Abstract
Multi-layered defense responses are activated in plants upon recognition of invading pathogens. Transmembrane receptors recognize conserved pathogen-associated molecular patterns (PAMPs) and activate MAP kinase cascades, which regulate changes in gene expression to produce appropriate immune responses. For example, Arabidopsis MAP kinase 4 (MPK4) regulates the expression of a subset of defense genes via at least one WRKY transcription factor. We report here that MPK4 is found in complexes in vivo with PAT1, a component of the mRNA decapping machinery. PAT1 is also phosphorylated by MPK4 and, upon flagellin PAMP treatment, PAT1 accumulates and localizes to cytoplasmic processing (P) bodies which are sites for mRNA decay. Pat1 mutants exhibit dwarfism and de-repressed immunity dependent on the immune receptor SUMM2. Since mRNA decapping is a critical step in mRNA turnover, linking MPK4 to mRNA decay via PAT1 provides another mechanism by which MPK4 may rapidly instigate immune responses.
Keywords: decapping, immunity, MAP kinases, mRNA decay, phosphorylation
Introduction
Plant innate immunity employs multilayered defense responses comprised of two overlapping mechanisms. In the first layer, plant pattern recognition receptors detect invading microorganisms by the presence of conserved pathogen-associated molecular patterns (PAMPs)(Boller & Felix, 2009). PAMP recognition is exemplified by the binding of the bacterial flagellin-derived flg22 peptide to the leucine-rich repeat-receptor-like kinase flagellin sensing 2 (FLS2) (Gomez-Gomez et al, 2001; Chinchilla et al, 2006). PAMP recognition initiates downstream signaling, including production of reactive oxygen species, calcium influx, MAP kinase activation and global changes in gene expression that induce PAMP-triggered immunity (PTI) (Chisholm et al, 2006; Zipfel, 2009). Adapted pathogens have evolved effector proteins that are delivered into host cells to compromise PTI by evading PAMP detection or suppressing defense responses. In the second layer of immunity, plant resistance (R) proteins have evolved to directly or indirectly recognize the activities of pathogen effectors (Jones & Dangl, 2006). In the best-studied examples, R proteins are found to guard host proteins or complexes (guardees) manipulated by specific pathogen effectors. Pathogen detection via R proteins leads to induction of strong defenses and to a form of host programmed cell death known as the hypersensitive response (HR) to sequester infections. These responses are collectively termed effector-triggered immunity (ETI) (Jones & Dangl, 2006).
Changes in phosphorylation are important regulatory mechanisms in cellular signaling. Activation of mitogen-activated protein (MAP) kinases occurs within 10 min of PAMP application (Asai et al, 2002; Boller & Felix, 2009) and relies on sequential phosphorylations between MAPKK-kinases (MEKK), MAPK kinases (MKK) and MAP kinases (MPK) (Pitzschke et al, 2009; Andreasson & Ellis, 2010; Rasmussen et al, 2012). In Arabidopsis, several MPKs are activated by PAMPs. A cascade comprising MEKK1-MKK4/5-MPK3/6 was initially found to be involved in PTI downstream of FLS2 (Asai et al, 2002; Droillard et al, 2004; Gao et al, 2008). Similarly, flg22 treatment activates another cascade including MEKK1, MKK1/2 and MPK4 (Petersen et al, 2000; Ichimura et al, 2006; Mészáros et al, 2006; Suarez-Rodriguez et al, 2007; Gao et al, 2008; Qiu et al, 2008a). Interestingly, Flg22-induced activation of MPK3, MPK4 and MPK6 is dependent on MKK1, while MPK3 and MPK6 are also activated by MKK4 (Mészáros et al, 2006). Thus, FLS2 activates two cascades, one with an unknown MEKK and MKK4/5-MPK3/6, the other with MEKK1-MKK1/2-MPK4. More recently, the closest homologue of MPK4, MPK11, was shown to also be activated by PAMPs (Bethke et al, 2012).
mpk4 mutants were originally found to exhibit autoimmunity, and MPK4 thus appeared to function genetically as a negative regulator of defense responses (Petersen et al, 2000; Droillard et al, 2004). However, MPK4 is activated in response to pathogens and PAMP elicitation (Droillard et al, 2004; Teige et al, 2004; Ichimura et al, 2006; Brader et al, 2007; Qiu et al, 2008a) which is counter-intuitive for a negative regulator. Subsequently, it was shown that activated MPK4 interacts with and phosphorylates MAP kinase substrate 1 (MKS1), bringing about the release of the transcription factor WRKY33 and induction of the expression of the PHYTOALEXIN DEFICIENT 3 (PAD3) gene required for biosynthesis of the antimicrobial camalexin (Andreasson et al, 2005; Qiu et al, 2008b). This illustrates how MPK4 functions as a positive regulator of PTI. Since it was recently reported that MPK4 is a target for manipulation by pathogen effectors (Zhang et al, 2007, 2012), and as mpk4 mutant phenotypes are partially suppressed by mutations in the R protein SUMM2 (suppressor or mkk1 mkk2) (Zhang et al, 2012), one model is that MPK4 is a PTI guardee whose absence triggers ETI.
Apart from regulation by transcription factors, mRNA translation and degradation also regulate gene expression, especially when they are reprogrammed to stabilize bulk mRNAs and favor mRNAs required for an appropriate stress response (Jiao et al, 2010; Munchel et al, 2011; Park et al, 2012; Ravet et al, 2012). mRNA is intrinsically unstable to facilitate rapid changes in mRNA abundance in response to stimuli. Eukaryotic mRNAs contain stability determinants including the 5′ 7-methylguanosine triphosphate cap (m7G) and the 3′ poly(A) tail. To achieve fine control of transcript abundance, mRNA is attacked by a decapping complex that breaks down RNA in a coordinated manner. mRNA decay is initiated by deadenylation, the removal of the 3′ poly(A) tail, whereafter mRNA can either be routed to 3′–5′ degradation by exosomal exonucleases, or 5′–3′ by the exoribonuclease activity of the decapping complex (Garneau et al, 2007). The decapping complex is highly conserved among eukaryotes (Garneau et al, 2007; Xu & Chua, 2011) and comprises the decapping enzyme DCP1/2 which removes the 5′ m7G cap, and the exoribonuclease XRN that degrades the monophosphorylated mRNA. Deadenylation and decapping are linked in eukaryotes by the decapping enhancer PAT1 (protein associated with topoisomerase II) (Hatfield et al, 1996; Wang et al, 1996; Tharun & Parker, 2001; Ozgur et al, 2010). In yeast, PAT1 promotes the interaction between mRNA and decapping enzyme, altering the messenger ribonucleoprotein (mRNP) organization from an arrangement favoring translation to one promoting decay. mRNA decay occurs in distinct cytoplasmic foci called processing bodies (PBs) in eukaryotes (Parker & Sheth, 2007; Balagopal & Parker, 2009), where mRNPs are sequestered for degradation, or from which they may re-enter polysomal complexes (Brengues, 2005). PBs can also harbor translationally arrested mRNPs and RNA silencing machinery (Balagopal & Parker, 2009; Xu & Chua, 2009; Thomas et al, 2011).
In plants, the decapping machinery includes the decapping enzyme DCP2 and its activators DCP1, DCP5, VARICOSE (VCS) (Deyholos et al, 2003; Xu et al, 2006; Goeres et al, 2007; Brodersen et al, 2008; Xu & Chua, 2009, 2011; Motomura et al, 2012) as well as the exoribonuclease XRN4 (Olmedo et al, 2006; Potuschak et al, 2006; Gregory et al, 2008; Rymarquis et al, 2011; Vogel et al, 2011). It is thought that DCP1, DHH1 and DCP5 form mRNPs for translational repression of target mRNA, which are then subject to decapping by recruitment of DCP2 and VCS and digestion by XRN4 (Xu & Chua, 2009, 2011). Importantly, although PAT1 functions have not previously been studied in plants, three homologues are encoded in the Arabidopsis genome, each of which contains a conserved C-terminal domain. The decapping activator Sm-like (LSM) proteins, which interact with PAT1 in eukaryotes (Salgado-Garrido et al, 1999; Bonnerot et al, 2000; Bouveret et al, 2000; Tharun et al, 2000; Tharun, 2009), have recently been characterized in Arabidopsis (Perea-Resa et al, 2012; Golisz et al, 2013). It was found that LSM1-7 proteins form a complex and lsm1 mutants accumulate capped mRNA. Furthermore, VCS, DHH1 and PAT1 homologs were identified in LSM1 immunoprecipitates (Golisz et al, 2013).
The decapping complex plays important roles in eukaryotic development. In contrast, links between mRNA decapping and stress signaling are just being uncovered (Jiao et al, 2010; Buchan et al, 2011; Munchel et al, 2011; Park et al, 2012), and how decapping may be involved in regulating immune systems is largely unknown. Here, we characterize the Arabidopsis homologue of the mRNA decay regulator PAT1. We show that PAT1 functions in decapping of mRNA. Furthermore, PAT1 is phosphorylated in response to flg22 and localizes to discrete, punctate foci in the cytosol. PAT1 also interacts with MPK4 and with the R protein SUMM2 in planta, and the absence of PAT1 triggers SUMM2 dependent immunity. This indicates that PAT1 is regulated by MPK4 in a pathway whose disruption leads to ETI via SUMM2.
Results
AtPAT1 is an ScPAT1 orthologue and interacts with MPK4 in planta
We previously conducted a yeast two-hybrid screen to identify Arabidopsis proteins that interact with MPK4 (Andreasson et al, 2005). In addition to the MPK4 substrate MKS1, we identified two clones encoding PAT1 (At1g79090), an mRNA decapping stimulator involved in post-transcriptional gene regulation (Coller & Parker, 2005). Two PAT1 homologs are encoded in the Arabidopsis genome (AtPAT1H1 At3g22270; AtPAT1H2 At4g14990, Supplementary Fig S1A). The steady-state expression level of PAT1 and the homologs compared to the housekeeping gene ACTIN8 (At1g49240) was analyzed by RT-PCR (Supplementary Fig S1B). We focused on PAT1 as the other two homologs were not identified by yeast two-hybrids. To confirm the PAT1-MPK4 interaction in planta, doubly transgenic Arabidopsis lines were generated in the Ler mpk4-1 background that expressed PAT1 with a C-terminal Myc tag and HA-tagged MPK4 under the control of their own promoters. Anti-Myc immunoprecipitation from either MPK4-HA or double transgenic MPK4-HA/Pat1-Myc tissue detected a 50 kDa band corresponding to MPK4-HA only in double transgenic lines (Fig1A). Thus, MPK4 and PAT1 can be found in complex in Arabidopsis.
Figure 1.
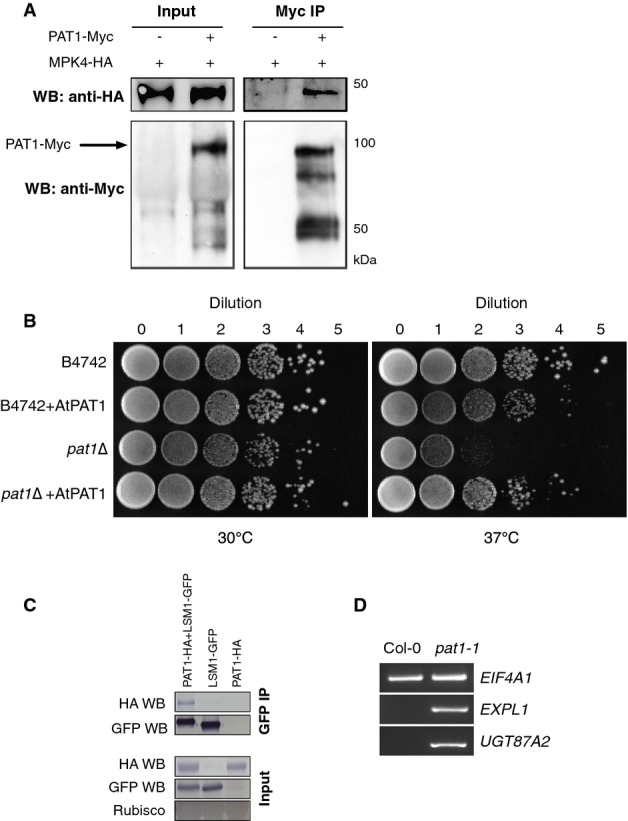
MPK4 and PAT1 interact in Arabidopsis
- A MPK4-HA is detected in PAT1-Myc immunoprecipitates from double transgenic Arabidopsis plants. Immunoblots of input and anti-Myc IPs probed with anti-HA and anti-Myc antibodies. Left panel, input; right panel, anti-Myc IP.
- B Yeast pat1Δ mutants are unable to grow at 37°C while wild-type (B4742) strain grows at 30°C (left panel) and 37°C (right). Growth at 37°C is restored in pat1Δ expressing AtPAT1. Serial dilutions of each yeast strain were plated on YPAD agar plates and grown at the indicated temperatures.
- C Co-IP between PAT1-HA and LSM1-GFP. Proteins were transiently co-expressed in N. benthamiana and tissue harvested 3 days post-infiltration. Immunoblots of GFP IPs (top panel) and inputs (20 μg each, bottom panel). Immunoblots were cut in half and probed with anti-HA antibodies and anti-GFP antibodies.
- D Agarose gel electrophoresis of 5′ RACE PCR to detect capped transcripts of EIF4A1, UGT87A2 and EXPL1 in 14-day-old Col-0 and pat1-1.
Source data are available online for this figure.
Yeast PAT1 engages with translating mRNPs and is involved in translational repression and decapping activation (Marnef & Standart, 2010). Since the function of PAT1 in Arabidopsis was unknown, we examined whether it functions similarly to yeast PAT1. To this end, a full-length Arabidopsis PAT1 cDNA was cloned from Col-0 (Supplementary Fig S1C and D) and transformed into wild-type yeast (B4742) and a yeast mutant (Y15797) in which yeast PAT1 was replaced with a G418 resistance cassette (BY4742 (YCR077c) pat1Δ::KanMX). In contrast to the wild-type, yeast lacking PAT1 (pat1Δ) display a temperature-sensitive phenotype and are impaired at 37°C but grow normally at 30°C (Tharun et al, 2005). This phenotype is reverted to wild-type in yeast containing Arabidopsis PAT1, as growth at 37°C was restored (Fig1B). As an additional control, we transformed Arabidopsis PAT1 into wild-type yeast (B4742/AtPAT1), and this grew similarly to wild-type at 30°C and almost as well at the wild-type at 37°C (Fig1B). The expression of Arabidopsis PAT1 in yeast was confirmed by anti-PAT1 immunoblotting of yeast protein extracts (Supplementary Fig S1E). This provides compelling evidence for the orthologous functions of these yeast and Arabidopsis PAT1 proteins. As PAT1 is found in complex with MPK4, these results provide a link between MPK4 and post-transcriptional regulation of mRNA stability.
We next analyzed the interaction between PAT1 and conserved components of mRNA decapping. PAT1-LSM1-7 complexes function in mRNA decapping and deadenylation (Bouveret et al, 2000; Tharun, 2009; Haas et al, 2010; Ozgur et al, 2010; Totaro et al, 2011). We therefore transiently expressed in Nicotiana benthamiana LSM1-GFP and PAT1-HA and then immunoprecipitated LSM1 with GFP Trap beads. PAT1-HA could be detected in LSM1 immunoprecipitates but did not adhere to GFP Trap beads in the absence of LSM1-GFP (Fig1C). This is consistent with the detection of peptides corresponding to PAT1 and its homologues in LSM1 immunoprecipitates (Golisz et al, 2013) and supports a role of PAT1 in mRNA decapping. In other organisms, interactions between PAT1 and LSM1 are robust, while those between PAT1 and other mRNA decapping proteins, including the DCP1-DCP2 complex and XRN1, are more transient (Bouveret et al, 2000; Nissan et al, 2010; Ozgur et al, 2010). This is consistent with our difficulty in detecting DCP1 in complex with PAT1 in Arabidopsis (Supplementary Fig S1F).
PAT1 is required for decapping of selected mRNAs
In order to determine whether PAT1 behaves as an activator of mRNA decapping, we used 5′ RACE to compare the levels of capped mRNAs in Col-0 and pat1 mutants. To this end, we identified an allele, pat1-1 (Salk_040660), with a T-DNA insertion in the last exon of PAT1 (Supplementary Fig S1C). We also generated an anti-PAT1 antibody against a C-terminal peptide (Supplementary Fig S1D). Immunoblotting of Col-0 protein extracts with this antibody detected a clear band around 90 kDa. In contrast, no protein could be detected in pat1-1 mutant extracts (Supplementary Fig S1G). This indicates that pat1-1 harbors either a truncated version of PAT1, no PAT1 protein, or levels of the protein that are below detection.
5′ RACE was performed on transcripts known to be degraded by the decapping complex (EXPL1; UGT87A2) (Perea-Resa et al, 2012), as well as a housekeeping transcript EIF4A1. We found that capped EXPL1 and UGT87A2 accumulated in pat1-1 mutants, while capped EIF4A1 mRNA was present in equal amounts in Col-0 and pat1-1 (Fig1D). This indicates that PAT1 plays a role in mRNA decay via decapping.
PAT1 is an MPK4 substrate
Since MPK4 and PAT1 are found in complexes in planta, we asked whether PAT1 is an MPK substrate. PAT1 contains 5 Ser-Pro (SP) motifs which are commonly phosphorylated by MPKs (Pearson et al, 2001; Ubersax & Ferrell, 2007) (Supplementary Fig S1D). To characterize PAT1 phosphorylation in vivo, we identified PAT1 phosphopeptides by mass spectrometry. Since MPK3/4/6 are activated by flagellin and by virulent strains of Pseudomonas syringae pv. tomato (Pto) DC3000 (Asai et al, 2002; Brader et al, 2007; Suarez-Rodriguez et al, 2007; Bethke et al, 2009, 2012; Rasmussen et al, 2012), PAT1 was immunoprecipitated from extracts of untreated control and flg22-treated wild-type Col-0 or PAT1-GFP transgenic lines. Bands corresponding to PAT1-GFP (130 kDa) were excised from the gel (Supplementary Fig S2), subjected to in-gel tryptic digestion, and peptides were extracted. Phosphopeptides were enriched by TiO2 chromatography and analyzed by liquid chromatography (RP-HPLC) coupled to electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS). This identified several phosphopeptides from PAT1-GFP IPs that were not detectable in the negative control (Col-0) (Table1; Supplementary Fig S2). The most abundant phosphopeptide, based on the extracted ion chromatogram from the LC-MS/MS analysis, revealed phosphorylation of Ser208 in an SP motif (Fig2A). Another peptide was identified with phosphorylation of Ser343. However, this site is not within an SP motif, making it a less likely site for phosphorylation by MPKs. Importantly, both these peptides have previously been detected in Arabidopsis by mass spectrometry (Phosphat Database, http://phosphat.mpimp-golm.mpg.de/). It should be noted that the PAT1 phosphopeptides were detected both whether or not the sample had been treated with flg22. Thus, under the conditions used here, PAT1 was phosphorylated and remained so after exposure to flg22.
Table 1.
Phosphopeptides identified in PAT1-GFP IPs by mass spectrometry analysis
| Sequence | Phospho-sitesa | m/z | Mascot score | |
|---|---|---|---|---|
| PAT1GFP water | PAT1GFP flg22 | |||
| SSFVSYPPPGSISPDQR | 1 (S208) | 950.93005 | 71 | 79 |
| SSFVSYPPPGSISPDQR | 2 (S208, S200) | 990.91333 | 46 | 64 |
| SSSGNYDGMLGFGDLR | 1 (S343) | 929.87323 | 70 | 114 |
| SSSGNYDGMLGFGDLR | 2 (S343, S342) | 969.85663 | 39 | 50 |
Phosphosites refers to the number and in brackets the location of the phosphorylation detected by MS analysis. Potential phosphorylation sites are indicated in bold letters. m/z refers to the mass-to-charge ratio of the tabulated peptides.
Figure 2.
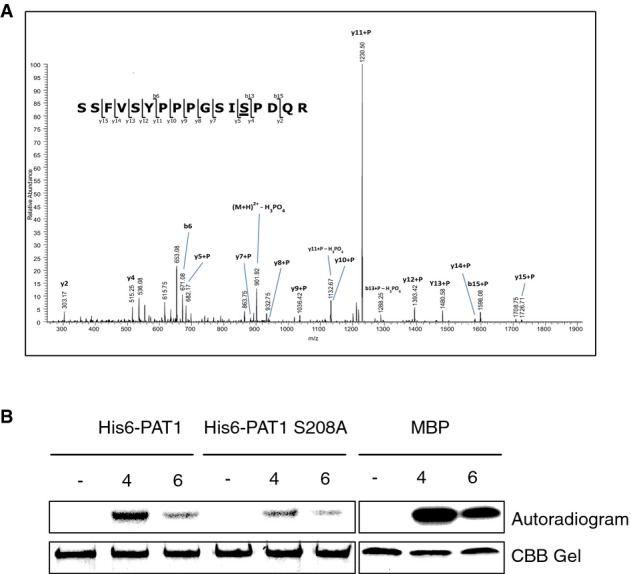
PAT1 is phosphorylated in planta and in vitro
- A Tandem MS spectrum of the phosphopeptide SSFVSYPPPGSISPDQR in which the underlined serine is phosphorylated. Y- and b-ions are indicated that localize phosphorylation to the SP site.
- B MPK4 and MPK6 were immunoprecipitated from extracts of Col-0 seedlings treated with 200 nM flg22 for 10 min. For the negative control (−), extracts were incubated with agarose beads without antibodies. IPs were incubated with His6-PAT1, His6-PAT1 S208A or MBP for 60 min at 37°C before boiling and SDS-PAGE. Autoradiogram (top panel), Coomassie-stained gel for loading control (bottom).
Source data are available online for this figure.
To determine whether PAT1 is a substrate of a specific MPK, we carried out in vitro kinase assays using immunoprecipitated, PAMP-activated MPKs with purified His6-PAT1 protein. Phosphorylated His6-PAT1 was detectable as a radioactive band around 95 kDa after incubation with flg22-activated MPK4 (Fig2B), while MPK6 caused only low levels of PAT1 phosphorylation and MPK3 did not significantly alter PAT1 phosphorylation (Supplementary Fig S3). Each MPK was also incubated with the generic MPK substrate myelin basic protein (MBP) to verify their activation (Fig2B; Supplementary Fig S3). This confirms that activated MPK4, and to a lesser extent, MPK6, is responsible for the phosphorylation of PAT1. A version of His6-PAT1 with Ser208 mutated to alanine (S208A) was also incubated with MPK4 and MPK6 IPs, and this mutant had significantly lower levels of phosphorylation (Fig2B). This supports the identification of S208 as a key phosphorylation site in PAT1.
pat1 mutants exhibit autoimmunity similar to mpk4
Given that MPK4 is an important immune regulator in Arabidopsis and that mpk4 mutants have de-repressed defense responses, we examined whether PAT1 may also be involved in immunity. The pat1-1 mutant has a distinct leaf serration phenotype and a slightly smaller rosette than Col-0 (Fig3). A similar rosette phenotype was seen in another T-DNA insertion line (pat1-2, WiscDsLox_734_D04 in Supplementary Figs S1C and S4), indicating that this phenotype is due to loss-of-function of PAT1. Importantly, however, the phenotype of pat1-1 is not as extreme as mpk4-2, which is much smaller and has more pronounced leaf curling and reduced fertility as well as constitutive defense gene expression (Petersen et al, 2000); Fig3).
Figure 3.

pat1 mutants have a serrated leaf, semi-dwarf phenotype
Plants photographed at 4 weeks of growth in short day conditions, genotypes as indicated. Col-0 and eds1-2 plants and pat1-1 and pat1-1/eds1-2, respectively, were placed in the middle of the same pots before the pictures were taken.
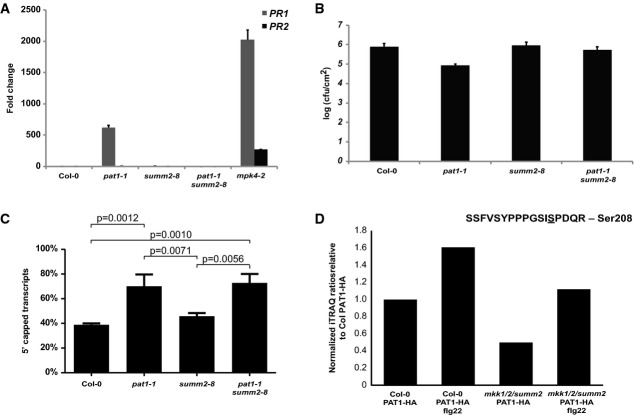
In order to determine whether pat1-1 is a constitutive defense mutant similar to mpk4, quantitative reverse-transcription PCR (qRT-PCR) was used to measure the steady-state level of the pathogenesis-related PR1 and PR2 genes in adult plants (Fig4A). Previous work showed that the enhanced defense response of mpk4 mutants is suppressed by mutations in EDS1 (Brodersen et al, 2006). Thus, eds1-2 was crossed with pat1-1 to explore the pat1 phenotype in the absence of this regulator (Fig3). Compared to Col-0, pat1-1 mutants accumulated 1,000-fold more PR1 and 150-fold more PR2 transcripts (Fig4A). In contrast, the levels of PR mRNAs in pat1-1 eds1-2 double mutants were similar to those in wild-type (Fig4A). Under the same conditions, mpk4-2 mutants accumulated 4,000-fold more PR1 and 800-fold more PR2 transcripts (Fig4A).
Figure 4.
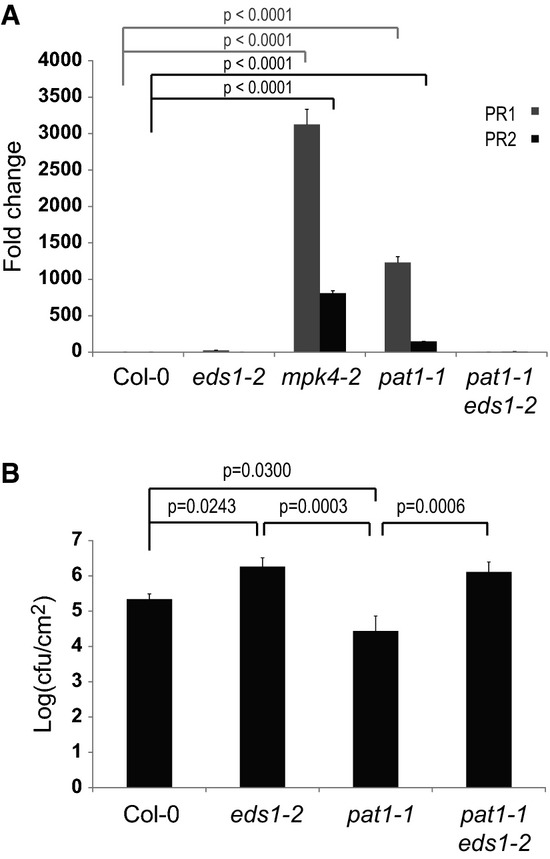
pat1 mutants display EDS1-dependent, constitutive defense responses
- A PR gene expression is elevated in mpk4-2 and pat1-1 mutants. Four-week-old plants were used for RNA extraction, followed by qRT-PCR. Fold-change in PR1 (gray bars) and PR2 (black) expression is relative to Col-0, normalized to UBQ10. Standard error of the mean is indicated by errors bars (n = 3). Statistical significance between the mean values was determined by ANOVA followed by Fisher's LSD test, P-values are only shown for mean values significantly different from Col-0.
- B pat1 is more resistant to colonization by P. syringae pv. tomato DC3000. Bacteria were syringe-infiltrated and samples taken 3 days post-infiltration. Data are shown as log10-transformed colony-forming units/cm2 leaf tissue (cfu/cm2). Standard error of the mean is indicated by errors bars (n = 4). Statistical significance between the mean values was determined by ANOVA followed by Fisher′s LSD test.
We found that the elevated PR gene expression in pat1-1 correlated to an enhanced resistance to infection by syringe-infiltrated Pto DC3000 when compared to Col-0 (Fig4B). While Pto DC3000 growth reached 5.5 cfu/cm2 in Col-0, pat1-1 mutants supported tenfold lower accumulation of Pto DC3000. In this experiment, eds1-2 mutants showed enhanced bacterial growth as expected for this mutant with compromised defense responses (Fig4B). pat1 disease resistance is EDS1 dependent, as bacterial growth pat1-1 eds1-2 double mutants was similar to that in eds1-2 (Fig4B). These findings indicate that, similar to mpk4, pat1 mutants express EDS1-dependent autoimmunity in the absence of microbes.
PAT1 detection in P-bodies is induced by PAMPs
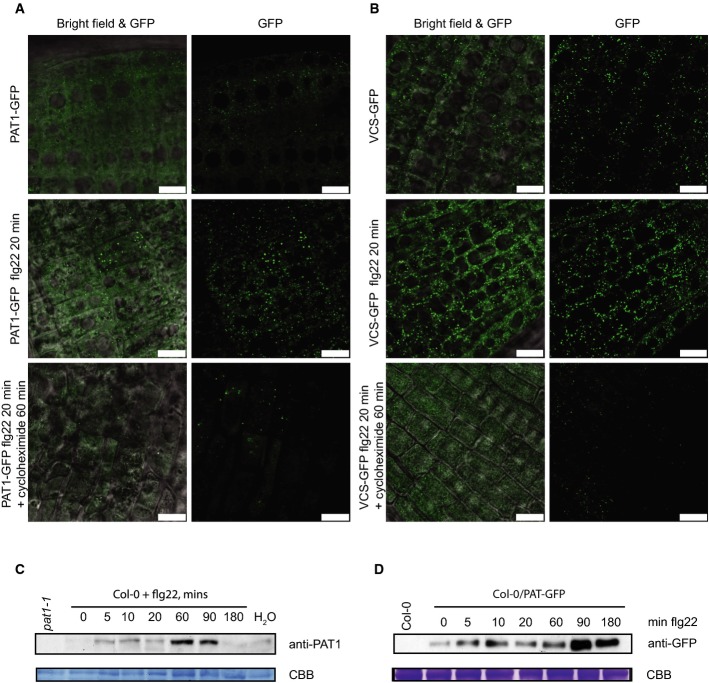
Processing bodies (PBs) are cytoplasmic granules involved in both mRNA decay and translational repression pathways (Kulkarni et al, 2010). PAT1 is a conserved PB component in yeast (Rodriguez-Cousino et al, 1995), C. elegans (Boag et al, 2008), Drosophila (Marnef et al, 2010) and mammals (Scheller et al, 2007) and PBs can be significantly induced in Arabidopsis by hypoxia and heat stress (Weber et al, 2008). To investigate whether PAT1 is found in PBs, we produced transgenic Arabidopsis lines that express PAT1-GFP from its native promoter complementing the pat1 phenotype (Supplementary Fig S5A and B). We detected GFP signal in small numbers of distinct foci by confocal microscopy in roots of young seedlings (Fig5A). Within 20 min of flg22 treatment, a significant increase in the number of GFP-positive foci could be seen in the root tips (Fig5A). To test whether these foci correspond to PBs, we treated the roots with cycloheximide in DMSO which is known to abrogate PB formation in plants (Goeres et al, 2007). This revealed that cycloheximide inhibited flg22-induced foci (Fig5A). Importantly, the control DMSO treatment did not reduce the number of foci (Supplementary Fig S6A and B). As a control, we also tracked the localization of the known decapping component VCS (Xu et al, 2006). Similar to PAT1-GFP, VCS-GFP was seen in flg22-induced PBs and absent when treated with cycloheximide (Fig5B). Interestingly, PAT1 protein, which is hardly detectable under steady-state conditions, also accumulated in response to flg22 treatment, with a peak by 60 min, and a return to normal levels after 2 h (Fig5C). PAT1-GFP mirrors this effect and was similarly up-regulated in response to flg22 treatment, as detected by anti-GFP Western blotting of seedling protein (Fig5D). Importantly, PAT1 transcript levels were not highly induced by flg22 treatment in Col-0 seedlings (Supplementary Fig S6C), suggesting that PAT1 induction occurs post-transcriptionally. These data indicate that activation of PTI leads to up-regulation of PAT1 protein levels by an unknown post-transcriptional mechanism, and this facilitates the detection of PAT1 in PBs. In addition, PTI could induce PB formation as part of cellular reprogramming, and PAT1 may localize to PBs to engage in this process.
Figure 5.
PAT1-GFP is present in P-bodies after PAMP treatment
- A Confocal microscopy with root elongation zones. Five-day-old Col-0/PAT1-GFP seedlings were treated with 1 μM flg22 on glass slides for 20 min (second panel) followed by treatment with 100 μg/ml cycloheximide for 60 min (bottom panel). The scale bar corresponds to 10 μm.
- B Confocal microscopy with 5-day-old VCS-GFP seedlings treated as in (A). The scale bar corresponds to 10 μm.
- C PAT1 protein is induced by PAMP treatment. Immunoblot detection of equal amounts of protein from Col-0 seedlings at times in minutes as indicated following vacuum infiltration with 1 μM flg22 or water (180 min after infiltration). Immunoblots were probed with anti-PAT1 antibodies. Negative control pat1-1 was loaded for comparison. Coomassie brilliant blue protein loading control is indicated by CBB.
- D Anti-GFP immunoblotting of proteins extracted from Col-0/PAT1-GFP seedlings treated over a time course with flg22 (top panel) and Coomassie brilliant blue (CBB)-stained PVDF loading control is shown (bottom panel). No PAT1-GFP was detected in negative control Col-0.
Source data are available online for this figure.
The MPK4 suppressor summ2 also suppresses the pat1 resistance phenotype
Autoimmunity caused by loss of different components of the MPK4 kinase cascade can be suppressed by mutations in the resistance protein SUMM2 (Zhang et al, 2012). An explanation for this is that SUMM2 keeps this PAMP responsive pathway under surveillance and that mutations in its components mimic the effects of microbial effectors that prevent phosphorylation within or below the cascade (Zhang et al, 2007). To further probe the connection between MPK4 and PAT1, we generated pat1-1 summ2-8 double mutants. These mutants retained the leaf serration phenotype of pat1-1 single mutants (Supplementary Fig S7A). However, double mutants no longer accumulated excessive PR1 and PR2 transcripts (Fig6A) and displayed summ2 levels of susceptibility to syringe-infiltrated Pto DC3000 (Fig6B). Importantly, the pat1 growth phenotype was not caused by overexpression of SUMM2 transcripts, as the level of SUMM2 in pat1-1 was similar to wild-type (Supplementary Fig S7B). To test whether the accumulation of 5′ capped transcripts in pat1-1 (Fig1D) was merely an effect of inappropriate activation of SUMM2, we tested the accumulation of 5′ capped UGT87A2 in an XRN1 sensitivity assay. XRN1 degrades all uncapped RNA leaving 5′ capped RNA intact (Blewett & Goldstrohm, 2012). The accumulation of 5′ capped versus uncapped UGT87A2 transcripts was at similar levels in pat1-1 and pat1-1 summ2-8 (Fig6C). Col-0 and summ2-8 plants had similarly reduced 5′ capped versus uncapped ratios but, importantly, these were much lower than the levels of pat1-1 and pat1-1 summ2-8 (Fig6C). Therefore, the accumulation of capped transcripts in pat1-1 mutants is not an artifact of defense activation but reflects a role for PAT1 in mRNA decay. In order to determine whether the SUMM2-mediated resistance in pat1 mutants is specific to pathogens of a specific class, we compared susceptibility to pathogens with different lifestyles. Pto DC300 is a hemi-biotrophic pathogen, which grows in living tissue during early infection. In contrast, the necrotrophic fungal pathogen Botrytis cinerea relies on dying tissue for its propagation in plants (Glazebrook, 2005). Infection of pat1-1 summ2 double mutants with B. cinerea resulted in similar growth of the fungus as detected in summ2-8 single mutants (Supplementary Fig S8).
Figure 6.
SUMM2 is required for the constitutive defense phenotype of pat1 mutants
- A Elevated PR gene expression in pat1 mutants is suppressed by summ2-8. Four-week-old plants were used for RNA extraction, followed by qRT-PCR. Fold-change in PR1 (gray bars) and PR2 (black bars) expression is relative to Col-0, normalized to UBQ10. Standard error of the mean is indicated by errors bars (n = 3).
- B pat1 resistance to P. syringae pv. tomato DC3000 is suppressed by summ2-8. Bacteria were syringe-infiltrated and samples taken 3 days post-infiltration. Data are log10-transformed colony-forming units/cm2 leaf tissue (cfu/cm2). Standard error of the mean is indicated by errors bars (n = 4).
- C 14-day-old seedlings grown on ms plates were used for RNA extraction. Next, 1 μg of RNA from each sample was treated with XRN1 (NEB) or mock treated before RT-qPCR. Data were normalized to ACT2, and transcript levels were compared between XRN1 and mock-treated RNA for each genotype. Standard error of the mean is indicated by error bars (n = 4). Statistical significance between the mean values was determined by ANOVA followed by Fisher's LSD test.
- D Phosphorylation of the PAT1 peptide SSFVSYPPPGSISPDQR, which include Ser208, in Col-0 and mkk1/1-summ2 before and after flg22 treatment. The phosphorylation stoichiometry is illustrated relative to Col PAT1 without flg22 treatment. The ratios were obtained using quantitative iTRAQ mass spectrometry.
To more accurately address to what extent PAT1 phosphorylation is MPK4 dependent and to measure PAT1 phosphorylation profiles before and after flg22 treatment, we generated PAT1-HA transgenic lines in the mkk1/2 summ2-8 background. The suppression of mpk4 by summ2-8 is only partial, but the autoimmunity phenotype is fully suppressed in mkk1/2 summ2 triple mutants. We next applied quantitative mass spectrometry (iTRAQ) to PAT1-HA IPs from Col-0 and mkk1/2-summ2 and this revealed increased phosphorylation stoichiometry of PAT1 at Ser208 in untreated Col-0 compared to mkk1/2 summ2 plants (Fig6D). Flg22 treatment leads to increased levels of PAT1 Ser208 in both genotypes, but the levels in Col-0 were higher than what we found for PAT1 in mkk1/2 summ2 (Fig6D). Since flg22 treatment augments PAT1 Ser208 phosphorylation in the absence of MKK1/2, MPK4 might be activated by other upstream kinases or PAT1 may be phosphorylated by MPK6. Nevertheless, these data indicate that flg22 treatment leads to increased phosphorylation of PAT1 at Ser208 and the MKK1/2 MPK4 pathway contributes to this phosphorylation.
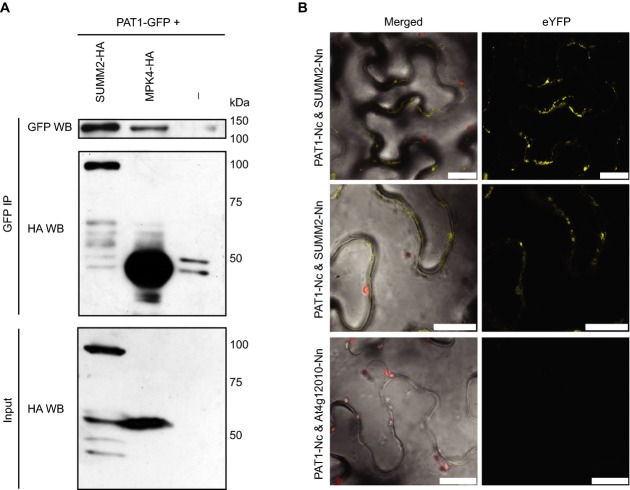
Our data indicate that PAT1 is part of the pathway including MPK4 and the SUMM2 R protein. SUMM2 and PAT1 might thus interact in planta. To test this, we transiently expressed and immunoprecipitated PAT1-GFP in N. benthamiana also transiently expressing MPK4-HA or SUMM2-HA. When PAT1-GFP was pulled down using GFP Trap beads, we could easily detect MPK4-HA and SUMM2-HA in the immunoprecipitates (Fig7A). To verify the association of MPK4, SUMM2 and LSM1, we also pulled down PAT1-HA and detected MPK4-GFP, SUMM2-GFP and LSM1-GFP in the immunoprecipitates but not Myc-GFP (Supplementary Fig S9). To further confirm the association of PAT1 and SUMM2, we used bimolecular fluorescence complementation (BiFC), which produces a fluorescent readout upon reconstruction of YFP. The BiFC assay showed detectable YFP signal in the cytoplasm when PAT1 and SUMM2 were co-expressed (Fig7B). No signals were observed when PAT1 was expressed with another R protein, At4g12010, implying that the association/reconstruction is specific (Fig7B, bottom panel). This indicates that PAT1 is in complex with SUMM2 in planta. Thus, MPK4 and PAT1 are both physically and genetically linked to SUMM2.
Figure 7.
PAT1 is associated with MPK4 and SUMM2 in N. benthamiana
- A Proteins were transiently co-expressed (PAT1-GFP + MPK4-HA or SUMM2-HA) in N. benthamiana and tissue harvested 2 days post-infiltration. Anti-HA immunoblots of inputs (20 μg each) are shown in the bottom panel. Immunoblots of GFP IPs were probed with anti-HA antibodies, then stripped and probed with anti-GFP antibodies. Molecular weights are shown in kDa on the right.
- B Confocal images of yellow fluorescent protein complementation. PAT1-Nc was transiently co-expressed with either SUMM2-Nn or At4g12010 in N. benthamiana. Confocal microscopy on leaf disks was conducted 2 days post-infiltration. Merged pictures show overlay of brightfield, chlorophyll and eYFP. The scale bar corresponds to 25 μm.
Source data are available online for this figure.
Discussion
In this study, we identify the Arabidopsis decapping component PAT1 and show it to be an interactor and substrate of MPK4 in plant innate immunity. Furthermore, we demonstrate that Arabidopsis PAT1 complements yeast pat1Δ mutants, indicating that the function of this conserved protein is maintained in plants. The accumulation of capped mRNAs in pat1 mutants is consistent with a role for PAT1 in the mRNA decapping pathway.
Pat1 mutants exhibit a distinct leaf serration phenotype (Supplementary Fig S4) resembling those of miRNA-loss-of-function mutants (Nikovics et al, 2006) such as abh1-8 (Gregory et al, 2008), serrate (Grigg et al, 2005) and hypomorphic alleles of ago1 (Morel, 2002). This suggests that PAT1 may have a role connected to microRNA activity. Arabidopsis decapping mutants such as dcp1, dcp2 and vcs accumulate lower levels of certain miRNAs (Motomura et al, 2012). Mutants with a pat1-like phenotype, such as vcs, suo6 and amp1, have revealed new components in miRNA-mediated translational repression (Brodersen et al, 2008; Yang et al, 2012; Li et al, 2013). In Drosophila, HPat interacts with components of the miRNA machinery including AGO1 and GW182 (Barišić-Jäger et al, 2013). The connection between the mRNA decay activator HPat and the miRNA effector complex may provide a link to promote the transition of mRNA from translation to degradation. Although PAT1 may regulate targets of miRNA-mediated translational repression, the mechanism by which translational repression occurs in Arabidopsis is still under investigation.
We also find here that PAT1 is an MPK4 substrate and that pat1 mutants exhibit autoimmunity as does mpk4. The connection between MPK4 and PAT1 is further supported by suppression of the pat1 constitutive defense phenotype by loss-of-function of the SUMM2 R protein (Fig6). However, SUMM2 deficiency only partially rescues mpk4 mutants (Zhang et al, 2012), thus it is possible that this partial rescue represents a SUMM2 PAT1 branch in the MPK4 pathway. Nevertheless, the pat1 constitutive defense phenotype is suppressed by summ2 such that pat1 summ2 mutants display a wild-type phenotype in response to biotrophic and necrotrophic pathogens (Fig6; Supplementary Fig S8). Since PAT1 and SUMM2 also interact in planta (Fig7), PAT1, or a PAT1-containing complex, is part of a pathway that includes SUMM2 as well as MPK4. It is therefore possible that PAT1 is under SUMM2 surveillance because it is an effector target with specific functions in immunity. Since pat1 summ2 mutants are not immunosuppressed, PAT1 and its homologues in Arabidopsis may function redundantly during PTI. An alternative explanation is that we simply have not yet tested a pathogen whose infection strategy could reveal a role of PAT1 in immunity.
mRNA decapping is not the only mRNA regulatory pathway characterized by constitutive defense responses. Indeed, mutants of nonsense-mediated decay including upf3-1, upf1-5 and smg7 mutants display autoimmune phenotypes (Jeong et al, 2011; Rayson et al, 2012a; Riehs-Kearnan et al, 2012; Shi et al, 2012). Their phenotypes are also suppressed by mutations in immune regulators such as eds1 and pad4 (Rayson et al, 2012b; Riehs-Kearnan et al, 2012), similar to what we find for pat1. This suggests that these components could also be under surveillance and may be suppressed by mutations in specific R genes. Recently, it was suggested that nonsense-mediated decay controls turnover of R gene mRNAs and cause autoimmunity in smg7 (Gloggnitzer et al, 2014). However, since autoimmunity in smg7 depends on a specific allele of the R protein RPS6, it is also possible that SMG7 is under surveillance. Most significantly, this suggests that plants have developed complex sensors to monitor the integrity of these pathways, which is consistent with the importance of differential gene expression in response to pathogen perception.
We detected PAT1 phosphorylation by PAMP-activated MPK4, and weakly also by MPK6 in vitro. We also identified PAT1 Ser208 and Ser343 as in planta phosphorylation sites by mass spectrometry. Since PAT1 phosphorylation was reduced in vitro when Ser208 was mutated to Ala, this site may be a key target of MPK4 (Fig2B). Interestingly, Ser208 is conserved in Physcomitrella patens (moss), rice, the Arabidopsis PAT1 homologues, and Xenopus PATL2, but not in human or yeast PAT1 (Supplementary Fig S10). Although Ser208 corresponds to an SP site in Xenopus PATL1/xPAT1b, PATL1 is known to be phosphorylated on Ser62 by an unknown kinase (Marnef et al, 2010). Thus, Ser208 may represent a plant-specific site or mechanism. While Ser208 is conserved in plant PAT1 orthologs, Ser342/3 is not. As Ser342/3 does not correspond to SP sites, they may be phosphorylated by kinases other than MPKs. Furthermore, we detected PAT1 phosphorylation in planta irrespective of PAMP treatment.
PAT1 phosphorylation by MPKs has not been shown in any system, although several phosphosites have been identified in human and yeast PAT1 proteins (PhosphoElm, http://phospho.elm.eu.org/ and Phosida http://www.phosida.com/databases). However, other decapping complex members are subject to stress-induced MPK-mediated phosphorylation. For example, human DCP1a is phosphorylated by c-Jun N-terminal kinase in response to stress (Rzeczkowski et al, 2011). Similarly, Ste20 phosphorylates yeast DCP2 upon glucose deprivation (Yoon et al, 2010). In both cases, phosphorylation seems only to be required for P-body formation and not for general decapping (Yoon et al, 2010; Rzeczkowski et al, 2011). Arabidopsis MPK6 was shown to specifically phosphorylate DCP1 in plants during dehydration stress (Xu & Chua, 2012). Thus, the regulation of mRNA decay machinery by MPKs during stress responses seems to be a key mechanism in plants and other organisms, although exactly how this affects mRNA turnover remains elusive.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used as a control. Seeds for T-DNA insertion lines were from NASC (Nottingham, UK). The T-DNA lines for At1g79090 (PAT1), both of which have insertions in the last exon, were Salk_040660 (here named pat1-1) and WiscDsLox437D04 (pat1-2). The T-DNA insertion in At1g12280 (SUMM2) summ2-8 (SAIL_1152A06), mkk1/2 (Zhang et al, 2012) and eds1-2 (Parker et al, 1996) have been described. Genotyping primers for newly described T-DNA lines are provided in Supplementary Table S1. Arabidopsis plants were grown in 9 × 9 cm pots at 22°C with a 8-h photoperiod, or on plates containing Murashige–Skoog (MS) salts medium (Duchefa), 1% sucrose and 1% agar with a 16-h photoperiod.
Cloning and transgenic lines
The genomic PAT1 (At1g79090) DNA sequence (without stop codon), plus 2 kb upstream from the start codon, was amplified from Col-0 genomic DNA and cloned into pENTR-D-TOPO (Invitrogen). The entry clone was recombined into pGWB513, pGWB517 and pGWB504 (Nakagawa et al, 2007) to obtain a C-terminal HA, Myc and GFP tags, respectively. The expression clones were transformed into Agrobacterium tumefaciens strain GV3101 and transformed into Col-0 plants by floral dipping. Transformants were selected on hygromycin (30 μg/ml) MS agar and survivors tested for protein expression by Western blotting.
The VCS promoter region was amplified and inserted into pGEM-T easy and next into the HindIII SalI in pBN:GFP to generate pBNpVCS:GFP. The VCS coding region was amplified and cloned into pGEM-T easy next inserted into SalI and KpnI in pBNpVCS:GFP and transformed into Agrobacterium LBA4404 and transformed into Col-0 plants by floral dipping. The cDNA of LSM1 (At3g14080) without stop codon was cloned into pENTR-D-TOPO (Invitrogen) and recombined into 35S promoter-containing pGWB505 (Nakagawa et al, 2007) to obtain C-terminal GFP-tagged LSM1. Genomic DNA of MPK4 and cDNA of SUMM2 were cloned into pENTR-D-TOPO and recombined into pGWB514 (Nakagawa et al, 2007) to obtain C-terminal HA-tagged constructs. The genomic DNA and promoter of DCP1 was cloned into pENTR-D-TOPO and recombined into pGWB513 to obtain C-terminal HA-tagged constructs under the control of the native promoter. Clones were transformed into A. tumefaciens strain GV3101 and used for transient expression in N. benthamiana or floral dipping in Arabidopsis. Genomic PAT1-GFP was transformed into the Col-0 background. DCP1-HA was dipped into the Col-0 background and T3 progeny were crossed with Col-0/PAT1-GFP T4 lines to obtain double transgenic lines. Genomic PAT1-Myc was transformed into Ler mpk4-1/MPK4-HA transgenic lines (described in Petersen et al, 2000) to obtain double transgenic lines.
The entry clones of PAT1 and SUMM2 were further recombined into pcYGW and pnYGW (Hino et al, 2011), respectively, obtaining the N- and C-terminal split YFP tags used for BiFC. At4g12010 clones were obtained by the same procedure. Clones were transformed into A. tumefaciens strain GV3101 and used for transient expression in N. benthamiana.
Mutagenesis of His6-PAT1
Site-directed mutagenesis of Ser208 to Ala was accomplished using pET15b-PAT1 as a template for PCR mutagenesis. The primers used were PAT1 S208A F&R (see Supplementary Table S1).
PAT1 protein purification and in vitro kinase assays
For in vitro experiments, PAT1 protein was purified from E. coli. The PAT1 cDNA was cloned into pET15b (for an N-terminal His fusion) and transformed into E. coli BL21 (pLysS). Protein expression was induced by overnight treatment with 0.5 mM IPTG at 18°C, added to cells at OD600 = 0.6. PAT1 protein was insoluble and thus purified from inclusion bodies using Bugbuster (Novagen). Proteins were solubilized in 6 M urea, 0.7% N-lauroylsarcosine, 100 mM Tris-HCl pH 8 and refolded overnight at 4°C in 0.88 M L-arginine, 55 mM Tris-HCl, 2 mM NaCl, 0.88 mM KCl, protease inhibitors. Protein was then dialyzed against 20 mM Tris-HCl pH 8, 100 mM NaCl using 3,500 MWCO dialysis tubing. Purified protein was concentrated using Centriprep 30K spin columns (Millipore) and then diluted with glycerol to a final concentration of 0.1 mg/ml.
For kinase assays, Col-0 plants were immersed in 200 nM flg22 for 10 min of to activate MAP kinases. MPK3, 4 and 6 were immunoprecipitated from 3 mg total protein extracted from flg22-treated tissue using 2 μg of each of their specific antibodies (Sigma) and 30 μl EZview protein A agarose beads (Sigma). Four microgram of purified myelin basic protein (MBP, Sigma) or 20 μg His6-PAT1 protein was incubated with washed MPK immunoprecipitates for 60 min at 37°C with 3 μCi γ-ATP in kinase buffer (62.5 μM ATP, 100 mM Tris pH 7.5, 150 mM NaCl, 150 mM MgCl2, 10 mM EGTA, 5 mM DTT, Phosstop inhibitor (Roche)). Kinase reactions were diluted with 4× SDS buffer and boiled for 5 min before loading on 12% SDS-PAGE gels. Following electrophoresis, gels were stained with Coomassie Brilliant Blue and incubated with gel drying buffer, followed by drying on a Bio-Rad gel dryer. Dried gels were exposed to a phosphor screen overnight.
Yeast transformation
Yeast strains pat1Δ (BY4742 (YCR077c) pat1Δ::KanMX) and B4742 were obtained from Euroscarf. PAT1 was cloned from A. thaliana Col-0 cDNA into pENTR-D-TOPO and recombined into yeast expression vector pVV215 (C-terminal HA tag, -URA selection; (Van Mullem et al, 2003) by Gateway recombination. Yeast pat1Δ and B4742 cells were transformed using lithium acetate/polyethylene glycol according to the Clontech Yeast Protocols handbook. Transformed yeast was selected on SD-URA agar plates and re-streaked after 3–4 days onto fresh selection plates. Pat1Δ and wild-type-transformed yeast was grown in liquid culture overnight, and protein was extracted from pelleted cells by vortexing with glass beads in 1× SDS extraction buffer. Boiled proteins were subjected to SDS-PAGE and anti-PAT1 immunoblotting. For temperature sensitivity assays, overnight cultures of wild-type, pat1Δ and PAT1-expressing transformants were plated on YPAD and grown at 30 and 37°C for 3 days.
Flg22 kinetics
Seedlings were grown on MS agar (Col-0) or MS agar containing 30 μg/ml hygromycin (Col-0/PAT1-GFP) for 5 days before being transferred to MS liquid medium in 24-well plates. After 10 days, seedlings were treated by the addition of flg22 to a final concentration of 100 nM (2 seedlings per well × 3 wells per treatment time point). Seedlings were harvested at the indicated times and immediately frozen in liquid nitrogen for later RNA or protein extraction.
Semi-quantitative and qRT-PCR
Total RNA was extracted from seedlings with Tri Reagent (Sigma). RNA samples were treated with DNase Turbo DNA-free (Ambion), quantified with a NanoDrop spectrophotometer (Thermo Scientific) and reverse-transcribed into cDNA with SuperScript III reverse transcriptase (Invitrogen). For semi-quantitative reverse-transcription PCR (RT-PCR), Col-0 seedling cDNA was used as a template for PCR with primers specific for ACTIN8, PAT1, PAT1H1 or PAT1H2 using Sigma Jumpstart REDTaq Readymix. PCR products (20 μl) were separated on 2% (w/v) agarose gels and visualized with ethidium bromide. Brilliant II SybrGreen master mix (Agilent) was used for qPCRs. The UBQ10 (At4g05320) gene was used for normalization. Gene expression of PR1, PR2, PAT1 and SUMM2 was measured by qPCR analysis, normalized to UBQ10 expression and plotted relative to Col-0 expression level. These experiments were repeated in three independent biological replicates, each with three technical replicates, and representative data are shown. Standard error is represented by error bars on figures, and statistical significance is indicated by letters above error bars. These are derived from one-way ANOVA with Tukey's multiple comparison test (GraphPad Prism).
RNA extraction and RACE PCR
RNA extraction used TRI reagent and 10 μg RNA was used for 5′ RACE according to instructions (First Choice RACE, Ambion). PCR was carried out on 1 μl of products from reverse transcription of capped RNA using DreamTaq polymerase (Fermentas) with 25 cycles. RACE PCR products (10 μl) were separated on 2% (w/v) agarose gels and visualized with ethidium bromide.
Quantification of capped versus uncapped transcripts
Total RNA was extracted from seedlings with NucleoSpin RNA columns (Machery-Nagel). To remove RNA with no 5′ cap structure, 1 μg of total RNA was incubated with 1 unit XRN1 (New England Biolabs) or no enzyme at 37°C for 1 h. Next RNA was reverse-transcribed into cDNA with SuperScript III reverse transcriptase (Invitrogen). UGT87A2 transcript accumulation was measured by qPCR using SybrGreen master mix (Agilent) and normalized to ACT2. Calculating 5′ capped versus uncapped transcripts was done by comparing transcript levels from XRN1 and mock-treated samples for the individual genotypes.
Confocal microscopy
Col-0/PAT1-GFP plants were grown on MS agar containing 30 μg/ml hygromycin for 5 days. Seedlings were placed on glass microscope slides with water, 1% DMSO or 100 nM flg22 for 20 min. For following cycloheximide and DMSO treatment, seedlings were removed from flg22 and placed on new glass microscope slides. Cycloheximide was used at 100 μg/ml in a 1% DMSO dilution, and the control was done with 1% DMSO. Imaging was done using a Leica SP5 inverted microscope.
Infection assays
Pseudomonas syringae pv. tomato DC3000 (Pto DC3000) strains were grown in overnight culture in Kings B medium supplemented with appropriate antibiotics. Cells were harvested by centrifugation and pellets resuspended in sterile water to OD600 = 0.002. Bacteria were infiltrated with a needleless 1-mm syringe into four leaves on four plants per genotype and maintained in growth chambers for 3 days. Samples were taken using a cork-borer (6.5 mm) to cut leaf disks from four leaves per plant and four plants per genotype. Leaf disks were ground in water, serially diluted and plated on Kings B with appropriate selection. Plates were incubated at 28°C and colonies counted 2–3 days later. These experiments were repeated in three independent biological replicates, and representative data are shown.
For drop inoculation of B. cinerea (strain B05.10), 10 μl of 2.5 × 105 spores/ml in Gamborg B5/2% sucrose (pH 5.6) was placed on the adaxial surface of fully expanded leaves of 4-week-old plants and sampled by harvesting 10–15 leaf disks per genotype were at each time point, namely 0, 2 and 3 days. Data are the average of 3 biological replicates. Disease severity was measured as accumulation of the B. cinerea CUTINASE A transcript by qPCR relative to A. thaliana α-SHAGGY KINASE (At5g26751) (primers are according to Gachon & Saindrenan, 2004).
Transient expression in Nicotiana benthamiana
Agrobacterium tumefaciens strains were grown in LB medium supplemented with appropriate antibiotics overnight. Cultures were spun down and resuspended in 10 mM MgCl2 to OD600 = 0.8. A. tumefaciens strains carrying PAT1-GFP and MPK4-HA or SUMM2-HA were mixed 1:1 and syringe-infiltrated into 3-week-old N. benthamiana leaves. Samples for protein extraction were harvested 2 days post-infiltration (dpi). Agrobacterium tumefaciens strains carrying either PAT1-HA, LSM1-GFP or PAT1-HA + LSM1-GFP were mixed 1:1 and syringe-infiltrated into 3-week-old N. benthamiana leaves. Samples for protein extraction were harvested 3 dpi. For BiFC, A. tumefaciens strains carrying PAT1 fused to the N-terminal part of YFP and SUMM2 or At4g12010 fused to the C-terminal part were mixed 1:1 and syringe-infiltrated into N. benthamiana leaves at an final OD600 = 0.8. Confocal microscopy on leaf disks was conducted 2 days post-infiltration under a Leica SP5 inverted microscope.
Protein extraction and immunoprecipitation in Nicotiana benthamiana
Leaves were ground in liquid nitrogen and extraction buffer [50 mM Tris-HCl pH 7.5; 150 mM NaCl; 10% (v/v) glycerol; 10 mM DTT; 10 mM EDTA; 1% (w/v) PVP; protease inhibitor cocktail (Roche); 0.1% (v/v) IGEPAL CA-630 (Sigma); Phosstop (Roche) added at 2 ml/g tissue powder. Samples were clarified by 20 min centrifugation at 4°C and 13,000 rpm. Supernatants (1.5 ml) were adjusted to 2 mg/ml protein and incubated 2 h at 4°C with 20 μl GFPTrap-A beads (Chromotek) or anti-HA antibodies (2 μg, Santa Cruz) and 30 μl EZview protein A agarose beads (Sigma). Following incubation, beads were washed four times with IP buffer, before adding 30 μl 2× SDS and heating at 95°C for 5 min.
Arabidopsis protein extraction and immunoprecipitation for mass spectrometry analysis
Leaves from adult plants were ground in liquid nitrogen and extraction buffer [50 mM Tris-HCl pH 7.5; 150 mM NaCl; 10% (v/v) glycerol; 5 mM DTT; 2 mM EDTA; protease inhibitor cocktail (Roche); 0,1% (v/v) IGEPAL CA-630 (Sigma) and Phosstop (Roche) added at 2 ml/g tissue powder. Samples were clarified by 20 min centrifugation at 4°C 13,000 rpm. Supernatants (45 ml) were adjusted to 3 mg/ml protein and incubated 4 h at 4°C with 50 μl GFPTrap-A beads (Chromotek) or anti-HA antibodies (4 μg, Santa Cruz) and 100 μl EZview protein A agarose beads (Sigma). Following incubation, beads were washed four times with IP buffer before adding 2× SDS and heating to 95°C for 5 min.
SDS-PAGE and immunoblotting
SDS-PAGE gels were prepared with either 8, 10 or 12% cross-linking. Proteins were loaded and gels run at 100–150 V for 1.5 h before electroblotting onto PVDF membrane (GE Healthcare). Membranes were rinsed in TBS and blocked for 1 h in 5% (w/v) non-fat milk in TBS-Tween (0.1% (v/v)). Antibodies were diluted in blocking solution to the following dilutions: anti-GFP (AMS Biotechnology (rabbit) 1:5,000 or Santa Cruz (mouse) 1:1,000), anti-HA 1:1,000 (Santa Cruz). Membranes were incubated with primary antibodies for 1 h to overnight. Membranes were washed 3 × 10 min in TBS-T (0.1%) before 1-h incubation in secondary antibodies, anti-rabbit or anti-mouse-HRP or anti-rabbit or anti-mouse AP conjugate (Promega; 1: 5,000). Chemiluminescent substrate (ECL Plus, Pierce) was applied before exposure to film (AGFA CP-BU). For AP-conjugated primary antibodies, membranes were incubated in NBT/BCIP (Roche) until bands were visible. For probing immunoblots with multiple antibodies, stripping was carried out using Restore Western Blot Stripping Buffer (Pierce) for 15–30 min, following by three washes with TBS-Tween.
Antibodies
Polyclonal anti-PAT1 antibodies were generated by immunizing rabbits with synthetic peptides derived from the N-terminus [EQRIPDRTKLYPEPQ] and C-terminus [KRSMLGSQKTEPVLS] of PAT1. Antibodies (final bleed) were affinity-purified against the C-terminal peptide (Eurogentec). Antibody specificity was verified by immunoblotting with plant extracts derived from Col-0 and pat1-1 tissue. Mouse anti-HA and anti-GFP antibodies were obtained from Santa Cruz. Rabbit anti-GFP antibodies were obtained from AMS Biotechnology. Anti-MPK3, anti-MPK-4 and anti-MPK-6 antibodies were obtained from Sigma. Secondary antibodies were obtained from Promega.
In-gel digestion, TiO2 chromatography and mass spectrometry
Bands excised from SDS-PAGE were chopped into small pieces and incubated for 1 h in 50 mM TEAB, acetonitrile (50:50) on a shaker at room temperature. After incubation, the sample was centrifuged shortly and the supernatant was discarded. Gel pieces were dried in a vacuum centrifuge for 15 min and subsequently 30 μl of trypsin (10 ng/μl) in 20 mM TEAB pH 7.5 was added to cover the dried gel pieces. The solution was incubated in 4°C for 30 min. After incubation, the trypsin solution was replaced with 30 μl 20 mM TEAB pH 7.5 and the tube was incubated at 37°C overnight. Peptides from the digestion solution after incubation were recovered in a low binding Eppendorf tube (Sorensen Bioscience), and the gel pieces washed with 50% acetonitrile for 15 min to extract more peptides. The washing solution was mixed with the recovered peptides, and the peptide solution was lyophilized. Phosphopeptides were purified by titanium dioxide (TiO2) chromatography (Larsen et al, 2005). Lyophilized peptides were redissolved in loading buffer for TiO2 chromatography (80% acetonitrile, 5% TFA, 1 M glycolic acid), and 0.3 mg TiO2 beads (GL Science, Japan) were added to the solution and incubated for 10 min. After incubation, the beads were pelleted by centrifugation and the supernatant was removed. The beads were washed once with 80% acetonitrile, 1% TFA and once with 10% acetonitrile, 0.1% TFA. Phosphopeptides were eluted using 1% ammonium hydroxide and desalted and concentrated prior to LC-MS/MS using a Poros Oligo R3 micro-column (Engholm-Keller et al, 2012).
For quantitative phosphopeptide analysis, the tryptic peptide mixtures after in-gel digestion were labeled with iTRAQ 4 plex (according to the manufacturer's protocol) and the samples were mixed prior to TiO2 enrichment as described above. The non-phosphorylated peptides were concentrated and desalted using a Poros Oligo R3 micro-column (Engholm-Keller et al, 2013). The non-phosphorylated peptides were used to normalize the PAT1 protein level.
LC-MS/MS analysis was performed using an EASY-LC system (Proxeon, Thermo Fischer Scientific) coupled to an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) as described previously (Engholm-Keller et al, 2012). Peptides were separated using a 20 min gradient from 0–34% B-buffer (A-buffer: 0.1% formic acid; B-buffer: 90% acetonitrile, 0.1% formic acid). The data-dependent analysis was performed using one MS full scan in the area 300–1,800 Da performed in the Orbitrap with 60,000 in resolution, followed by the five most intense ions selected for MSMS (collision induced dissociation) performed in the linear ion trap.
Raw data from the LTQ-Orbitrap-XL were processed using the Proteome Discoverer (PD) program (Thermo Fisher Scientific, Bremen, Germany). The data were searched against the Arabidopsis database (33,596 sequences; 13,487,687 residues) using the Mascot 2.2 version database. The parameters for the database search were as follows: Enzyme—trypsin; missed cleavages—2; MS mass accuracy—10 ppm; MSMS mass accuracy—0.8 Da; Variable modifications—Phosphorylation (S, T, Y), Oxidation (met), Propionamide (C). The identified peptides were filtered for 1% false discovery rate using the ‘Fixed Value PSM Validator’ in PD. All identified phosphopeptides were manually validated. For iTRAQ-labeled phosphopeptides, the PD quantitation node was used and the data were normalized based on the non-phosphorylated peptides.
Acknowledgments
We thank Yuelin Zhang for providing summ2-8 and mkk1/2 summ2-8 seeds. This work was supported by the Danish Research Council for independent Research, Natural Sciences Grant #10-084139 (M.P.) & Postdoctoral Fellowship #11-116368 (M.E.R.) & Technology and Production Sciences #11-106302. KP was supported by an EMBO Long-Term Fellowship and a Marie Curie International Incoming Fellowship. We thank Suksawad Vonvisuttikun for technical help and Dr. Tsuyoshi Nakagawa (Shimane University) for providing Gateway binary pGWB vectors. All confocal work was done at Center for Advanced Bioimaging. MRL was supported by the Lundbeck Foundation (Junior Group Leader Fellowship).
Author contributions
MER, MWR, KP, JM and MP conceived and designed the experiments. MER, MWR KP, SL, MRL, JG, AMR, LS, WZ and GB performed experiments. MER, MWR, JM and MP analyzed the data. MER, MWR, JM and MP wrote the manuscript.
Conflict of interest
The authors declare that they have no conflicts of interest.
Supporting Information
Supplementary Figures S1–S10
Supplementary Table S1
Supplementary Legends
Review Process File
Source Data for Figure 1A
Source Data for Figure 1C
Source Data for Figure 2B
Source Data for Figure 5C
Source Data for Figure 5D
Source Data for Figure 7A
References
- Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NH, Zhu S, Qiu J-L, Micheelsen P, Rocher A, Petersen M, Newman M-A, Nielsen HB, Hirt H, Somssich IE, Mattsson O, Mundy J. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 2005;24:2579–2589. doi: 10.1038/sj.emboj.7600737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E, Ellis B. Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 2010;15:106–113. doi: 10.1016/j.tplants.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gómez-Gómez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barišić-Jäger E, Kręcioch I, Hosiner S, Antic S, Dorner S. HPat a decapping activator interacting with the miRNA effector complex. PLoS One. 2013;8:e71860. doi: 10.1371/journal.pone.0071860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G, Unthan T, Uhrig JF, Poeschl Y, Gust AA, Scheel D, Lee J. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. PNAS. 2009;106:674. doi: 10.1073/pnas.0810206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G, Pecher P, Eschen-Lippold L, Tsuda K, Katagiri F, Glazebrook J, Scheel D, Lee J. Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol Plant Microbe Interact. 2012;25:471–480. doi: 10.1094/MPMI-11-11-0281. [DOI] [PubMed] [Google Scholar]
- Blewett NH, Goldstrohm AC. A eukaryotic translation initiation factor 4E-binding protein promotes mRNA decapping and is required for PUF repression. Mol Cell Biol. 2012;32:4181–4194. doi: 10.1128/MCB.00483-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boag PR, Atalay A, Robida S, Reinke V, Blackwell TK. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J Cell Biol. 2008;182:543–557. doi: 10.1083/jcb.200801183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Bonnerot C, Boeck R, Lapeyre B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol Cell Biol. 2000;20:5939–5946. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brader G, Djamei A, Teige M, Palva ET, Hirt H. The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis. Mol Plant Microbe Interact. 2007;20:589–596. doi: 10.1094/MPMI-20-5-0589. [DOI] [PubMed] [Google Scholar]
- Brengues M. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Nielsen H, Zhu S, Newman M-A, Shokat KM, Rietz S, Parker J, Mundy J. Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 2006;47:532–546. doi: 10.1111/j.1365-313X.2006.02806.x. [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakcarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Yoon J-H, Parker R. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci. 2011;124:228–239. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006;18:465–476. doi: 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos MK, Cavaness GF, Hall B, King E, Punwani J, van Norman J, Sieburth L. VARICOSE, a WD-domain protein, is required for leaf blade development. Development. 2003;130:6577–6588. doi: 10.1242/dev.00909. [DOI] [PubMed] [Google Scholar]
- Droillard M-J, Boudsocq M, Barbier-Brygoo H, Laurière C. Involvement of MPK4 in osmotic stress response pathways in cell suspensions and plantlets of Arabidopsis thaliana: activation by hypoosmolarity and negative role in hyperosmolarity tolerance. FEBS Lett. 2004;574:42–48. doi: 10.1016/j.febslet.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Engholm-Keller K, Birck P, Størling J, Pociot F, Mandrup-Poulsen T, Larsen MR. TiSH–a robust and sensitive global phosphoproteomics strategy employing a combination of TiO2, SIMAC, and HILIC. J Proteomics. 2012;75:5749–5761. doi: 10.1016/j.jprot.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Gachon C, Saindrenan P. Real-time PCR monitoring of fungal development in Arabidopsis thaliana infected by Alternaria brassicicola and Botrytis cinerea. Plant Physiol Biochem. 2004;42:367–371. doi: 10.1016/j.plaphy.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, Zhang Y. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008;18:1190–1198. doi: 10.1038/cr.2008.300. [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Gloggnitzer J, Akimcheva S, Srinivasan A, Kusenda B, Riehs N, Stampfl H, Bautor J, Dekrout B, Jonak C, Jimenez-Gomez JM, Parker JE, Riha K. Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe. 2014;16:376–390. doi: 10.1016/j.chom.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Goeres D, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth L. Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell. 2007;19:1549–1564. doi: 10.1105/tpc.106.047621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golisz A, Sikorski PJ, Kruszka K, Kufel J. Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res. 2013;41:6232–6249. doi: 10.1093/nar/gkt296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L, Bauer Z, Boller T. Both the extracellular leucine-rich repeat domain and the kinase activity of FLS2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell. 2001;13:1155–1163. [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, Lister R, OMalley R, Urich MA, Tonti-Phillipini J, Chen H, Millar AH, Ecker JR. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Grigg SP, Canales C, Hay A, Tsiantis M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005;437:1022–1026. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- Haas G, Braun JE, Tritschler F, Nishihara T, Izaurralde E. HPat provides a link between deadenylation and decapping in metazoa. J Cell Biol. 2010;189:289–302. doi: 10.1083/jcb.200910141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield L, Beelman CA, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino T, Tanaka Y, Kawamukai M, Nishimura K, Mano S, Nakagawa T. Two Sec13p homologs, AtSec13A and AtSec13B, redundantly contribute to the formation of COPII transport vesicles in Arabidopsis thaliana. Biosci Biotechnol Biochem. 2011;75:1848–1852. doi: 10.1271/bbb.110331. [DOI] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K. MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J Biol Chem. 2006;281:36969–36976. doi: 10.1074/jbc.M605319200. [DOI] [PubMed] [Google Scholar]
- Jeong H-J, Kim YJ, Kim SH, Kim Y-H, Lee I-J, Kim YK. Nonsense-mediated mRNA decay factors, UPF1 and UPF3, contribute to plant defense. Plant Cell Physiol. 2011;52:2147–2156. doi: 10.1093/pcp/pcr144. [DOI] [PubMed] [Google Scholar]
- Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjan M. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature. 2010;467:608–611. doi: 10.1038/nature09338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochem Soc Trans. 2010;38:242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jørgensen TJD. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, Zhang F, Raikhel N, Jiang L, Chen X. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell. 2013;153:562–574. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A, Maldonado M, Buguaut A, Balasubramanian S, Kress M, Weil D, Standart N. Distinct functions of maternal and somatic Pat1 protein paralogs. RNA. 2010;16:2094–2107. doi: 10.1261/rna.2295410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A, Standart N. Pat1 proteins: a life in translation, translation repression and mRNA decay. Biochem Soc Trans. 2010;38:1602–1607. doi: 10.1042/BST0381602. [DOI] [PubMed] [Google Scholar]
- Mészáros T, Helfer A, Hatzimasoura E, Magyar Z, Serazetdinova L, Rios G, Bardóczy V, Teige M, Koncz C, Peck S, Bögre L. The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 2006;48:485–498. doi: 10.1111/j.1365-313X.2006.02888.x. [DOI] [PubMed] [Google Scholar]
- Morel JB. Fertile hypomorphic ARGONAUTEago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomura K, Le QT, Kumakura N, Fukaya T, Takeda A, Watanabe Y. The role of decapping proteins in the miRNA accumulation in Arabidopsis thaliana. RNA Biol. 2012;9:644–652. doi: 10.4161/rna.19877. [DOI] [PubMed] [Google Scholar]
- Munchel SE, Shultzaberger RK, Takizawa N, Weis K. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol Biol Cell. 2011;22:2787–2795. doi: 10.1091/mbc.E11-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo G, Guo H, Gregory BD, Nourizadeh SD, Aguilar-Henonin L, Li H, An F, Guzman P, Ecker JR. ETHYLENE-INSENSITIVE5 encodes a 5 ‘-> 3 ‘ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. PNAS. 2006;103:13286–13293. doi: 10.1073/pnas.0605528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur S, Chekulaeva M, Stoecklin G. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol Cell Biol. 2010;30:4308–4323. doi: 10.1128/MCB.00429-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-H, Chung PJ, Juntawong P, Bailey-Serres J, Kim YS, Jung H, Bang SW, Kim Y-K, Do Choi Y, Kim J-K. Posttranscriptional control of photosynthetic mRNA decay under stress conditions requires 3′ and 5′ untranslated regions and correlates with differential polysome association in rice. Plant Physiol. 2012;159:1111–1124. doi: 10.1104/pp.112.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell. 1996;8:2033–2046. doi: 10.1105/tpc.8.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pearson G, Robinson F, Gibson TB, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- Perea-Resa C, Hernández-Verdeja T, López-Cobollo R, del Mar Castellano M, Salinas J. LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell. 2012;24:4930–4947. doi: 10.1105/tpc.112.103697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, Sharma SB, Klessig DF, Martienssen R, Mattsson O, Jensen AB, Mundy J. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H. MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol. 2009;12:421–426. doi: 10.1016/j.pbi.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Potuschak T, Vansiri A, Binder BM, Vierstra RD, Genschik P. The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell. 2006;18:3047–3057. doi: 10.1105/tpc.106.046508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J-L, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, Thorgrimsen S, Palma K, Suarez-Rodriguez MC, Sandbech-Clausen S, Lichota J, Brodersen P, Grasser KD, Mattsson O, Glazebrook J, Mundy J, Petersen M. Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 2008a;27:2214–2221. doi: 10.1038/emboj.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J-L, Zhou L, Yun B-W, Nielsen HB, Fiil BK, Petersen K, MacKinlay J, Loake GJ, Mundy J, Morris PC. Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 2008b;148:212–222. doi: 10.1104/pp.108.120006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen MW, Roux M, Petersen M, Mundy J. MAP kinase cascades in Arabidopsis innate immunity. Front Plant Sci. 2012;3:169. doi: 10.3389/fpls.2012.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravet K, Reyt G, Arnaud N, Krouk G, Djouani E, Boucherez J, Briat J, Gaymard F. Iron and ROS control of the DownSTream mRNA decay pathway is essential for plant fitness. EMBO J. 2012;31:175–186. doi: 10.1038/emboj.2011.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayson S, Arciga-Reyes L, Wooton L, de Torres Zabala M, Truman W, Grant M, Davies B. A role for nonsense-mediated mRNA decay in plants: pathogen responses are induced in Arabidopsis thaliana NMD mutants. PLoS One. 2012a;7:e31917. doi: 10.1371/journal.pone.0031917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayson S, Ashworth M, de Torres Zabala M, Grant M, Davies B. The salicylic acid dependent and independent effects of NMD in plants. Plant Signal Behav. 2012b;7:1434–1437. doi: 10.4161/psb.21960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehs-Kearnan N, Gloggnitzer J, Dekrout B, Jonak C, Riha K. Aberrant growth and lethality of Arabidopsis deficient in nonsense-mediated RNA decay factors is caused by autoimmune-like response. Nucleic Acids Res. 2012;40:5615–5624. doi: 10.1093/nar/gks195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Cousino N, Lill R, Neupert W, Court D. Identification and initial characterization of the cytosolic protein Ycr77p. Yeast. 1995;11:581–585. doi: 10.1002/yea.320110608. [DOI] [PubMed] [Google Scholar]
- Rymarquis LA, Souret FF, Green PJ. Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA. 2011;17:501–511. doi: 10.1261/rna.2467911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzeczkowski K, Beuerlein K, Müller H, Dittrich-Breiholz O, Schneider H, Kettner-Buhrow D, Holtmann H, Kracht M. c-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J Cell Biol. 2011;194:581–596. doi: 10.1083/jcb.201006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Seraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller N, Resa-Infante P, la Luna de S, Galao RP, Albrecht M, Kaestner L, Lipp P, Lengauer T, Meyerhans A, Díez J. Identification of PatL1, a human homolog to yeast P body component Pat1. Biochim Biophys Acta. 2007;1773:1786–1792. doi: 10.1016/j.bbamcr.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Shi C, Baldwin I, Wu J. Arabidopsis plants having defects in nonsense-mediated mRNA decay factors UPF1, UPF2, and UPF3 show photoperiod-dependent phenotypes in development and stress responses. J Integr Plant Biol. 2012;54:99–114. doi: 10.1111/j.1744-7909.2012.01093.x. [DOI] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su S-H, Jester PJ, Zhang S, Bent AF, Krysan PJ. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 2007;143:661–669. doi: 10.1104/pp.106.091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, Doczi F, Ichimura K, Shinozaki K, Dangl JL, Hirt H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell. 2004;15:141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Tharun S, He WH, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Tharun S, Parker R. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- Tharun S, Muhlrad D, Chowdhury A, Parker R. Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3′ end protection. Genetics. 2005;170:33–46. doi: 10.1534/genetics.104.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S. Lsm1-7-Pat1 complex. RNA Biol. 2009;6:228–232. doi: 10.4161/rna.6.3.8282. [DOI] [PubMed] [Google Scholar]
- Thomas M, Loschi M, Desbats M, Boccaccio G. RNA granules: the good, the bad and the ugly. Cell Signal. 2011;23:324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A, Renzi F, La Fata G, Mattioli C, Raabe M, Urlaub H, Achsel T. The human Pat1b protein: a novel mRNA deadenylation factor identified by a new immunoprecipitation technique. Nucleic Acids Res. 2011;39:635–647. doi: 10.1093/nar/gkq797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- Van Mullem V, Wery M, De Bolle X, Vandenhaute J. Construction of a set of Saccharomyces cerevisiae vectors designed for recombinational cloning. Yeast. 2003;20:739–746. doi: 10.1002/yea.999. [DOI] [PubMed] [Google Scholar]
- Vogel F, Hofius D, Paulus KE, Jungkunz I, Sonnewald U. The second face of a known player: Arabidopsis silencing suppressor AtXRN4 acts organ-specifically. New Phytol. 2011;189:484–493. doi: 10.1111/j.1469-8137.2010.03482.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Watt P, Louis E, Borts RH, Hickson I. Pat1: a topoisomerase II-associated protein required for faithful chromosome transmission in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:4791–4797. doi: 10.1093/nar/24.23.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C, Nover L, Fauth M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 2008;56:517–530. doi: 10.1111/j.1365-313X.2008.03623.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Yang J-Y, Niu Q-W, Chua N-H. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18:3386–3398. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua NH. Arabidopsis decapping 5 is required for mRNA decapping, P-Body formation, and translational repression during postembryonic development. Plant Cell. 2009;21:3270–3279. doi: 10.1105/tpc.109.070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua N-H. Processing bodies and plant development. Curr Opin Plant Biol. 2011;14:88–93. doi: 10.1016/j.pbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua NH. Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J. 2012;31:1975–1984. doi: 10.1038/emboj.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wu G, Poethig RS. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. PNAS. 2012;109:315–320. doi: 10.1073/pnas.1114673109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J-H, Choi E-J, Parker R. Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J Cell Biol. 2010;189:813–827. doi: 10.1083/jcb.200912019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, Chai J, Chen S, Tang X, Zhou J-M. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe. 2007;1:175–185. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe. 2012;11:253–263. doi: 10.1016/j.chom.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Zipfel C. Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol. 2009;12:414–420. doi: 10.1016/j.pbi.2009.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S10
Supplementary Table S1
Supplementary Legends
Review Process File
Source Data for Figure 1A
Source Data for Figure 1C
Source Data for Figure 2B
Source Data for Figure 5C
Source Data for Figure 5D
Source Data for Figure 7A