per bone surface in fra-1-transgenic mice and increased activity of ALP in primary transgenic osteoblast cultures (Jochum et al, 2000). Unexpectedly, osteoblast surface per bone surface was not reduced and the activity of ALP was not affected in osteoblasts of fra-1Δ/Δ mice. The expression of osteoblast differentiation markers runx2 and bsp was also comparable to control osteoblasts in vitro and in vivo. This suggests that Fra-1 is dispensable for the initial stages of osteoblast differentiation but can promote osteoblast differentiation when ectopically expressed in mice. A late differentiation defect of fra-1Δ/Δ osteoblasts cannot be excluded, since expression of oc, a late marker of osteoblast differentiation, was indeed reduced. It is conceivable that other AP-1 proteins compensate for the absence of Fra-1 in osteoblasts. One candidate for such a functional overlap is ΔFosB, a splice variant of FosB, because mice that ectopically express ΔFosB develop a bone phenotype that is highly reminiscent of that observed in fra-1-transgenic animals (Sabatakos et al, 2000). A second candidate is the potential Fra-1 dimerizing partner ATF4, which has recently been implicated in Coffin–Lowry syndrome (CLS), a disease associated with skeletal abnormalities (Yang et al, 2004). The phenotype of ATF4-deficient mice is strikingly similar to that of fra-1Δ/Δ mice with reduced bone mass, decreased expression of oc and reduced collagen synthesis. Moreover, expression of early differentiation markers such as Runx2 and Osterix is also not affected (Yang et al, 2004), suggesting that both transcription factors may control the activity of osteoblasts by a similar mechanism.
The most likely mechanism that can account for the low bone mass phenotype of fra-1Δ/Δ mice is a defect in the late maturation of osteoblasts to bone matrix-producing cells, accompanied by insufficient induction of the bone matrix components oc and col1a2. In addition, reduced levels of mgp, which is mainly expressed in proliferating and hypertrophic chondrocytes of long bones (Luo et al, 1995), might contribute to the low bone mass phenotype of fra-1Δ/Δ mice. Whereas reduced expression of col1a2, a major component of bone collagen, and lower levels of mgp can clearly contribute to the low bone mass phenotype of fra-1Δ/Δ mice, the role of oc is controversial. Loss of oc expression in knockout mice leads to increased bone mass (Ducy et al, 1996). This phenotype, however, becomes first noticeable at 6 months of age (Ducy et al, 1996), which is much later than the osteopenia in fra-1Δ/Δ mice. In addition, oc and mgp belong to the Gla (γ carboxyglutamic acid)-rich proteins that can bind mineral via carboxyl groups and are implicated in extracellular matrix mineralization. Whereas the absence of oc in knockout mice did not affect bone mineralization (Ducy et al, 1996), deletion of mgp leads osteopenia (Luo et al, 1997) and to increased mineralization of the growth plate and the arteries (Luo et al, 1997). A mineralization defect was, however, not observed in the long bones of fra-1Δ/Δ mice. It is likely that the reduction of mgp expression is insufficient to cause a significant change in the mineralization rate. In addition, von Kossa staining of arterial walls revealed no calcification within the elastic lamellae (data not shown). This is consistent with the bone-specific downregulation of mgp in fra-1Δ/Δ mice and supports recent data that Mgp regulates matrix mineralization locally but not systemically (Murshed et al, 2004).
How is the expression of oc, col1a2 and mgp regulated? It is possible that fra-1Δ/Δ osteoblasts are defective in an anabolic signaling pathway that is essential for bone matrix production (Harada and Rodan, 2003). Several osteopenic or osteoporotic mice with mutations in signaling pathways show osteoblast dysfunctions, such as knockout mice for runx2 (Ducy et al, 1997), c-abl (Li et al, 2000), lrp-5 (Kato et al, 2002), membrane-bound matrix metalloproteinase mt-1-mmp (Holmbeck et al, 1999), insulin receptor substrate-1 (Ogata et al, 2000) and insulin receptor substrate-2 (Akune et al, 2002). However, none of these genes was deregulated in fra-1Δ/Δ mice. In addition, the TGF-β signaling pathway is positively implicated in bone matrix production (Thiebaud et al, 1994). However, the expression of TGF-β 1, 2, 3 as well as SMAD3, an essential mediator of TGF-β signaling to the nucleus, was analyzed by RPA and was found to be unchanged in fra-1Δ/Δ mice. This suggests that Fra-1 does not regulate these essential components of the TGF-β signaling pathway (data not shown). Alternatively, it is possible that Fra-1 is downstream of an anabolic signaling pathway and can directly activate the expression of the matrix genes. This is supported by the presence of AP-1-binding sites in the human promoters of oc (Schule et al, 1990), col1a2 (Chung et al, 1996) and mgp (Farzaneh-Far et al, 2001). However, the upregulation of oc and col1a2 expression was less pronounced in the bones of fra-1-transgenic mice than that of mgp. This suggests that mgp might be a specific target gene of Fra-1, whereas oc and col1a2 are regulated indirectly by Fra-1.
Several reports have demonstrated that ectopic expression of Fra-1 can potentiate osteoclastogenesis (Owens et al, 1999; Jochum et al, 2000; Matsuo et al, 2000). Consistently, differentiation of fra-1Δ/Δ osteoclasts was reduced in vitro. This defect could not be rescued on wild-type, fra-1Δ/Δ or fra-1-transgenic osteoblast feeder layers, suggesting that the altered expression of matrix molecules on fra-1Δ/Δ and fra-1-transgenic osteoblasts had no effect on osteoclast differentiation. In particular, we have also tested the effect of Mgp on osteoclast differentiation. Addition of mouse serum from ApoE-Mgp-transgenic mice (Murshed et al, 2004), which was enriched for Mgp, rescued the differentiation of fra-1Δ/Δ osteoclasts. However, serum from non-transgenic control mice also rescued the defect, suggesting that a soluble factor different from Mgp can rescue osteoclast differentiation. In addition, plating the fra-1Δ/Δ osteoclast precursors on bone slices also abolished the differentiation defect. This might account for the unchanged osteoclast surface per bone surface in fra-1Δ/Δ mice, because osteoclasts differentiate in close proximity to the bone surface in vivo. The molecular nature of the signal(s) from the bone surface or the serum that rescues osteoclast differentiation and activity has not yet been identified.
In summary, these results demonstrate that Fra-1 is an important transcription factor and novel regulator of bone mass by affecting bone matrix production and maintaining osteoblast activity. Other Fos proteins such as ΔFosB might have related functions, which could explain the similarity between the osteosclerotic phenotypes of fra-1- and Δfosb-transgenic mice (Jochum et al, 2000; Sabatakos et al, 2000). Therefore, Fos proteins could be potential drug targets for the treatment of low bone mass diseases.
Materials and methods
Generation of mice with conditional fra-1 alleles
For the fra-1 targeting vector, a 6 kb SpeI/NotI fragment containing exons 3 and 4 of the fra-1 gene (Schreiber et al, 1997) was cloned into an artificial MCS of pBluescript II. The upper loxP sequence (followed by an artificial HindIII restriction site) was cloned into the EcoRI site of intron 3. The lower loxP sequence was part of a separately cloned DNA fragment that was inserted into the AflII site downstream of exon 4. This fragment contained a pGK-Neo selection cassette followed by the loxP sequence, a 0.2 kb splice acceptor sequence (splice acceptor of exon 3 of fra-1) and an EGFP reporter gene. For negative selection, the gene for diphtheria toxin was used. The targeting vector was used for homologous recombination in HM-1 embryonic stem cells. Positive clones were identified by PCR and Southern analysis and used for blastocyst injection.
Animals
Mice carrying the floxed fra-1 allele (fra-1f) were crossed to MORE-cre knock-in mice (Tallquist and Soriano, 2000). The genetic background of this intercross was C57Bl/6 × 129Sv. Mutant mice were intercrossed at least five times before bone histomorphometry to get a recombinant inbred strain. To avoid sex-specific differences of bone formation, only male mice were used in this study.
Southern, Northern and Western blot analysis
For Southern blots, 10 μg of genomic DNA was digested with EcoRV/HindIII yielding a 12.5 kb fragment for the wild-type fra-1 allele, a 4.5 kb fragment for the floxed fra-1 allele and an 11.5 kb fragment for the deleted fra-1 allele. For detection of the bands, a 0.8 kb AflII fragment downstream of exon 4 was used as a probe. For Northern blot experiments, 20 μg of total RNA was loaded and mRNA bands for mgp and tubulin were detected with labeled PCR products that have been amplified using cDNA from long bones. The PCR product for detection of mgp was cloned and sequenced before hybridization. Western blot analysis was performed according to standard procedures using anti-GFP and anti-actin antibodies (Roche).
RNase protection assay
Total RNA was isolated with the TRIZOL protocol (Sigma) and 10 μg was used for each RPA reaction. Long bones and calvariae were pulverized in liquid nitrogen prior to RNA isolation. RPA was performed using the RiboQuant multi-probe RPA systems mJun/Fos and mCK3b (PharMingen) according to the manufacturer's instructions.
Histology and in situ hybridization analysis
For histological analysis and ISH, bones were fixed overnight with neutral buffered 4% PFA at 4°C. The tissues were then decalcified for 10 days in 0.5 M EDTA and embedded in paraffin. Sections (5 μm) were stained with hematoxylin and eosin (H&E). For ISH analysis, sections were deparaffinized and mRNAs for runx2, oc, col1a2 and mgp were detected with a probe according to standard procedures.
TRAP staining and osteoclast activity assays
TRAP staining of osteoclasts was performed on deparaffinized bone sections using the leukocyte acid phosphatase kit (Sigma) according to the manufacturer's instructions. The activity of osteoclasts was assayed in vitro after differentiation on bovine cortical bone slices. The area of the resorption pits on the slices was quantified using MetaMorph software. Osteoclast activity in vivo was measured in urine samples of mice using Metra DPD EIA and Metra Creatinine Assay Kits (Quidel) according to the manufacturer's instructions.
PCR and real-time PCR analysis
PCR for genotyping of fra-1Δ/Δ mice was performed with primers P1 (gaaatggctccgtgggtaaaggta), P2 (gacagggttcatcttcatagttct) and P3 (tgtaccggacgcttgtcatctcat) giving rise to wild-type (308 bp), floxed (354 bp) and deleted (408 bp) bands. Deletion of fra-1 in fra-1Δ/Δ mice was analyzed by semiquantitative RT–PCR using the primers fra-1up (cccccgcaagctcaggcacagac) and fra-1do (gcagatggggcgatgggcttcc). For real-time PCR, light cycler Fast start DNA Master SYBR Green (Roche Diagnostics) was used. The following primers for osteoblast and osteoclast genes were used: RANK (420 bp) up: gcttacctgcccagtctcatcgtt, down: gggccggtccgtgtactcatcct; c-src (435 bp) up: gaaggcaggcaccaaactcagc, down: ccccggtaatcccccagcatca; runx2 (399 bp) up: cggagcggacgaggcaagagtttc, down: agacagcggcgtggtggagtggat; collagen1a1 (528 bp) up: ggtgcccccggtcttcag, down: agggccagggggtccagcatttc; mgp (309 bp) up: gcgagctaaagcccaaaagaga, down: tcaacccgcagaaggaagga; biglycan (407 bp) up: tgcccttatcctctggtgttctct, down: tagttgccccatatttgtttgtg; decorin (393 bp) up: gcccctaccgatgccagtgt, down: ggttgccgcccagttctatga; mt-1 (396 bp) up: tgagggtttccacggcgacagta, down: taggcggggtttttgggcttat; lrp5 (380 bp) up: ggcgctgtgacggcttccctgagt, down: gtagaccccgcccatgacgaagag; irs-1 (336 bp) up: cttggcgcagttacctcgtccttc, down: tgccgccacttcttctcgttctc; irs-2 (360 bp) up: ctgcaaccccgccacaacctatc, down: tgggcggctcatcacctcctc; c-abl (421 bp) up: ccccagacggcagcctaaatgaag, down: gaggccgaacaagaacgcaggaag; fosB (440 bp) up: catcccggtggccttcttgctca, down: ctgggggtggggtttgggattagg. The primer pairs for TRAP, cathepsin K, carbonic anhydrase II, MMP-9, tubulin, ALP, bone sialoprotein, osteocalcin, RANKL, OPG and collagen1a2 have been described previously (Kenner et al, 2004). The expression levels of RNA transcripts were calculated with the comparative CT method. The individual RNA levels were normalized for tubulin and depicted as relative expression levels with the corresponding controls (fra-1f/Δ) set to 1.
X-ray analysis, histology and histomorphometry
Mice were killed at 1.5 and 3 months of age. After whole animal contact radiography (Faxitron X-ray cabinet, Faxitron Co.) and autopsy, bones were dissected out and fixed in 3.7% PBS-buffered formaldehyde for 18 h at 4°C. After dehydration, the undecalcified tibiae and lumbar spines were embedded in methylmethacrylate, and 5 μm sections were cut in a sagittal plane on a rotation microtome (Cut 4060E, MicroTech) as previously described (Amling et al, 1999). Sections were stained with toluidine blue and modified von Kossa/van Gieson and were evaluated microscopically. For assessment of dynamic histomorphometric parameters, 12-μm-thick sections were mounted unstained in Fluoromount (Electron Microscopy Sciences) to permit evaluation by fluorescent microscopy.
Microcomputed tomography analysis
For three-dimensional histomorphometry and visualization of the vertebral bone structure, lumbar vertebrae L6 of 3-month-old mice was dissected out and scanned in a microCT 40 (Scanco Medical) at a resolution of 6 μm. The raw data were manually segmented and analyzed with μCT Evaluation Program V4.4A (Scanco Medical). For visualization, the segmented data were imported and displayed in μCT Ray V3.0 (Scanco Medical).
Quantitative histomorphometry
Quantitative histomorphometry was performed on toluidine blue-stained, undecalcified proximal tibia and lumbar vertebra sections. Experiments were performed in a blinded fashion. An analysis of bone histomorphometry parameters was carried out according to standardized protocols of the American Society for Bone and Mineral Research (Parfitt et al, 1987) using the Osteomeasure histomorphometry system (Osteometrix). For assessment of dynamic histomorphometric indices, mice were injected with calcein according to a standard double labeling protocol (Amling et al, 1999). Fluorochrome measurements were made on two nonconsecutive 12-μm-thick unstained sections per animal. Statistical analysis was performed using Student's t-test and P<0.05 was accepted as significant; error bars represent s.d.
Osteoblast and osteoclast cultures
Primary osteoblasts and osteoclasts were isolated and differentiated in vitro as described previously (Kenner et al, 2004). For ALP activity measurement, cells were washed with PBS and sonicated in 10 mM Tris–HCl buffer (pH 8.0). ALP activity in the osteoblast lysate was measured using a colorimetric assay (Sigma). Nodules of mineralized extracellular matrix were identified morphologically by alizarin red staining (Sigma). Osteoclast differentiation was evaluated by TRAP staining (leukocyte acid phosphatase kit from Sigma). To test resorption activity, co-cultures were performed on bovine cortical bone slices, and resorption pits were stained with toluidine blue. For co-culture experiments, primary osteoblasts (105 per well) were co-cultured with bone marrow cells (106 per well) in MEM containing 10% FCS, 10−8 M 1,25-dihydroxyvitamin D3 and 10−7 M dexamethasone in 24-well plates.
Acknowledgments
We thank Hans Christian Theussl for blastocyst injections, Hannes Tkadletz for help with the illustrations, Jean Pierre David for helpful discussions, Matthias Priemel for help with histomorphometry and Oskar Hoffmann for providing the bovine bone slices. We thank Phil Soriano for providing the MORE-cre mouse. We also thank Christine Hartmann, Koichi Matsuo and Tim Chambers for critical reading of the manuscript. We especially thank Monzur Murshed and Gerard Karsenty for providing the ApoE-Mgp mouse serum. The IMP is funded by Boehringer Ingelheim and this work was supported by the Austrian Industrial Research Promotion Fund.
References
- Akune T, Ogata N, Hoshi K, Kubota N, Terauchi Y, Tobe K, Takagi H, Azuma Y, Kadowaki T, Nakamura K, Kawaguchi H (2002) Insulin receptor substrate-2 maintains predominance of anabolic function over catabolic function of osteoblasts. J Cell Biol 159: 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amling M, Priemel M, Holzmann T, Chapin K, Rueger JM, Baron R, Demay MB (1999) Rescue of the skeletal phenotype of vitamin D receptor-ablated mice in the setting of normal mineral ion homeostasis: formal histomorphometric and biomechanical analyses. Endocrinology 140: 4982–4987 [DOI] [PubMed] [Google Scholar]
- Behrens A, Haigh J, Mechta-Grigoriou F, Nagy A, Yaniv M, Wagner EF (2003) Impaired intervertebral disc formation in the absence of Jun. Development 130: 103–109 [DOI] [PubMed] [Google Scholar]
- Boyle WJ, Simonet WS, Lacey DL (2003) Osteoclast differentiation and activation. Nature 423: 337–342 [DOI] [PubMed] [Google Scholar]
- Chung KY, Agarwal A, Uitto J, Mauviel A (1996) An AP-1 binding sequence is essential for regulation of the human alpha2(I) collagen (COL1A2) promoter activity by transforming growth factor-beta. J Biol Chem 271: 3272–3278 [DOI] [PubMed] [Google Scholar]
- Ducy P, Desbois C, Boyce B, Pinero G, Story B, Dunstan C, Smith E, Bonadio J, Goldstein S, Gundberg C, Bradley A, Karsenty G (1996) Increased bone formation in osteocalcin-deficient mice. Nature 382: 448–452 [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89: 747–754 [DOI] [PubMed] [Google Scholar]
- Eferl R, Wagner EF (2003) AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 3: 859–868 [DOI] [PubMed] [Google Scholar]
- Farzaneh-Far A, Davies JD, Braam LA, Spronk HM, Proudfoot D, Chan SW, O'Shaughnessy KM, Weissberg PL, Vermeer C, Shanahan CM (2001) A polymorphism of the human matrix gamma-carboxyglutamic acid protein promoter alters binding of an activating protein-1 complex and is associated with altered transcription and serum levels. J Biol Chem 276: 32466–32473 [DOI] [PubMed] [Google Scholar]
- Fleischmann A, Hafezi F, Elliott C, Reme CE, Ruther U, Wagner EF (2000) Fra-1 replaces c-Fos-dependent functions in mice. Genes Dev 14: 2695–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia T, Roman-Roman S, Jackson A, Theilhaber J, Connolly T, Spinella-Jaegle S, Kawai S, Courtois B, Bushnell S, Auberval M, Call K, Baron R (2002) Behavior of osteoblast, adipocyte, and myoblast markers in genome-wide expression analysis of mouse calvaria primary osteoblasts in vitro. Bone 31: 205–211 [DOI] [PubMed] [Google Scholar]
- Grigoriadis AE, Schellander K, Wang ZQ, Wagner EF (1993) Osteoblasts are target cells for transformation in c-fos transgenic mice. J Cell Biol 122: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriadis AE, Wang ZQ, Cecchini MG, Hofstetter W, Felix R, Fleisch HA, Wagner EF (1994) c-Fos: a key regulator of osteoclast–macrophage lineage determination and bone remodeling. Science 266: 443–448 [DOI] [PubMed] [Google Scholar]
- Harada S, Rodan GA (2003) Control of osteoblast function and regulation of bone mass. Nature 423: 349–355 [DOI] [PubMed] [Google Scholar]
- Holmbeck K, Bianco P, Caterina J, Yamada S, Kromer M, Kuznetsov SA, Mankani M, Robey PG, Poole AR, Pidoux I, Ward JM, Birkedal-Hansen H (1999) MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell 99: 81–92 [DOI] [PubMed] [Google Scholar]
- Jochum W, David JP, Elliott C, Wutz A, Plenk H Jr, Matsuo K, Wagner EF (2000) Increased bone formation and osteosclerosis in mice overexpressing the transcription factor Fra-1. Nat Med 6: 980–984 [DOI] [PubMed] [Google Scholar]
- Karsenty G, Wagner EF (2002) Reaching a genetic and molecular understanding of skeletal development. Dev Cell 2: 389–406 [DOI] [PubMed] [Google Scholar]
- Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA II, Hartmann C, Li L, Hwang TH, Brayton CF, Lang RA, Karsenty G, Chan L (2002) Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol 157: 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenner L, Hoebertz A, Beil T, Keon N, Karreth F, Eferl R, Scheuch H, Szremska A, Amling M, Schorpp-Kistner M, Angel P, Wagner EF (2004) Mice lacking JunB are osteopenic due to cell-autonomous osteoblast and osteoclast defects. J Cell Biol 164: 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Boast S, de los Santos K, Schieren I, Quiroz M, Teitelbaum SL, Tondravi MM, Goff SP (2000) Mice deficient in Abl are osteoporotic and have defects in osteoblast maturation. Nat Genet 24: 304–308 [DOI] [PubMed] [Google Scholar]
- Luo G, D'Souza R, Hogue D, Karsenty G (1995) The matrix Gla protein gene is a marker of the chondrogenesis cell lineage during mouse development. J Bone Miner Res 10: 325–334 [DOI] [PubMed] [Google Scholar]
- Luo G, Ducy P, McKee MD, Pinero GJ, Loyer E, Behringer RR, Karsenty G (1997) Spontaneous calcification of arteries and cartilage in mice lacking matrix GLA protein. Nature 386: 78–81 [DOI] [PubMed] [Google Scholar]
- Matsuo K, Owens JM, Tonko M, Elliott C, Chambers TJ, Wagner EF (2000) Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat Genet 24: 184–187 [DOI] [PubMed] [Google Scholar]
- McCabe LR, Banerjee C, Kundu R, Harrison RJ, Dobner PR, Stein JL, Lian JB, Stein GS (1996) Developmental expression and activities of specific fos and jun proteins are functionally related to osteoblast maturation: role of Fra-2 and Jun D during differentiation. Endocrinology 137: 4398–4408 [DOI] [PubMed] [Google Scholar]
- McCabe LR, Kockx M, Lian J, Stein J, Stein G (1995) Selective expression of fos- and jun-related genes during osteoblast proliferation and differentiation. Exp Cell Res 218: 255–262 [DOI] [PubMed] [Google Scholar]
- Murshed M, Schinke T, McKee MD, Karsenty G (2004) Extracellular matrix mineralization is regulated locally; different roles of two gla-containing proteins. J Cell Biol 165: 625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata N, Chikazu D, Kubota N, Terauchi Y, Tobe K, Azuma Y, Ohta T, Kadowaki T, Nakamura K, Kawaguchi H (2000) Insulin receptor substrate-1 in osteoblast is indispensable for maintaining bone turnover. J Clin Invest 105: 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens JM, Matsuo K, Nicholson GC, Wagner EF, Chambers TJ (1999) Fra-1 potentiates osteoclastic differentiation in osteoclast–macrophage precursor cell lines. J Cell Physiol 179: 170–178 [DOI] [PubMed] [Google Scholar]
- Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR (1987) Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res 2: 595–610 [DOI] [PubMed] [Google Scholar]
- Sabatakos G, Sims NA, Chen J, Aoki K, Kelz MB, Amling M, Bouali Y, Mukhopadhyay K, Ford K, Nestler EJ, Baron R (2000) Overexpression of DeltaFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat Med 6: 985–990 [DOI] [PubMed] [Google Scholar]
- Schreiber M, Poirier C, Franchi A, Kurzbauer R, Guenet JL, Carle GF, Wagner EF (1997) Structure and chromosomal assignment of the mouse fra-1 gene, and its exclusion as a candidate gene for oc (osteosclerosis). Oncogene 15: 1171–1178 [DOI] [PubMed] [Google Scholar]
- Schreiber M, Wang ZQ, Jochum W, Fetka I, Elliott C, Wagner EF (2000) Placental vascularisation requires the AP-1 component fra1. Development 127: 4937–4948 [DOI] [PubMed] [Google Scholar]
- Schule R, Umesono K, Mangelsdorf DJ, Bolado J, Pike JW, Evans RM (1990) Jun–Fos and receptors for vitamins A and D recognize a common response element in the human osteocalcin gene. Cell 61: 497–504 [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P (2000) Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis 26: 113–115 [DOI] [PubMed] [Google Scholar]
- Teitelbaum SL, Ross FP (2003) Genetic regulation of osteoclast development and function. Nat Rev Genet 4: 638–649 [DOI] [PubMed] [Google Scholar]
- Thiebaud D, Guenther HL, Porret A, Burckhardt P, Fleisch H, Hofstetter W (1994) Regulation of collagen type I and biglycan mRNA levels by hormones and growth factors in normal and immortalized osteoblastic cell lines. J Bone Miner Res 9: 1347–1354 [DOI] [PubMed] [Google Scholar]
- Vidal F, Sage J, Cuzin F, Rassoulzadegan M (1998) Cre expression in primary spermatocytes: a tool for genetic engineering of the germ line. Mol Reprod Dev 51: 274–280 [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Ovitt C, Grigoriadis AE, Mohle-Steinlein U, Ruther U, Wagner EF (1992) Bone and haematopoietic defects in mice lacking c-fos. Nature 360: 741–745 [DOI] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G (2004) ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology: implication for Coffin–Lowry syndrome. Cell 117: 387–398 [DOI] [PubMed] [Google Scholar]


