Abstract
The human genome has been referred to as the blueprint of human biology. In this review we consider an essential but largely ignored overlay to that blueprint, the human microbiome, which is composed of those microbes that live in and on our bodies. The human microbiome is a source of genetic diversity, a modifier of disease, an essential component of immunity, and a functional entity that influences metabolism and modulates drug interactions. Characterization and analysis of the human microbiome have been greatly catalyzed by advances in genomic technologies. We discuss how these technologies have shaped this emerging field of study and advanced our understanding of the human microbiome. We also identify future challenges, many of which are common to human genetic studies, and predict that in the future, analyzing genetic variation and risk of human disease will sometimes necessitate the integration of human and microbial genomic data sets.
Keywords: bacteria, microbiota, metagenomics, 16S rRNA
INTRODUCTION
A logical step following the completion of the human genome sequence was the identification and functional validation of variation in the human genome as well as dissection of its association with disease. An important aspect to be considered in parallel to these studies is now emerging: the contribution of the human microbiome to health and disease. The term microbiome was coined by Joshua Lederberg to “signify the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space and have been all but ignored as determinants of health and disease” (44). And although the microbiota of our bodies have largely been overlooked (aside from attempts at suppression and eradication), they constitute 90% of the total number of cells associated with our bodies; only the remaining 10% are human cells (78). Despite the extensive demonization that has ensued in this age of antibiotics and antimicrobials, the microbes living in and on our bodies are largely commensal and provide us with genetic variation and gene functions that human cells have not had to evolve on their own. In this review, we discuss (a) the genomic technology that instigated and continues to advance human microbiome studies; (b) how the human microbiome causes, contributes to, and modulates health and disease, including conditions that once were thought to be genetically encoded purely by the 23 human chromosomes; and (c) the challenges and pitfalls associated with this area of research, which is still in its infancy.
A HISTORICAL PERSPECTIVE
The cultivation and isolation of bacteria have long been the gold standard for the identification and characterization of microbes, beginning in the late 1800s when Robert Koch developed techniques to isolate the agent responsible for anthrax. After the isolation of single colonies, bacterial identification was achieved by direct observation of the bacterial cells and their morphology, biochemical testing, differential staining, and the use of enrichment cultures. On several occasions, it was noted that the number and diversity of cells observed microscopically far exceeded those of cells grown in culture (2, 90). It soon became apparent that culture-based approaches introduced bias, selecting for those microbes that thrive in isolation and under specific laboratory conditions, and that the full diversity of microorganisms had remained largely unexplored.
The elucidation of bacterial phylogeny based on the well-conserved small-subunit 16S ribosomal RNA (rRNA) sequence—the seminal work of Woese and colleagues (105, 106)—set the stage for genomic identification and analysis of microbial communities. Soon after the framework for bacterial phylogeny was established, Pace and colleagues developed a method to circumvent culture-based approaches in identifying bacteria (43, 89) based on isolation [via polymerase chain reaction (PCR)] of rRNA genes from bulk DNA extracted from an environmental sample. The 16S rRNA gene, which is approximately 1,500 base pairs in length, contains species-specific hypervariable regions but is conserved enough for PCR amplification using broad-range primers (36). The 16S rRNA gene sequences are compared with the phylogenetic reference tree to assign taxonomy. Since the original 16S rRNA–based phylogeny was described from an initial group of 11 bacterial phyla in 1987, reference databases have exploded with sequence data (105). As of February 2012, the Ribosomal Database Project (RDP) (version 28) contained more than 2 million 16S rRNA sequences and 35 phyla (13).
TAKING CUES FROM THE ENVIRONMENT AND ECOLOGY
Much of the technical and computational methodology for human microbiome research and analysis was developed to investigate environmental/ ecological communities. This included the interrogation of organisms and their functions from acid mine drainage (AMD) biofilms, activated sludge, Yellowstone hot springs, surface water of the Sargasso Sea, agricultural soil, and deep-sea whale skeletons (8, 37, 93, 96, 98). The first studies to couple nextgeneration sequencing technology with the microbiome were also environmental, including metagenomic analysis of a Minnesotan iron mine and a 16S rRNA tag sequencing survey of the deep sea to reveal the rare biosphere (23, 86).
Notably, AMD biofilms proved an ideal system for analyzing microbial community function using cultivation-independent approaches. AMD sites are low in microbial species richness and complexity owing to low pH, high metal concentrations, and limited resource availability. The results of such analyses uncovered species of bacteria and archaea that resist cultivation (22, 81). Shotgun sequencing of small-insert libraries constructed from AMD biofilms allowed the reconstruction of ~12 near-complete composite bacterial and archaeal genomes and opened the field of proteogenomic analysis (18). These studies, in addition to others, have demonstrated the tractability of the low-complexity AMD system to resolve the ecological and functional roles of microbial diversity and provide insights into interactions among bacteria, archaea, phages, and viruses. The utility of low-complexity environments such as AMD sites to provide insights into more complex, heterogeneous, and dynamic microbial populations is analogous to the utility of early human genetic studies that focused primarily on traits inherited in a monogenic, Mendelian fashion (e.g., hemophilia, sickle cell anemia, and cystic fibrosis).
Studies of host-microbe mutualism also gained momentum beginning with insights gleaned from ecological communities. A striking example is the southern pine beetle, which leverages a bacterium to protect its fungal food source from a competitor fungus. 16S rRNA gene sequencing revealed a novel actinomycete that colonizes the beetle and produces an antibiotic that selectively suppresses the antagonistic fungi (82). This example illustrates one of many ways in which a host is able to maintain beneficial microbes while suppressing hostile invaders.
GENOMIC TECHNOLOGY CATALYZES HUMAN MICROBIOME RESEARCH
Amplicon-Based Surveys of Human Microbial Communities
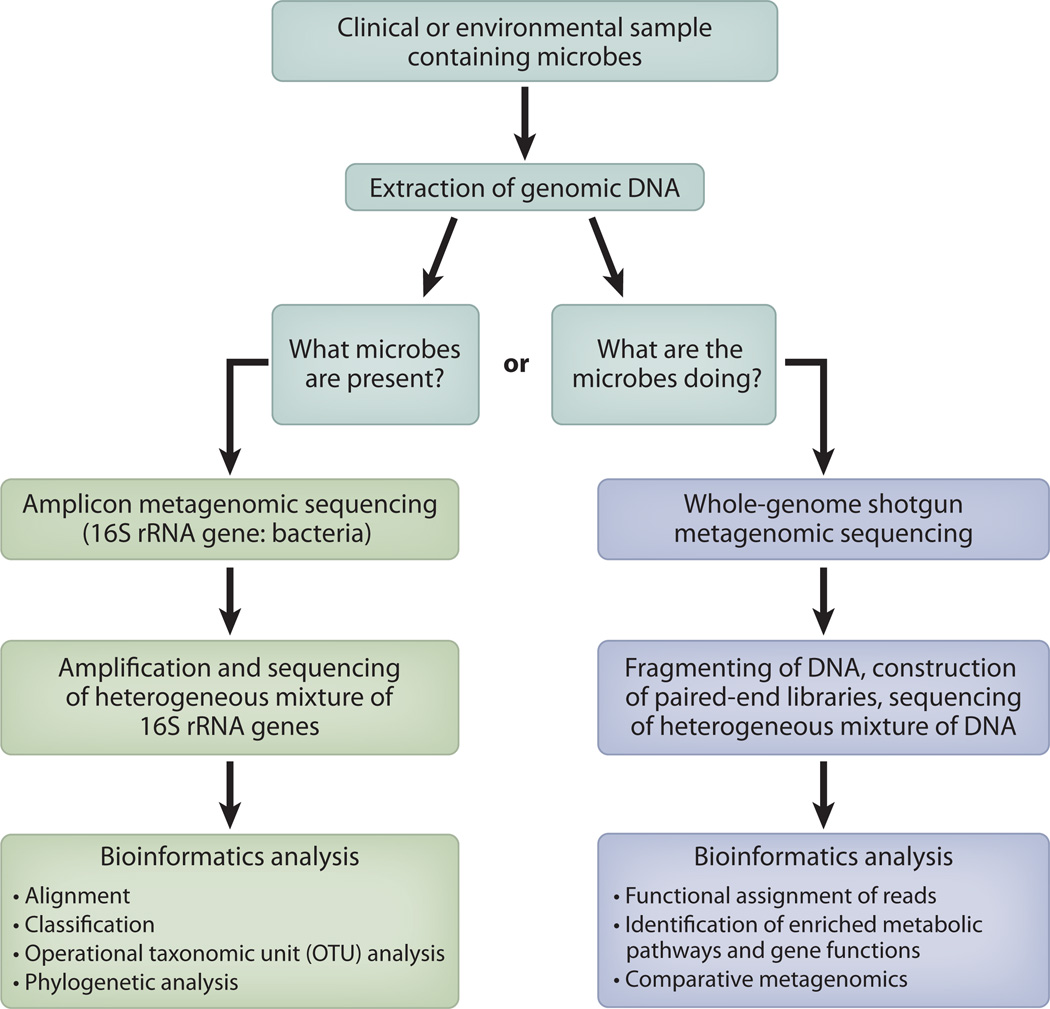
High-throughput surveys of microbial communities have been greatly enabled by culture-independent identification methods coupled with advancement in genomic technologies, especially DNA sequencing technology (Figure 1). The earliest human microbial surveys were based upon fingerprinting techniques, which relied upon physical separation of 16S rRNA genes by denaturing gradient gel electrophoresis and terminal restriction fragment length polymorphism analyses (49, 56). Greater specificity was gained with Sanger sequencing of the amplified and cloned 16S rRNA gene. The longer read lengths produced by Sanger sequencing are still a benefit to studies in the microbial community discovery phase. However, the advent and commercialization of highly parallel DNA sequencing technology have revolutionized our ability to quickly and precisely characterize microbial populations at a much lower cost and a greater depth than Sanger sequencing can provide.
Figure 1.
Workflow for metagenomic sequencing and analysis projects.
At present, many researchers in the field rely upon the Roche/454 pyrosequencing platform for 16S rRNA gene sequencing, which produces ~1 million ~400-nucleotide reads per run. Incorporating bar codes into the 5′ primer sequence multiplexes samples into a single sequencing run and enables researchers to produce thousands of 16S rRNA sequences per sample. If the goal of the study is to distinguish species within a genus (e.g., Staphylococcus epidermidis versus the pathogenic S. aureus), then longer read lengths (> 300 nucleotides) are necessary. As read lengths of the 16S rRNA gene decrease, so does taxonomic precision, and thus the ability to distinguish between strains, species, and even genera. However, for some studies researchers have preferred the greater sampling depth offered by Illumina platforms, and have explored paired-end sequencing to mitigate the shorter read length. Caporaso et al. (10) sequenced 1,967 microbiome samples across six lanes in a single run of the Illumina Genome Analyzer IIx. Balancing sequence length and sampling depth is one of the moving targets for the field, and the appropriate balance for a particular study is clearly shaped by the overall objectives. In general, the final decision of which sequencing platform to utilize depends upon the question being posed.
In addition to the choice of sequencing platform, investigators must consider many associated pitfalls when embarking on a bacterial survey study. Although the next-generation sequencing technologies eliminate cloning biases associated with Sanger sequencing, 16S rRNA gene diversity surveys still rely upon limited PCR amplification, which can potentially introduce bias. Known bias can derive from the design of the PCR primers or from PCR conditions that can cause amplification bias and chimera formation (102). Many of these study design issues and their analysis outcomes were recently reviewed by the Knight and Schloss groups (41, 79).
Microbial Genome Sequencing
Clearly, the use of next-generation sequencing has revolutionized the way that microbial diversity surveys are performed, but this technology has also had a tremendous impact on deciphering individual microbial genomes. In 1995, Haemophilus influenzae became the first bacterial genome sequenced with Sanger sequencing, introducing whole-genome random sequencing (85). At 1.8 Mb, the H. influenzae genome was 10 times larger than previously sequenced viral genomes. However, generating this genome assembly required the equivalent of three years of Sanger sequencing. (It was in fact completed by 14 machines over three months.) Then, with the advent of nextgeneration sequencing, generating this amount of data required only one instrument and one week. The ability to bypass cloning of DNA into bacteria for shotgun sequencing was also a major turning point in obtaining better coverage. Promoter regions of bacterial genes are to some degree uncloneable, and as a result these regions are typically underrepresented in initial data sets generated from cloning-based Sanger sequencing. As of February 2012, the Genomes Online Database (GOLD; http://www.genomesonline.org) provided nearly 3,000 finished bacterial genomes, with many thousands more underway.
Technological advances now make sequencing a single bacterial genome seem trivial and the potential to sequence thousands realistic. One of the initiatives of the National Institutes of Health (NIH) Common Fund’s Human Microbiome Project (http://commonfund.nih.gov/hmp) is to sequence 3,000 cultivated and uncultivated bacterial reference strains. This catalog of reference genomes is intended as a scaffold for the assembly of metagenomic sequences and as a reference for 16S rRNA gene sequences (69). In May 2010, 178 bacterial genomes were published, with the great majority isolated from the gastrointestinal (GI) tract (58). From this work and others, it is clear that only the tip of the iceberg of species diversity has been uncovered.
Targeted sequencing projects have focused on the pan-genome—the collection of genes found across all members of a species—as a useful framework for describing genomic diversity within a taxon. For example, Tettelin, Fraser-Liggett, and colleagues (92) characterized the pan-genome of Streptococcus agalactiae, an important pathogen for newborn infants, and found that ~20% of any given genome is made up of genes that are shared only partially with other strains. Furthermore, they found that sequencing additional genomes was predicted to increase the size of the pan-genome, indicating that S. agalactiae has an open genome. This type of analysis has been used to characterize a number of bacterial species as well as genera and has been used to estimate the pan-genome size of all bacteria.
Recent studies foreshadow a transition in the near future to sequencing technologies that can provide even faster turnaround for microbial genomics and microbiome studies. During the recent Escherichia coli outbreak in Germany, the Ion Torrent Personal Genome Machine proved its utility when a draft genome of the outbreak strain was produced in three days, furthering efforts to determine the evolutionary origins and pathogenic potential associated with the strain (76).
Whole-Genome Shotgun Metagenomic Sequencing
Highly parallelized DNA sequencing, such as that offered by the Illumina HiSeq, has paved the way for whole-genome shotgun (WGS) metagenomic analysis of microbial communities (Figure 1). Metagenomics provides the potential to analyze microbial communities from the viewpoint of functionality while circumventing both PCR and cloning bias. WGS metagenomics is a powerful alternative to sequencing 16S rRNA genes because it can in part answer not only the question “Who is there?” but also “What can they do?” The general strategy for a metagenomic study (Figure 1) is to directly extract DNA from a clinical sample (such as feces) and then construct a library of inserts that are multiplexed and sequenced. Tens of millions of Illumina reads are typically generated to articulate the full genetic potential of a sample. Unassembled reads that match signature genes (such as 16S rRNA) can be classified by searching against databases (such as greengenes or RDP-II) to determine the taxonomic makeup of the metagenomic data set. 16S rRNA read counts are used to calculate the frequency of microbial species, and differences between samples are a measure of relative microbial abundances.
However, individual reads are too short to generate the sequence of a complete gene and thus predict a protein-coding function. Reads are therefore assembled into contiguous DNA fragments to provide maximum information. However, there are often multiple highly similar genomes present in a WGS metagenomic data set. The sheer volume of sequence data generated combined with incomplete catalogs of sequenced reference genomes (bacterial, viral, archaeal, eukaryotic) makes WGS metagenomics an extremely computationally intensive undertaking. Sequence reads can be assembled as much as possible using tools such as SOAPdenovo (47), as longer reads provide more accurate gene annotation and phylogeny prediction. To identify putative protein-coding genes, a BLASTX search can be performed against either a nonredundant database or a database of microbial genome sequences specific to the environment/site of interest (e.g., the gut).
Functional data can then be inferred from the predicted proteins by searching against the KEGG (Kyoto Encyclopedia of Genes and Genomes) (38) and COG (Clusters of Orthologous Groups) (91) pathway databases. MG-RAST (Metagenomics Rapid Annotation Using Subsystem Technology), a Web-based metagenomics analysis server, has implemented many of these functions into an open-source metagenome analysis pipeline that combines normalization, alignment, functional and taxonomic assignment, and subsystem reconstruction into an automated workflow (53). MEGAN (Metagenome Analyzer) is a similar tool that performs taxonomic classification, functional analysis using KEGG or SEED classification, and computational comparison of metagenomes, all in an interactive analysis and visualization platform (55) (Table 1). It should be noted that reference databases (e.g., phylogenetic, pathway, and functional databases) are highly biased toward those organisms that are readily cultivable and those genes with known functions.
Table 1.
Selected examples of freely available bioinformatics tools for the analysis of 16S ribosomal RNA (rRNA) sequence data sets and metagenomic data sets
| Analysis tool | Web site | Reference |
|---|---|---|
Ribosomal Database Project:
|
http://rdp.cme.msu.edu | 13 |
mothur:
|
http://www.mothur.org | 80 |
QIIME (Quantitative Insights into Microbial Ecology):
|
http://www.qiime.org | 9 |
LEfSe (LDA Effect Size):
|
http://www.huttenhower.org/galaxy | 83 |
Metastats:
|
http://metastats.cbcb.umd.edu | 103 |
MG-RAST (Metagenomics Rapid Annotation Using Subsystem Technology):
|
http://metagenomics.anl.gov | 53 |
MEGAN (Metagenome Analyzer):
|
http://ab.inf.uni-tuebingen.de/software/megan | 55 |
CULTIVATING, SEQUENCING, AND DECIPHERING THE UNCULTIVABLE
One of the greatest obstacles to analyzing WGS metagenomic sequence data is the lack of reference genome sequence. To generate reference genome databases, it is crucial that new methods be developed to cultivate and isolate previously uncultivable and/or highly fastidious organisms. One recently developed technology is an ichip (isolation chip) composed of hundreds of miniature diffusion chambers, each inoculated with a single microbial cell (59). This method is able to recover numerous novel phylotypes not recovered by cultivation, but will work only when the environment is aqueous and capable of diffusion (e.g., seawater). For human samples, dissociation into single nonadherent bacterial cells would be a prerequisite to employ this method.
Some technologies are circumventing the cultivation step and leveraging whole-genome amplification to sequence single-cell genomes. Microfluidic isolation of bacteria from the subgingival crevice followed by single-cell genome amplification and sequencing resulted in the first glimpse into the phylum TM7, for which no isolate or sequence data were previously available (50). Woyke et al. (107) reported the first complete genome isolated from a single cell, in this case from a polyploid bacterium, Sulcia muelleri, isolated from the green sharpshooter. Isolation of single cells via fluorescence-activated cell sorting is also possible, and Chisholm’s group (75) employed this method followed by amplification of single-cell genomes on hundreds of bacteria simultaneously while virtually eliminating contaminating nontarget DNA. Multiple displacement amplification procedures leveraging the high-fidelity bacteriophage ϕ29 DNA polymerase are typically preferred for singlecell genome amplification because only minute quantities of DNA are required to amplify by several orders of magnitude (17). However, this method introduces significant biases in genome coverage, which some groups have attempted to mitigate through postamplification normalization procedures (75). Genomic methods to query single microbial cells could have an immense impact on the study of microbes, making previously uncultivable organisms and their genes readily accessible.
Once reference genome sequence is available, another significant obstacle is the functional annotation of putative open reading frames. Several efforts have been targeted at deciphering the function of hypothetical proteins. One such effort is COMBREX(Computational Bridges to Experiments), a consortium of closely collaborating experimental and computational biologists. Based on a concept put forth by Roberts (74), bioinformatic approaches are used to predict protein function and prioritize prime targets. The list of these targets is then open to experimentalists to functionally test using their experimental knowledge and reagents. The Joint Center for Structural Genomics has developed and integrated high-throughput structural biology methods to solve more than 1,000 protein structures, many of unknown function. The Web-based annotation initiative TOPSAN (The Open Protein Structure Annotation Network) facilitates collaborative efforts to annotate and investigate such structures (24). Collaborative efforts such as these demonstrate the value of open dialogue and partnership between computational biologists and experimental biologists. Progress toward deciphering the role of the human microbiome in health and disease will continue to rely upon such alliances.
THE HUMAN MICROBIOME AS A COMPOSITE OF MANY MICROBIOMES
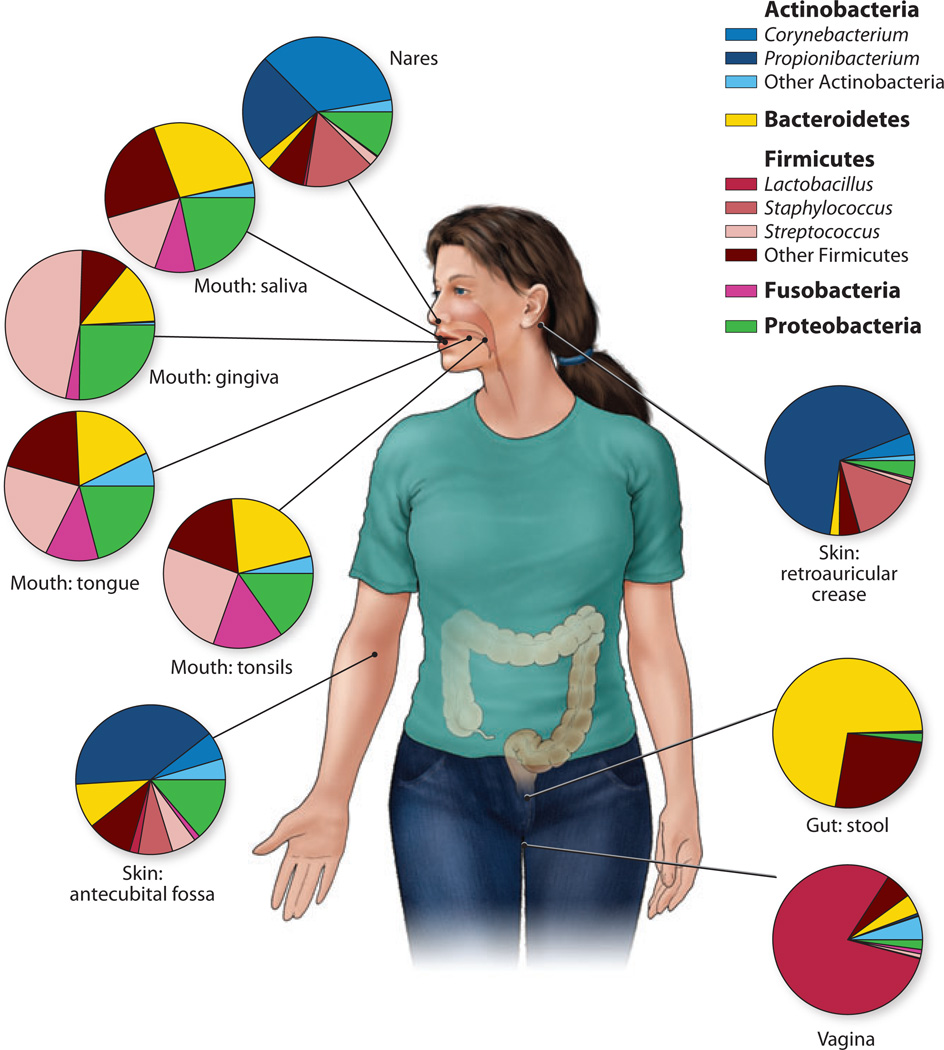
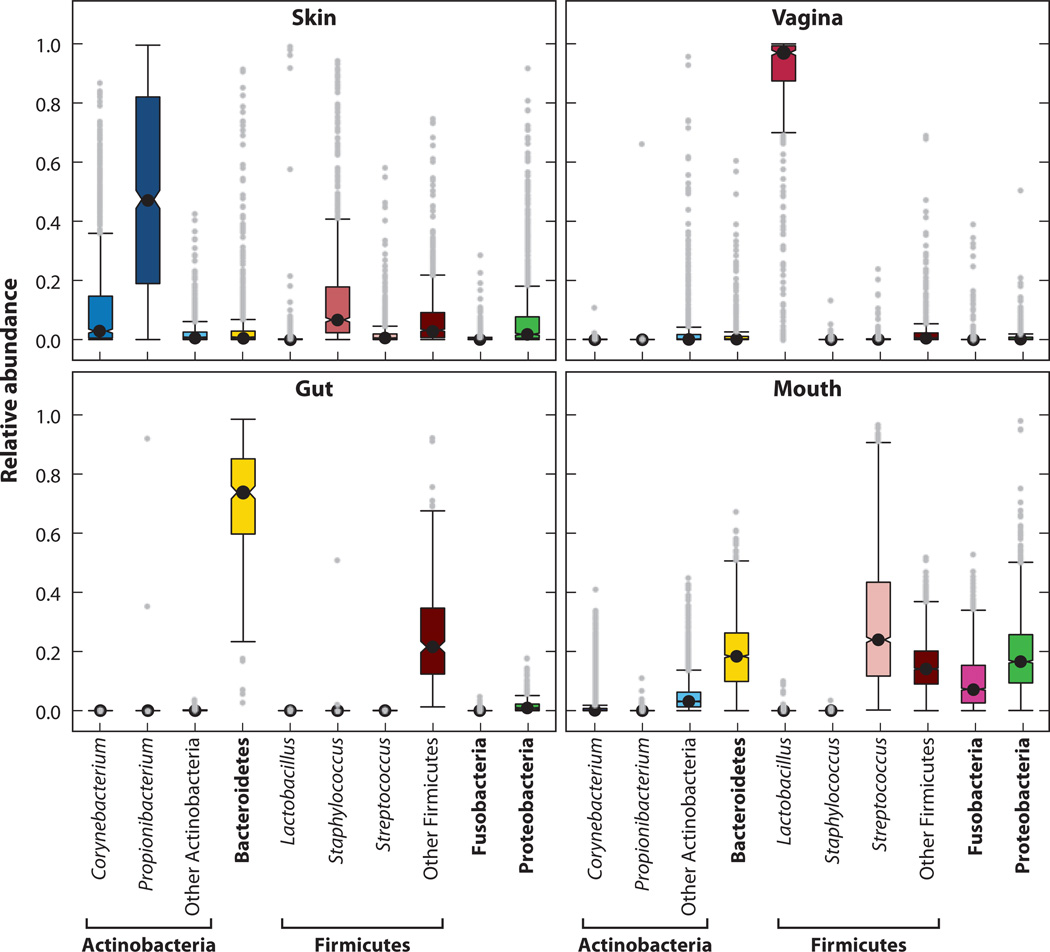
One of the major goals of the five-year, NIH-funded Human Microbiome Project was to define the healthy human adult microbiome at multiple body sites in a large cohort (n = 242) (65). The body sites sampled included the oral cavity, skin, and GI tract as well as the vagina in females. Both 16S rRNA and WGS metagenomic data sets were generated from this large number of clinical samples. Not surprisingly, this study confirmed that each body habitat harbors dominant signature taxa, something that had been shown by individually focused studies (14, 33, 70, 71, 94). For example, the sebaceous area behind the ear, the retroauricular crease, is consistently dominated by the lipophilic Propionibacterium; Lactobacillus dominates the vagina; and Bacteroidetes and Firmicutes dominate the gut (Figure 2). Body habitat was also noted as a strong determinant of microbial co-occurrence and coexclusion. On the one hand, with its large cohort, this study underscored the findings of multiple previous studies in which interpersonal variation was significantly greater than intrapersonal variation (Figure 3). On the other hand, the relative abundances of metabolic and functional pathways in the metagenomic data were much more stable than organismal abundances as measured by 16S rRNA sequences. Surprisingly, actively pathogenic organisms were rarely present in the microbial communities of these individuals (Hum. Microbiome Proj. Consort., manuscripts in review).
Figure 2.
Genus- and phylum-level classification of bacteria colonizing a composite subject, showing that human microbiome diversity is dependent on the site sampled. Sites in the oral cavity share greater similarity than other types of sites, such as the skin, vagina, and gut. Data derived from the NIH Human Microbiome Project study (http://commonfund.nih.gov/hmp).
Figure 3.
Interpersonal variation in levels of bacteria (classified at the genus and phylum levels) of the skin, vagina, gut, and mouth. The median relative abundance of each bacteria is indicated by a central point, the boxes extend from the first to third quartiles, and the whiskers extend to the highest and lowest data points no farther than 1.5 times the interquartile range from the box. Gray dots represent individual samples that lie outside this range. Each body habitat harbors dominant signature taxa. Actintobacteria (Corynebacterium and Propionibacterium), Firmicutes (Staphylococcus, Streptococcus, and others), and Proteobacteria predominate on the skin, with interindividual variation displayed; Lactobacillus predominates in the vagina; Bacteriodetes and Firmicutes predominate in the gut; and Bacteriodetes, Firmicutes (Streptococcus), Fusobacteria, and Proteobacteria predominate in the mouth. Data derived from the NIH Human Microbiome Project study (http://commonfund.nih.gov/hmp).
As was clear from the Human Microbiome Project study and individual site-specific studies, each body site is a highly specialized niche characterized by its own microbial consortia, community dynamics, and interaction with host tissue. Next, we address several of these niches, highlighting salient and unique findings that have advanced our understanding of the microbiome’s role in human health and disease.
The Gastrointestinal Tract: Readily Accessible with Abundant Microbiota
The GI tract contains the bulk of microbiota associated with the human body and has been one of its most thoroughly examined ecosystems. Fecal samples are commonly used for these analyses because they are easy to obtain and contain a large amount of biomass. The gut microbiome is quite low in diversity at higher phylogenetic levels, comprising primarily the bacterial phyla Firmicutes and Bacteroidetes (21), but contains great diversity at lower phylogenetic levels (species, strains) (45). One study suggested that more than 5,000 bacterial taxa may reside in the gut (19).
Initial microbial colonization of the gut in infants appears to be dependent on delivery mode; vaginally delivered babies acquire microbiota similar to those of their mother’s vagina (i.e., dominated by Lactobacillus and Prevotella), and babies delivered via Caesarian section acquire microbiota similar to those typically associated with the skin (i.e., Propionibacterium, Staphylococcus, and Corynebacterium) (20). Furthermore, there were significant differences between the gut microbiota of formula-fed and breast-fed infants, with some of those bacteria common in formula-fed babies being associated with a higher prevalence of antibiotic use, hospitalization, and prematurity (64). The gut microbiota eventually converge toward an adultlike profile during the first year of life (61).
Even more abundant than bacteria are the viruses that infect them—the bacteriophages. In their lysogenic phase, bacteriophages can integrate into the bacterial genome and provide additional genetic diversity. Virome analysis of the gut revealed that the major type of variation was interindividual, but significant changes were observed when the host was placed on a defined diet (54). The diet-induced changes in the gut virome covaried with changes in the gut bacterial community, and gut virus populations converged in individuals placed on similar diets. This is in sharp contrast to another study that found the gut virome to be stable over time (72). It is unclear what accounts for the differences observed between these two studies, but this example highlights the need for standardization in study design and result reporting.
The GI tract was one of the first human ecosystems to be examined by WGS metagenomic analysis. Early WGS metagenomic sequencing and analysis of the fecal matter of two individuals identified the metabolic potential of the gut microbiota by assigning function to Sanger sequence reads (31). The MetaHIT (Metagenomics of the Human Intestinal Tract) Consortium, funded by the European Commission and comprising 13 academic and industry partners, has been a key leader in the gut microbiome and metagenomics arena. Qin et al. (70) described a WGS metagenomic analysis of 124 Europeans that used Illumina Genome Analyzer technology and generated over 576 Gb of metagenomic sequence reads. Using this data set, they described the core gut metagenome, which consists of genes essential for host-microbe interactions such as those that degrade complex polysaccharides and those that synthesize short-chain fatty acids, vitamins, and amino acids. Even so, it should be noted that only ~12% of the genes derived from this sequence data set mapped to reference genomes. As more reference genomes are sequenced—for example, as part of the MetaHIT Consortium and NIH Human Microbiome Project initiatives—these numbers should improve. WGS metagenomic analysis of this data set and others subsequently demonstrated the existence of three distinct enterotypes—i.e., groups of individuals defined by the composition of their gut microbiota (3). The enterotypes were neither nation nor continent specific and could not be explained by body mass index, age, or gender. This study provided the first indication that the composition of the human gut microbiota is stratified and not continuous.
Because a fecal sample is theoretically a composite of bacteria collected throughout the length of the GI tract, one challenge lies in the analysis of spatial microbial diversity in the GI tract, for which there is very little data. One of the first genomic analyses of gut microbiota utilized 16S rRNA gene sequencing to begin to define the intrapersonal (spatial) and interpersonal variation of the adherent mucosal and fecal microbiota (21). Much of the sequence diversity along theGI tract was novel and revealed spatial differences not captured by a fecal sample. This is to be expected, as the host tissue varies greatly throughout the length of the GI tract, dictating distinct microenvironments depending on dominant cell types and functions.
This leads to the question of what determines the microbiota that colonize the gut. In addition to host- and tissue-specific factors, there are certainly environmental factors that must play some role in selecting for those microbes that constitute the gut microbiota. Wu and colleagues (108) demonstrated that gut enterotypes are strongly correlated with long-term dietary patterns, especially high-fat, high-protein diets as compared with high-carbohydrate diets. Gordon’s group (26) definitively demonstrated a relationship between both the total abundance and the relative levels of bacteria evoked by diet perturbations in mice colonized with defined bacterial communities. Genetic factors are also implicated in microbial gut colonization, with initial studies carried out in animal models. For example, studies in advanced intercross mice demonstrated that host genotype had a large influence on gut microbiota composition, independent of litter and cohort effects (5). In fact, a relative abundance of gut microbial taxa was associated by genome-wide linkage with 18 quantitative trait loci, which demonstrates the power of the host genotype in shaping microbial diversity.
Interesting features of the gut microbiome and its role in disease are beginning to emerge, especially its role in obesity, diabetes, and metabolic disease. Perhaps the best-known example is the contribution of gut microbiota to obesity. Although human geneticists continue to search for host variation tied to obesity, it is now appreciated that the gut microbiota of obese individuals are significantly altered and carry a greater capacity for energy harvest (95). Animal models have proven especially useful in demonstrating mechanistic and functional linkages of the gut microbiota to disease. An animal model deficient in Toll-like receptor 5 (Tlr5)— an important component of innate immunity and infection control in the gut—exhibits hallmarks of metabolic syndrome (adiposity, insulin resistance, hypertension, hyperlipidemia) and altered gut microbiota as compared with healthy control mice (99). Additionally, when transferred to germ-free mice, the gut microbiota of Tlr5-deficient mice were sufficient to recapitulate features of metabolic syndrome.
Interaction of the innate immune system with the gut microbiota was demonstrated to be a critical epigenetic factor governing the development of type 1 diabetes in nonobese diabetic (NOD) mice. Specific-pathogen-free NOD mice deficient in MyD88—an adaptor protein essential to multiple innate immune receptors and signaling pathways—do not develop diabetes. This effect appears to be dependent on the gut microbiota, as germ-free Myd88−/− NOD mice develop robust diabetes. However, diabetes was attenuated in germ-free Myd88−/− NOD mice colonized with defined microbiota from donor specific-pathogen-free Myd88−/− NODmice (101). This is yet another example of how interactions between the microbiota and the host govern predisposition to disease.
Studies in Drosophila have provided insight into the complex interactions between host and microbe in the gut. In Drosophila, the homeobox gene Caudal represses the NF-κβ-dependent signaling of antimicrobial peptides, which in turn regulates host-microbe mutualism (77). RNA interference knockdown of Caudal results in altered gut microbiota and eventually gut cell apoptosis and death. In a separate study, the commensal bacterium Acetobacter pomorum proved to be a key link in the regulation of Drosophila development, body size, metabolism, and intestinal stem cell activity (84). A. pomorum–derived, pyrroloquinoline quinone–dependent alcohol dehydrogenase activity modulates Drosophila insulin/insulin-like growth factor signaling.
In humans, beyond aiding with digestion, bacteria were recently found to play a role in cancer. Colorectal carcinoma is associated with enriched abundance of Fusobacterium (11, 40), an invasive anaerobic bacterium that was previously associated with inflammatory bowel disease but is a rare constituent of the healthy gut microbiota. Though the exact role of Fusobacterium in colorectal carcinoma remains unclear, it is capable of eliciting a host inflammatory response, a known risk factor for colorectal cancer (52, 109). Additionally, Fusobacterium abundance was positively associated with lymph node metastasis (11). More work is needed to define the mechanism by which Fusobacterium is related to colorectal carcinoma, but a more immediate value may be in exploiting it as a marker for colorectal cancer presence, risk, or prognosis.
Finally, the role of the gut microbiota through their metabolic potential extends far beyond the intestine to the metabolism of systemic drugs and disease manifestation in other organ systems. Nicholson’s group (12) deciphered the first example of metabolomics intersecting with the microbiome, showing a clear link between an individual’s ability to metabolize acetaminophen (most commonly known in the United States under the brand name Tylenol) and bacterial metabolic state. Acetaminophen has been one of the most commonly used nonprescription medicines for decades, so one might think that its toxicology and metabolism would be well understood. However, Nicholson’s study showed a novel and striking association between an individual’s predose metabolite profile and his or her specific metabolism and excretion of acetaminophen. As these pathways described for acetaminophen impact the metabolism of many drugs, the gut bacterial metabolism might have a large influence on both drug-induced responses and disease development.
Microbiome profiling clearly has a role in personalized medicine that extends beyond the variation in drug-metabolizing enzymes. Hazen’s group (100) made the first link between diet, gut bacteria, liver metabolism, and atherosclerosis leading to cardiovascular disease. Their initial experiments screened for plasma metabolites in patients who had experienced a heart attack or stroke. While testing this metabolic cascade in animal models, they found dramatic differences in mice treated with broad-spectrum antibiotics. They teased apart this mechanism by examining atherosclerosis development in mouse models of human metabolic disorders that were placed on restricted diets and then treated with antibiotics. The hunt is now on to find a probiotic approach to selectively eliminate the gut microbes that process metabolites derived from high-fat foods before they are delivered to the liver and ultimately deposited in blood vessels. These studies clearly show the vast unexplored role of the gut microbiota in regulating human health beyond simply intestinal disorders.
The Oral Microbiome
In general, the dominant bacteria of the oral cavity are streptococcal species, with common representation also from Veillonella, Gamella, Rothia, Fusobacterium, and Neisseria (1, 6). Many diseases of the oral cavity, such as dental caries (cavities) and periodontitis (gum disease), have long been suspected to be caused at least in part by microbes. A metagenomic analysis of dental caries demonstrated that consortia of microbiota inhabit the caries and that these microbes differ functionally from those inhabiting the healthy oral cavity (4). Strikingly, those who never suffered from caries were colonized with microbiota enriched for genes encoding antimicrobial peptides and quorum-sensing molecules. In periodontitis, Porphyromonas gingivalis, an anaerobic bacterium, has historically been the suspected etiological agent. Mechanistic work in germ-free mice demonstrated that only minute quantities of P. gingivalis induce quantitative and qualitative changes in the oral microbiota, not directly but rather via exploitation of the complement cascade to cause periodontal bone loss (34).
Bacteriophages have been another focus of oral microbiome research and have provided the greater microbiome community with scientific insight. Pride et al. (67) recently analyzed the salivary viromes of five healthy individuals and found that the vast majority of human oral viruses are bacteriophages. Many of the viral genes encoded virulence factors, and thus the oral virome may be a reservoir for oral pathogenicity factors. In this same study, comparisons of the salivary virome with gut and respiratory viromes showed that habitat is an important selection factor for the virome.
Genome-encoded clustered regularly interspaced short palindromic repeats (CRISPRs) are a bacterial defense mechanism against mobile genetic elements such as bacteriophages and conjugative plasmids. CRISPRs are acquired from the invading element, forming a heritable, adaptive record of prior infection. Bacterial cells express CRISPR RNA to interfere with invading nucleic acids (51). An analysis of streptococcal CRISPR sequences in the oral cavity revealed great diversity within individuals, suggesting that each individual was exposed to unique viral populations (68). Although much remains to be learned about CRISPR elements, the historical perspective they provide about bacteria strains may be useful for ecological and epidemiological studies and eventually personalized medicine.
Epidemiological studies have suggested a correlation between periodontitis and diseases that affect seemingly unrelated organ systems, e.g., diabetes and atherosclerosis. A common link that could explain this apparent association is the human microbiota, their priming of the immune system, or their long-term effects on inflammation. Metagenomic sequence analysis of the oral cavity revealed enrichment of Streptococcus mitis bacteriophage SM1–derived genes encoding platelet-binding factors, which are key virulence factors in endocardium infection (104). Koren et al. (39) demonstrated not only that bacteria were present in the atherosclerotic plaques, but also that the types and abundance of those bacteria correlated with the abundance of those same bacteria in the oral cavity. Furthermore, the abundance of several bacterial taxa in the oral cavity and the gut showed correlation with plasma cholesterol levels. These studies also illustrate the potential of the microbiome as a disease marker.
The Reproductive Tract Microbiome
Even before genomics arrived on the scene, it was suspected that the vaginal microbiota play a key role in the prevention of multiple diseases, including bacterial vaginosis (BV), yeast infections, sexually transmitted diseases, urinary tract infections, and human immunodeficiency virus. In a study of 396 reproductive-aged women, 16S rRNA gene sequence analysis revealed that vaginal bacteria profiles generally fall into one of five clusters. Four of the clusters were dominated by Lactobacillus species, whereas the fifth group had a higher proportion of anaerobic species and overall greater bacterial diversity (71). This group was also associated with a higher vaginal pH and a higher Nugent score, the latter of which is a cellular morphological indicator of BV. Several independent analyses of women with BV compared with healthy women showed the same trend of greater vaginal microbiota diversity associated with BV (48, 60, 88). A longitudinal study of the vaginal microbiota indicated that levels of lactobacilli fluctuate with menses, with a notable increase in Gardnerella vaginalis during menstruation; the authors hypothesized that this is due to increased iron availability because of the presence of vaginal blood during menses (88). Longitudinal analysis of women with BV during a treatment time course demonstrated that BV-associated bacteria were eradicated by treatment with the antibiotic metronidazole but tended to reappear following treatment.
The male genitourinary tract microbiome does not get the same attention as the vagina’s, but could be a very important factor in sexually transmitted infections. In an analysis of the male urine microbiota, anaerobic, fastidious bacteria were associated with sexually transmitted infections (57). In an analysis of pre- and postcircumcision microbial diversity in 12 African men, circumcision was associated with a decrease in anaerobic bacteria and overall decreased bacterial diversity of the coronal sulci of the penis (66).
The Skin Microbiome
Genomic approaches characterizing skin bacteria have revealed a much greater diversity of organisms than was previously apparent from culture-based methods (14, 27, 29, 32, 33). Our group and others have demonstrated that the physiology of the skin site is a strong determinant of the dominant colonizing bacteria, with specific bacteria associated with moist, dry, and oily microenvironments (14, 33). In general, bacterial diversity appears to be lowest in oily sites (such as the back and face) and highest in dry, exposed sites (such as the arms and legs). In general, intrapersonal variation in microbial community membership and structure between symmetric skin sites is lower than interpersonal variation, as determined by 16S rRNA gene sequencing (14, 29, 33).
There is a particular need to develop better methods for typing fungal and other microeukaryotic species, as these organisms are known to thrive on the skin and in some cases are associated with skin disorders. Analysis of 18S rRNA gene sequences has shown that the vast majority of fungal organisms residing on the healthy skin resemble Malassezia species, closely mirroring culture-based data, but the identity of more rare species may still prove important to understanding human disorders such as toenail infections and athlete’s foot (28, 62, 63). Demodex mites are also considered part of the normal skin microflora, residing in the sebaceous glands and hair follicles of the facial skin and increasing in prevalence with age (25, 35). Molecular methods for typing Demodex do not exist, but there is evidence suggesting a role for Demodex in some skin disorders such as rosacea (30, 42, 46).
WGS metagenomic data for skin communities would provide a fuller articulation of gene content and function to address basic questions such as the potentially beneficial role of skin bacteria to the human host. But although skin holds tremendous advantages for ease of sampling, the clear disadvantage lies in the difficulty of obtaining the critical threshold of starting material required for WGS metagenomic sequencing. Swabbing a 1-cm2 area of skin typically yields only tens of nanograms of DNA, which is at the very low end of the amount required to make a next-generation sequencing library. And although skin cells undergo a linear program of terminal differentiation that limits the amount of human DNA present at the surface, unbiased whole-genome amplification needs to be further developed before skin WGS metagenomic sequencing can be tractable on a large scale. It seems that beyond the diseases that affect an organ system, every microbial community comes with its own technical advantages and limitations for genomic analysis.
ANTIBIOTICS
The prevalence of antibiotics in our society has effectively transformed a fraction of their intended targets into superbugs, selecting for those mutants and strains of bacteria with the capacity to survive large doses of antibiotic drugs. Blaser & Falkow (7) go further to hypothesize that the recent increase in allergic and other diseases without any obvious explanation (e.g., asthma and metabolic diseases) are a consequence of the disappearing microbiota. Cleaner water, smaller families, delivery by Caesarian section, and widespread antibiotic use (especially in young children) have all contributed to the disappearance of our microbiota, which have evolved and coexisted with their hosts for millions of years. Is it possible that our cleanliness is making us sick?
In moving toward answers to these types of questions, genomic approaches have been useful in determining the effects of antibiotics on the human microbiota. In a longitudinal study of the gut microbiota in three antibiotictreated adults, ciprofloxacin treatment was associated with decreased bacterial diversity (19). Most bacterial taxa recovered after four weeks, but a handful of taxa did not recover even after six months, demonstrating the persistent effects of antibiotic treatment on the gut microbiota. In mice, antibiotic treatment completely displaced the normal gut microbiota, allowing exogenously administrated vancomycin-resistant Enterococcus to completely invade the gut (97). Parallel analysis in patients undergoing allogeneic hematopoietic stem cell transplantation showed that these drug-resistant strains dominate the gut microbiota prior to bloodstream infection, suggesting the prognostic value of microbiome markers.
The emergence of multidrug-resistant bacterial strains has focused attention on identifying reservoirs for antibiotic resistance genes that may be available to clinically relevant pathogens. Lateral gene transfer events are largely responsible for the dissemination of antibiotic resistance genes as well as virulence genes. Lateral gene transfer can occur by bacteriophage-mediated transduction, direct uptake of environmental DNA (such as transformation by plasmids), transposition, or conjugation. The soil has also been identified as a rich source of antibiotic resistance genes (15, 16, 73), which is not surprising because many clinically relevant antibiotics are derived from soil actinomycetes. Culture-independent functional characterization of the antibiotic resistance reservoir of human gut and saliva microbiota revealed that most resistance genes have not been previously identified and were evolutionarily distant from known resistance genes (87). Furthermore, the majority of resistance genes identified in cultured isolates of the gut microbiota were identical to known resistance genes in clinically relevant pathogens. These findings raise many questions about the commensal human microbiota as reservoirs for antibiotic resistance genes.
THE CHALLENGES THAT LIE AHEAD
Because we are only beginning to realize the full potential of the human microbiome and its significance to health and disease, the road ahead is neither easy nor straightforward. The same questions were asked in ecology and environmental microbiology for years prior to the realization that the human microbiome is also an ecosystem. Integrating strategies and findings from environmental ecosystems into human microbiome studies is a logical and expedient way forward.
One of the main challenges ahead lies in the sheer amount of data generated by increasingly cheap DNA sequencing. Generating the sequence data is probably the easiest step in these studies, but the computational capacity and bioinformatic expertise to process and analyze these data are hard to come by. Furthermore, obtaining biologically relevant samples, with carefully annotated metadata and meticulous clinical phenotyping associated with them, is essential to creating meaningful microbiome data sets. Wise use of sequencing resources in carefully designed studies will produce the greatest advances in answering questions about the human microbiome. Another overarching challenge for the field is how to visualize the complex, multitiered analyses from these microbial studies.
In addition to these challenges, many questions remain. We are beginning to glimpse the effect of the microbiome on multiple organ systems. For example, asthma and hay fever are often preceded by skin eczema, referred to as the atopic march. Is it possible that exposure early in life to microbes associated with eczema predisposes an individual to other allergic/ atopic diseases? In the same way, how does the microbiome educate the immune system so that it distinguishes the pathogenic from the commensal, and therefore repress any damaging and unnecessary host responses? In this way, one can envision the value of the microbiome in its prognostic and predictive potential.
Although it is unlikely that our modern society will give up its love affair with everything antibiotic and antimicrobial, is there a way that we can leverage commensal microbes such that they can repress pathogens and host response to pathogens? This is a critical question to address as an increasing number of pathogenic microbes become multidrug-resistant superbugs. Another challenge will be the regulatory landscape for prebiotics and probiotics as the United States and other countries roll out guidance on whether microbial organisms will be considered drugs, natural products, or food additives.
Finally, we envision that eventually metagenomic and human genetic data sets will be integrated, so that in addition to genetic markers (such as single-nucleotide polymorphisms), metagenomic markers will also be queried. Just as in genome-wide association data, assigning meaning to these markers when an association is observed will require functional studies to move beyond association to causation.
ACKNOWLEDGMENTS
We thank Sean Conlan and Daryl Leja for assistance with figure preparation.
Glossary
- Microbiota
microorganisms—such as bacteria, fungi, viruses, and archaea—present in a community
- Microbiome
all of the genetic material of a microbial community sequenced together
- Metagenomics
genomic analysis of DNA extracted directly from a clinical or environmental sample; it can assess community diversity (through 16S rRNA metagenomics) or fuller complexity (through whole-genome shotgun metagenomics)
- NIH Common Fund’s Human Microbiome Project
study aimed at characterizing the microbial communities found at several different sites on the human body and analyzing the role of these microbes in human health and disease
- Pan-genome
the collection of genes found across all members of a species
- Bacteriophage
a virus that infects a bacterium
- MetaHIT (Metagenomics of the Human Intestinal Tract) Consortium
consortium comprising 13 academic and industry partners funded by the European Commission to intensively study the gut microbiome’s role in health and disease
- Germ-free mice
mice born and reared in sterile environments from which no microbiota can be cultured
- Metadata
clinical or environmental data associated with samples collected
Footnotes
This is a work of the US Government and is not subject to copyright protection in the United States.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simon-Soro A, et al. The oral metagenome in health and disease. ISME J. 2012;6:45–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bond PL, Hugenholtz P, Keller J, Blackall LL. Bacterial community structures of phosphateremoving and non-phosphate-removing activated sludges from sequencing batch reactors. Appl. Environ. Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. USA. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dantas G, Sommer MO, Oluwasegun RD, Church GM. Bacteria subsisting on antibiotics. Science. 2008;320:100–103. doi: 10.1126/science.1155157. [DOI] [PubMed] [Google Scholar]
- 16.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 17.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denef VJ, Mueller RS, Banfield JF. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J. 2010;4:599–610. doi: 10.1038/ismej.2009.158. [DOI] [PubMed] [Google Scholar]
- 19.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA. 107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards KJ, Bond PL, Gihring TM, Banfield JF. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science. 2000;287:1796–1799. doi: 10.1126/science.287.5459.1796. [DOI] [PubMed] [Google Scholar]
- 23.Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, et al. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics. 2006;7:57. doi: 10.1186/1471-2164-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellrott K, Zmasek CM, Weekes D, Sri Krishna S, Bakolitsa C, et al. TOPSAN: a dynamic web database for structural genomics. Nucleic Acids Res. 2011;39:D494–D496. doi: 10.1093/nar/gkq902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elston DM. Demodex mites: facts and controversies. Clin. Dermatol. 2010;28:502–504. doi: 10.1016/j.clindermatol.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Z, Perez-Perez GI, Chen Y, Blaser MJ. Quantitation of major human cutaneous bacterial and fungal populations. J. Clin. Microbiol. 2010;48:3575–3581. doi: 10.1128/JCM.00597-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georgala S, Katoulis AC, Kylafis GD, Koumantaki-Mathioudaki E, Georgala C, Aroni K. Increased density of Demodex folliculorum and evidence of delayed hypersensitivity reaction in subjects with papulopustular rosacea. J. Eur. Acad. Dermatol. Venereol. 2001;15:441–444. doi: 10.1046/j.1468-3083.2001.00331.x. [DOI] [PubMed] [Google Scholar]
- 31.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grice EA, Kong HH, Renaud G, Young AC, Bouffard GG, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hay R. Demodex and skin infection: fact or fiction. Curr. Opin. Infect. Dis. 2010;23:103–105. doi: 10.1097/QCO.0b013e3283360a18. [DOI] [PubMed] [Google Scholar]
- 36.Hugenholtz P, Pace NR. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 1996;14:190–197. doi: 10.1016/0167-7799(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 37.Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koren O, Spor A, Felin J, Fak F, Stombaugh J, et al. Human oral gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl. 1):4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, et al. Experimental and analytical tools for studying the human microbiome. Nat. Rev. Genet. 2012;13:47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lacey N, Delaney S, Kavanagh K, Powell FC. Mite-related bacterial antigens stimulate inflammatory cells in rosacea. Br. J. Dermatol. 2007;157:474–481. doi: 10.1111/j.1365-2133.2007.08028.x. [DOI] [PubMed] [Google Scholar]
- 43.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lederberg J, McCray AT. ’Ome sweet ’omics—a genealogical treasury of words. Scientist. 2001;15:8. [Google Scholar]
- 45.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 46.Li J, O’Reilly N, Sheha H, Katz R, Raju VK, et al. Correlation between ocular Demodex infestation and serum immunoreactivity to Bacillus proteins in patients with facial rosacea. Ophthalmology. 2010;117:870–877. doi: 10.1016/j.ophtha.2009.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K. SNP detection for massively parallel wholegenome resequencing. Genome Res. 2009;19:1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ling Z, Kong J, Liu F, Zhu H, Chen X, et al. Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC Genomics. 2010;11:488. doi: 10.1186/1471-2164-11-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu WT, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. USA. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLean MH, Murray GI, Stewart KN, Norrie G, Mayer C, et al. The inflammatory microenvironment in colorectal neoplasia. PLoS ONE. 2011;6:e15366. doi: 10.1371/journal.pone.0015366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinforma. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitra S, Rupek P, Richter DC, Urich T, Gilbert JA, et al. Functional analysis of metagenomes and metatranscriptomes using SEED and KEGG. BMC Bioinforma. 2011;12(Suppl. 1):S21. doi: 10.1186/1471-2105-12-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muyzer G. DGGE/TGGE a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 1999;2:317–322. doi: 10.1016/S1369-5274(99)80055-1. [DOI] [PubMed] [Google Scholar]
- 57.Nelson DE, Van Der Pol B, Dong Q, Revanna KV, Fan B, et al. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS ONE. 2010;5:e14116. doi: 10.1371/journal.pone.0014116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nichols D, Cahoon N, Trakhtenberg EM, Pham L, Mehta A, et al. Use of ichip for highthroughput in situ cultivation of ”uncultivable” microbial species. Appl. Environ. Microbiol. 2010;76:2445–2450. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oakley BB, Fiedler TL, Marrazzo JM, Fredricks DN. Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl. Environ. Microbiol. 2008;74:4898–4909. doi: 10.1128/AEM.02884-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paulino LC, Tseng CH, Blaser MJ. Analysis of Malassezia microbiota in healthy superficial human skin and in psoriatic lesions by multiplex real-time PCR. FEMS Yeast Res. 2008;8:460–471. doi: 10.1111/j.1567-1364.2008.00359.x. [DOI] [PubMed] [Google Scholar]
- 63.Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J. Clin. Microbiol. 2006;44:2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 65.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Price LB, Liu CM, Johnson KE, Aziz M, Lau MK, et al. The effects of circumcision on the penis microbiome. PLoS ONE. 2010;5:e8422. doi: 10.1371/journal.pone.0008422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pride DT, Salzman J, Haynes M, Rohwer F, Davis-Long C, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012;6:915–926. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pride DT, Sun CL, Salzman J, Rao N, Loomer P, et al. Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects over time. Genome Res. 2011;21:126–136. doi: 10.1101/gr.111732.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Proctor LM. The human microbiome project in 2011 and beyond. Cell Host Microbe. 2011;10:287–291. doi: 10.1016/j.chom.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 70.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl. 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Riesenfeld CS, Goodman RM, Handelsman J. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ. Microbiol. 2004;6:981–989. doi: 10.1111/j.1462-2920.2004.00664.x. [DOI] [PubMed] [Google Scholar]
- 74.Roberts RJ. Identifying protein function—a call for community action. PLoS Biol. 2004;2:e42. doi: 10.1371/journal.pbio.0020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rodrigue S, Malmstrom RR, Berlin AM, Birren BW, Henn MR, Chisholm SW. Whole genome amplification and de novo assembly of single bacterial cells. PLoS ONE. 2009;4:e6864. doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rohde H, Qin J, Cui Y, Li D, Loman NJ, et al. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N. EnglJMed. 2011;365:718–724. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- 77.Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, et al. Innate immune homeostasis by the homeobox gene Caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 78.Savage DC. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 79.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE. 2012;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. Introducing mothur: opensource, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schrenk MO, Edwards KJ, Goodman RM, Hamers RJ, Banfield JF. Distribution of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans: implications for generation of acid mine drainage. Science. 1998;279:1519–1522. doi: 10.1126/science.279.5356.1519. [DOI] [PubMed] [Google Scholar]
- 82.Scott JJ, Oh DC, Yuceer MC, Klepzig KD, Clardy J, Currie CR. Bacterial protection of beetlefungus mutualism. Science. 2008;322:63. doi: 10.1126/science.1160423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shin SC, Kim SH, You H, Kim B, Kim AC, et al. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 85.Smith HO, Tomb JF, Dougherty BA, Fleischmann RD, Venter JC. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenza Rd genome. Science. 1995;269:538–540. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 86.Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Srinivasan S, Liu C, Mitchell CM, Fiedler TL, Thomas KK, et al. Temporal variability of human vaginal bacteria and relationship with bacterial vaginosis. PLoS ONE. 2010;5:e10197. doi: 10.1371/journal.pone.0010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stahl DA, Lane DJ, Olsen GJ, Pace NR. Analysis of hydrothermal vent-associated symbionts by ribosomal RNA sequences. Science. 1984;224:409–411. doi: 10.1126/science.224.4647.409. [DOI] [PubMed] [Google Scholar]
- 90.Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 91.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, et al. The COG database: an updated version includes eukaryotes. BMC Bioinforma. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 94.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 96.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 97.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Investig. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 99.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weng L, Rubin EM, Bristow J. Application of sequence-based methods in human microbial ecology. Genome Res. 2006;16:316–322. doi: 10.1101/gr.3676406. [DOI] [PubMed] [Google Scholar]
- 103.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Willner D, Furlan M, Schmieder R, Grasis JA, Pride DT, et al. Metagenomic detection of phageencoded platelet-binding factors in the human oral cavity. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl. 1):4547–4553. doi: 10.1073/pnas.1000089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Woese CR. Bacterial evolution. Microbiol. Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Woyke T, Tighe D, Mavromatis K, Clum A, Copeland A, et al. One bacterial cell, one complete genome. PLoS ONE. 2010;5:e10314. doi: 10.1371/journal.pone.0010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]