Abstract
Degradation of mRNAs is usually initiated by deadenylation, the shortening of long poly(A) tails to oligo(A) tails of 12–15 As. Deadenylation leads to decapping and to subsequent 5′ to 3′ degradation by XRN proteins, or alternatively 3′ to 5′ degradation by the exosome. Decapping can also be induced by uridylation as shown for the non-polyadenylated histone mRNAs in humans and for several mRNAs in Schizosaccharomyces pombe and Aspergillus nidulans. Here we report a novel role for uridylation in preventing 3′ trimming of oligoadenylated mRNAs in Arabidopsis. We show that oligo(A)-tailed mRNAs are uridylated by the cytosolic UTP:RNA uridylyltransferase URT1 and that URT1 has no major impact on mRNA degradation rates. However, in absence of uridylation, oligo(A) tails are trimmed, indicating that uridylation protects oligoadenylated mRNAs from 3′ ribonucleolytic attacks. This conclusion is further supported by an increase in 3′ truncated transcripts detected in urt1 mutants. We propose that preventing 3′ trimming of oligo(A)-tailed mRNAs by uridylation participates in establishing the 5′ to 3′ directionality of mRNA degradation. Importantly, uridylation prevents 3′ shortening of mRNAs associated with polysomes, suggesting that a key biological function of uridylation is to confer 5′ to 3′ polarity in case of co-translational mRNA decay.
INTRODUCTION
Uridylation participates in the control of RNA stability in various eukaryotes, from Schizosaccharomyces pombe to nematodes, plants or man. The broad spectrum of RNAs subjected to uridylation includes U6 snRNA, mRNAs, RNA-induced silencing complex (RISC)-cleaved fragments, small RNAs and their precursors (1–5). Uridylation has diverse effects on the fate of non-coding RNAs. For instance, oligouridylation destabilizes human let-7 microRNA (miRNA) precursors in human embryonic cells and cancer cells, whereas monouridylation of let-7 miRNA precursors favours let-7 miRNA maturation by Dicer in different cell types (6). Uridylation can therefore promote or prevent let-7 miRNA production depending on the cellular context. In addition to miRNA precursors, mature small RNAs can also be uridylated, with different consequences on their stability. In mammals, uridylation can abrogate miRNA activity without affecting miRNA stability (7,8). However, uridylation can also trigger the degradation of miRNA and siRNA as shown in Caenorhabditis elegans, zebrafish, Chlamydomonas reinhardtii or Arabidopsis (9–13). In Arabidopsis, the 3′ nucleotide of siRNAs and miRNAs is methylated by the methyltransferase HEN1 (14). Methylation stabilizes small RNA and prevents their uridylation by the terminal uridylyltransferase HESO1 (12–15). Mutations in HESO1 partially stabilize small RNAs in a hen1 background, revealing that methylation and uridylation have opposing effects on small RNA stability in Arabidopsis (12,13).
Other RNA targets of terminal uridylyltransferases include RISC-cleaved transcripts and mRNAs, and for both RNA types, uridylation enhances decapping followed by 5′ to 3′ degradation (16–22). For instance, oligouridylation of histone mRNAs in humans was shown to favour the binding of the Lsm1-7 complex, which in turn promotes decapping by Dcp2 and subsequent 5′ to 3′ degradation by Xrn1 (19). Uridylation of histone mRNAs triggers also 3′ to 5′ degradation by the exosome (19) and by the Eri1 exoribonuclease, which is recruited on binding of the Lsm1 complex to the uridylated 3′ terminal stem-loop (16). To date, the demonstration of uridylation-mediated mRNA degradation is restricted to non-polyadenylated histone mRNAs in mammals. However, uridylation of several mRNAs by the nucleotidyl transferase Cid1 in S. pombe triggers decapping and subsequent 5′ to 3′ degradation, revealing that uridylation can be an integral step of bulk mRNA decay (20). Interestingly, uridylation-induced decapping is independent of deadenylation in S. pombe (20). By contrast, deadenylation precedes addition of U/C-rich sequences to gdhA mRNAs in Aspergillus nidulans, which also triggers decapping (21). Deadenylation before 3′ U/C tagging is not required for all mRNA types in A. nidulans because it can be bypassed for transcripts containing premature stop codons that are substrates of the non-sense–mediated decay pathway (22). Collectively, these data reveal an emerging role of uridylation in controlling the stability of coding and non-coding RNAs in eukaryotes (4).
Two lines of evidence argue in favour of mRNA uridylation playing a role in mRNA metabolism in Arabidopsis. First, the uridylation of CCR2 mRNAs was recently reported (22). However, the biological significance of this post-transcriptional modification remains to be determined. Second, the Arabidopsis Cid1-related protein encoded by At2g45620 catalyses the addition of uridines when artificially tethered to a non-adenylated reporter mRNA ectopically expressed in Xenopus oocytes (23). Here, we describe the function of UTP:RNA uridylyltransferase 1 (URT1), the At2g45620 gene product. We show that URT1 targets oligoadenylated mRNAs in vivo and has little impact on mRNA degradation rates. More importantly, in urt1 mutants lacking mRNA uridylation, we observed the 3′ trimming of oligo(A)-tailed transcripts for all mRNAs tested, indicating that uridylation prevents 3′ to 5′ ribonucleolytic attacks. Importantly, uridylation can prevent the 3′ trimming of mRNAs still engaged on polysomes. We propose that mRNA uridylation participates in establishing the 5′ to 3′ polarity of mRNA degradation, which could be crucial in case of co-translational decay.
MATERIALS AND METHODS
Materials
Arabidopsis thaliana plants, Columbia ecotype (Col-0), were grown on soil with 16 h light/8 h darkness cycles. The characterization of urt1-1 (Salk_087647C) and urt1-2 (WISCDSLOXHS208_08D) T-DNA insertion lines is shown in Supplementary Figure S1. xrn4-3 (SALK_014209) has been described previously (24). Sequences of primers are shown in Supplementary Table S1.
Subcellular localization of URT1
The coding sequence of URT1 (At2g45620), DCP1 (At1g08370) and RBP47 (At1g19130) was fused 5′ to GFP or RFP sequences using the Gateway® destination vectors pB7FWG2 or pH7RWG2, respectively. Biolistic transformation of tobacco BY2 cells and analysis by confocal microscopy were performed by standard methods.
Expression and purification of recombinant proteins
Recombinant His-tagged GST-URT1 or the catalytically inactive mutant GST-URT1D491/3A were produced in BL21(DE3) cells grown at 17°C. Cells were disrupted by sonication in 20 mM MOPS, pH 7.5, 250 mM KCl, 15% (v/v) glycerol, 1 mM DTT, 0.1% (v/v) Tween 20 in presence of protease inhibitors (Roche). Recombinant proteins were purified by classical Immobilized Metal Affinity Chromatography on Ni-NTA resin followed by glutathion affinity chromatography. Purified proteins were dialysed against 20 mM MOPS, pH 7.5, 100 mM NaCl, 15% (v/v) glycerol, 0.1% (v/v) Tween 20. Aliquots were snap frozen in liquid nitrogen and stored at −80°C.
Activity assays
URT1 and Cid1 activities were compared by using identical amounts of arbitrary units defined by measuring trichloroacetic acid-precipitable incorporation of [α-32P]-UTP into tRNAs and reaction conditions based on (25). In vitro assays shown in Figure 1A contained 100 nM of GST-URT1 or GST-URT1D491/3A or 0.1 unit/μl of Cid1 (NEB), 25 mM potassium phosphate, pH 7, 5 mM MgCl2, 5 mM DTT, 10% (v/v) glycerol, 0.1 μg/μl bovine serum albumin (BSA), 0.1 mM UTP, GTP, CTP or ATP and 5 nM of a 21-nucleotide synthetic RNA 5′-end labelled by T4 Polynucleotide Kinase (NEB) and [γ-32P]-ATP. For experiments shown in Figure 1B, the reaction buffer was 20 mM MOPS, pH 7.5, 100 mM NaCl, 15% (v/v) glycerol, 0.1% (v/v) Tween 20, 1 mM MgCl2, 0.5 µg/µl BSA, 1 mM UTP. Reaction products were separated by denaturing 17% (w/v) polyacrylamide gel electrophoresis before autoradiography. To determine the composition of the tails synthesized by URT1 in the presence of the four ribonucleotides, the latter reaction conditions were used except that the concentration for each ribonucleotide was 1 mM. Nucleotide composition was determined by 3′ Rapid Amplification of cDNA ends (RACE) polymerase chain reaction (PCR), cloning and sequencing as detailed below.
Figure 1.
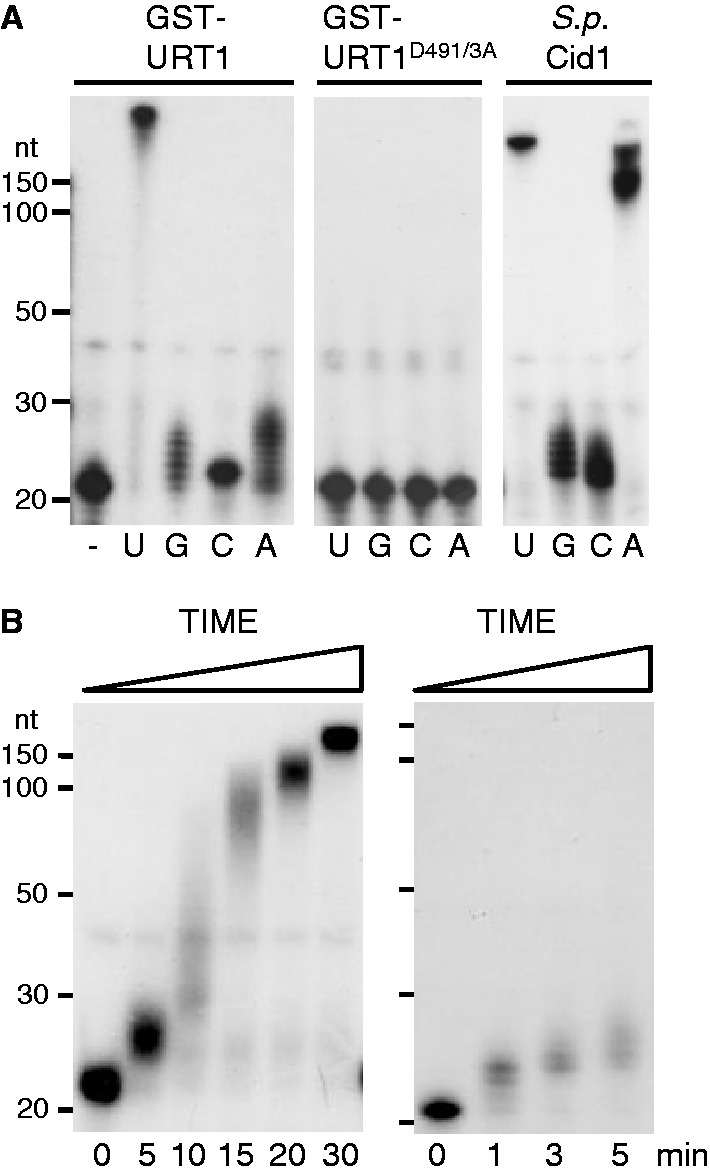
Catalytic properties of URT1. (A) URT1 is a UTP:RNA uridylyltransferase. GST-URT1 or the catalytically inactive mutant GST-URT1D491/3A were incubated for 30 min with a 21-nucleotide 5′ [32P]-labelled synthetic RNA without (−) or in the presence of each ribonucleotide as indicated. Reaction products were separated on 17% denaturing polyacrylamide gels before autoradiography. The migration of size standards (150, 100, 50, 30 and 20 nt) is indicated on the left. For comparison, the catalytic activity of S. pombe Cid1 is shown. (B) URT1’s activity is distributive for the first added nucleotides as shown by time-course analysis. Incubation time is indicated in minutes at the bottom of panels.
3′ RACE and circular reverse transcriptase-polymerase chain reaction (cRT-PCR) analyses
The 3′ RACE PCR protocol used to detect uridylated mRNAs is detailed in (26). Briefly, 5 µg of total RNA was purified with Nucleospin® RNA plant columns (Macherey Nagel), dephosphorylated with calf intestinal phosphatase (CIP, NEB) before ligation to the RNA primer (5′P-CUAGAUGAGACCGUCGACAUGAAUUC-3′NH2) with T4 RNA ligase (Fermentas). This sequence was used as an anchor to initiate cDNA synthesis by SuperScript® III reverse transcriptase (Invitrogen) and the anchor rv1 primer (Supplementary Table S1). Two nested PCR amplification of 30 cycles were performed using anchor rv1 and rv2 primers in combination with gene-specific sense primers (Supplementary Table S1).
Circular reverse transcriptase (cRT)-PCR analysis was used to characterize the 5′ and 3′ extremities of capped and uncapped LOM1 transcripts. To detect capped transcripts, 5 µg of total RNA was first treated with 5 units of CIP (NEB) to dephosphorylate uncapped RNAs and then with 2.5 units of tobacco acid pyrophosphatase (TAP) (Epicentre) to generate 5′ phosphorylated transcripts from capped mRNAs. The 5′ phosphorylated transcripts were then circularized by 20 units of T4 RNA ligase at 18°C for 16 h in a final volume of 200 µl. Uncapped transcripts were detected following mRNA circularization from total RNA that was neither treated with CIP nor with TAP. Negative controls were treated only with CIP because neither dephosphorylated nor capped mRNAs can be circularized by T4 RNA ligase.
3′ RACE and cRT PCR products were cloned in pGEM®-T Easy (Promega) before sequence analysis.
Polysome purification
Polysome purification was performed according to (27) except that 400 mg of tissue powder was loaded on 11 ml of 15–60% (w/v) sucrose gradients. A puromycin treatment abolished the typical UV polysome profiles and, as expected, dramatically reduced the RNA content in heavy fractions of the gradients, preventing reliable 3′ RACE analysis of puromycin controls.
Additional methods
Northern blot analyses, mRNA half-life measurements, microarray analysis and statistical analysis of microarray data are described in Supplementary Methods.
RESULTS
Nucleotide specificity of URT1
To characterize the nucleotidyltransferase activity of URT1, we produced recombinant proteins corresponding either to the wild-type protein or to a mutated version in which two conserved aspartate residues essential for catalysis of nucleotidyltransferases (D491 and D493 in URT1) were mutated to alanines. The nucleotidyltransferase activity of both proteins was tested using a 5′ [32P]-labelled oligoribonucleotide substrate and conditions previously used to describe Cid1’s activity (25). No activity was observed for the mutant protein URT1D491/3A, whereas URT1 showed a robust uridylation activity (Figure 1A). Under these assay conditions, URT1 synthesizes tails of >200 Us, while only 1–9 of the three other ribonucleotides are appended to the RNA substrate in 30 min. To further test that URT1 preferentially incorporates uridines, URT1 was incubated with the unlabelled oligoribonucleotide substrate and the four ribonucleotides ATP, GTP, CTP and UTP. The composition of tails synthesized by URT1 was then determined by sequence analysis of 12 cloned reaction products. URT1-synthesized tails contained 97.8% (947/967 nucleotides) uridines, confirming the strong uridine selectivity of URT1. In line with these in vitro results, the residues involved in uridine selectivity in Cid1 (D330, E333, H336) (28–30) are conserved in URT1 (D708, E711, H714). Interestingly, under the in vitro conditions used, URT1 better discriminates UTP from ATP as compared with Cid1 (Figure 1A).
Testing the stringency of URT1’s nucleotide specificity in vitro entailed the use of reaction conditions that maximize activity. As a result, large U-tails are polymerized, which unlikely reflect the in vivo situation. In fact, a restricted number of Us (1–10) are added to almost all RNA substrate molecules on short incubation times, indicating that the activity of URT1 is distributive for the first added nucleotides (Figure 1B). Following the addition of ∼10 uridines, URT1’s activity seems to become more processive. This switch from distributivity to processivity is not due to the short size of the initial oligoribonucleotide substrate (21 nt) because it was also observed for two RNA substrates of 31 nt (Supplementary Figure S2). The distributivity for the first added nucleotides appears therefore an inherent feature of URT1.
Taken together, these data show that Arabidopsis URT1 is a UTP:RNA uridylyltransferase with a marked preference for uridine polymerization and a distributive activity for the first added nucleotides. In line with this conclusion, the in vivo RNA substrates of URT1 are modified by the addition of a limited number of uridines as detailed below.
URT1 uridylates oligoadenylated mRNAs
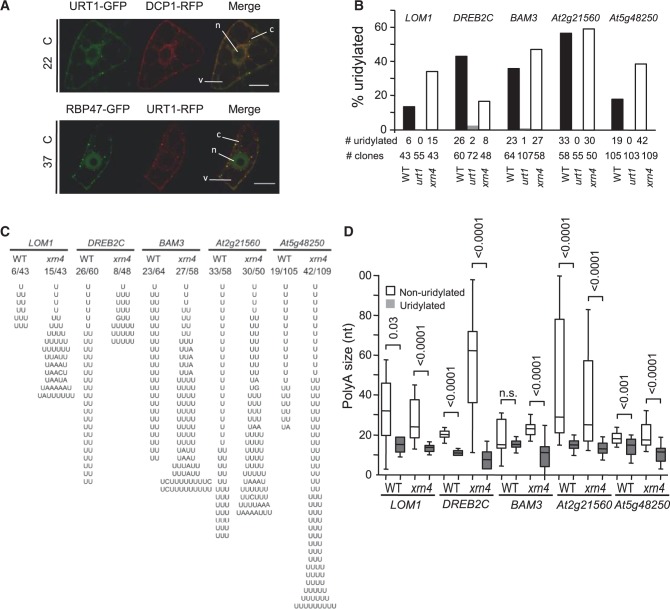
Expressing fluorescent URT1 fusion proteins revealed a diffuse cytosolic pattern with foci (Figure 2A). These foci were observed in all cells examined. URT1-GFP and URT1-RFP partially co-localized with known markers of processing bodies (P-Bodies) and stress granules, DCP1-RFP or RBP47-GFP, respectively (31,32), suggesting that URT1 can associate with cytosolic ribonucleoprotein (RNP) granules (Figure 2A). These results also indicate that URT1 targets cytosolic RNAs. To identify such targets, we investigated mRNA uridylation by nested 3′ RACE PCR experiments using gene-specific forward primers and reverse primers complementary to a 3′ ligated RNA anchor sequence as detailed in (26). Initial tests on mRNAs selected on different criteria such as short half-lives or misregulation in urt1 mutants revealed that any mRNA investigated is a substrate of URT1. Detailed features of mRNA uridylation are presented for five mRNAs: LOM1 (At2g45160), DREB2C (At2g40340), BAM3 (At4g20270), At2g21560 and At5g48250. Of note, DREB2C and BAM3 are among the shortest-lived mRNAs in Arabidopsis (33) and At2g21560 and At5g48250 were identified among genes misregulated in urt1 (Supplementary Table S2). mRNA uridylation was first assessed in WT plants. Cloning and sequence analysis of 3′ RACE products revealed that 14% (6/43) of LOM1, 43% (26/60) of DREB2C, 36% (23/64) of BAM3, 57% (33/58) of At2g21560 and 18% (19/105) of At5g48250 clones corresponded to uridylated mRNAs (Figure 2B). Uridine extensions were short in WT, with 1 or 2 uridines detected for 92% (98/107) of the clones corresponding to uridylated mRNAs (Figure 2C). Overall, uridylated transcripts had significant shorter poly(A) tails than non-uridylated ones (Figure 2D). The average size of uridylated oligo(A) tails for the 5 mRNAs analysed in WT was 14 (n = 106). This size is similar to the length recently reported for oligo(A) tails of uridylated CCR2 mRNAs (22) and is in the range for the known oligo(A) size of deadenylated mRNAs in eukaryotes. Taken together, these data indicate that oligoadenylated mRNAs can be uridylated in Arabidopsis.
Figure 2.
Deadenylation precedes uridylation. (A) URT1 is a cytosolic protein. URT1 fused to GFP (upper panel) or RFP (lower panel) was transiently expressed in tobacco BY-2 cells. Markers of P-bodies and stress granules were DCP1-RFP and RBP47-GFP, respectively. Co-localization of URT1-GFP with DCP1-RFP and of URT1-RFP with RBP47-GFP was observed for 27 of 35 and 39 of 50 foci, respectively, as measured from two independent samples. Cells shown in the lower panel were incubated 30 min at 37°C before observation. n, nucleus; v, vacuole; c, cytosol. Scale bars are 30 µm (top) and 15 µm (bottom). (B) URT1 uridylates mRNAs. Percentage of uridylated mRNAs determined by 3′ RACE PCR in WT, urt1 and xrn4 for LOM1 (At2g45160), DREB2C (At2g40340), BAM3 (At4g20270), At2g21560 and At5g48250 (n = 2). Number of clones (# clones) and number of clones corresponding to uridylated mRNAs (# uridylated) are indicated for each genotype. (C) Composition and length of U-tails detected at the 3′-ends of LOM1, DREB2C, BAM3, At2g21560 and At5g48250 mRNAs in WT and xrn4. The number of clones corresponding to uridylated mRNAs on the total number of clones is indicated. (D) Deadenylation precedes uridylation. Box plot analysis of poly(A) tail length distribution for non-uridylated (white) and uridylated (grey) LOM1, DREB2C, BAM3, At2g21560 and At5g48250 mRNAs in WT and xrn4 as indicated. The second and third quartiles are shown by the box edges, the median is indicated by a horizontal bar, whiskers show the range of data except far-outliers. P-values (Mann–Whitney test) are indicated for each pair, except when non-significant (n.s.).
Next, we analysed the uridylation status of the five candidate mRNAs in urt1 mutants. Interestingly, we did not detect uridylated transcripts for LOM1 (0/55), At2g21560 (0/55) and At5g48250 (0/103) and only 2/72 DREB2C clones and 1/107 BAM3 clones corresponded to uridylated transcripts in urt1 (Figure 2B). URT1 is therefore the nucleotidyltransferase activity that is mainly responsible for uridylating mRNAs in Arabidopsis.
mRNA uridylation was also determined in a xrn4 mutant because the Arabidopsis 5′ to 3′ cytosolic exoribonuclease XRN4 (34) is a reasonable candidate for degrading uridylated mRNAs by analogy with data reported for humans and S. pombe (19,20). As in WT, uridylated mRNAs were detected in xrn4 for the five candidate mRNAs (Figure 2B), and uridylated mRNAs had significant shorter oligo(A) tails than non-uridylated ones in xrn4 (Figure 2D). However, both composition and length of the U tails differ between WT and xrn4 (Figure 2C). Whereas most U tails of WT mRNAs are 1-2 Us in length, the majority of oligo(U) tails (68/122) in xrn4 were at least 3 nucleotide long and could reach up to 11 nucleotides (Figure 2C). In addition, the oligo(U)-rich tails in xrn4 included up to 10% As (37/381), whereas the short tails in WT were almost exclusively composed of Us (180/181 nucleotides). The fact that both length and composition of U-tails are modified in absence of XRN4 suggests that uridylated mRNAs are substrates of XRN4.
Collectively, these data support the idea that URT1 uridylates mRNAs, that a deadenylation step precedes uridylation in Arabidopsis and that uridylated mRNAs are substrates of XRN4.
Uridylation marks uncapped, 5′ shortened LOM1 transcripts
Uridylation was reported to enhance decapping of mRNAs in S. pombe and A. nidulans and of non-polyadenylated histone mRNAs in humans (19–21). We therefore compared the proportion of uridylated transcripts between capped and uncapped transcripts by cRT-PCR using LOM1 as a model substrate. Capped LOM1 transcripts were detected in WT, urt1 and xrn4, whereas uncapped LOM1 mRNAs accumulated to detectable levels only in xrn4 (Figure 3A). This is in line with the idea that uncapped transcripts are degraded by XRN4. The 3′ extremities of both capped and uncapped transcripts in xrn4 mapped between positions +2074 to +2102 (Figure 3B). For the uncapped samples, 42 out of 70 clones were uridylated and only 2 out of 70 were not. For the rest of the clones (26/70), sequence analysis of cRT-PCR products cannot discriminate between uridines that could be either genomically encoded at the 5′ or added at the 3′ extremities. We favour the latter possibility because >90% of the uridylated LOM1 mRNAs had shortened poly(A) tails and half of them correspond to 5′ truncated transcripts (see below). Therefore up to 98% of the clones in the uncapped sample correspond to uridylated transcripts. By contrast, only 1 clone out of 73 corresponding to capped transcripts was uridylated. In addition, 95% (69/73) of 5′ extremities of capped transcripts map at or within 27-nt downstream of the known LOM1 5′-ends. By contrast, the 5′ extremities of 49% (34/70) of uncapped transcripts map within the coding sequence, indicating that a 5′ shortening occurs even in absence of the known cytosolic 5′ to 3′ exoribonuclease XRN4 (Figure 3B). Whether these 5′ shortened transcripts in xrn4 are generated by an endoribonuclease activity or by a cytosolic 5′ to 3′ exoribonuclease distinct from XRN4 is presently unknown. Interestingly, the uncapped, 5′ shortened transcripts that are detected in xrn4 mutants share the same 3′ extremities as capped transcripts. This indicates that even in xrn4 where 5′ to 3′ RNA degradation is compromised, uridylated LOM1 transcripts are still prone to 5′ to 3′ degradation rather than to 3′ attacks.
Figure 3.
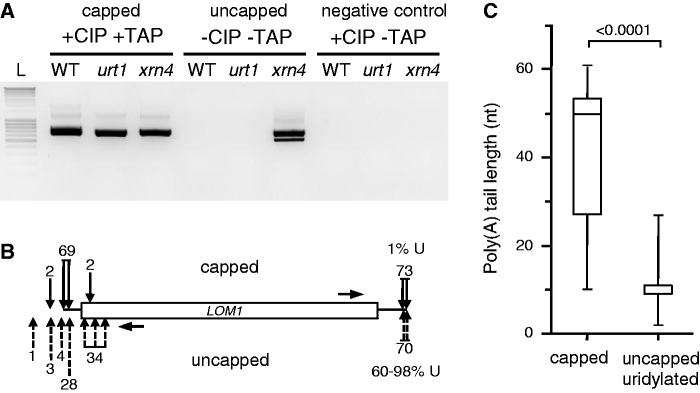
Uridylation marks 5′ shortened uncapped LOM1 mRNAs. (A) Detection of capped and uncapped LOM1 mRNAs by cRT-PCR analysis following combinations of CIP and TAP treatments as indicated. Capped mRNAs were detected following successive treatments with CIP and TAP. Both CIP and TAP treatments were omitted to detect uncapped transcripts. Negative controls were treated with CIP only (see details in ‘Materials and Methods’ section). PCR products were run alongside a DNA ladder (L) with 100-bp size increments. The negative image of an ethidium bromide–stained agarose gel is shown. (B) Most uncapped LOM1 mRNAs are uridylated. Positions of 5′- and 3′-ends of capped (black arrows above diagram) and uncapped (dashed arrows below diagram) LOM1 mRNAs in xrn4. Seventy-three and 70 clones were analysed for capped and uncapped mRNAs, respectively. The numbers of clones is indicated for each position. Horizontal arrows indicate the position of primers used for cRT-PCR. The percentage of clones corresponding to uridylated mRNAs is shown at the 3′-ends. (C) Uncapped uridylated LOM1 mRNAs have shorter poly(A) tails in xrn4 as compared with capped transcripts. Box plot analysis as in Figure 2.
The cRT-PCR experiments also confirmed that uridylated LOM1 transcripts have a decreased poly(A) size as compared with non-uridylated capped transcripts (Figure 3C). In xrn4, the LOM1 poly(A) tail distribution had a median of 50 As for capped transcripts and 11 As for uridylated uncapped transcripts (P < 0.0001, Mann–Whitney test). The prevalence of uridylated transcripts in the uncapped fraction, the 5′ shortening of uridylated transcripts and the fact that uridylated transcripts have short poly(A) tails suggest that mRNA uridylation in Arabidopsis is linked to decapping and 5′ to 3′ degradation, as previously reported in humans, S. pombe and A. nidulans. However, it is difficult at present to estimate whether uridylation facilitates decapping in Arabidopsis because uridylation marks oligoadenylated mRNAs, and deadenylation alone can promote decapping. Importantly, none of the phenotypes linked with compromised decapping or 5′ to 3′ RNA degradation in Arabidopsis were observed in urt1 mutants, suggesting that decapping is not adversely compromised in absence of URT1. For instance, urt1 mutants do not show the ethylene-insensitive phenotype of xrn4 seedlings (35,36) (Supplementary Figure S3) and we did not observe abnormalities in embryo-to-seedling transition, a developmental step strongly affected by the loss of decapping activity (31,37).
Uridylation has no major impact on mRNA degradation rates
The genome-wide impact of URT1 on mRNA steady-state levels was determined by microarray analysis. One hundred sixty-four genes showed significant different expression levels between WT and urt1 (65 up, 99 down) (see Supplementary Table S2 for a list of misregulated genes and gene ontology analysis, and Supplementary Figure S4A for the validation of three misregulated genes in urt1). The number of differentially expressed genes is comparable with those observed for xrn4 (38,39). However, there is no significant overlap between urt1 and xrn4 data sets because only one gene (At5g49280) is misregulated both in urt1 and xrn4. To test a potential impact of URT1 on mRNA degradation rates, half-lives of five short-lived transcripts (i.e. half-lives ranging between 20 and 40 min) were determined following inhibition of transcription by cordycepin or actinomycin D. The degradation rates of the five mRNAs were not significantly affected regardless of their respective up-regulated, down-regulated or unchanged expression level in urt1 (Supplementary Figure S4B and C). Therefore, the variation of steady-state levels of these mRNAs in urt1 is rather due to indirect effects or multifactorial causes that remain to be identified. Although we cannot exclude that the mRNA half-lives determined following transcription inhibition by cordycepin or actinomycin D could not faithfully reproduce the actual in vivo half-lives, these data argue that URT1 has no major impact on mRNA degradation rates.
Oligoadenylated mRNAs are trimmed in urt1 mutants
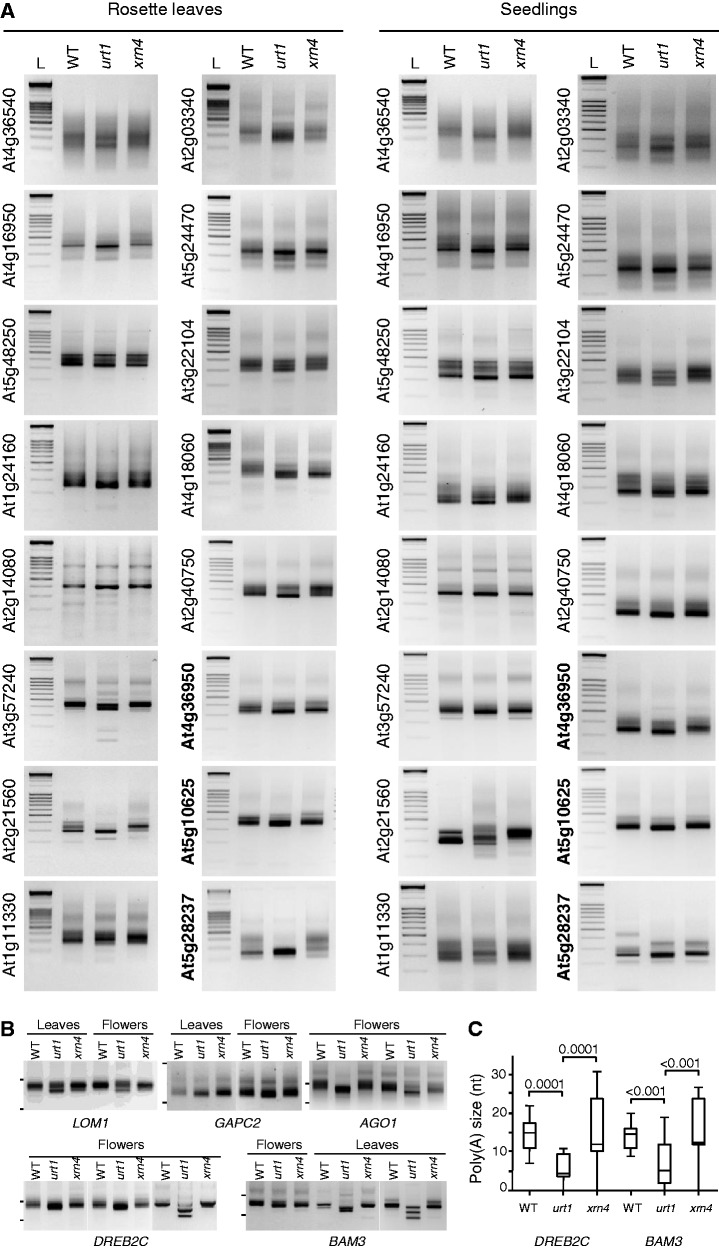
A new role for mRNA uridylation in Arabidopsis was determined following the observation of a slight but systematic reduction in the size of the main 3′ RACE PCR products for 16 of the most up- or down-regulated mRNAs in urt1 as well as for the short-lived transcripts DREB2C and BAM3 or any other mRNAs investigated (Figures 4A and B, Supplementary Figure S1C and S5B). This size shift in urt1 is not only detected for all mRNAs tested but is also observed for the different developmental stages analysed: rosette leaves, seedlings and flowers (Figures 4A and B, Supplementary Figure S5B) showing the robustness of this molecular phenotype in urt1 mutants.
Figure 4.
Uridylation protects from mRNA 3′ shortening. (A) URT1 prevents 3′ shortening of mRNAs irrespective of their misregulation in urt1. Thirteen down- and 3 up-regulated transcripts (regular and bold AGI numbers, respectively) in urt1 were analysed by 3′ RACE analysis in WT, urt1 and xrn4 at two developmental stages, rosette leaves and seedlings. All panels correspond to negative images of ethidium bromide–stained 2% agarose gels. 3′ RACE PCR products were run alongside a DNA ladder (L) with 100-bp size increments. Complex patterns reflect multiple polyadenylation sites frequently observed for Arabidopsis mRNAs. (B) 3′ shortening in urt1 detected by 3′ RACE PCR for 5 mRNAs. Genotypes, tissues and gene identities are indicated for all panels corresponding to negative images of ethidium bromide–stained 2% agarose gels. The migration of 0.5 and 0.2 kb size standards are shown on the left. (C) Increased deadenylation of DREB2C and BAM3 mRNAs in urt1. Box plot analysis as in Figure 2.
To understand the nature of the size shift, we cloned size-selected amplification products corresponding to the main band for DREB2C and BAM3. The distribution of poly(A) sites was similar between WT, urt1 and xrn4. However, the cloned poly(A) tails were shorter in urt1 (Mann–Whitney test P < 0.001) (Figure 4C). Enhanced deadenylation is therefore responsible for the size shift observed by 3′ RACE PCR in urt1. Because 3′ RACE PCR preferentially detects shorter over long poly(A) tails, we checked whether only oligo(A) tails rather than full-length poly(A) tails were shortened in urt1 mutants, a rationale hypothesis because URT1’s RNA substrates correspond to oligoadenylated mRNAs. To this end, we chose the At5g08040 mRNAs which, unlike DREB2C and BAM3, are expressed to sufficient levels to be detected by northern analysis and are well resolved on acrylamide gels owing to their short size of 371 nt [excluding poly(A) tails]. A northern analysis was performed using RNA samples from WT, urt1 and xrn4 flowers that were either treated with RNase H and oligo(dT) to remove poly(A) tails, or not treated with RNase H. This analysis revealed that similar long poly(A) tails of a maximal size of ∼100 nt are observed for At5g08040 mRNAs in the three genotypes (Supplementary Figure S5A). The slight size shift for RNA species with shorter tails is more difficult to observe on northern blots but is convincingly detected by 3′ RACE PCR in flowers, as well as in seedlings (Supplementary Figure S5B).
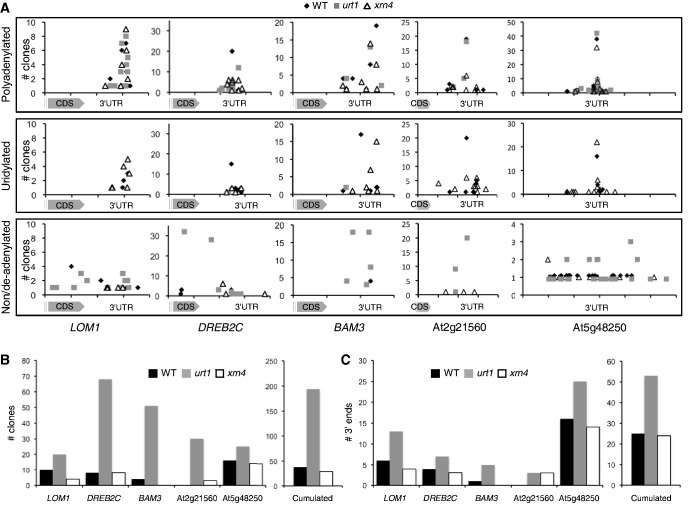
A second molecular phenotype was observed in urt1. As shown for several experiments for BAM3 and DREB2C, additional 3′ RACE PCR products with reduced sizes were occasionally amplified in urt1 mutants (Figure 4B). To understand the nature of these heterogeneous patterns, whole PCR reactions from two to three independent experiments were cloned. The corresponding sequence analysis is presented for LOM1, DREB2C, BAM3, At2g21560 and At5g48250 (Figure 5). Cloned 3′ RACE PCR products were split into three categories: (i) polyadenylated transcripts, (ii) uridylated transcripts and (iii) non-polyadenylated or deadenylated transcripts. Transcripts were considered deadenylated when the poly(A) tail length was <10, a size inferior to the minimal poly(A) tail length required for the binding of a poly(A) binding protein (40). Plotting the clone numbers against the position of 3′-ends on the respective mRNAs shows that multiple polyadenylation sites are detected for the five genes (Figure 5A). This observation is coherent with the intrinsic 3′ heterogeneity described for mRNA 3′-ends in Arabidopsis (41). The overall distribution of polyadenylation sites appeared unchanged in WT, urt1 and xrn4 (Figure 5A). Of note, the distribution of uridylated ends in WT and xrn4 coincides perfectly with the positions of polyadenylation sites (Figure 5A). In fact, all uridylated mRNAs detected in the course of this study in WT correspond to adenylated mRNAs [with a strong bias toward oligo(A) tails]. Therefore, although we do not formally exclude the possible uridylation of truncated and/or non-polyadenylated mRNAs in WT, our data indicate that oligoadenylated mRNAs are the main substrates of URT1. Importantly, the number and the diversity of clones corresponding to deadenylated or non-polyadenylated (from 0 to 10 As) transcripts were increased in urt1 as compared with WT and xrn4 (Figure 5A). Because clones having identical 3′-ends could correspond either to sister amplification or reflect the amplification of preferential degradation intermediates, we considered both the number of clones and the number of distinct 3′-ends detected for non/de-adenylated mRNAs. Overall, both the total number of clones and the diversity of the 3′-ends were similar in WT and xrn4 but markedly increased in urt1 (Figures 5B and C, respectively). For LOM1 and DREB2C, the 3′ extremities of deadenylated or non-polyadenylated transcripts covered the whole range of the sequence investigated by 3′ RACE PCR with some 3′-ends mapping even within the CDS (Figure 5A). This analysis revealed that the occasional shorter PCR products detected by 3′ RACE correspond to 3′ truncated transcripts and that the occurrence of 3′ truncated transcripts is increased in urt1.
Figure 5.
3′ truncated transcripts accumulate in urt1. (A) Number of clones (n = 141 for LOM1, n = 180 for DREB2C, n = 229 for BAM3, n = 163 for At2g21560, n = 314 for At5g48250) from at least two independent experiments and corresponding to polyadenylated, uridylated, non/deadenylated (from 0 to 10 As) mRNAs were plotted against the 3′ region of each respective mRNA as indicated. One interval on abscissas is 100 nt. CDS, coding sequence. (B) Individual and cumulated numbers of non/deadenylated clones for LOM1, DREB2C, BAM3, At2g21560 and At5g48250 in WT, urt1 and xrn4. (C) Individual and cumulated numbers of distinct 3′-ends of non/deadenylated clones for LOM1, DREB2C, BAM3, At2g21560 and At5g48250 in WT, urt1 and xrn4.
Taken together, these results show that the major phenotype observed in urt1 mutants corresponds to a 3′ shortening of oligoadenylated mRNAs. Because this shortening was systematically observed and uridylation was detected for any mRNA investigated, we propose that mRNA uridylation is a frequent process in Arabidopsis and that uridylation prevents complete deadenylation of mRNAs. This is further supported by the increased occurrence of 3′ truncated transcripts in urt1 mutants that likely results from 3′ exoribonucleolytic attacks in the mRNA body once the mRNA has been fully deadenylated (see ‘Discussion’ section).
Uridylation can prevent 3′ trimming of polysomal mRNAs
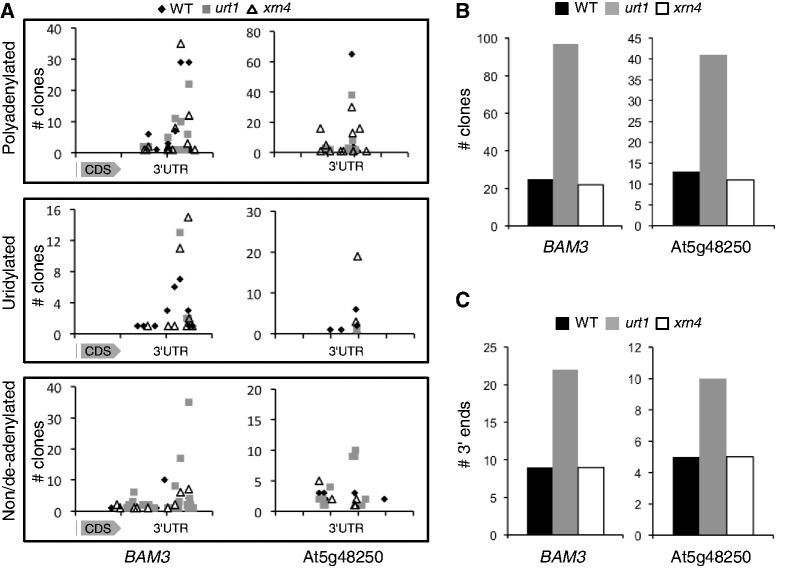
The occurrence of co-translational mRNA decay was reported in S. cerevisiae (42) and recently, uridylation was shown to tag polysomal mRNAs in another fungus, A. nidulans (22). In this species, uridylation tags oligoadenylated transcripts in a similar way as for Arabidopsis mRNAs (22). We therefore tested whether uridylation could prevent the occurrence of 3′ shortened mRNAs associated with polysomes. Polysomes were isolated from seedlings, and typical UV profiles were observed for WT, urt1 and xrn4 samples, indicating no major effects of the urt1 mutation on polysome assembly (Figure 6A). RNA extracted from polysomal fractions was used to determine 3′ RACE PCR profiles for four transcripts, and the characteristic 3′ shortening was observed in urt1 (Figure 6B). Two experiments are shown for BAM3 and At5g48250 to show that the 3′ shortening is systematically observed in urt1 even though the PCR profiles may vary slightly. In addition, shorter amplicons corresponding to 3′ truncated transcripts were occasionally detected in urt1 polysomal fractions as shown for BAM3. Cloning and sequence analysis of BAM3 and At5g48250 samples revealed the presence of uridylated mRNAs in polysomal fractions in WT and xrn4 and confirmed that both the number of clones corresponding to non- or de-adenylated transcripts and the number of distinct 3′-ends are increased in urt1 (Figure 7). These results indicate that uridylation can prevent 3′ trimming of mRNAs associated with polysomes.
Figure 6.
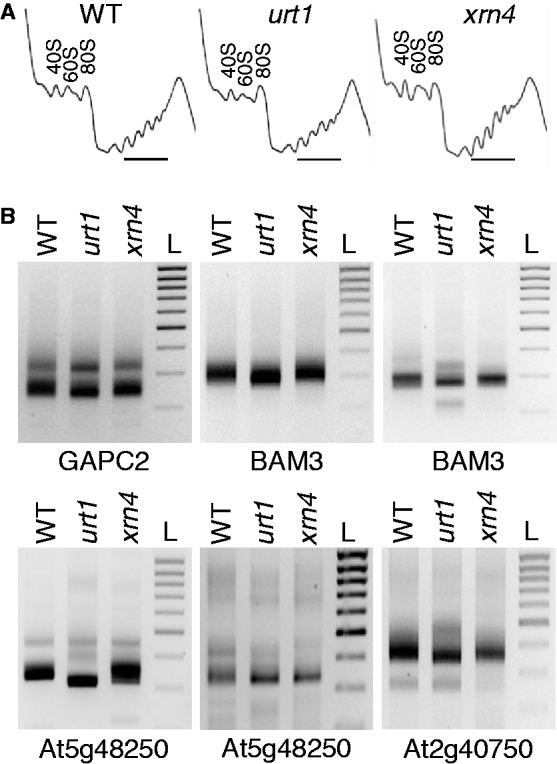
Uridylated mRNAs are present in polysomal fractions. (A) Polysome profiles in WT, urt1 and xrn4. The position of the 40S, 60S and 80S peaks is indicated on each panel. Polysomal fractions analysed by 3′ RACE PCR are marked by a horizontal line. (B) 3′ shortening of mRNAs in urt1 is detected in polysomal fractions by 3′ RACE PCR. Genotypes and identity of mRNAs are indicated on all panels corresponding to negative images of ethidium bromide–stained agarose gels. The migration of a DNA ladder (L) (1 to 0.2 kb, 0.1 kb size increment) is shown on the right of each panel. Two experiments are shown for BAM3 and At5g48250 to show that the PCR profiles may vary but the 3′ shortening is systematically observed in urt1.
Figure 7.
3′ truncated transcripts accumulate in urt1 polysomal fractions. (A) Number of clones (n = 423 for BAM3, n = 326 for At5g48250) corresponding to polyadenylated, uridylated, non/deadenylated (from 0 to 10 As) mRNAs plotted against position alongside the 3′ region of each respective mRNA as indicated. One interval on abscissas is 100 nt. CDS, coding sequence. (B) Numbers of non/deadenylated clones for BAM3 and At5g48250 in WT, urt1 and xrn4. (C) Numbers of distinct 3′-ends of non/deadenylated clones for BAM3 and At5g48250 in WT, urt1 and xrn4.
DISCUSSION
Uridylation of mRNAs was recently reported to promote 5′ to 3′ degradation through decapping (19,20). Here we show that in addition, uridylation prevents the formation of 3′ trimmed mRNAs in Arabidopsis, revealing a novel role for uridylation. We propose that uridylation favours the 5′ to 3′ directionality of mRNA decay, likely by protecting the 3′-ends of oligoadenylated mRNAs that are committed for 5′ to 3′ degradation as discussed below. 5′ to 3′ polarity is crucial in case of co-translational decay (42) and, interestingly, uridylation limits the occurrence of 3′ shortened mRNAs engaged in polysomes, highlighting one role for mRNA uridylation in Arabidopsis.
Our results indicate that uridylation marks oligoadenylated mRNAs in Arabidopsis. The exact proportion of a given mRNA to be uridylated remains to be determined because the PCR-based technique used to detect uridylated mRNAs favours the amplification of shorter versus longer poly(A) tails (Supplementary Figure S5) and uridylated transcripts have shorter tails than non-uridylated ones. The ratio of clones corresponding to uridylated mRNAs is therefore likely overestimated. Despite this technical limitation, the 3′ RACE PCR strategy has the advantages of being gene-specific, sensitive and allows the detection of a single uridine present at the 3′-end of a poly(A) tail. This method is therefore well suited for detecting uridylated RNAs and comparing their occurrence between genotypes. To date, uridylation was systematically detected for any mRNA investigated and is therefore likely to be widespread in Arabidopsis.
The absence of a significant impact of URT1 on mRNA degradation rates is coherent with urt1 mutants displaying none of the phenotypes linked with compromised decapping or 5′ to 3′ RNA degradation. A likely interpretation of these observations is that the mRNAs that are marked by uridylation are already committed to degradation by the deadenylation step. Deadenylation is a rate-limiting step in mRNA degradation and is sufficient to trigger decapping. Both of these features could explain why mRNA degradation is not adversely compromised in urt1 mutants.
The most important finding of this study is that uridylation is required to limit the occurrence of 3′ trimmed and truncated mRNAs. Three alternative interpretations could explain this observation. First, the 5′ to 3′ RNA degradation pathway could be impaired in urt1, thereby promoting 3′ to 5′ attacks of oligoadenylated mRNAs. Second, uridylation could be required to trigger the degradation of 3′ trimmed and truncated transcripts. Third, uridylation of oligoadenylated mRNAs could prevent 3′ trimming by protecting their 3′-ends. We do not favour the two first hypotheses for the following reasons. Several lines of evidence support the idea that Arabidopsis uridylated mRNAs are substrates of the 5′ to 3′ pathway as previously reported for humans and S. pombe (19,20): uridylation marks decapped mRNAs as shown for LOM1, these transcripts are 5′ shortened (Figure 3) and both the composition and length of U-tails are modified in xrn4 (Figure 2C). However, we never observed 3′ trimming in xrn4 for all mRNAs tested (Figures 4A and B, Figure 6, Supplementary Figure S1C and S5B), and truncated transcripts do not accumulate in xrn4 as compared with WT (Figures 5B and C and 7B and C). Both observations indicate that 3′ trimmed and truncated transcripts are not merely generated by favouring 3′ to 5′ attacks when 5′ to 3′ degradation is impaired.
Importantly, we never detected 3′ truncated transcripts that were uridylated, neither in WT nor in xrn4, even though the PCR strategy used here is sensitive enough to detect even low amounts of truncated transcripts in WT and xrn4 (Figures 5 and 7). Although we cannot rule out that truncated transcripts could be uridylated by URT1 at low levels, we have no indication that URT1 is required for the degradation of truncated transcripts. In addition, all uridylated mRNAs detected in WT and all but 2 clones in xrn4 (387 of 389 clones when combining data presented in Figures 3, 5 and 7) correspond to adenylated mRNAs and only 2 of 389 clones are deadenylated mRNAs in xrn4. This observation strongly supports the idea that the main mRNA substrates of URT1 are mRNAs that are still adenylated, albeit with short poly(A) tails.
By contrast, several lines of evidence support the idea that uridylation could prevent 3′ to 5′ attacks of mRNAs that have been deadenylated to ∼10–14 As. As described above, the main substrates of URT1 are mRNAs with short poly(A) tails. In addition, when we characterized uncapped uridylated LOM1 transcripts in xrn4, we observed that these RNAs are 5′ shortened but showed no sign of 3′ trimming (Figure 3). This indicates that uridylated mRNAs are prone to 5′ rather than 3′ attacks, even in a xrn4 genetic background. A similar observation was made in wild-type plants for uridylated RISC-cleaved transcripts that are 5′ but not 3′ shortened (17). Uridylation also prevents 3′ to 5′ exoribonucleolytic decay in mammalian cell extracts, in addition to promote decapping (18). Both studies concluded that uridylation favours the 5′ to 3′ directionality of RNA decay and supports the idea that uridylation could stabilize 3′-ends (17,18). In fact, U-tracts that terminate the transcription of RNA polymerase III precursors are well known to prevent 3′ to 5′ degradation by promoting the binding of the La protein and the Lsm2-8 heptameric complex in the nucleus (43). Interestingly, a link between uridylation and binding of the cytosolic Lsm1-7 complex also exists. The Lsm1-7 complex recognizes preferentially oligo(A) versus poly(A) tails, but the binding of the Lsm1-7 complex is enhanced by uridylation (18). In line with this, the Lsm1-7 complex is an integral component of the uridylation-mediated decapping pathway (19,20). The current model of this pathway is that uridylation enhances binding of the Lsm1-7 complex (19,20), which recruits the decapping complex via Pat1 (44–48). Importantly, in the context of our study, the Lsm1-7 complex also protects the 3′-ends of oligoadenylated mRNAs from ribonucleolytic attacks (49,50). In yeast lsm mutants, mRNAs are trimmed by 10–20 nucleotides at their 3′-ends, a phenotype similar to the one observed for urt1 mutants. Although the trimming reaction itself is not performed by the exosome, 3′ trimming enhances exosome-mediated 3′ to 5′ decay (50,51). The Lsm1-7 complex could therefore play a dual role in RNA decay, by promoting 5′ to 3′ degradation but also by playing a direct role in inhibiting 3′ ribonucleolytic attacks. Both functions of the Lsm1-7 complex are likely conserved in Arabidopsis. However, the presence of the Lsm1-7 complex is not sufficient to ensure protection of the 3′-ends of oligoadenylated mRNAs in Arabidopsis urt1 mutants, and the uridylation activity of URT1 is mandatory. This observation implies that preventing the formation of 3′ trimmed and truncated transcripts in Arabidopsis requires an active process, uridylation, highlighting its biological significance.
Degradation is commonly preceded by mRNAs exiting translation (45,48) but mRNA degradation can also occur on polysomes in budding yeast (42). This finding led to the hypothesis that the 5′ to 3′ directionality of RNA decay evolved to allow the last translocating ribosome to complete translation. Our data indicate that uridylation definitely participates in the establishment of 5′ to 3′ polarity of RNA degradation in Arabidopsis by preventing the formation of 3′ trimmed and truncated transcripts. Detecting uridylated mRNAs in polysomal fractions strongly suggests that protection by uridylation operates on polysomes. It is unclear at present what proportion of uridylated mRNAs is associated to polysomes. However, even if this proportion is low, protection of mRNA 3′-end by uridylation could be advantageous to limit the occurrence of truncated transcripts in polysomes. Further work is also required to identify the ribonuclease(s) involved in nibbling the unprotected oligoadenylated mRNAs in urt1. In yeast lsm mutants, the nibbling enzyme is also unknown but the exosome was shown to degrade 3′ trimmed mRNAs (50). The 3′ truncated transcripts in urt1 mutants could also become substrate of the 3′ to 5′ degradation pathway, either of the exosome or other cytosolic 3′ to 5′ exoribonucleolytic activities such as SUPPRESSOR OF VARICOSE, a member of the RNase II family associated with RNP granules or RRP6L3, a cytosolic RRP6-like exoribonuclease (52,53). Irrespective of the exact fate of 3′ truncated mRNAs in urt1 mutants, the most important consequence of our data is that the preventive action of URT1 ensures that such aberrant transcripts are not, or to a much lesser extent, produced in WT plants. Whereas mechanisms of RNA quality control have evolved to detect and eliminate defective transcripts, uridylation is rather part of a preventive process that prevents the occurrence of 3′ trimmed and truncated transcripts.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–2, Supplementary Figures 1–5, Supplementary Methods and Supplementary References [54–61].
FUNDING
Centre National de la Recherche Scientifique (CNRS, France); Agence Nationale de la Recherche (ANR, France) [ANR-2010-BLAN-1707 to D.G. and J.M.D.], [ANR-2010-LABX-36 to D.G.] in the frame of the programme d'investissements d'avenir]. Funding for open access charge: Centre National de la Recherche Scientifique (CNRS), France.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Benjamin Stupfler and Barbara Class for technical help.
REFERENCES
- 1.Trippe R, Sandrock B, Benecke BJ. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 1998;26:3119–3126. doi: 10.1093/nar/26.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilusz CJ, Wilusz J. New ways to meet your (3′) end oligouridylation as a step on the path to destruction. Genes Dev. 2008;22:1–7. doi: 10.1101/gad.1634508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marzluff W. A new way to initiate mRNA degradation. Nat. Struct. Mol. Biol. 2009;16:613–614. doi: 10.1038/nsmb0609-613. [DOI] [PubMed] [Google Scholar]
- 4.Norbury CJ. 3′ Uridylation and the regulation of RNA function in the cytoplasm. Biochem. Soc. Trans. 2010;38:1150–1153. doi: 10.1042/BST0381150. [DOI] [PubMed] [Google Scholar]
- 5.Morozov IY, Caddick MX. Cytoplasmic mRNA 3′ tagging in eukaryotes: does it spell the end? Biochem. Soc. Trans. 2012;40:810–814. doi: 10.1042/BST20120068. [DOI] [PubMed] [Google Scholar]
- 6.Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-MicroRNA as a key step in the biogenesis of group II let-7 MicroRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 7.Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, Wolf DA, Mizgerd JP. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol. 2009;11:1157–1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones MR, Blahna MT, Kozlowski E, Matsuura KY, Ferrari JD, Morris SA, Powers JT, Daley GQ, Quinton LJ, Mizgerd JP. Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival. PLoS Genet. 2012;8:e1003105. doi: 10.1371/journal.pgen.1003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc. Natl Acad. Sci. USA. 2010;107:3906–3911. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJ, Roovers EF, Ladurner P, Berezikov E, Ketting RF. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 2010;29:3688–3700. doi: 10.1038/emboj.2010.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren G, Chen X, Yu B. Uridylation of miRNAs by HEN1 SUPPRESSOR1 in Arabidopsis. Curr. Biol. 2012;22:695–700. doi: 10.1016/j.cub.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X. The arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 2012;22:689–694. doi: 10.1016/j.cub.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, Kremmer E, Ansel KM, Heissmeyer V. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat. Struct. Mol. Biol. 2012;20:73–81. doi: 10.1038/nsmb.2450. [DOI] [PubMed] [Google Scholar]
- 17.Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 18.Song MG, Kiledjian M. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA. 2007;13:2356–2365. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol. 2009;16:616–623. doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morozov IY, Jones MG, Razak AA, Rigden DJ, Caddick MX. CUCU modification of mRNA promotes decapping and transcript degradation in Aspergillus nidulans. Mol. Cell. Biol. 2010;30:460–469. doi: 10.1128/MCB.00997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morozov IY, Jones MG, Gould PD, Crome V, Wilson JB, Hall AJ, Rigden DJ, Caddick MX. mRNA 3′ tagging is induced by nonsense-mediated decay and promotes ribosome dissociation. Mol. Cell. Biol. 2012;32:2585–2595. doi: 10.1128/MCB.00316-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwak JE, Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 25.Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sement FM, Gagliardi D. Polydenylation: Methods and Protocols. In: Rorbach J, editor. Methods In Molecular Biology. Springer Science: Humana Press; 2013. in press. [Google Scholar]
- 27.Mustroph A, Juntawong P, Bailey-Serres J. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol. Biol. 2009;553:109–126. doi: 10.1007/978-1-60327-563-7_6. [DOI] [PubMed] [Google Scholar]
- 28.Munoz-Tello P, Gabus C, Thore S. Functional implications from the Cid1 poly(U) polymerase crystal structure. Structure. 2012;20:977–986. doi: 10.1016/j.str.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 29.Lunde BM, Magler I, Meinhart A. Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity. Nucleic Acids Res. 2012;40:9815–9824. doi: 10.1093/nar/gks740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yates LA, Fleurdepine S, Rissland OS, De Colibus L, Harlos K, Norbury CJ, Gilbert RJ. Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase. Nat. Struct. Mol. Biol. 2012;19:782–787. doi: 10.1038/nsmb.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, Yang JY, Niu QW, Chua NH. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18:3386–3398. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber C, Nover L, Fauth M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 2008;56:517–530. doi: 10.1111/j.1365-313X.2008.03623.x. [DOI] [PubMed] [Google Scholar]
- 33.Narsai R, Howell KA, Millar AH, O'Toole N, Small I, Whelan J. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell. 2007;19:3418–3436. doi: 10.1105/tpc.107.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kastenmayer JP, Green PJ. Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl Acad. Sci. USA. 2000;97:13985–13990. doi: 10.1073/pnas.97.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potuschak T, Vansiri A, Binder BM, Lechner E, Vierstra RD, Genschik P. The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell. 2006;18:3047–3057. doi: 10.1105/tpc.106.046508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olmedo G, Guo H, Gregory BD, Nourizadeh SD, Aguilar-Henonin L, Li H, An F, Guzman P, Ecker JR. ETHYLENE-INSENSITIVE5 encodes a 5′–>3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc. Natl Acad. Sci. USA. 2006;103:13286–13293. doi: 10.1073/pnas.0605528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goeres DC, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth LE. Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell. 2007;19:1549–1564. doi: 10.1105/tpc.106.047621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rymarquis LA, Souret FF, Green PJ. Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA. 2011;17:501–511. doi: 10.1261/rna.2467911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gregory BD, O'Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherstnev A, Duc C, Cole C, Zacharaki V, Hornyik C, Ozsolak F, Milos PM, Barton GJ, Simpson GG. Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat. Struct. Mol .Biol. 2012;19:845–852. doi: 10.1038/nsmb.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beggs JD. Lsm proteins and RNA processing. Biochem. Soc. Trans. 2005;33:433–438. doi: 10.1042/BST0330433. [DOI] [PubMed] [Google Scholar]
- 44.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tharun S. Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int. Rev. Cell. Mol. Biol. 2009;272:149–189. doi: 10.1016/S1937-6448(08)01604-3. [DOI] [PubMed] [Google Scholar]
- 46.Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E. HPat provides a link between deadenylation and decapping in metazoa. J. Cell Biol. 2010;189:289–302. doi: 10.1083/jcb.200910141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozgur S, Chekulaeva M, Stoecklin G. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol. Cell. Biol. 2010;30:4308–4323. doi: 10.1128/MCB.00429-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boeck R, Lapeyre B, Brown CE, Sachs AB. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He W, Parker R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics. 2001;158:1445–1455. doi: 10.1093/genetics/158.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tharun S. Lsm1-7-Pat1 complex: a link between 3′ and 5′-ends in mRNA decay? RNA Biol. 2009;6:228–232. doi: 10.4161/rna.6.3.8282. [DOI] [PubMed] [Google Scholar]
- 52.Lange H, Holec S, Cognat V, Pieuchot L, Le Ret M, Canaday J, Gagliardi D. Degradation of a polyadenylated rRNA maturation by-product involves one of the three RRP6-like proteins in Arabidopsis thaliana. Mol. Cell. Biol. 2008;28:3038–3044. doi: 10.1128/MCB.02064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang W, Murphy C, Sieburth LE. Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proc. Natl Acad. Sci. USA. 2010;107:15981–15985. doi: 10.1073/pnas.1007060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 58.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Provart N, Zhu T. A Browser-based functional classification superviewer for Arabidopsis genomics. Curr. Comput. Mol. Biol. 2003;2003:271–272. [Google Scholar]
- 60.Maere S, Heymans K, Kuiper M. BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 61.Gagnot S, Tamby JP, Martin-Magniette ML, Bitton F, Taconnat L, Balzergue S, Aubourg S, Renou JP, Lecharny A, Brunaud V. CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res. 2008;36:D986–D990. doi: 10.1093/nar/gkm757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.