Summary
Cellular plasticity contributes to the regenerative capacity of plants, invertebrates, teleost fishes, and amphibians. In vertebrates, differentiated cells are known to revert into replicating progenitors, but these cells do not persist as stable stem cells. We now present evidence that differentiated airway epithelial cells can revert into stable and functional stem cells in vivo. Following the ablation of airway stem cells, we observed a surprising increase in the proliferation of committed secretory cells. Subsequent lineage tracing demonstrated that the luminal secretory cells had dedifferentiated into basal stem cells. Dedifferentiated cells were morphologically indistinguishable from stem cells and they functioned as well as their endogenous counterparts to repair epithelial injury. Indeed, single secretory cells clonally dedifferentiated into multipotent stem cells when they were cultured ex vivo without basal stem cells. In contrast, direct contact with a single basal stem cell was sufficient to prevent secretory cell dedifferentiation. In analogy to classical descriptions of amphibian nuclear reprogramming, the propensity of committed cells to dedifferentiate was inversely correlated to their state of maturity. This capacity of committed cells to dedifferentiate into stem cells may play a more general role in the regeneration of many tissues and in multiple disease states, notably cancer.
The term dedifferentiation was first coined to describe the process in which cells of the Urodele retinal pigment epithelium lose their differentiated properties to replace extirpated lens cells1. Although not formally demonstrated, the term was used to suggest that differentiated epithelial cells reverted to a prior developmental stage before their subsequent differentiation into an alternative cell fate. Dedifferentiation has since been explored in plants, invertebrates, teleost fishes and amphibians2–17. In vertebrates, quiescent differentiated cells can revert into replicating progenitor cells5–7,11,12,14 to replace lost cells, but these progenitor cells do not persist as stable stem cells11. Indeed, in murine hair follicle regeneration, the immediate differentiated progeny of epithelial stem cells are already resistant to dedifferentiation17. On the other hand, the undifferentiated secretory progenitors of the intestine that are the immediate progeny of intestinal stem cells are able to dedifferentiate into stem cells after injury13, mimicking the capacity for dedifferentiation of the immediate progeny of drosophila germline stem cells3,15,16. Recently, airway epithelial cells have been shown to be more plastic than previously recognized using stringent lineage tracing strategies18 and differentiated secretory cells have been shown to give rise to very rare cells (0.34±0.09%) that express basal cell markers after severe injury, but the properties of these rare basal-like cells were not studied and their functional capacity was not assessed19. Here, we specifically sought to determine whether stably committed luminal cells could dedifferentiate into functional stem cells.
Secretory cells replicate after stem cell ablation
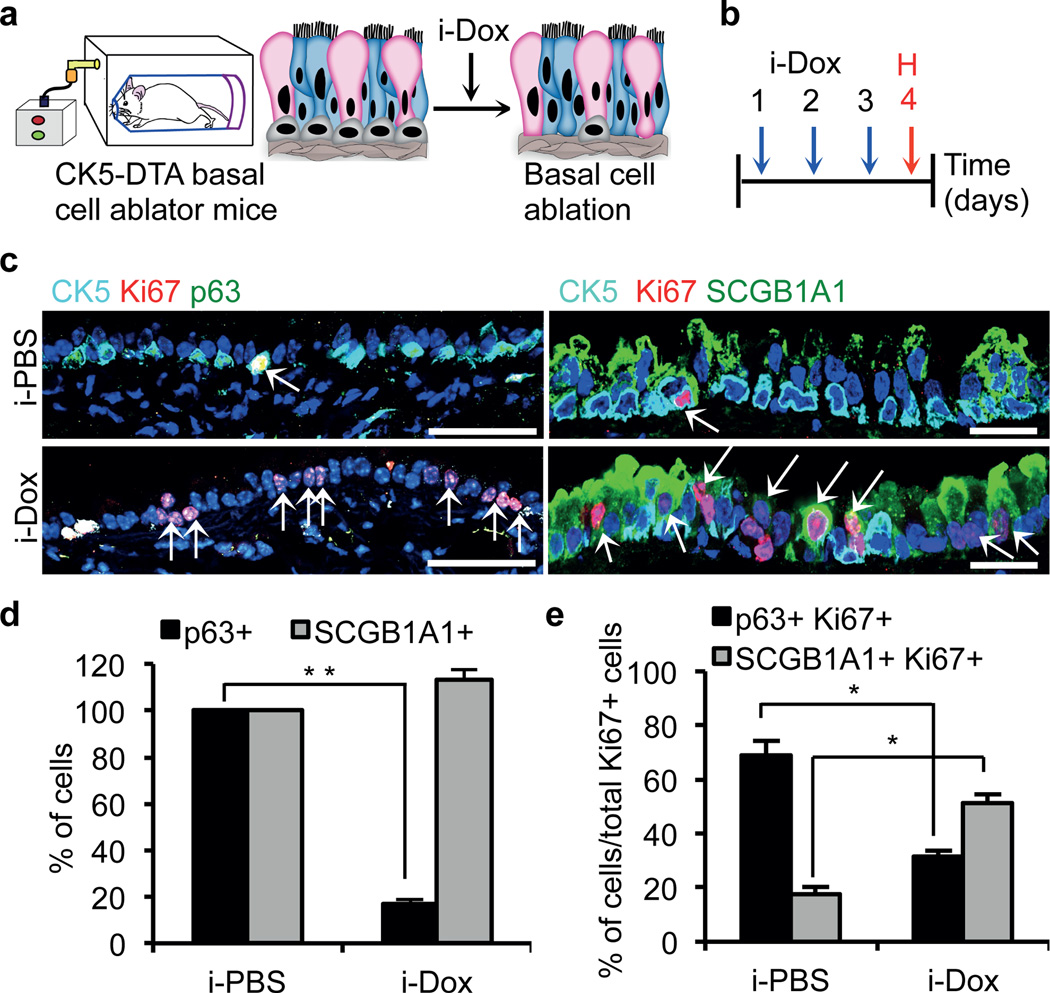
Airway basal stem cells have been shown to self-renew and differentiate into multiple airway epithelial cell types using genetic lineage tracing20,21. Secretory cells are differentiated luminal cells that have both secretory and detoxifying functions. Secretory cells can also further differentiate into ciliated cells19. To test whether secretory cells can dedifferentiate into stem cells, we ablated basal stem cells of the airway epithelium and simultaneously lineage traced the secretory cells of the same mouse (Extended Data Fig. 1). To ablate the airway basal stem cells, we generated a CK5-rtTA/tet(O)DTA22,23 (hereafter referred to as CK5-DTA) mouse, which expresses active subunit of the diphtheria toxin (DTA) in CK5+ airway basal stem cells following doxycycline administration. CK5 expression is, however, not restricted to the basal stem cells of the airway epithelium and is expressed in many others epithelial tissues20,22. Therefore, the ablation of CK5-expressing cells using systemic doxycycline administration through drinking water or by intraperitoneal injection is lethal in the adult mouse (data not shown). To circumvent this problem, we designed a method for the inhalational delivery of doxycycline24 as a means to induce DTA transgene expression exclusively in the airway epithelium. CK5-DTA mice were exposed to either inhaled PBS (i-PBS) or inhaled doxycycline (i-Dox) for two or three consecutive days (Fig. 1a, b). Tracheas were isolated 24 hours after the final dose of i-Dox. Immunofluorescence analysis using the basal stem cell markers p63, CK5, NGFR and T1alpha (T1α) demonstrated a specific dose dependent ablation of basal cells and a preservation of normal secretory cell numbers (Fig. 1c–d and Extended Data Fig. 2a–c). Approximately 80% (n=3) of the airway basal cells were ablated after three doses of inhaled doxycycline (an 81.0±3.4% decrease in the number of CK5+ cells and a 78.8±2.4% decrease in the number of p63+ cells) (Extended Data Fig. 2d). Immunofluorescence staining of the proliferation marker Ki67 in combination with cell type specific markers demonstrated that residual CK5+ and p63+ basal cells do proliferate, but there are actually fewer numbers of proliferating basal cells relative to the total population of replicating cells following ablation (Fig. 1c and e). Surprisingly, there was a 2-fold increase in the numbers of replicating secretory cells (SCGB1A1+ Ki67+) in i-Dox treated animals (51.29±3.02%) as compared to i-PBS treated animals (17.7±2.68%) (Fig. 1c and e and Extended Data Fig. 3a). Consistent with the increased proliferation of differentiated secretory cells, Ki67 staining was specifically increased in the CK8+ suprabasal layer of the airway epithelium (Extended Data Fig. 3b). Thus, secretory cells are the predominant cells that replicate after stem cell ablation. Interestingly, occasional cells expressed both the basal cell marker CK5 and the secretory cell marker SCGB1A1 in i-Dox treated mice (Extended Data Fig. 3c).
Figure 1. Secretory cells proliferate after basal cell ablation.
a, Schematic representation of the ablation of CK5-expressing basal cells of the trachea. Secretory, ciliated and basal stem cells are shown in pink, blue and grey colors respectively. b, Schematic of the timeline of i-dox or i-PBS administration and tissue harvest. c, Immunostaining for basal (p63 (green) and CK5 (cyan)) and secretory cells (SCGB1A1 (green)) in combination with Ki67 (red) on either i-PBS (upper panels) or i-Dox (lower panels) treated mice (n=6). White arrows, Ki67+ cells. d, Quantification of the percentage of p63+ and SCGB1A1+ cells per total DAPI+ cells in i-PBS or i-Dox n=3. e, Percentage of p63+Ki67+ and SCGB1A1+Ki67+ cells relative to total Ki67+ cells in i-PBS and i-Dox (n=3) treated CK5-DTA mice. i-Dox, inhaled doxycycline; i-PBS, inhaled PBS. Nuclei, DAPI (blue). *- p<0.05 and ** - p<0.01; n=3 (3 mice per condition). Error bars, average±s.e.m. P-values, two tailed and paired t-test. Scale bar, 20µm.
Secretory cells dedifferentiate in vivo
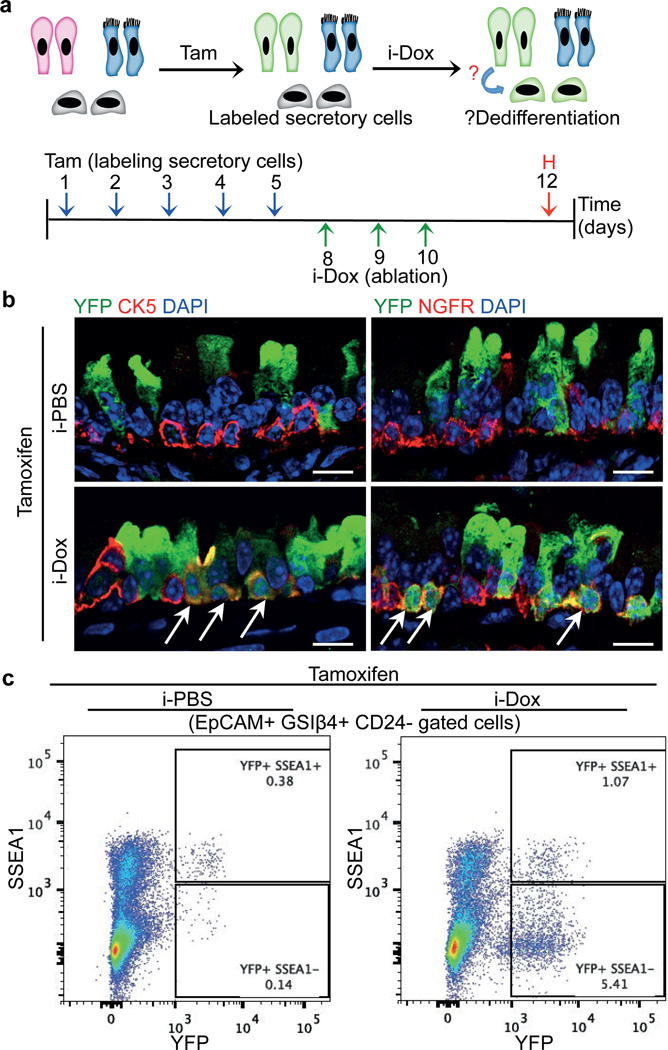
To lineage label secretory cells prior to stem cell ablation, we generated quadruple transgenic mice: Scgb1a1-CreER/LSL-YFP::CK5-rtTA-tet(O)DTA (hereafter referred to as Scgb1a1-YFP/CK5-DTA mice). Administration of tamoxifen to induce the CreER-mediated expression of the YFP label in secretory cells was followed by 3 doses of i-Dox to induce basal cell ablation (Fig. 2a). Lineage labeled YFP+ secretory cells demonstrated increased rates of proliferation in i-Dox treated animals as compared to i-PBS treated controls (Extended Data Fig. 3d–e). We identified YFP+ secretory cell-derived cells that were morphologically indistinguishable from basal stem cells (Fig. 2b). In addition, we found that a subset of lineage labeled cells expressed a suite of basal cell markers including CK5, NGFR, p63 and T1α (Fig. 2b and Extended Data Fig. 3f). Quantification revealed that 7.9±2.08% of basal cells (585 CK5+ YFP+ cells out of 7320 total CK5+ cells in i-Dox treated animals, n=6 mice) expressed a YFP lineage label demonstrating that dedifferentiated basal-like cells comprised a substantial fraction of the total stem cell pool. Dedifferentiated cells did not appear in PBS-treated controls (3 CK5+ YFP+ cells out of 7558 total CK5+ cells counted (0.041±0.028%; n=6 mice). Consistently, when the entire basal cell population is purified by flow cytometry, the YFP lineage labeled basal-like cells have lost the secretory cell surface marker SSEA1 (Fig. 2c). Thus, dedifferentiating cells lose markers of secretory cell differentiation as they acquire markers of stem cells.
Figure 2. Luminal secretory cells dedifferentiate into basal stem cells after stem cell ablation.
a, Schematic representation of tamoxifen (Tam) and i-dox administration to Scgb1a1-YFP/CK5-DTA mice followed by tissue harvest (H). b, Immunostaining for CK5 (red) (left panels) and NGFR (red) (right panels) in combination with YFP (green) in i-PBS (upper panels) or i-Dox (lower panels) treated mice (n=3). White arrows indicate double positive cells. c, Flowcytometric analysis of dedifferentiated cells. EpCAM+GSIβ4+CD24- cells were analyzed for expression of SSEA1 and YFP in either i-Dox and i-PBS treated mice. i-Dox, inhaled doxycycline; i-PBS, inhaled PBS. Nuclei - DAPI (blue). n=3 (at least 2 mice each per condition). Scale bar, 20µm.
Secretory cells dedifferentiate ex vivo
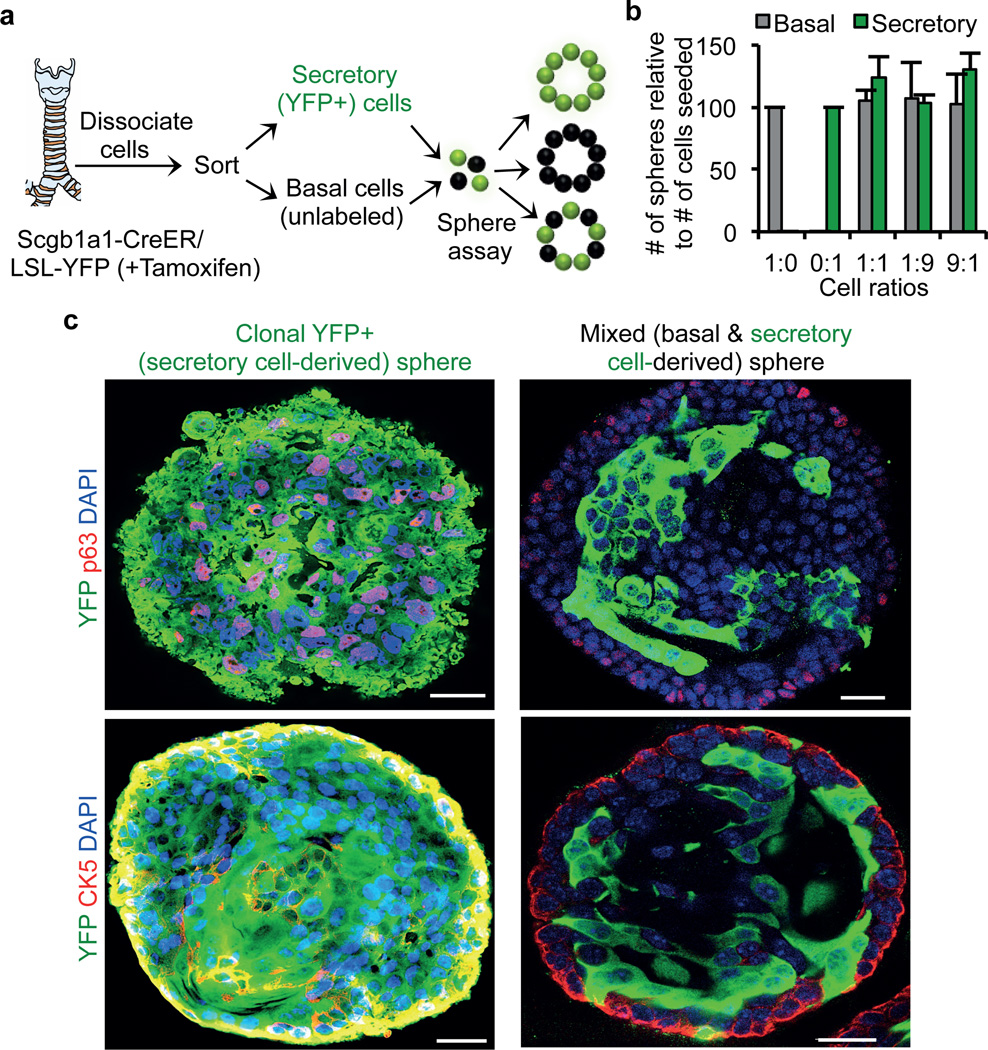
Since our in vivo experiments demonstrated that secretory cells are stimulated to dedifferentiate by the ablation of basal stem cells, we wondered whether secretory cell dedifferentiation could be induced ex vivo when secretory cells were cultured in the absence of basal stem cells. We reasoned that such an assay would provide a platform for further determining whether the dedifferentiation process is actively suppressed by the presence of co-cultured basal stem cells. To assess this possibility, we isolated and sorted unlabeled basal stem cells and YFP+ secretory cells from Scgb1a1-CreER/LSL-YFP mice after tamoxifen injection (Extended Data Fig. 4). We then performed sphere-forming assays with these pure YFP labeled secretory cells alone or in combination with pure sorted GSIβ4+ unlabeled basal cells in varying proportions. The formation of three possible types of spheres is predicted to occur including spheres that are comprised of (1) exclusively YFP+ secretory cell-derived cells, (2) exclusively unlabeled basal cell-derived cells, or (3) chimeric spheres containing YFP+ secretory cell-derived and unlabeled basal cell-derived cells in the same sphere (Fig. 3a). Using our mixed cell populations, we identified a seeding density at which 99.14%±0.18 (n=3) of spheres were either entirely YFP+ or entirely unlabeled, thus establishing a clonal origin of the spheres. Regardless of the relative ratio of input secretory and basal stem cells cultured together in a well, the aggregate clonal sphere-forming efficiency of each cell type remained constant (Fig. 3b). Immunofluorescence analysis demonstrated that in many spheres originating from a single YFP+ secretory cell, some YFP+ cells expressed the basal stem cell markers CK5 and p63, indicating that secretory cell dedifferentiation occurs ex vivo (even in the presence of nearby purely stem cell-derived spheres; Fig. 3c). Thus, we established an ex vivo clonal dedifferentiation assay. Of the total spheres derived clonally from YFP+ secretory cells 88% showed evidence of dedifferentiation, and in 70.58% of these spheres, greater than 10% of cells expressed the stem cell marker CK5. Even when the input proportion of basal cells to secretory cells plated in matrigel was 100:1, no inhibition of dedifferentiation was observed in the spheres that were clonally derived from secretory cells (data not shown). This suggested the possibility that basal cells in one sphere do not provide a secreted factor to suppress the dedifferentiation of secretory cells in another sphere and that direct contact is required to suppress secretory cell dedifferentiation. Indeed, we observed that in the small fraction (0.86%±0.09; n=3) of chimerically derived spheres, not a single YFP+ labeled secretory cell dedifferentiated and went on to express basal cell markers (Fig. 3c). Therefore a single basal stem cell in direct contact with a secretory cell prevents dedifferentiation. Of note, these findings cannot entirely exclude the possibility that a secreted stem cell-derived growth factor can locally suppress secretory cell dedifferentiation. However, they do suggest that an intimate association of basal cells and secretory cells would be required to generate sufficient concentrations of a putative secreted dedifferentiation suppressing activity.
Figure 3. Secretory cells dedifferentiate in the absence of basal cells in an ex vivo sphere forming assay.
a, Schematic representation of lineage labeling, sorting and ex vivo sphere forming assay. Schematic representation of different type of spheres anticipated from basal and secretory cell mixing assay. b, Quantification of the number of spheres that are either basal cell derived (grey bars) and secretory cell (green bars) derived. x-axis, the ratios (1:0, 0:1, 1:1, 1:9 and 9:1) of basal to secretory cells seeded. y-axis, number of spheres formed relative to the number of cells seeded. c, Immunostaining for p63 (red) (upper panels) or CK5 (red) (lower panels) in combination with YFP (green). Nuclei - DAPI (blue). n=3 (2 replicates per condition). Error bars, average±s.e.m. Scale bar 20µm.
To provide further confirmation that secretory cells can dedifferentiate into basal cells ex vivo, we used mice carrying a doxycycline-inducible basal cell-specific reporter allele (CK5-rtTA/tet(O)H2BGFP) and sorted SSEA1+/H2BGFP- secretory cells from their trachea (Extended Data Fig. 5a and b) and performed sphere forming assay. We verified that no GFP+ cells were present prior to induction by doxycycline. When doxycycline was administered during the course of sphere formation, the first appearance of H2BGFP occurred at day 3 of culture, indicating that the secretory cells had been converted into basal-like cells. Immunofluorescence analysis revealed that the resulting H2BGFP+ cells also expressed p63 and CK5 (Extended Data Fig. 5c, left panels). The same results were obtained when the sorted SSEA1+/H2BGFP- cells were grown on transwells (Extended Data Fig. 5c, right panels). Additionally, secretory cells also dedifferentiated into basal-like cells when we cultured lineage-labeled YFP+ cells of Scgb1a1-CreER/LSL-YFP mice on transwell membranes or as spheres (Extended Data Fig. 6a–d). These ex vivo dedifferentiated cells could be serially passaged 5 times in transwell culture or as spheres and retain their expression of basal stem cell markers and their ability to self-renew (Extended Data Fig. 6d–e). Similarly, cells that had undergone dedifferentiation in vivo could also be passaged as stable stem cells (Extended Data Fig. 6f–g). Thus, dedifferentiated secretory cells can stably self-renew.
Mature secretory cells resist dedifferentiation
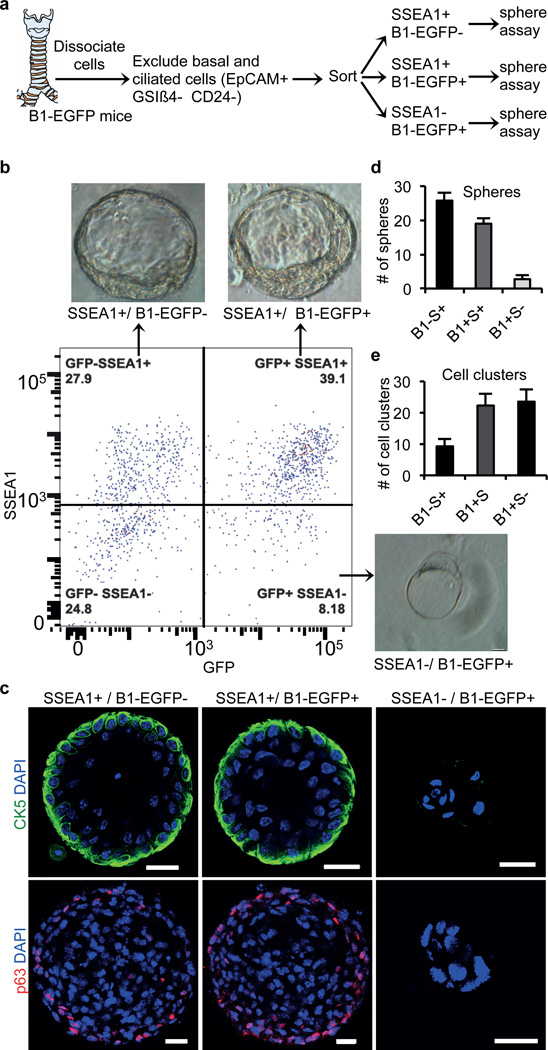
To determine whether all secretory cells have the potential to dedifferentiate or whether only a subset of secretory cells are endowed with this capacity, we attempted to subset this class of epithelial cells. To do so we made use of a transgenic mouse strain that expresses EGFP specifically in secretory cells of the airway epithelium driven by the promoter of the B1 subunit of vacuolar H(1)-ATPase (which we reasoned would be associated with mature secretory cells and hereafter refer to as B1-EGFP)25,26. Co-immunostaining for SSEA1 and GFP demonstrated the existence of 3 subpopulations of secretory cells SSEA1+/GFP-, SSEA1+/GFP+, and SSEA1-/GFP+ (Extended Data Fig. 7a). Of note, all GFP+ cells are SCGB1A1+ secretory cells and none are CK5+ basal cells (Extended Data Fig. 7a). To define the cellular hierarchy of these three subsets of cells, we exposed the airway epithelium of B1-EGFP mice to sulfur dioxide injury. In this injury model, sulfur dioxide causes the complete sloughing of only the suprabasal differentiated cells. The remaining basal stem cells are left intact and start replicating within 24 hours to give rise to a mature epithelium within 14 days. We found that single-positive SSEA1+ cells appeared first on day 4 and then matured into double-positive SSEA1+/GFP+ on day 6 (Extended Data Fig. 7b) prior to the formation of any fully mature single-positive B1-EGFP+ cells evident in the fully mature homeostatic epithelium (Extended Data Fig. 7a, lower panel, arrowheads). Using B1-EGFP mice; we performed sphere-forming assays with each of the three subsets of secretory cells (Fig. 4a). Intriguingly, all three subsets of cells formed similar large spheres and all these spheres contained basal-like cells (Fig. 4b–d). Interestingly, the sphere forming ability of the three populations was inversely proportional to the relative maturity of the secretory cell subsets (Fig. 4d). Of note, the majority of the cell aggregates produced from the most mature SSEA1-/GFP+ secretory cell subset occurred as small cell clusters instead of spheres (Fig. 4c, e). Furthermore, these cell clusters did not contain CK5 and p63 expressing basal stem cells (Fig. 4c).
Figure 4. Dedifferentiation potential of secretory cells is inversely proportional to their maturity.
a, Schematic representation of dissociation and sorting of 3 subsets of secretory cells based on expression of SSEA and GFP. b, Sorting of secretory cell subsets followed by sphere forming assay. Representative images of the predominant type of cell aggregates (spheres or cell clusters) from secretory cell subsets. c, Immunostaining for CK5 (green) and p63 (red) on cell aggregates. d-e, Quantification of the number of spheres (d) or cell clusters (e) from secretory cell subsets. Nuclei - DAPI (blue). n=3 (2 replicates per condition). Error bars, average±s.e.m. Scale bar 20µm.
Dedifferentiated cells stably persist
To assess whether dedifferentiated stem cell-like cells have the ability to self renew and persist in vivo, we generated Scgb1a1-YFP/CK5-DTA mice that possessed lineage-labeled dedifferentiated basal-like cells as above and these mice were then maintained for 2 months prior to sacrifice (Extended Data Fig. 8a). Dedifferentiated YFP+CK5+ cells persisted and continued to represent a sizeable fraction of the stem cell pool (9.15%±0.41; n=3). Indeed, the relative pool size of dedifferentiated basal-like cells remained stable over the course of 2 months (dedifferentiated basal cells represented 8% of the stem cell pool immediately after dedifferentiation). Additionally, triple immunostaining for CK5, YFP and Ki67 revealed that YFP+CK5+ dedifferentiated basal-like cells have the same self-renewal rates as do their normal YFP-CK5+ basal stem cell counterparts (Extended Data Fig. 8b–c).
Dedifferentiated cells are functional stem cells
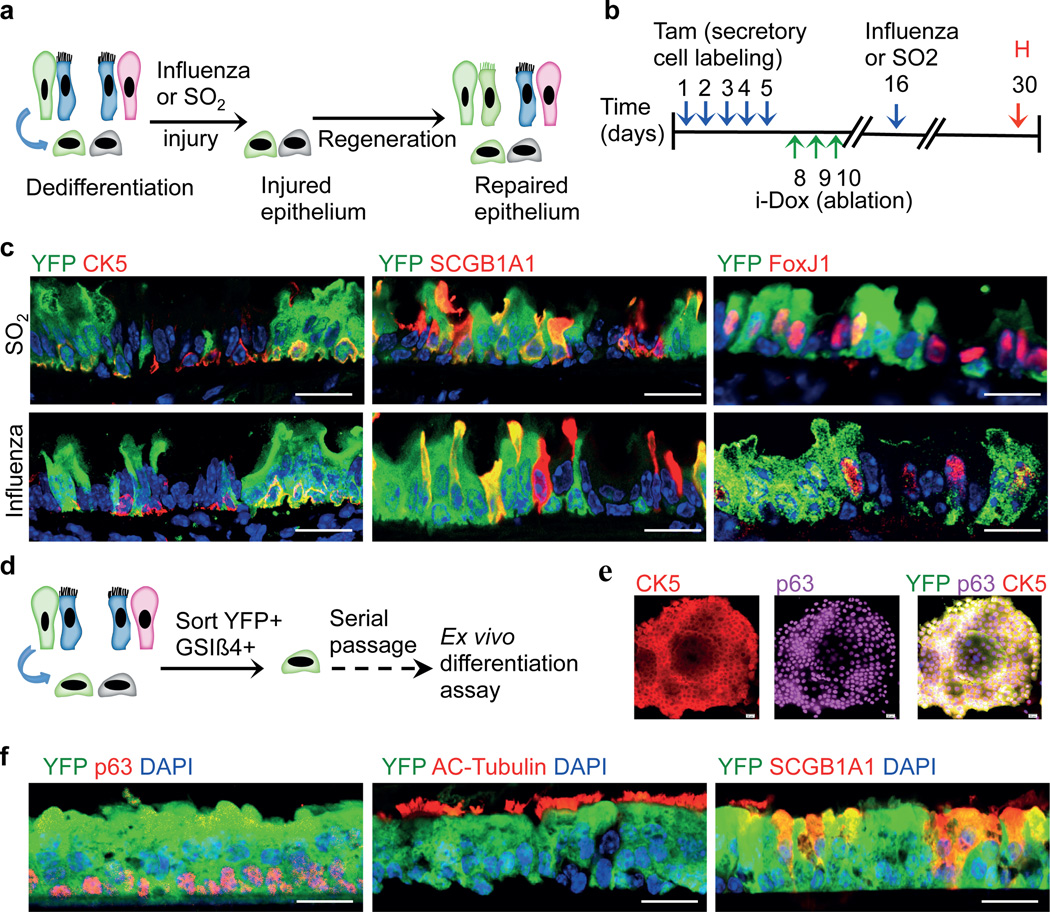
To assess the functional stem cell capacity of dedifferentiated basal-like cells, we generated Scgb1a1-YFP/CK5-DTA mice that possessed lineage labeled dedifferentiated basal-like cells and then exposed these animals to two forms of physiologic airway injury (Fig. 5a and b). First, a toxin-induced airway injury with inhaled sulfur dioxide was used to efficiently denude suprabasal cells from the airway epithelium, leaving behind a single layer of basal cells, some of which were derived from labeled secretory cells that had dedifferentiated (marked by YFP) (Extended Data Fig. 9a). The epithelium fully regenerated in 14 days as expected and immunofluorescence analysis for YFP in combination with CK5 (basal cell), SCGB1A1 (secretory cell), and FoxJ1 (ciliated cell) revealed that YFP+ cells contributed to all three epithelial cell lineages in the form of scattered YFP+ patches (Fig. 5c, upper panels). In order to further scrutinize the functional potential of our dedifferentiated basal-like cells, we used influenza viral infection as a second physiologic injury model (Extended Data Fig. 9b)27. We again observed that dedifferentiated basal-like cells participate in regeneration by giving rise to all three epithelial cell types of the airway (Fig. 5c, lower panels). Similarly, sorted dedifferentiated cells that were produced either in vivo or ex vivo could be serially passaged in culture and differentiated into mature airway epithelium (Fig. 5d–f and Extended Data Fig. 10a–c).
Figure 5. Dedifferentiated cells are functional stem cells both in vivo and ex vivo.
a, Schematic representation of the regeneration of epithelium from dedifferentiated cells after SO2 or influenza induced injury. b, Timeline for the induction of dedifferentiation prior to infectious or toxic injury followed by tissue harvest. c, Co-labeling of YFP (green) with CK5 (red; left panels), SCGB1A1 (red; middle panels) and FoxJ1 (red; right panels) after SO2 (upper panels) or influenza induced injury (lower panels). d. Ex vivo expansion, and differentiation of sorted dedifferentiated cells. e, Immunostaining for CK5(red), p63 (magenta) and YFP (green) on colonies from sorted dedifferentiated cells. f, Co-labeling of YFP (green) with p63 or Acetylated-Tubulin or SCGB1A1 (red). i-Dox, inhaled doxycycline; Tam, tamoxifen; H, harvest. Nuclei - DAPI (blue). n=3 (2 replicates/mice per condition). Scale bar 20µm.
Furthermore, we asked if individual dedifferentiated basal-like cells are multipotent (ie, able to give rise to ciliated, secretory, and basal cells) or unipotent (ie, able to give rise to only one of the cell types). To address this issue, we cultured individual dedifferentiated cells and then performed an air-liquid interface culture using these clonally derived stem cells. Immunofluorescence analyses for basal, secretory and ciliated cell markers revealed that majority of the clonally derived basal-like cells (11 out of 13 clones) are multipotent (Extended Data Fig. 10d). Intriguingly rare clones give rise to only ciliated (1 out of 13 clones) or secretory cells (1 out of 13 clones). The potential of single dedifferentiated cells to act as multipotent stem cells suggests that some or most basal stem cells in vivo may analogously serve as multipotent stem cells. However, the rare lineage restricted clones we observe may reflect a heterogeneity in the basal cell population in vivo that warrants future scrutiny.
Discussion
Here, we have presented evidence using lineage tracing that fully differentiated cells can revert into stable and functional basal stem cells. In contrast, in the hair follicle, the committed progeny of skin stem cells do not revert into stem cells when the skin stem cells are ablated17. However, in the murine intestinal epithelium, undifferentiated secretory progenitors that are the immediate Villin-negative progeny of stem cells can revert back to a stem cell state after injury13. Notably, the capacity of fully differentiated Villin-positive intestinal secretory cells to revert into stem cells was not assessed. Here we show that fully committed secretory cells respond to stem cell ablation by proliferating and converting into functional epithelial stem cells. Our study points to an alternative cellular mechanism through which tissues can regenerate after stem cell loss. The existence of multiple cellular reservoirs of regenerative capacity may allow a more effective reparative response when one or the other cell type is damaged by a toxic or infectious insult.
The ability of basal stem cells to prevent the dedifferentiation of secretory cells has many implications for tissue biology in general since stem cells and their progeny can now be seen to reciprocally modulate one another to regulate their relative ratios and thereby overall tissue architecture. In our example, the prevention of secretory cell dedifferentiation occurs through direct contact with even a single stem cell. This mechanism ensures a precise and local control of epithelial architecture. More generally, it suggests that the reciprocal interactions of stem and committed cells may have been “designed” to ensure a robust self-organizing property in diverse tissue types.
Furthermore, the ability of differentiated cells to acquire stem cell properties appears to be inversely proportional to the degree of the maturity of the differentiated cells. This is analogous to the results seen in amphibian nuclear reprogramming in which nuclei from more mature cells were less easily reprogrammed than those of their immature counterparts28,29. The capacity to segregate secretory cells according to their maturity and associated ability to resist dedifferentiation provides an ideal in vivo experimental model for dissecting the molecular mechanisms through which cells might lock their identity as they mature. Finally, our findings have broad implications for cancer biology since our results point to an underlying physiologic form of cell plasticity that could be co-opted in the process of tumorogenesis30–34. Indeed, some lung cancers appear to be able to resist chemotherapy by employing a lineage conversion into a different tumor subtype35.
Methods summary
CK5-rtTA22, Scgb1a1-CreER19, and tet(O)DTA23, and B1-EGFP25,26 mice were previously described. Corn oil or Tamoxifen (2mg/20gms body weight) were intraperitoneally injected for five consecutive days. Aerosolized PBS or Doxycycline was administered by inhalation24. SO2 injury models have been previously reported19. Mice were anesthetized and infected with a sub-lethal dose of influenza by intranasal inhalation as previously described27. Sphere culture and staining was performed as described previously20. Immunofluorescence and cell sorting were performed using standard protocols.
Methods
Mouse models
CK5-rtTA22, Scgb1a1-CreER19, tet(O)DTA23, and B1-EGFP25,26 mice were previously described. Rosa26R-eYFP (Gt(Rosa)26Sortm1(eYFP)Cos/J) mice (stock number 006148) and Tg(tet(O)HIST1H2BJ/GFP)47Efu/J; (stock #005104) were purchased from The Jackson Laboratory (Bar Harbor, ME). CK5-rtTA females were crossed to tet(O)DTA males to generate a double-transgenic mouse (CK5-DTA) that expresses DTA protein upon doxycycline administration. Aerosolized doxycycline or PBS was administered as described previously24. For secretory cell lineage tracing after basal cell ablation, we crossed male Scgb1a1-CreER/Rosa26R-YFP to CK5-DTA female mice to generate quadruple (Scgb1a1-YFP/CK5-DTA) transgenic mice. To label secretory cells, we injected tamoxifen intraperitoneally (2mg/20gms body weight) for five consecutive days to induce the Cre mediated excision of a stop codon and subsequent expression of YFP. Both male and female mice were used for experiments. 6–12 week old mice were used for experiments. Similar aged mice were used for both control and treated animals. We analyzed at least 3 mice per condition in each experiment. The MGH Subcommittee on Research Animal Care approved animal protocols in accordance with NIH guidelines.
SO2 and Influenza infection induced injury
SO2 injury models have been previously reported19. Briefly, mice were exposed to 500ppm of SO2 for 3-hour 40 mins. For influenza experiments, influenza A/Puerto Rico/8/34 (PR8) was obtained from Charles River Laboratories International (Wilmington, MA). Mice were anesthetized and infected with a sub-lethal dose of influenza by intranasal inhalation as previously described27. Mice were sacrificed at day 3 or 14 post-infection, and the tracheas were removed and placed in 4% paraformaldehyde.
Immunofluorescence, microscopy and cell counting
Trachea was dissected and fixed in 4% PFA for 2 hours at 4°C followed by two washes in PBS, and then embedded in OCT. Cryosections (6 µm) were permeabilized with 0.1% Triton X-100 in PBS, blocked in 1% BSA for 30 minutes at room temperature, incubated with primary antibodies for 1 hour at room temperature, washed, incubated with appropriate secondary antibodies diluted in blocking buffer for 1 hour at room temperature, washed, and counterstained with DAPI. Spheres were fixed with 4% PFA on day 9 after plating and washed with PBS and stained as described above.
The following primary antibodies used were: chicken anti-green fluorescent protein (1:500; GFP-1020, Aves Labs); rabbit anti-Ki67 (1:200; ab15580, Abcam); rat anti-Ki67 (1:200; 14-5698-82, eBioscience); goat anti-SCGB1A1 (1:500; kindly provided by Barry Stripp); mouse anti-p63 (1:100; sc-56188, Santa Cruz); mouse anti- tubulin, acetylated (1:100; T6793, Sigma); mouse anti-FoxJ1 (1:500; 14-9965, eBioscience), rabbit anti-NGFR (1:200, ab8875, Abcam); Mouse IgM anti-SSEA1 (1:100; 14-8813-82, eBioscience); Hamster anti-T1alpha (1:50, DSHB) and rabbit anti-cytokeratin 5 (1:1000; ab53121, Abcam). All secondary antibodies were Alexa Fluor conjugates (488, 594 and 647) and used at 1:500 dilution (Life Technologies).
Images were obtained using Olympus IX81 Inverted microscope (Olympus, Center Valley, PA). Confocal images were obtained with a Nikon A1 confocal laser-scanning microscope with a 40X or 60X oil objective (Nikon CFI Plan APO VC 40X or 60X Oil). Cells were manually counted based on immunofluorescence staining of markers for each of the respective cell types. Cartilage 1 to 9 was used as reference points in all the tracheal samples to count specific cell types based on immunostaining. Serial sections were stained for the antibodies tested and randomly selected slides were used for cell counting. Percentage of basal cells per sphere was calculated based on CK5 immunostaining. Spheres with less than 2% CK5+ cells were not included in the quantification.
Cell dissociation and sorting
Airway epithelial cells from trachea were dissociated using papain solution. Longitudinal haves of the trachea were cut into 5 pieces and incubated in Papain dissociation solution and incubated at 37°C for 2 hours. After incubation, dissociated tissues were passed through a cell strainer and centrifuged and pelleted at 500×g for 5 mins. Cell pellets were dispersed and incubated with Ovo-mucoid protease inhibitor (Worthington biochemical Corporation, cat. # LK003182) to inactivate residual papain activity by incubating on a rocker at 4°C for 20 minutes. Cells were then pelleted and stained with EpCAM-PECy7 (1:50; 25-5791-80, eBiosciences) or EpCAM-APC (1:50; 17-5791, eBiosciences); GSIβ4 (Griffonia Simplicifolia Isolectin beta4)-Biotin (L2120, Sigma); SSEA1 eFluor 650NC (1:75, 95-8813-41, eBiosciences); PE anti-mouse CD24 (1:100, 553262, BD Pharmingen); for 30 min in 2.5% FBS in PBS on ice. After washing, cells were sorted on BD FACS Aria (BD, San Jose, CA) using FACS Diva software and analysis was performed using FlowJo (version 10) software.
Sphere forming assays and transwell cultures
Cells were cultured and expanded in complete SGAM medium (Lonza, CC-3118) containing TGFβ/BMP4/WNT antagonist cocktails and 5 uM Rock inhibitor Y-27632 (Selleckbio, S1049). To initiate air-liquid interface cultures, airway basal stem cells were dissociated and seeded onto transwell membranes. After confluence, media was removed from the upper chamber. Mucocilary differentiation was performed with PneumaCult™-ALI Medium (StemCell, 05001). Differentiation of airway basal stem cells on an air-liquid interface was followed by directly visualizing beating cilia in real time after 10–14 days. For clonal culture assays, dedifferentiated basal-like cells (GSIβ4+ YFP+) were sorted and plated on collagen-coated plates at low cell density to obtain individual colonies. Individual colonies were isolated using trypsin and directly visualized aspiration on single colonies. Individual colony-derived cells were maintained and expanded separately and used for air-liquid interface culture. Sphere culture was performed as described previously20. Briefly, 50 µl of 1:4 cold matrigel / MTEC-plus medium was layered on an 8-well chamber slide (Thermo Scientific, Cat.# 177402) and incubated at 37°C for 10 mins to solidify the matrigel. Sorted cells were mixed in 2% matrigel in MTEC-plus and plated on pre-coated 8-well chamber slide at a density of 4000 cells per well. For mixing assays, sorted cells were seeded at a density of 6000 cells per well. In each experiment, three independent wells were used for each condition tested. Medium was changed every other day for 9 days. For transwells cultures, cells were suspended in MTEC-plus medium and plated on transwell inserts at a cell density of 6000 cells per well. Medium was changed every day for 7 days. For basal cell reporter assays, cells were treated with 1µg /mL doxycycline either at 24 hours after plating or just 24 hours before harvest as described in the results section. For serial passaging of spheres, medium from culture wells was aspirated, washed with PBS, and then treated with trypsin-EDTA (0.25%) for 2 minutes. Trypsin was inactivated and dissociated cells were collected and centrifuged at 350xg for 3 minutes at 4°C. Cells were re-seeded at a 1:20 dilution in matrigel for the next round of sphere culture.
Statistical Analysis
The standard error of the mean was calculated from the average of at least 3 independent tracheal samples unless otherwise mentioned. Data was compared among groups using the Student’s t-test (unpaired, two-tailed). A p-value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgements
The work in this manuscript was supported by a Harvard Stem Cell Institute Seed Grant and an NIH-NHLBI Early Career Research New Faculty (P30) award (5P30HL101287-02) and a Harvard Stem Cell Institute (HSCI) Junior Investigator Grant to JR. JR is a New York Stem Cell Foundation - Robertson Investigator. We wish to extend our thanks to William Anderson, Yuval Dor, Qiao Zhou, Andrew Brack, Jenna Galloway and all the members of the Rajagopal laboratory for their constructive criticism. We thank the members of the HSCI Flow cytometry core facility for helping us with cell sorting.
Footnotes
Supplementary Information is linked to the online version of the paper at http://www.nature.com/nature.
Author Contributions
P.R.T. designed and performed experiments and wrote the manuscript; H.M., A.P., R.Z., M.P., B.L., V.V. performed ex vivo experiments; J.L.C. performed influenza infection experiments; A.S. provided tet(O)DTA mice and edited the manuscript; S.B. provided B1-EGFP mice; B.D.M. reviewed the manuscript; J.R. suggested and co-designed the study and co-wrote the manuscript with P.R.T.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Wolff G. Entwicklungsphysiologische Studien. I. Die Regeneration der Urodelenlinse. Wilhelm Roux Arch. Entwickl. -Mech. Org. 1895;1:380–390. [Google Scholar]
- 2.Brockes JP, Kumar A. Plasticity and reprogramming of differentiated cells in amphibian regeneration. Nat. Rev. Mol. Cell Biol. 2002;3:566–574. doi: 10.1038/nrm881. [DOI] [PubMed] [Google Scholar]
- 3.Kai T, Spradling A. Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature. 2004;428:564–569. doi: 10.1038/nature02436. [DOI] [PubMed] [Google Scholar]
- 4.Slack JMW. Metaplasia and transdifferentiation: from pure biology to the clinic. Nat. Rev. Mol. Cell Biol. 2007;8:369–378. doi: 10.1038/nrm2146. [DOI] [PubMed] [Google Scholar]
- 5.Kragl M, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 6.Lehoczky JA, Robert B, Tabin CJ. Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20609–20614. doi: 10.1073/pnas.1118017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476:409–413. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugimoto K, Gordon SP, Meyerowitz EM. Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 2011;21:212–218. doi: 10.1016/j.tcb.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 11.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 13.Van Es JH, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dabeva MD, et al. Liver regeneration and alpha-fetoprotein messenger RNA expression in the retrorsine model for hepatocyte transplantation. Cancer Res. 1998;58:5825–5834. [PubMed] [Google Scholar]
- 15.Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304:1331–1334. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- 16.Cheng J, et al. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu Y-C, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song H, et al. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 2012;109:17531–17536. doi: 10.1073/pnas.1207238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rawlins EL, et al. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rock JR, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rock JR, Hogan BLM. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol. 2011;27:493–512. doi: 10.1146/annurev-cellbio-100109-104040. [DOI] [PubMed] [Google Scholar]
- 22.Diamond I, Owolabi T, Marco M, Lam C, Glick A. Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J. Invest. Dermatol. 2000;115:788–794. doi: 10.1046/j.1523-1747.2000.00144.x. [DOI] [PubMed] [Google Scholar]
- 23.Weber T, et al. Inducible gene expression in GFAP+ progenitor cells of the SGZ and the dorsal wall of the SVZ-A novel tool to manipulate and trace adult neurogenesis. Glia. 2011;59:615–626. doi: 10.1002/glia.21129. [DOI] [PubMed] [Google Scholar]
- 24.Tata PR, et al. Airway Specific Inducible Transgene Expression Using Aerosolized Doxycycline. Am. J. Respir. Cell Mol. Biol. 2013 doi: 10.1165/rcmb.2012-0412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller RL, et al. V-ATPase B1-subunit promoter drives expression of EGFP in intercalated cells of kidney, clear cells of epididymis and airway cells of lung in transgenic mice. Am. J. Physiol. Cell Physiol. 2005;288:C1134–C1144. doi: 10.1152/ajpcell.00084.2004. [DOI] [PubMed] [Google Scholar]
- 26.Kim JK, et al. In vivo imaging of tracheal epithelial cells in mice during airway regeneration. Am. J. Respir. Cell Mol. Biol. 2012;47:864–868. doi: 10.1165/rcmb.2012-0164OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho JL, et al. Enhanced Tim3 activity improves survival after influenza infection. J. Immunol. 2012;189:2879–2889. doi: 10.4049/jimmunol.1102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briggs R, King TJ. Transplantation of Living Nuclei From Blastula Cells into Enucleated Frogs’ Eggs. Proc. Natl. Acad. Sci. U.S.A. 1952;38:455–463. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- 30.Blanpain C. Tracing the cellular origin of cancer. Nat. Cell Biol. 2012;15:126–134. doi: 10.1038/ncb2657. [DOI] [PubMed] [Google Scholar]
- 31.Schwitalla S, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Friedmann-Morvinski D, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 34.Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329:568–571. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.