Abstract
mRNA turnover in eukaryotes involves the removal of m7GDP from the 5′ end. This decapping reaction is mediated by a protein complex well characterized in yeast and human but not in plants. The function of the decapping complex in the development of multicellular organisms is also poorly understood. Here, we show that Arabidopsis thaliana DCP2 can generate from capped mRNAs, m7GDP, and 5′-phosphorylated mRNAs in vitro and that this decapping activity requires an active Nudix domain. DCP2 interacts in vitro and in vivo with DCP1 and VARICOSE (VCS), an Arabidopsis homolog of human Hedls/Ge-1. Moreover, the interacting proteins stimulate DCP2 activity, suggesting that the three proteins operate as a decapping complex. Consistent with their role in mRNA decay, DCP1, DCP2, and VCS colocalize in cytoplasmic foci, which are putative Arabidopsis processing bodies. Compared with the wild type, null mutants of DCP1, DCP2, and VCS accumulate capped mRNAs with a reduced degradation rate. These mutants also share a similar lethal phenotype at the seedling cotyledon stage, with disorganized veins, swollen root hairs, and altered epidermal cell morphology. We conclude that mRNA turnover mediated by the decapping complex is required for postembryonic development in Arabidopsis.
INTRODUCTION
Cytoplasmic processing bodies (P bodies) are dynamic RNA protein aggregates found in yeast and mammalian cells with critical roles in mRNA decapping (Dunckley and Parker, 1999; Dunckley et al., 2001; Lykke-Andersen, 2002; Sheth and Parker, 2003; Coller and Parker, 2004; Fenger-Gron et al., 2005), translation and translation suppression (Coller and Parker, 2005; Liu et al., 2005b), RNA interference (Liu et al., 2005a, 2005b; Rehwinkel et al., 2005; Sen and Blau, 2005), RNA virus replication, and innate cellular immunity against retroviruses (Beliakova-Bethell et al., 2006; Wichroski et al., 2006). It is believed that the P bodies are evolutionarily conserved among eukaryotes, because several P body components, such as the decapping complex, are found in both yeast and mammalian cells (Coller and Parker, 2004). However, plant P bodies have not yet been described.
The decapping of mRNAs, which occurs inside P bodies, represents a critical step in mRNA turnover because it is highly regulated. In yeast, the decapping complex/machinery consists of DCP2, DCP1, EDC3, EDC2, EDC1, DHH1, PAT1, and LSM1-7 (Coller and Parker, 2004). Whereas EDC3, EDC2, EDC1, DHH1, PAT1, and LSM1-7 are regulatory factors, DCP2 and DCP1 are directly associated with decapping activity both in vivo and in vitro (Dunckley and Parker, 1999; Tharun and Parker, 1999; Zhang et al., 1999; Steiger et al., 2003; Sakuno et al., 2004). However, only recombinant DCP2 protein alone showed decapping activity in vitro, and structural analysis of DCP2 (Sakuno et al., 2004; She et al., 2004, 2005) revealed that the DCP2 Nudix domain is responsible for the decapping activity. In mammalian cells, the decapping complex contains hDCP2, hDCP1, hEDC3, Rck/p54 (a DHH1 homolog), and Hedls/Ge-1 (Fenger-Gron et al., 2005; Yu et al., 2005). Whereas hDCP2 displays robust decapping activity in vitro, hDCP1 is devoid of any decapping activity (Lykke-Andersen, 2002; van Dijk et al., 2002; Wang et al., 2002). Hedls/Ge-1 has no obvious homolog in yeast; nevertheless, it appears to be the central component of the mammalian decapping complex because small interfering RNA knockdown of Ge-1 but not hDCP2 resulted in P body disassembly (Yu et al., 2005).
Phenotypic analysis provided some evidence that mRNA decapping complex may be required for cell growth and development. In Saccharomyces cerevisiae, dcp1-1 mutants could not grow at 36°C (Beelman et al., 1996; Hatfield et al., 1996) owing to a complete inhibition of decapping in vivo, whereas a dcp2 deletion mutant grew extremely slowly at all temperatures correlating with the decreased RNA turnover rate (Dunckley and Parker, 1999). Sp DCP1-deficient mutants of Schizosaccharomyces pombe are nonviable as well (Sakuno et al., 2004). A Drosophila mutant disrupted in the dDCP1 gene shows the embryo abdominal deletion phenotype resulting from a reduced degradation of maternal RNAs and mislocalization of the oskar messenger ribonucleoprotein complex (Lin et al., 2006). The pleiotropic phenotypes displayed by these mutants suggest a critical role of RNA degradation in cell growth and development. A phenotype associated with a deficiency of Hedls/Ge-1, another P body component, has not yet been described.
In plants, nucleotide pyrophosphatase activity, which may include decapping activity, was previously detected in potato (Solanum tuberosum) extracts (Kole et al., 1976). Gazzani et al. (2004) reported that knockout of XRN4 (5′ to 3′ exonuclease) promotes transgene silencing in an xrn4 null mutant. This result, which suggests that decapped mRNAs serve as substrates for RNA-dependent RNA polymerase, provides a link between mRNA turnover and posttranscriptional gene silencing. Moreover, the role of transcript decapping in the development of multicellular organisms is poorly understood. For these reasons, we have decided to identify and characterize protein components responsible for mRNA decapping in plants and to explore the function of these proteins in plant development.
We show here that the Arabidopsis thaliana decapping activity is performed by a complex containing DCP2, DCP1, and VARICOSE (VCS). Recombinant DCP2 is enzymatically active in vitro, generating from capped mRNAs, m7GDP, and 5′-phosphorylated mRNAs that are substrates of 5′ to 3′ exonuclease. Mutational analyses revealed that the Nudix domain is required for this activity. Interactions among DCP2, DCP1, and VCS in vitro and in vivo demonstrate that the three proteins form a decapping complex. DCP2, DCP1, and VCS were found to colocalize in cytoplasm foci resembling P bodies, consistent with their role in mRNA decay. In dcp1-1, dcp2-1, and vcs-6 mutants, cell differentiation is arrested at postembryonic growth. Accumulation of transcripts in the capped form results in their decreased degradation rate and prolonged half-life in decapping-deficient mutants. We conclude that the decapping complex plays a critical role in postembryonic development in Arabidopsis.
RESULTS
Arabidopsis DCP2, DCP1, and VCS Are Homologs of hDCP2, hDCP1a, and Hedls/Ge-1, Respectively
We performed a BLAST search to identify proteins that may be associated with mRNA turnover in Arabidopsis. In yeast, DCP1 and DCP2 form a complex catalyzing the removal of m7GDP caps from mRNAs. DCP2 shares overall 45 and 52% conserved amino acids with yeast DCP2 and human DCP2, respectively (Figure 1A). Moreover, the N-terminal domain and the Nudix domain, which is required for the catalytic activity, are highly conserved among these proteins. Similarly, DCP1 appears to be the homolog of yeast DCP1 (30% homology) and human DCP1 (40% homology) at the amino acid level (Figure 1B). Both genes encoding DCP2 and DCP1 are present in single copy in the Arabidopsis genome.
Figure 1.
Schemes of Arabidopsis (At) DCP2, DCP1, VCS, and VCR Aligned with Their Homologs in S. cerevisiae (Sc) and Homo sapiens (h).
Conserved domains are boxed and labeled. The number of amino acid (aa) residues in each protein is shown along with the degree of sequence similarity.
(A) DCP2. Gray bar, N-terminal domain; black bar, Nudix domain.
(B) DCP1. Gray bar, EVH1/WH1, a member of the PH domain superfamily (She et al., 2004).
(C) VCS and VCR. WD40, nuclear localization signal (NLS), S domain, and ψ(X2-3) repeats are indicated below (Yu et al., 2005).
In human cells, Hedls/Ge-1, DCP1a, and DCP2 form a decapping complex. Arabidopsis VCS and VARICOSE-RELATED (VCR) share 40% amino acid similarity over the entire sequence of Hedls/Ge-1 (Figure 1C). Nevertheless, VCS and VCR retain some of the features of the Hedls/Ge-1 protein structure, such as the WD40 domain and the ψ(X2-3) repeats. Genes for VCS and VCR are closely located on Arabidopsis chromosome 3. Mutants vcs-1 and its four alleles show a pleiotropic phenotype, including disorganized vascular bundles, temperature sensitivity, and reduced shoot apical meristem size. However, a vcr null mutant is indistinguishable from the wild type, suggesting that VCR may not be functional (Deyholos et al., 2003). Therefore, we chose VCS for further analysis.
Subcellular Localization of DCP1, DCP2, and VCS
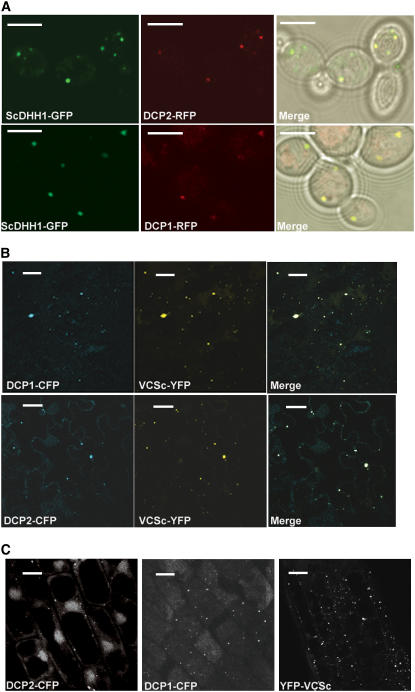
Because yeast P bodies have been well characterized, we first assessed the localization of DCP1–red fluorescent protein (RFP) and DCP2-RFP in yeast (strain yRP1736) using Sc DHH1–green fluorescent protein (GFP) as a yeast P body marker (Sheth and Parker, 2003). Indeed, in all of the cells with appropriate expression levels, both Arabidopsis fusion proteins localized to cytoplasmic foci containing Sc DHH1-GFP (Figure 2A). Those foci disappeared 10 min after cycloheximide (100 μg/mL) treatment. As negative controls, three other Arabidopsis proteins (At eIF4E, At eIF4Eiso, and At RPL18) did not localize to yeast cytoplasmic foci. We conclude that DCP2 and DCP1 specifically localized to yeast P bodies.
Figure 2.
Subcellular Localization of DCP2, DCP1, and VCSc.
Single focal planes for each sample are shown.
(A) Subcellular localization of DCP2-RFP and DCP1-RFP compared with Sc DHH1-GFP in yeast P bodies. RFP fluorescence is pseudocolored in red, and GFP is pseudocolored in green. Bars = 5 μm.
(B) Subcellular localization of DCP1-CFP and DCP2-CFP with YFP-VCSc in Nicotiana epidermal cells. CFP fluorescence is pseudocolored in blue, and YFP is pseudocolored in yellow. Bars = 20 μm.
(C) Subcellular localization of DCP2-CFP, DCP1-CFP, and YFP-VCSc in root cells of Arabidopsis transgenic plants 3 d after germination carrying individual transgenes. Bars = 20 μm.
Using the transient expression system in Nicotiana benthamiana leaves, we found that yellow fluorescent protein (YFP)–DCP2 and DCP1–cyan fluorescent protein (CFP) localized to cytoplasmic foci in leaf epidermal cells. By contrast, At eIF4E-CFP, At eIF4Eiso-CFP, and YFP-At RPL18 contributed evenly throughout the cell. Coexpression of YFP-DCP2 and DCP1-CFP confirmed that the two proteins localized to the same cytoplasmic foci (data not shown). Similar experiments with CFP-VCS (CFP fused to the N terminus of full-length VCS [1344 amino acids]) in N. benthamiana showed that the fusion protein localized to cytoplasmic foci as well. Because VCS is a relatively large protein, we tested its deletion derivatives to define which region is responsible for the localization. Three YFP fusion constructs were made, encoding VCSn (amino acids 1 to 401), VCSm (amino acids 402 to 995), and VCSc (amino acids 996 to 1344) (Figure 3B). YFP-VCSn and YFP-VCSm did not colocalize with full-length CFP-VCS. By contrast, YFP-VCSc colocalized with CFP-VCS (data not shown). Therefore, the C-terminal region (VCSc) of VCS is necessary and sufficient for VCS localization to cytoplasmic foci. Moreover, the YFP-VCSc cytoplasmic foci overlapped with the DCP2-CFP and DCP1-CFP foci (Figure 2B). As Ge-1, the human homolog of Arabidopsis VCS, colocalized with hDCP2 and hDCP1a to P bodies in human cell lines (Yu et al., 2005), we conclude that the DCP2/DCP1/VCS foci in Arabidopsis are likely to be P bodies as well.
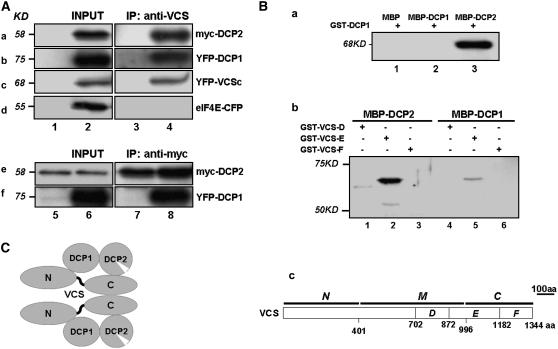
Figure 3.
Protein Interactions among DCP2, DCP1, and VCS.
(A) Self-association of VCS and its interaction with DCP2 and DCP1 in vivo. Coimmunoprecipitation (IP) was performed with extracts prepared from the wild type (lanes 1 and 3) and various Arabidopsis transgenic lines (lanes 2 and 4). Panels a and b, 35S-myc-DCP2 and XVE-YFP-DCP1 coexpression lines. Panel c, 35S-YFP-VCSc line. Panel d, 35S-eIF4E-CFP line. Extracts were immunoprecipitated with anti-VCS antibody (a to d). Immunoprecipitates were then analyzed by protein gel blotting using antibodies against myc (a and e) and YFP/CFP (b to d and f). For panels e and f, transgenic plants carrying 35S-myc-DCP2 and XVE-YFP-DCP1 were used. Plants were either not treated (lanes 5 and 7) or induced using β-estradiol (20 μM) for 6 d (lanes 6 and 8). Extracts were immunoprecipitated with anti-myc antibody.
(B) In vitro interactions between DCP2 and DCP1 (a) and interactions between DCP2 and various VCS domains (b). Proteins were purified from E. coli, and amylose resin was used in the pull-down assay. Proteins input are indicated above each lane (a and b), and different portions of VCS used in the assays are illustrated in panel c. The presence of GST fusion proteins was detected by anti-GST monoclonal antibody on protein gel blots.
(C) A proposed model of the DCP2/DCP1/VCS complex based on in vivo and in vitro interaction data. aa, amino acids.
Based on sequence homology, we have identified a putative Arabidopsis homolog (At3g61240) of DHH1 that shares 80 and 79% similarity to DHH1 in yeast and Rck/p54 in mammalian cells, respectively. Both DHH1 and Rck/p54 are components of P bodies in these organisms (Sheth and Parker, 2003; Andrei et al., 2005; Coller and Parker, 2005). By transient expression assays, we found that YFP-At3g61240 was localized to DCP2 foci (see supplemental Figure 1 online), supporting the notion that the latter are putative plant P bodies.
By a transgenic approach, DCP2, DCP1, and VCSc were shown to localize to cytoplasmic foci in transgenic Arabidopsis plants (Figure 2C). Because of the localization of DCP1/2 in yeast P bodies, these cytoplasmic foci are likely to be P bodies in Arabidopsis. Although the number of foci per cell (10 ± 3 in 35S-DCP2-CFP, 11 ± 5 in 35S-DCP1-CFP, and 11 ± 6 in 35S-YFP-VCSc cells; n = 30) was comparable, the brightness and size of the foci differed. Because P bodies are dynamic foci, these differences may reflect differential incorporation of the three exogenous proteins into endogenous P bodies.
Interactions among DCP2, DCP1, and VCS
The colocalization of DCP1, DCP2, and VCS prompted us to test whether they interact with each other. Using transgenic Arabidopsis plants expressing Myc-DCP2 and YFP-DCP1 recombinant proteins, we investigated protein interactions by coimmunoprecipitation followed by protein gel blot analysis. Interactions could be observed between (1) VCS and Myc-DCP2, (2) VCS and YFP-DCP1, and (3) VCS and YFP-VCSc, a truncated form of the VCS C terminus (Figure 3A). These data suggest that VCS may form dimers/multimers through its C-terminal region; in addition, the results show that VCS forms a complex with DCP2 and DCP1 in vivo (Figure 3A). Myc-DCP2 can pull-down YFP-DCP1 in the coexpression line, suggesting that they form a heterodimer in vivo.
To test whether the three proteins interact directly with each other in vitro, Maltose Binding Protein (MBP)–DCP2, MBP-DCP1, GST-DCP1, and several C-terminal fragments of VCS (GST-VCSD, GST-VCSE, and GST-VCSF) were expressed in Escherichia coli and purified (see Methods). In some cases, as seen with MBP-DCP2 and MBP-DCP1, the full-length proteins were contaminated by shorter fragments resulting from premature termination and/or degradation. However, we found that MBP-DCP2 specifically pulled down GST-DCP1 and the E portion of C-terminal VCS (Figure 3B). Neither the D nor the F portion of the VCS C-terminal region was pulled down by MBP-DCP1; on the other hand, the E portion interacted weakly with MBP-DCP1. These results suggest that the VCS N-terminal region is primarily responsible for interaction with DCP1, with minor contributions from the E segment of the C-terminal region. Unfortunately, the inability to express the N-terminal region of VCS in E. coli precluded us from a direct confirmation of this result in vitro.
Figure 3C shows a proposed model of the interactions among components of the DCP2/DCP1/VCS complex. The direct interactions among these proteins support the notion that they colocalize in putative P bodies.
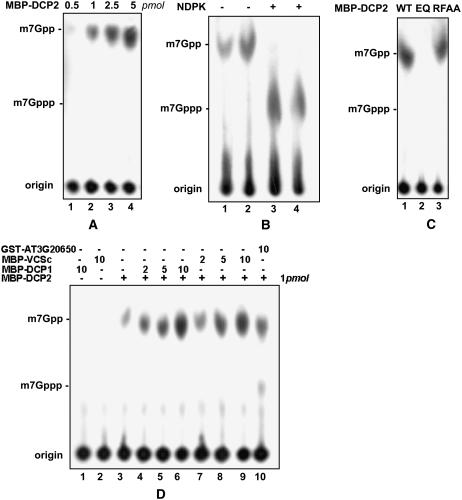
Decapping Activity of DCP2 in Vitro
In assays for decapping activity, only MBP-DCP2 was active—not MBP-DCP1 and MBP-VCSc (Figure 4A). As described with hDCP2 (van Dijk et al., 2002), this activity was insensitive to an RNase inhibitor or excess uncapped tRNA. To provide evidence that the product of the reaction catalyzed by DCP2 contains two phosphates, we performed parallel reactions with MBP-hDCP2, which served as a positive control, and incubated the reaction products with nucleotide diphosphate kinase (NDPK) in the presence of ATP. NDPK converted the product into species comigrating with m7GTP, confirming that the decapping activity generated m7GDP (Figure 4B).
Figure 4.
Decapping Activity of DCP2 in Vitro.
(A) Decapping assay of the MBP-DCP2 fusion protein. Amounts of protein in each reaction mix are shown at top. Migration positions of m7Gppp and m7Gpp are indicated at left.
(B) Comparison of decapping products generated from MBP-DCP2 (lanes 1 and 3) and MBP-hDCP2 (lanes 2 and 4) with and without NDPK treatment.
(C) Decapping activity of MBP-DCP2 and its mutants. EQ, mutant MBP-DCP2 bearing the E158Q mutation in the Nudix domain; RFAA, mutant MBP-DCP2 bearing the R30A and F31A mutations.
(D) Decapping activity of MBP-DCP2 with or without the presence of MBP-DCP1 (lanes 1 and 4 to 6) and MBP-VCSc (lanes 2 and 7 to 9). In lane 10, GST-At3g20650 (10 pmol) was added as a negative control. Amounts of protein added are indicated above each lane.
DCP2 contains a Nudix domain found in a variety of proteins, which has conserved residues that participate in the formation of a catalytic center (Dunckley and Parker, 1999; She et al., 2005). We constructed and purified two mutant proteins: (1) MBP-DCP2E158Q has a mutation changing Glu (E) to Gln (Q) at the 158th amino acid, a conserved residue in the Nudix domain; and (2) MBP-DCP2R30AF31A has two mutations in the N-terminal domain: Arg (R) to Ala (A) at the 30th amino acid and Phe (F) to Ala (A) at the 31st amino acid. Mutations in MBP-DCP2E158Q but not MBP-DCP2R30AF31A blocked the decapping activity, suggesting that an intact Nudix domain is required for the decapping activity of DCP2 (Figure 4C). At a low enzyme:substrate ratio, the decapping activity of MBP-DCP2 was stimulated at least fivefold by MBP-DCP1 or MBP-VCSc (Figure 4D). This led us to hypothesize that the latter two proteins activate and/or stabilize DCP2 in the decapping complex in vivo.
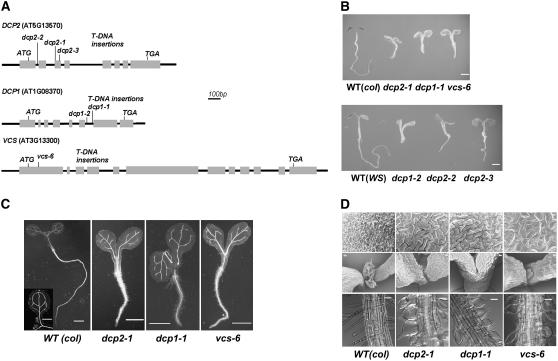
Characterization of Decapping Complex–Deficient Mutants
To access the function of the decapping complex in Arabidopsis, we obtained and analyzed T-DNA insertion mutants with deficiency in DCP2, DCP1, and VCS. The positions of all of the insertions were confirmed by sequencing the PCR products obtained with primers specific to the T-DNA left border and the gene locus (Figure 5A). T-DNA insertion sites in dcp1-1 and dcp1-2 are located 126 bp apart in the sixth intron of DCP1. dcp2-1 and dcp2-3 are mutant alleles with T-DNAs inserted into the third exon of DCP2, whereas the exact insertion site in dcp2-3 is 80 bp away from the insertion site in dcp2-1. In another mutant, dcp2-2, the T-DNA was inserted in the first intron of DCP2. vcs-6 contains a T-DNA in the VCS first exon, 123 bp downstream of the first ATG.
Figure 5.
Phenotypical Analysis of Mutants Deficient in DCP2, DCP1, and VCS.
(A) Schemes of T-DNA insertion lines and alleles of each locus. Gray boxes indicate sequences corresponding to mRNA sequences. Locus numbers, open reading frames (first ATG and stop), and allele names are shown.
(B) Six-day-old seedlings of the wild type and various mutant alleles (listed below). col, Columbia; WS, Wassilewskija. Bars = 1 mm.
(C) Light microscopy images of 6-d-old wild-type and three mutant seedlings after clearing. Bars = 1 mm.
(D) Detailed morphological phenotypes of 6-d-old wild-type and three mutant plants. Top panel, scanning electron microscopic images of cotyledon epidermis. Bars = 20 μm. Middle panel, scanning electron microscopic images of shoot apical meristems. Bars = 50 μm. Bottom panel, light microscopy images of root hairs. Bars = 50 μm.
Adult plants of these mutants were heterozygous for the T-DNA insertions, indicating that homozygous mutants were lethal and died before the developmental stage we sampled. Careful examination of early seedlings showed that the growth of these mutants was severely perturbed at the cotyledon stage (Figure 5B). PCR confirmed that these growth-arrested seedlings were homozygous mutants. The mutants were null, as we could not detect wild-type DCP1 or DCP2 transcripts (Figure 6B). Although RT-PCR confirmed that vcs-6 is a null mutant as well (Figure 6C), RNA gel blot analysis revealed detectable transcripts in vcs-6 (Figure 6B). These transcripts were presumably derived from VCR (Deyholos et al., 2003), which has high sequence similarity to VCS transcripts.
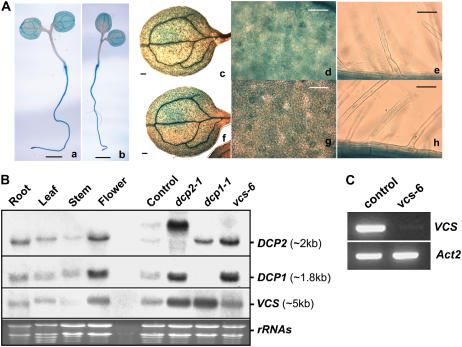
Figure 6.
Expression Profiles of DCP2, DCP1, and VCS in Arabidopsis Tissues and Different Mutants.
(A) GUS staining of 6-d-old seedlings of ProDCP2-GUS (panels a, c, d, and e) and ProDCP1-GUS (panels b, f, g, and h) transgenic plants. Vascular cells (panels a, b, c, and f; bars = 1 mm), guard cells and epidermal cells (panels d and g; bars = 50 μm) on leaves and root hairs (panels e and h; bars = 50 μm) are shown.
(B) RNA gel blot analysis of DCP2, DCP1, and VCS expression in Arabidopsis tissues and in different organs of wild-type plants and in decapping-deficient mutants. Specific bands are indicated at right along with estimated sizes. Ethidium bromide–stained RNA gel lanes are shown at bottom as loading controls. The seedling samples were collected from progeny of the DCP2+/− heterozygote line that did not display a phenotype, which includes wild-type seedlings as well as DCP2+/− heterozygotes. Note that the high molecular mass RNA species detected in dcp2-1 were attributable to the insertion of T-DNA (see Figure 5A). This band was also detected in the control sample because of the presence of DCP2+/− heterozygotes.
(C) Transcript levels of VCS (top panel) and Actin2 (bottom panel) in a control sample and vcs-6 as determined by RT-PCR.
Mutant Phenotypes of dcp2-1, dcp1-1, and vcs-6
The phenotype of dcp2-1 became apparent at 6 d after germination. Offspring of DCP2+/− heterozygotes segregated at 3:1 (820:273; P < 0.05), consistent with the observation that there were no visible differences during embryogenesis, suggesting that dcp2-1 is not embryo-lethal. Six-day-old seedlings of dcp2-1 lacked visible true leaves. Their hypocotyls and roots were short compared with those of the wild type, suggesting that cell elongation and growth were severely arrested (Figure 5C). The veins in dcp2-1 cotyledons were disrupted and did not form a closed loop as in wild-type cotyledons. Development of cotyledon epidermal cells was arrested, losing the typical lob-neck interlock structure commonly seen in the wild type, resulting in misshapen and uneven cotyledons in the mutant (Figure 5C). The density of stomata in dcp2-1 was at least two times less than that in the wild type, but it was the same as observed with vcs-1 grown at 29°C (Deyholos et al., 2003). Although true leaves and trichomes cannot be seen in shoot tips, shoot apical meristems were present in dcp2-1 (Figure 5D). After 3 weeks, the mutant meristem, which still remained green and continued to proliferate, accumulated extra cells, suggesting the continuation of stem cell division. Root hair cells in dcp2-1 were swollen, a phenotype also observed in vcs-1 (Figure 5D). However, root hair cell initiation was not affected, suggesting that cell growth terminated during differentiation. The growth of highly differentiated cell types, including vascular cells, stomata cells, epidermal cells, and root hairs, was perturbed in dcp2-1, leading us to conclude that DCP2 plays a critical role in postembryonic development.
The morphological phenotype of dcp2-1 was also seen in two other mutant alleles, dcp2-2 and dcp2-3 (Figure 5B), confirming that the phenotype is indeed correlated to a deficiency of DCP2. However, in addition to the dcp2-1 null phenotype, dcp2-2 and dcp2-3 mutants also produced small true leaves 2 weeks after germination, suggesting that they are weaker alleles. The dcp2-1 morphological phenotypes were shared by dcp1-1, dcp1-2, and vcs-6 (Figures 5B and 5C). dcp1-2 phenocopies dcp1-1, indicating that the phenotype is correlated to a deficiency of DCP1. Moreover, the phenotype similarities among all mutants deficient in DCP2, DCP1, and VCS suggest that the three proteins form a decapping complex in vivo and that each component is required for mRNA decapping during postembryonic development.
Expression Pattern of DCP2 and DCP1
Using promoter-GUS fusion constructs, we investigated the expression pattern of DCP1 and DCP2 in transgenic plants. Both ProDCP2-GUS and ProDCP1-GUS showed strong expression in 6-d-old seedlings. Although there was DCP1 and DCP2 expression in almost all cells, expression was enhanced in root tip, root hair, and the vascular system (Figure 6A), which were the cells/organs affected in the decapping-deficient mutants. DCP2, DCP1, and VCS transcripts can be detected in roots, leaves, stems, and flowers (Figure 6B), suggesting that they are generally expressed in a number of organs. This is consistent with the role of the decapping complex, which is required for cell growth. Interestingly, we observed increased accumulation of DCP1 and VCS transcripts in dcp2-1, DCP2 and VCS transcripts in dcp1-1, and DCP2 and DCP1 transcripts in vcs-6. These results suggest that an inhibition of mRNA decapping in these mutants leads to a general effect on mRNA accumulation.
Accumulation of Capped mRNA and Prolongation of mRNA Stability in dcp2-1, dcp1-1, and vcs-6
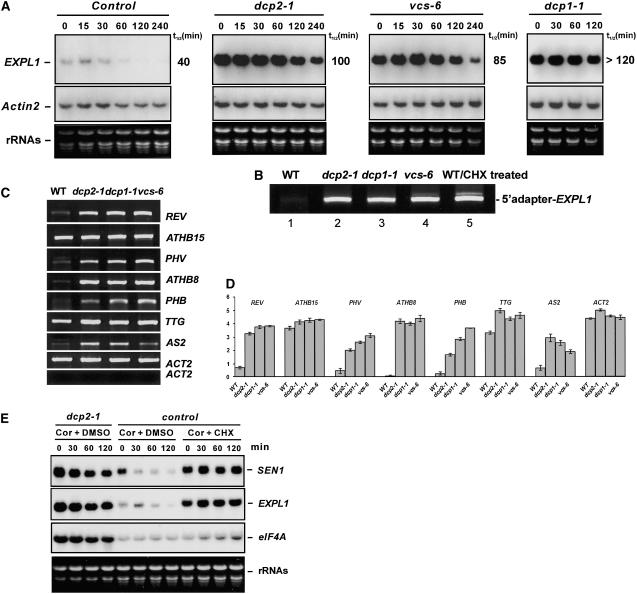
We performed RNA gel blot analysis to test whether mRNA turnover is altered in the decapping-deficient mutants dcp2-1, dcp1-1, and vcs-6. Expansin-Like1 (EXPL1) transcript levels were >10-fold higher in dcp2-1, dcp1-1, and vcs-6 mutants than in the control (Figure 7A). Moreover, the estimated half-life of EXPL1 transcripts was ∼100 min in dcp2-1, 120 min in dcp1-1, and 85 min in vcs-6. These values were at least two times longer than the half-life of 40 min found in control samples (Figure 7A). A similar increase in half-life was observed with another transcript, SEN1, suggesting that decapping may precede the degradation of many transcripts.
Figure 7.
Transcript Accumulation in Three Decapping-Deficient Mutants.
Six-day-old seedlings of each mutant were sampled, and seedlings without a phenotype from progeny of DCP2+/− heterozygotes served as the control.
(A) Transcript levels of EXPL1 (top panels) and Actin2 (middle panels) in control and decapping-deficient mutants at different time points after cordycepin treatment. The estimated half-life (t1/2) is shown at right. Ethidium bromide–stained RNA gel lanes (bottom panels) are shown as loading controls.
(B) RACE-PCR (see Methods) shows accumulation of capped EXPL1 transcript in decapping mutants and in the wild type treated with cycloheximide (CHX).
(C) Semiquantitative RT-PCR to examine transcript abundance in 6-d-old seedlings of the wild type and decapping-deficient mutants. Ethidium bromide–stained cDNA bands corresponding to specific transcripts (listed at right) were confirmed by sequencing. Using templates from reaction mix without the RT step during cDNA synthesis produced no bands, suggesting no genomic DNA contamination in the samples used for RT-PCR (bottom panel).
(D) Quantitative representation of the RT-PCR results from (C). Each column represents the mean value from three independent experiments normalized with respect to the background of the gel. se bars are shown.
(E) Accumulation of SEN1, EXPL1, and eIF4A transcripts after cordycepin (Cor) and cycloheximide (CHX) treatments. Specific bands are labeled at right, and an ethidium bromide–stained RNA gel (bottom panel) is shown as a loading control. Samples and time course after cycloheximide treatment are shown above each lane.
Using modified rapid amplification of cDNA ends (RACE)–PCR, we found that EXPL1 accumulated in the capped form in all three decapping-deficient mutants compared with the control (Figure 7C). Sequence analysis of the PCR products confirmed that the cDNA bands were indeed derived from the transcripts. Moreover, the existence of the adapter sequence verified that the bands were products of capped rather than uncapped mRNAs. Accumulation of capped transcripts suggests that these mutants are deficient in mRNA decapping in vivo.
As many transcription factors were found to be involved in cell differentiation, we tested whether accumulation of their transcripts was altered in the three decapping-deficient mutants. Using semiquantitative RT-PCR, we found transcripts with differential accumulation patterns between mutants and the wild type, which were seen consistently in all three mutants. Steady state transcript levels of PHB, ATHB8, PHV, and REV are at least fivefold higher in the three mutants compared with the control. AS2 and TTG transcripts are increased by approximately twofold in the mutants, whereas ATHB15 and Actin2 transcript levels are not affected significantly (Figures 7C and 7D).
We further tested the other RNA species transcribed by RNA polymerase II, transcripts containing premature termination code (PTC+ transcripts) and small nucleolar RNAs (snoRNAs). For PTC+ transcripts, we tested At5g07910, At3g63340, At2g45670, and At1g72050, according to Hori and Watanabe (2005) and Yoine et al. (2006). Semiquantitative RT-PCR and subsequent sequencing results show that these transcripts, including PTC+ and PTC-free forms, were not significantly different in the three decapping-deficient mutants compared with the wild type (data not shown). For snoRNAs, we tested At U14 and At U51 (Brown et al., 2001). Semiquantitative RT-PCR using primers specific to each transcript shows that they are accumulated at the same level in the three decapping-deficient mutants as in the wild type (data not shown).
Cycloheximide Effect on mRNA Stability
It has been proposed that cycloheximide blocks translational elongation and stabilizes mRNAs by freezing them on polysome. We treated 6-d-old seedlings with cycloheximide to arrest translation and also with cordycepin to stop the synthesis of new polyadenylated RNAs and then examined the decay rates of EXPL1 and SEN1 transcripts. SEN1 transcripts, which had a half-life of 27 min in the control sample, were stabilized by cycloheximide such that no transcript decay was detected in 120 min (Figure 7E). By contrast, in the absence of cycloheximide, the estimated half-life of SEN1 transcripts was increased to 70 min in dcp2-1. Analysis of EXPL1 transcripts showed a similar stabilization effect by cycloheximide. Moreover, the transcripts accumulated in the capped form, as assayed by RACE-PCR (Figure 7B). These results support the notion that blocking translation elongation stabilizes mRNA by preventing decapping.
DISCUSSION
A Decapping Complex in Arabidopsis
A decapping complex comprising DCP1/2 was first described in yeast (Beelman et al., 1996; Dunckley and Parker, 1999) and subsequently in human cells (Lykke-Andersen, 2002; van Dijk et al., 2002; Wang et al., 2002). Homologs of these two proteins are found in Caenorhabditis elegans (Lall et al., 2005) as well as in Drosophila melanogaster (Lin et al., 2006), but only the yeast and human decapping complexes have been biochemically characterized. The human decapping complex also contains a WD-40 domain protein, called Hedls/Ge-1, which appears not to be present in the yeast genome (Fenger-Gron et al., 2005; Yu et al., 2005). Available evidence suggests that DCP2 is the only component in the complex that possesses catalytic activity, although this activity can be modulated by DCP1 (Steiger et al., 2003) and Hedls (Fenger-Gron et al., 2005).
We have identified from the Arabidopsis genome homologs of DCP1/2. Here, we show that DCP1/2 together with VCS (Deyholos et al., 2003), an Arabidopsis homolog of Hedls/Ge-1 (Fenger-Gron et al., 2005; Yu et al., 2005), form a decapping complex in vitro and in vivo. This notion is supported by the following lines of evidence. (1) DCP1, DCP2, and VCS can associate with each other in vitro. The VCS-E region containing a putative coiled-coil motif (amino acids 1147 to 1175) is involved in this interaction. (2) Recombinant DCP2, but not DCP1 and VCS, displays decapping activity in vitro, and the decapping activity is stimulated by DCP1 and VCS. Moreover, the E158Q mutation in the DCP2 Nudix domain abolished the decapping activity (3) Coimmunoprecipitation experiments show that the three proteins are present in the same complex in vivo. (4) The three proteins were found to be concentrated in cytoplasmic foci, which likely represent P bodies in plants.
Recombinant DCP2 from E. coli is the third protein known to be able to remove m7GDP caps from capped mRNAs in vitro, after their counterparts in yeast (Steiger et al., 2003) and human (van Dijk et al., 2002) cells. The absence of an obvious VCS homolog in yeast and the stimulation of DCP2 activity by DCP1 and VCS suggest that the plant decapping complex is more closely related to the human decapping complex (Fenger-Gron et al., 2005).
dcp2-1, dcp1-1, and vcs-6 Are Defective in Decapping in Vivo
Analysis of several unstable transcripts and transcripts encoding transcription factors in the three decapping mutants showed a decreased degradation rate and a corresponding increase in transcript half-life. Moreover, these transcripts accumulated in the capped form. These results support the view that DCP2, DCP1, and VCS are needed for transcript decapping in vivo. Treatment of wild-type plants with cycloheximide, which disassembles P bodies (Sheth and Parker, 2003), also produced the same effects, suggesting that decapping is a translation-linked mRNA process residing in P bodies.
In all three decapping-deficient mutants, the vascular tissues are abnormally initiated and disrupted, resulting in varicose veins. Because ectopic expression of ATHB8 was reported to significantly accelerate and stimulate the formation of vascular tissues (Baima et al., 2001), we analyzed expression levels of ATHB8 and its related mRNAs. Among members of the small homeodomain-leucine zipper (HD-ZipIII) family, PHB, PHV, REV, and ATHB8 transcripts accumulated consistently in all three decapping mutants, whereas ATHB15 transcripts levels were not significantly different from those in the wild type. Based on the results of Baima et al. (2001), it is reasonable to assume that the thickened and varicose veins result from the accumulation of ATHB8 and related mRNAs. Thus, it is likely that the accumulation of many mRNAs, whose degradation depends on mRNA decapping in P bodies, may be responsible for the pleiotropic phenotype of the decapping-deficient mutants.
All five HD-ZipIII family mRNAs are subject to microRNA (miRNA) regulation. However, the observation that non-miRNA targets also accumulate in the mutants suggests that the decapping process is not specifically related to the miRNA pathway. No significant accumulation of ATHB15 was seen in the mutants, suggesting no direct linkage between miRNA regulation and decapping for this transcript in Arabidopsis. Whether the Arabidopsis decapping complex is also required for miRNA-mediated gene silencing, as in Drosophila (Rehwinkel et al., 2005; Behm-Ansmant et al., 2006), remains to be elucidated.
To determine whether decapping is required for the degradation of all capped transcripts, we also analyzed PTC+ transcripts and snoRNAs. In mammalian cells, downregulating the DCP2 decapping protein significantly increases the abundance of steady state nonsense-containing but not nonsense-free transcripts (Lejeune et al., 2003). However, we observed no preferential accumulation of PTC+ transcript among the three decapping mutants and the wild type, or among their corresponding nonsense-free transcripts. This finding suggests the possible existence of a different nonsense-mediated decay pathway for these plant transcripts. The accumulation of several snoRNAs tested in the mutants is not significantly different from that in the wild type, suggesting that decapping is not required for the turnover of these snoRNAs.
Eukaryotes deploy both the 5′→3′ pathway and the 3′→5′ pathway for mRNA turnover. The decapping process is the first step of the 5′→3′ pathway (Coller and Parker, 2004; Meyer et al., 2004). We found that degradation of Act2 transcripts, transcripts targeted by specific miRNAs, PTC+ transcripts, and snoRNAs was unaffected by defective decapping, suggesting that these RNA species are likely degraded by the 3′→5′ pathway. Some of the components of the exosome were found and confirmed to be functional in Arabidopsis (Chekanova et al., 2002). It will be interesting to investigate the interplay between the two pathways.
Decapping-Deficient Mutants Are Defective in Postembryonic Development
The observation that all three decapping mutants have a similar lethality phenotype with growth arrest at the postembryonic stage provides additional insights into the function of the decapping complex in vivo. First, we consider it somewhat surprising that dcp1-1 and vcs-6 should have the same phenotype as dcp2-1. The dcp1-1 mutant should continue to produce DCP2 and VCS, whereas vcs-6 should continue to produce DCP2 and DCP1. Because DCP2 is the catalytic subunit, it is surprising that dcp1-1 and vcs-6 have no decapping activity in vivo. One reasonable hypothesis is that DCP2 produced in dcp1-1 and vcs-6 is either unstable or has little or no in vivo decapping activity, because DCP1 and VCS are needed for its activation or stabilization. Indeed, in human cells, hDCP2 accumulates to ∼10- to 20-fold higher levels in the presence of coexpressed Hedls (Fenger-Gron et al., 2005). In this regard, we have found that VCS is able to form dimers/multimers and DCP1 and DCP2 can form heterodimers in vivo. Second, the high similarity in phenotype allows us to conclude that each of the three proteins executes its function largely through the activity of the decapping complex. The function of Hedls in decapping in vivo has not yet been reported. Our molecular analysis of the Arabidopsis vcs-6 provides direct evidence that the main function of VCS, a homolog of Hedls, is to modulate decapping. However, other possible roles of VCS cannot be excluded (e.g., the VCS N-terminal WD-40 domains [Deyholos et al., 2003] may interact with other proteins to link transcript decapping with other signaling pathways).
In Drosophila oocytes, the dDCP1 gene is required for the degradation and localization of the oskar messenger ribonucleoprotein complex during oogenesis (Lin et al., 2006), providing evidence that dDCP1 may have specialized functions in distinct developmental processes. By contrast, microscopic observations and analysis of the segregation ratio of heterozygotes indicate that gametogenesis and embryogenesis are not affected in any of the Arabidopsis decapping mutants. However, postembryonic differentiation is severely arrested at the stage of cotyledonary seedlings. Hypocotyl and root elongation in decapping mutants is arrested, and the vascular system is disrupted and disorganized. Although neither meristem activities nor root hair initiation is affected, root hairs are swollen and cotyledon epidermal cells no longer assume the typical lob-neck structure, indicating an impairment in polar cell growth (Kost et al., 1999; Fu et al., 2005). This loss of polar growth is also consistent with the abnormal development of vcs trichomes at restrictive temperatures reported previously (Deyholos et al., 2003). The pleiotropic mutant phenotype suggests that some important steps in postembryonic development require decapping-mediated mRNA turnover. Because little is known regarding genes or mutants specifically involved in postembryonic development, we hypothesize that the lethality might be caused by the accumulation of a number of different transcripts.
The arrested cell differentiation phenotype in mutant seedlings not only suggests a critical role of decapping in development but also provides an opportunity to investigate how decapping as well as mRNA turnover affect this process, and how related pathways are regulated at the whole organism level. Because decapping is an important aspect of P body function, the phenotype of these decapping-deficient mutants suggests that P bodies may be required for cell development as well in other multicellular eukaryotes.
METHODS
Plant and Yeast Strains
Arabidopsis thaliana dcp1-1 (844B03) was obtained from GABI-Kat (Rosso et al., 2003), and dcp2-1 (SALK_00519) and vcs-6 (SAIL_831_D08) (Sessions et al., 2002) were from the ABRC. These three accessions are in the Columbia ecotype. vcs-6 is an allele to and named after vcs-1 to vcs-5 (Deyholos et al., 2003). dcp1-2 (FLAG_563G05), dcp2-2 (FLAG_453E05), and dcp2-3 (FLAG_427G03) were Institut National de la Recherche Agronomique public lines (ecotype Wassilewskija) (Samson et al., 2002). Plants were grown on sterile Murashige and Skoog (MS) medium for 18 d before being transferred to a greenhouse under similar conditions (22°C, 16-/8-h photoperiod cycle). Agrobacterium tumefaciens–mediated genetic transformation of Arabidopsis was by the floral-dip method (Clough and Bent, 1998).
Yeast strain yRP1736 MATa leu2-3,112 trp1 ura3-52 his4 cup1∷LEU2/PGK1pG/MFA2pG dcp1∷URA3 DHH1-GFP (Sheth and Parker, 2003) was used for transformation. Transformants were selected on SD medium (BD Bioscience) without Trp supplement.
Transformation and Expression Vectors
To express fusion proteins in Arabidopsis and Nicotiana benthamiana, the coding sequences of DCP2, DCP1, At3g61240, and VCSc were amplified by PCR from reverse-transcribed products of Arabidopsis total RNA, digested, and ligated into pENT3C (Invitrogen). These entry clones were then cloned into destination vectors by LR clonase (Invitrogen).
To express recombinant proteins in Saccharomyces cerevisiae, pESC-Trp (Stratagene) was modified to pESC-GW-RFP(-Trp), in which a gateway cassette and a mRFP sequence (Campbell et al., 2002) were inserted downstream of the GAL10 promoter. pESC-DCP2-RFP and pESC-DCP1-RFP were produced by the LR recombination system.
To express GUS under the control of Arabidopsis native promoters, 2435 bp of 5′ upstream sequence of DCP2 (the first nucleotide of the first ATG being +1) and 3068 bp of 5′ upstream sequence of DCP1 were amplified from the relevant genomic regions. These promoter fragments were enzymatically ligated to a GUS coding sequence, giving ProDCP2-GUS and ProDCP1-GUS. GUS staining was performed by incubating transgenic plants in a 5-bromo-4-chloro-3-indolyl-β-glucuronic acid staining solution for 2 h at 37°C.
Microscopic Analysis
Transient expression of fusion proteins in Nicotiana leaves was assayed 2 d after agroinfiltration. Roots of 3-d-old transgenic Arabidopsis seedlings were incubated in 5% glycerol and imaged for subcellular localization of fusion proteins. Yeast cells were grown in SD medium at 30°C to OD600 = 0.3 and transferred to synthetic galactose medium for 2 h before being imaged in PBS. Tissues for scanning electron microscopy were prepared as described by Deyholos et al. (2003). Confocal laser scanning microscopy images were collected with an Axiovert 200 microscope (Zeiss) with LSM510 META laser scanning module and processed with an LSM image browser.
Plasmids and Expression of Recombinant Proteins
pMBP-DCP2, pMBP-DCP1, pMBP-VCSc, and pMBP-hDCP2 were generated by the LR recombination system. pGEX-DCP1 was from direct cloning of the PCR product into pGEX-4T1 (GE Bioscience). The plasmids pMBP-DCP2E158Q and pMBP-DCP2R30AF31A containing mutations in the DCP2 coding sequence were generated by the QuickChange XL mutagenesis system according to the manufacturer's instructions (Stratagene). Insertions and changes in plasmids were confirmed by sequencing.
Recombinant proteins were purified and then dialyzed against dialysis buffer (20 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 150 mM NaCl, 1 mM DTT, and 10% glycerol).
Coimmunoprecipitation and Pull-Down Assays
Polyclonal antibody against VCS in rabbits was prepared commercially (Cocalico Biologicals). Rabbit polyclonal antibody against c-Myc and mouse monoclonal antibody against GST and GFP were purchased from Santa Cruz Biotechnology.
For coimmunoprecipitation, proteins from 6-d-old Arabidopsis seedlings carrying overexpression constructs were extracted in binding buffer (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 1 mM DTT, 0.2% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride, and protease inhibitors). After incubation with the appropriate antibody for 3 h at 4°C, protein A–agarose was added to the lysate and further incubated for 3 h at 4°C. After six washes with the binding buffer, protein A–agarose was collected and proteins released were analyzed by protein gel blotting.
For in vitro pull-down assay, purified proteins (2 μg each) were incubated with 2 μg of each of the target proteins at room temperature for 2 h in 1 mL of binding buffer (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 0.2% glycerol, 0.6% Triton X-100, and 0.5 mM β-mercaptoethanol). Amylose resin beads were added and the incubation was continued under the same conditions. The beads were washed six times with the washing buffer (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, and 0.6% Triton X-100). Pulled-down proteins were resolved by 10% SDS-PAGE and analyzed by protein gel blotting using the appropriate antibody.
In Vitro Decapping Assay
RNAs (121nt-G16) containing a 16-guanine track at the 3′ end were transcribed with the MAXIscript T7 kit (Ambion) from a DNA fragment of At2g38280 corresponding to the 5′ untranslated region (121 nucleotides) of the transcribed mRNA. Cap-labeled RNAs generated by guanylyltransferase (Ambion) were gel-purified before use.
Decapping assays were performed at 37°C for 30 min with cap-labeled RNA (10,000 cpm) and the indicated amounts of purified proteins. Reaction products were resolved by thin layer chromatography as described by Zhang et al. (1999). The reactions were performed in IVDA buffer (10 mM Tris-Cl, pH 7.5, 100 mM KAc, 2 mM MgAc2, 0.5 mM MnCl2, 2 mM DTT, 0.1 mM spermine, and 25 μg/mL yeast tRNA), modified from Piccirillo et al. (2003). Unlabeled standards were loaded alongside the samples and visualized by UV light shadowing. The NDPK reactions were performed as described (Wang and Kiledjian, 2001).
RT-PCR and RACE-PCR
Total RNA was isolated from 6-d-old seedlings grown on MS medium using the RNeasy plant mini kit (Qiagen). Reverse transcription was performed using oligo(dT) primers and 5 μg of each total RNA after DNase treatment. Detection of RNA 5′ ends was performed using the RLM-RACE kit (Ambion), with calf intestinal phosphatase and tobacco acid pyrophosphatase treatments, modified as described (Gazzani et al., 2004). Sequences of primers used for the detection of specific transcripts will be provided upon request.
Cordycepin and Cycloheximide Treatments and RNA Gel Blot Analysis
Six-day-old seedlings were incubated in MS medium with cordycepin (3′-deoxyadenosine; Sigma-Aldrich) and/or cycloheximide added. Total RNA was extracted from samples harvested at various time points using Trizol reagent (Invitrogen). For RNA gel blot analysis, 15 μg of total RNA was fractionated on a 1.2% (w/v) agarose gel and then transferred to a Hybond-XL membrane. DNA probes were labeled with [α-32P]dCTP using the random prime labeling system (GE Biosciences).
Accession Numbers
Representative sequences for genes mentioned in this article can be found in the GenBank data library under the following accession numbers: DCP2 (AK119112); DCP1 (NM100710); VCS (NM180245); VCR (NM112174); eIF4E (NM117914); eIF4Eiso (NM203123); RPL18 (NM111432); EXPL1 (NM114466); SEN1 (NM119743); and ACTIN2 (U41998).
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Colocalization of At3g61240 and DCP2 in Tobacco Leaf Epidermal Cells.
Supplementary Material
Acknowledgments
We thank Roy Parker for yeast strains containing P body markers, Jens Lykke-Andersen for plasmid containing hDCP2, Leslie E. Sieburth for the vcs-1 mutant, Barbara Mazur, Enno Krebbers, Carl Falco, and Richard Broglie for discussions and comments, and Alison North and Eleana C. Sphicas for assistance with microscopy. This work was supported in part by a grant from DuPont to N.-H.C.; J.-Y.Y. was supported by a Taiwan Merit Scholarship from the Ministry of Education, the Council for Economic Planning and Development, and the National Science Council, Taiwan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Nam-Hai Chua (chua@mail.rockefeller.edu).
Online version contains Web-only data.
References
- Andrei, M.A., Ingelfinger, D., Heintzmann, R., Achsel, T., Rivera-Pomar, R., and Luhrmann, R. (2005). A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M.M., Ruberti, I., and Morelli, G. (2001). The Arabidopsis ATHB-8 HD-Zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman, C.A., Stevens, A., Caponigro, G., LaGrandeur, T.E., Hatfield, L., Fortner, D.M., and Parker, R. (1996). An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382 642–646. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant, I., Rehwinkel, J., Doerks, T., Stark, A., Bork, P., and Izaurralde, E. (2006). mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20 1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliakova-Bethell, N., Beckham, C., Giddings, T.H.J., Winey, M., Parker, R.O.Y., and Sandmeyer, S. (2006). Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.W., Clark, G.P., Leader, D.J., Simpson, C.G., and Lowe, T. (2001). Multiple snoRNA gene clusters from Arabidopsis. RNA 7 1817–1832. [PMC free article] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova, J.A., Dutko, J.A., Mian, I.S., and Belostotsky, D.A. (2002). Arabidopsis thaliana exosome subunit AtRrp4p is a hydrolytic 3′→5′ exonuclease containing S1 and KH RNA-binding domains. Nucleic Acids Res. 30 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Coller, J., and Parker, R. (2004). Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73 861–890. [DOI] [PubMed] [Google Scholar]
- Coller, J., and Parker, R. (2005). General translational repression by activators of mRNA decapping. Cell 122 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos, M.K., Cavaness, G.F., Hall, B., King, E., Punwani, J., Van Norman, J., and Sieburth, L.E. (2003). VARICOSE, a WD-domain protein, is required for leaf blade development. Development 130 6577–6588. [DOI] [PubMed] [Google Scholar]
- Dunckley, T., and Parker, R. (1999). The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18 5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley, T., Tucker, M., and Parker, R. (2001). Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics 157 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron, M., Fillman, C., Norrild, B., and Lykke-Andersen, J. (2005). Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 20 905–915. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Gu, Y., Zheng, Z., Wasteneys, G., and Yang, Z. (2005). Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120 687–700. [DOI] [PubMed] [Google Scholar]
- Gazzani, S., Lawrenson, T., Woodward, C., Headon, D., and Sablowski, R. (2004). A link between mRNA turnover and RNA interference in Arabidopsis. Science 306 1046–1048. [DOI] [PubMed] [Google Scholar]
- Hatfield, L., Beelman, C.A., Stevens, A., and Parker, R. (1996). Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 16 5830–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, K., and Watanabe, Y. (2005). UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 43 530–540. [DOI] [PubMed] [Google Scholar]
- Kole, R., Sierakowska, H., and Shugar, D. (1976). Novel activity of potato nucleotide pyrophosphatase. Biochim. Biophys. Acta 438 540–550. [DOI] [PubMed] [Google Scholar]
- Kost, B., Mathur, J., and Chua, N.-H. (1999). Cytoskeleton in plant development. Curr. Opin. Plant Biol. 2 462–470. [DOI] [PubMed] [Google Scholar]
- Lall, S., Piano, F., and Davis, R.E. (2005). Caenorhabditis elegans decapping proteins: Localization and functional analysis of Dcp1, Dcp2, and DcpS during embryogenesis. Mol. Biol. Cell 16 5880–5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune, F., Li, X., and Maquat, L.E. (2003). Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12 675–687. [DOI] [PubMed] [Google Scholar]
- Lin, M.-D., Fan, S.-J., Hsu, W.-S., and Chou, T.-B. (2006). Drosophila Decapping Protein 1, dDcp1, is a component of the oskar mRNP complex and directs its posterior localization in the oocyte. Dev. Cell 10 601–613. [DOI] [PubMed] [Google Scholar]
- Liu, J., Rivas, F.V., Wohlschlegel, J., Yates, J.R., Parker, R., and Hannon, G.J. (2005. a). A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7 1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Valencia-Sanchez, M.A., Hannon, G.J., and Parker, R. (2005. b). MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J. (2002). Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22 8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, S., Temme, C., and Wahle, E. (2004). Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 39 197–216. [DOI] [PubMed] [Google Scholar]
- Piccirillo, C., Khanna, R., and Kiledjian, M. (2003). Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA 9 1138–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel, J.A.N., Behm-Ansmant, I., Gatfield, D., and Izaurralde, E. (2005). A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11 1640–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Sakuno, T., Araki, Y., Ohya, Y., Kofuji, S., Takahashi, S., Hoshino, S.-i., and Katada, T. (2004). Decapping reaction of mRNA requires Dcp1 in fission yeast: Its characterization in different species from yeast to human. J. Biochem. (Tokyo) 136 805–812. [DOI] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Balzergue, S., Dubreucq, B., Lepiniec, L., Pelletier, G., Caboche, M., and Lecharny, A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, G.L., and Blau, H.M. (2005). Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7 633–636. [DOI] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She, M., Decker, C.J., Chen, N., Tumati, S., Parker, R., and Song, H. (2006). Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 13 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She, M., Decker, C.J., Sundramurthy, K., Liu, Y., Chen, N., Parker, R., and Song, H. (2004). Crystal structure of Dcp1p and its functional implications in mRNA decapping. Nat. Struct. Mol. Biol. 11 249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U., and Parker, R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger, M., Carr-Schmid, A., Schwartz, D.C., Kiledjian, M., and Parker, R.O.Y. (2003). Analysis of recombinant yeast decapping enzyme. RNA 9 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun, S., and Parker, R. (1999). Analysis of mutations in the yeast mRNA decapping enzyme. Genetics 151 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk, E., Cougot, N., Meyer, S., Babajko, S., Wahle, E., and Seraphin, B. (2002). Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21 6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Jiao, X., Carr-Schmid, A., and Kiledjian, M. (2002). From the cover. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. USA 99 12663–12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., and Kiledjian, M. (2001). Functional link between the mammalian exosome and mRNA decapping. Cell 107 751–762. [DOI] [PubMed] [Google Scholar]
- Wichroski, M.J., Robb, G.B., and Rana, T.M. (2006). Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2 e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoine, M., Ohto, M.-a., Onai, K., Mita, S., and Nakamura, K. (2006). The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 47 49–62. [DOI] [PubMed] [Google Scholar]
- Yu, J.H., Yang, W.-H., Gulick, T.O.D., Bloch, K.D., and Bloch, D.B. (2005). Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 11 1795–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Williams, C.J., Wormington, M., Stevens, A., and Peltz, S.W. (1999). Monitoring mRNA decapping activity. Methods 17 46–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.