Abstract
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) emerged in December 20191,2 and is responsible for the COVID-19 pandemic3. Vaccines are an essential countermeasure urgently needed to control the pandemic4. Here, we show that the adenovirus-vectored vaccine ChAdOx1 nCoV-19, encoding the spike protein of SARS-CoV-2, is immunogenic in mice, eliciting a robust humoral and cell-mediated response. This response was predominantly Th1, as demonstrated by IgG subclass and cytokine expression profiling. Vaccination with ChAdOx1 nCoV-19 (prime-only and prime-boost regimen) induced a balanced Th1/Th2 humoral and cellular immune response in rhesus macaques. We observed a significantly reduced viral load in bronchoalveolar lavage fluid and lower respiratory tract tissue of vaccinated rhesus macaques challenged with SARS-CoV-2 compared with control animals, and no pneumonia was observed in vaccinated animals. However, there was no difference in nasal shedding between vaccinated and control animals. Importantly, no evidence of immune-enhanced disease following viral challenge in vaccinated animals was observed. Safety, immunogenicity and efficacy of ChAdOx1 nCoV-19 against symptomatic PCR-positive COVID-19 disease will now be assessed in randomised controlled human clinical trials.
ChAdOx1 is a replication-deficient simian adenoviral vector derived from isolate Y25. The seroprevalence of antibodies to Y25 in the human population was found to be 0% in the United Kingdom and 9% in the Gambia5. We previously demonstrated that a single dose of ChAdOx1 MERS, encoding the spike protein of MERS-CoV, protected non-human primates (NHPs) against MERS-CoV-induced disease6. Upon the emergence of SARS-CoV-2, we designed a similar ChAdOx1-vectored vaccine encoding a codon-optimized full-length SARS-CoV-2 spike protein.
Immunogenicity in mice
Two mouse strains (BALB/c, N=5 and outbred CD1, N=8) were vaccinated intramuscularly (IM) with ChAdOx1 nCoV-19 or ChAdOx1 GFP, a control vaccine encoding green fluorescent protein. Humoral and cellular immunity were studied 9–14 days later. Total IgG titers were detected against spike protein in all vaccinated mice (Figure 1a). Profiling of the IgG subclasses showed a predominantly Th1 response post vaccination (Extended Data Figure 1a) and induction of IgM antibodies (Extended Data Figure 2a). Virus-specific neutralising antibodies were detected in all mice vaccinated with ChAdOx1 nCoV-19, whereas no neutralisation was detected in serum from mice vaccinated with ChAdOx1 GFP (Figure 1b). Splenic T-cell responses measured by IFN-γ ELISpot and intracellular cytokine staining (ICS) were detected against peptides spanning the full length of the spike construct (Figure 1c). Again, a strong Th1-type response was detected post vaccination as supported by high levels of IFN-γ and TNF-α, and low levels of IL-4 and IL-10 (Figure 1d & Extended Data Figure 1b–c).
Figure 1: Humoral and cellular immune responses to ChAdOx1 nCoV-19 vaccination in mice.
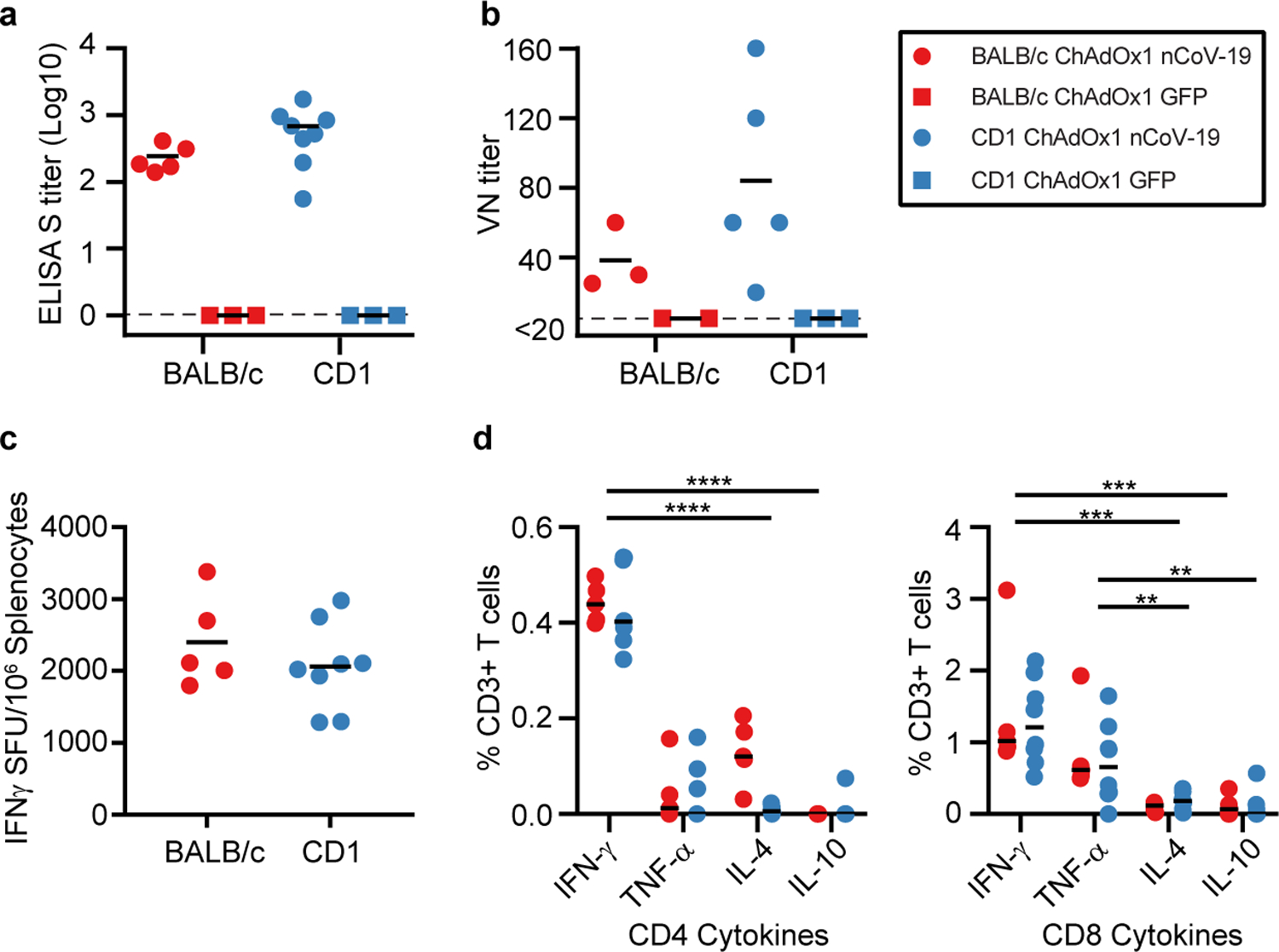
a. End point titer of serum IgG detected against S protein at 14 days post vaccination. No positive responses were detected in the control group. n=5, 3, 8, and 3 animals respectively examined over 1 independent experiment. b. Virus neutralizing titer in serum at 9 days post vaccination. n=3, 2, 5, and 3 animals respectively examined over 1 independent experiment. c. Summed IFN-γ ELISpot responses in splenocytes toward peptides spanning the spike protein at 14 days post vaccination. Control mice had low (<100 SFU) responses. n=5 and 8 animals respectively examined over 1 independent experiment. d. Summed frequency of spike-specific cytokine positive CD3+ T cells at 14 days post vaccination. P-value left panel: <0.0001. P-value right panel 0.0002 (IFN-γ−IL-4); 0.0001 (IFN-γ-IL-10); 0.0054 (TNF-α-IL-4); 0.0022 (TNF-α-IL-10). n=5 and 8 animals respectively examined over 1 independent experiment. BALB/c = red; CD1 = blue; vaccinated = circle; control = square; dotted line = limit of detection; line = mean; SFU = spot-forming units; Spl. = splenocytes; * = p<0.05. Statistical significance determined via 2-way ANOVA (repeated measure) and post-hoc positive test.
Immunogenicity in rhesus macaques
We next evaluated the efficacy of ChAdOx1 nCoV-19 in rhesus macaques, a non-human primate model that displays robust infection of upper and lower respiratory tract and virus shedding upon inoculation with SARS-CoV-27. Six animals per group were vaccinated via a prime-only regimen (28 days before challenge) or a prime-boost regimen (56 and 28 days before challenge) intramuscularly with 2.5 x 1010 ChAdOx1 nCoV-19 virus particles each. As a control, six animals were vaccinated via the same route with the same dose of ChAdOx1 GFP (one animal 56 and 28 days before challenge, five animals 28 days before challenge, Figure 2a). No adverse events were observed upon vaccination. Spike-specific antibodies were present as early as 14 days post vaccination and were significantly boosted upon the second immunization (two-tailed Wilcoxon test). Endpoint IgG titers of 400–6400 (prime) and 400–19,200 (prime-boost) were measured on the day of challenge (Figure 2b). Virus-specific neutralising antibodies were also significantly boosted upon secondary immunization (two-tailed Wilcoxon test) and detectable in all vaccinated animals before challenge (5–40 (prime) and 10–160 (prime-boost)), whereas no virus-specific neutralising antibodies were detected in control animals (Figure 2c). IgM antibodies were present in serum post vaccination at challenge in 6 out of 6 prime-boost and 2 out of 6 prime only animals (Extended Data Figure 2b). SARS-CoV-2 spike specific T-cell responses were detected at 0 DPI by IFN-γ ELISpot assay upon stimulation of peripheral blood mononuclear cells (PBMCs) with a peptide library spanning the full length of the spike protein. No statistically significant difference in the magnitude of the response was found between the prime-boost and prime-only group (Mann Whitney test, p-value = 0.3723, Figure 2d). As reported previously6, vaccination with ChAdOx1 nCoV-19 resulted in the induction of neutralizing antibodies against the vaccine vector itself within 28 days after vaccination (Extended Data Figure 3). Nonetheless, a boost vaccination with ChAdOx1 nCoV-19 resulted in a significant increase in binding and neutralizing antibodies in NHPs (Figure 2b,c) and increase of SARS-CoV-2 VN titer was not significantly correlated to ChAdOx1 VN titer (two-tailed Pearson correlation, r2=0.6493 p=0.0529).
Figure 2. Humoral and cellular immune responses to ChAdOx1 nCoV-19 vaccination in rhesus macaques.

a. Study schedule for NHPs. V = vaccination; E = exam; C = exam and challenge; N = exam and necropsy. Violin plots of b. endpoint IgG titer in serum against trimeric spike protein, c. VN titer in serum and d. Summed IFN-γ ELISpot responses in PBMCs collected at 0 DPI toward peptides spanning the spike protein. Red circles = prime-only vaccine; blue squares = prime-boost vaccine; green triangle = controls; dotted line = limit of detection; SFU = spot-forming units; VN= virus-neutralizing; * = p-value=0.0313. Statistical significance determined via two-tailed Wilcoxon test.
Clinical signs
Upon challenge with 2.6 x 106 TCID50 SARS-CoV-2 to both the upper and lower respiratory tract the average clinical score of control animals was higher compared to ChAdOx1 nCoV-19 vaccinated animals. This was significantly different as determined via Mann-Whitney’s rank’s test on 4–7 days post infection (DPI, Figure 3a).
Figure 3. Clinical signs and viral load in rhesus macaques inoculated with SARS-CoV-2 after vaccination with ChAdOx1 nCoV-19.
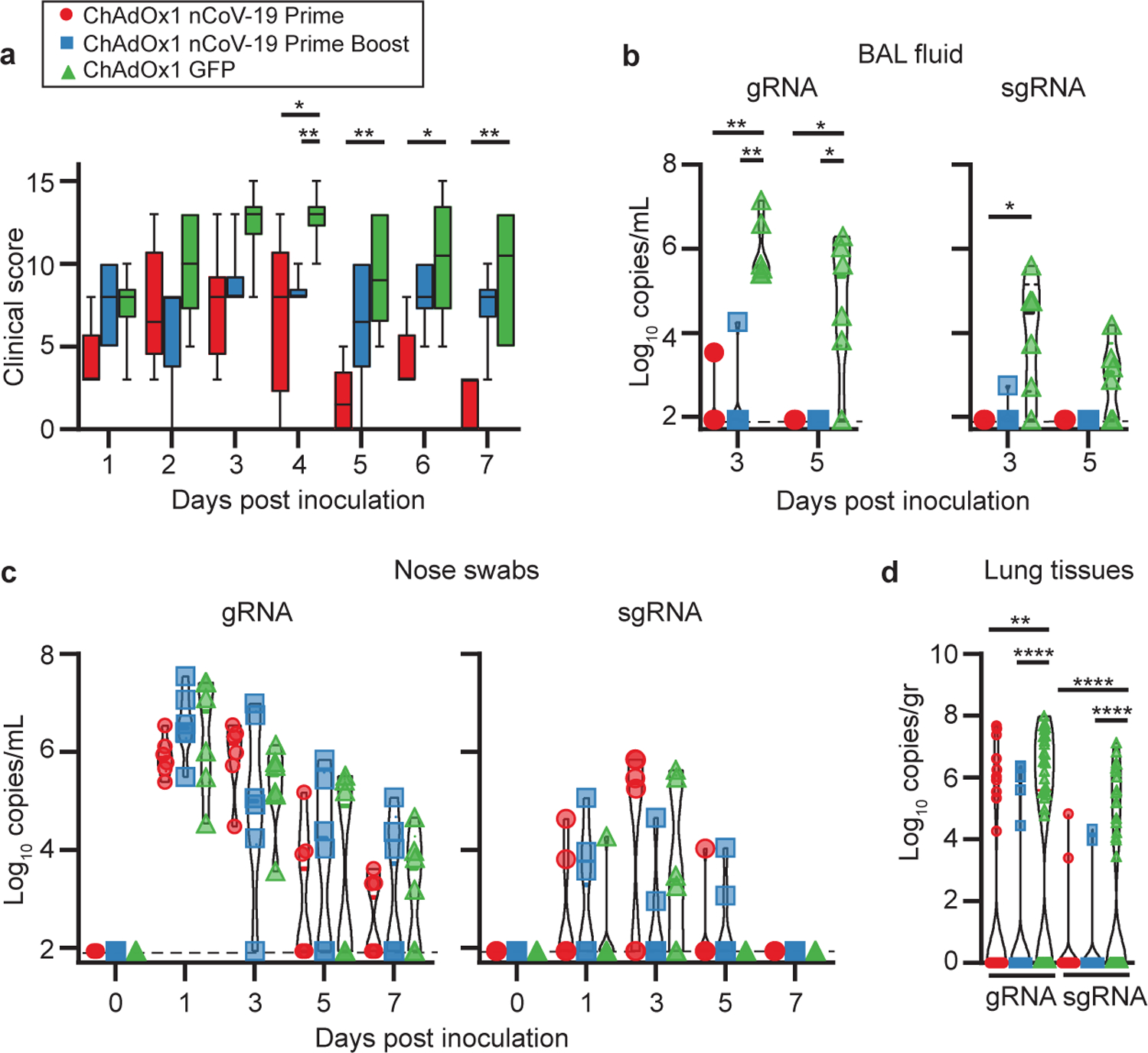
a. Boxplot of 25th to 75th percentile with median as centre and whiskers of 5th to 95th percentile clinical score in NHPs. n=6 animals per group examined over 2 independent experiments. P-values = 0.0455 (Prime-control D3); 0.0238 (Prime-control D4); 0.0043 (Prime-boost-control D4); 0.0043 (Prime-control D5); 0.0152 (Prime-control D6); 0.0022 (Prime-control D7). Violin plot of viral load in b. BAL fluid (* = p-value 0.0152; ** = p-value 0.0022) and c. nose swabs obtained from rhesus macaques. d. Violin plot of viral load in lung tissue. n=6 lung lobes of 6 animals per group examined over 2 independent experiments. **=p-value=0.0011; ****=p-value<0.0001. Dotted line = limit of detection. Statistical significance determined via two-tailed Mann-Whitney test.
Viral load in respiratory tract samples
In BAL fluid obtained from control animals, viral genomic RNA (gRNA) and subgenomic RNA (sgRNA), indicative of virus replication, was detected on all days. In contrast, viral gRNA and sgRNA was detected in only two vaccinated animals on 3 DPI, and viral load was significantly lower (Figure 3b). Viral gRNA was detected in nose swabs from all animals and no difference was found on any day between vaccinated and control animals. Viral sgRNA was detected in a minority of animals, with no difference between groups (Figure 3c). Infectious virus could only be detected at 1 and 3 DPI in prime-only vaccinated and control animals, and 1 DPI in prime-boost vaccinated animals (Extended Data Table 1).
Cytokine response
Cytokines in serum were analysed after challenge to monitor immune responses. Following vaccination of NHPs the level of Th1 (IFN-γ and IL-2) or Th2 (IL-4, IL-5, or IL-13) cytokines in the serum were low and no evidence of a dominant Th2 response was detected (Extended Data Figure 4). We observed a significant upregulation in IFN-γ at 1 DPI in ChAdOx1 nCoV-19 prime-only vaccinated animals, but not in prime-boost vaccinated or control animals. IL-10 and IL-13 were significantly upregulated in control animals compared to prime-boost animals on 1 and 7 DPI (IL-13 only), but not compared to prime-only animals. No significant differences were observed between ChAdOx1 nCoV-19 and control animals for TNF-α, IL-2, IL-4, IL-5 and IL-6 (Extended Data Figure 4); this is in line with a previously observed lack of upregulation of cytokines and chemokines in rhesus macaques upon infection with SARS-CoV-2.
Pulmonary pathology and viral load
At 7 days post inoculation, all animals were euthanized, and tissues were collected. None of the vaccinated monkeys developed pulmonary pathology after inoculation with SARS-CoV-2. All lungs were histologically normal and no evidence of viral pneumonia nor immune-enhanced inflammatory disease was observed. In addition, no SARS-CoV-2 antigen was detected by immunohistochemistry in the lungs of any of the vaccinated animals. Three out of 6 control animals developed some degree of viral interstitial pneumonia. Lesions were widely separated and characterized by thickening of alveolar septae by small amounts of edema fluid and few macrophages and lymphocytes. Alveoli contained small numbers of pulmonary macrophages and, rarely, edema. Type II pneumocyte hyperplasia was observed. Multifocally, perivascular infiltrates of small numbers of lymphocytes forming perivascular cuffs were observed. Immunohistochemistry demonstrated viral antigen in type I and II pneumocytes, as well as in alveolar macrophages, in 5 out of 6 control animals (Figure 4). We were unable to detect any lesions or IHC-positive cells in nasal mucosa in any animals.
Figure 4. Histological changes in lungs of rhesus macaques on 7 DPI.
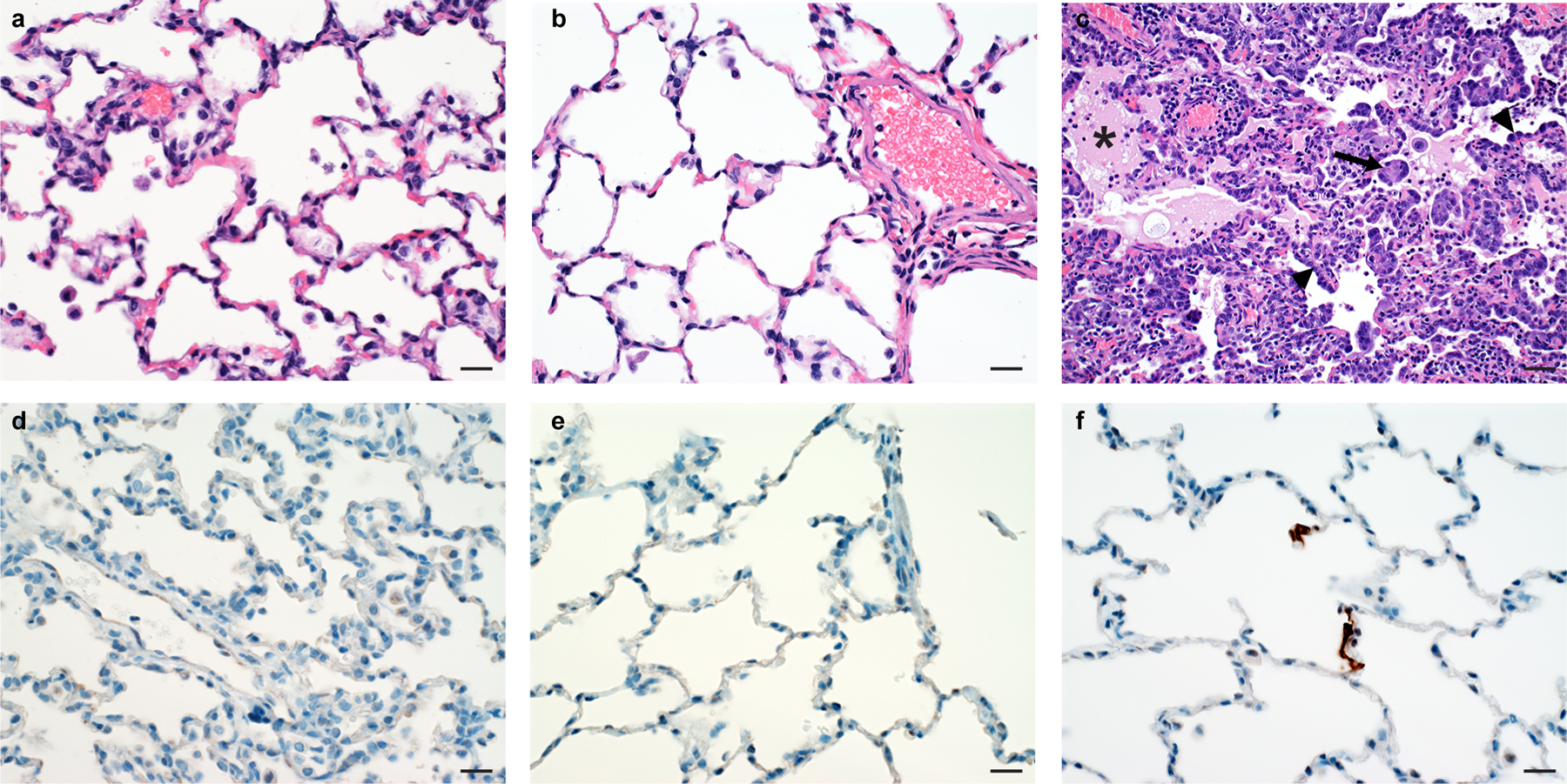
No histological changes were observed in the lungs of ChAdOx1 nCoV-19 prime (a) and prime-boost (b) vaccinated animals. c) Interstitial pneumonia with edema (asterisk), type II pneumocyte hyperplasia (arrowhead) and syncytial cells (arrow) in control animals. No SARS-CoV-2 antigen was detected by immunohistochemistry in the lungs of ChAdOx1 nCoV-19 prime (d) and prime-boost (e) vaccinated animals. f) SARS-CoV-2 antigen (visible as red-brown staining) was detected by immunohistochemistry in type I and type II pneumocytes in the lungs of control animals. 18 sections, taken from 6 different lung lobes are evaluated for each animal; a representative lesion from each group was selected for the figure. Magnification: 400x, scale bar = 20 µm.
Viral gRNA load was high in lung tissue of all control animals and viral sgRNA was detected in 5 out of 6 control animals (Figure 3d). In the prime-only vaccinated group, the viral gRNA load was significantly lower in lung tissue as determined via Mann-Whitney’s rank test, and below limit of detection in 2 of 6 vaccinated animals; viral sgRNA was only detected in lung tissue of one animal (Figure 3d, Extended Data Figure 5). In the prime-boost vaccinated group, viral gRNA was detected in 2 out of 6 animals; one animal was weakly positive in one lung lobe and one animal which mounted a limited response to vaccination was positive in four lung lobes. Viral sgRNA could only be detected in lung tissue from the animal with a lower immune response (Figure 3d, Extended Data Figure 5). We did not detect infectious virus in any lung tissue.
Extra-pulmonary pathology and viral load
No lesions were observed in gastro-intestinal tissues of any animals. As reported previously7, SARS-CoV-2 antigen could be detected in lymphocytes and macrophages in the lamina propria of the intestinal tract of all control animals. This phenomenon was also observed in 6 out of 6 prime vaccinated animals and 3 out of 6 prime-boost vaccinated animals. There were no histological differences between lymphoid tissues of vaccinated or control animals.
Viral gRNA could be detected in extra-respiratory tissues but was predominantly low in all animals and not associated with detection of sgRNA. Viral gRNA in intestinal tissues of prime-boost vaccinated animals was higher than that measured in control and prime-only vaccinated animals and was associated with the detection of sgRNA. However, no infection of intestinal tissue was observed by immunohistochemistry, nor were we able to detect infectious virus in intestinal tissue (Extended Data Figure 6).
Discussion
Here, we show that a single vaccination with ChAdOx1 nCoV-19 is effective in preventing damage to the lungs upon high dose challenge to both upper and lower respiratory tract with SARS-CoV-2, and a prime-boost regimen significantly increased humoral immune responses. We did not see an increase in cellular responses following a second dose of ChAdOx1 nCoV-19, in line with previous results reported by Bliss et al. using a homologous prime-boost regimen with malaria vaccine candidate ChAd63 ME-TRAP8. A small decrease in S and vector-specific antibody titers was observed between D-14 and D0 in the prime-boost group. Longitudinal preclinical and clinical studies will investigate whether this decline in antibody titer is significant over time.
Two recently published SARS-CoV-2 vaccine studies in NHPs showed similar results: Gao et al. used a three dose vaccination regimen of a high dose of whole inactivated SARS-CoV-2 which protected rhesus macaques from SARS-CoV-2 pneumonia9 and Yu et al. used a two dose vaccination regimen with a DNA vaccine encoding spike protein which significantly reduced viral RNA presence in BAL fluid and nasal swabs10. The three studies were conducted at different locations using different protocols, and thus direct comparison is difficult. Animals were challenged with 1 x 106 TCID50 by the intratracheal route9, 1.1 x 104 PFU via the intratracheal and intranasal route10, or here, animals were challenged with 2.6 x 106 TCID50 via the intratracheal, intranasal, ocular and oral route. VN titers induced in vaccinated animals were similar between studies; vaccination with two doses of ChAdOx1 nCoV-19, three doses of 6 µg inactivated SARS-CoV-2, or two doses of spike-encoding DNA resulted in median VN titres of 80, 50, and 74, respectively. A prime vaccination with ChAdOx1 nCoV-19 resulted in VN titers similar to those obtained after inoculation of rhesus macaques with SARS-CoV-27. Upon vaccination with one or two doses of ChAdOx1 nCoV-19, viral load in BAL fluid and lung tissue of vaccinated animals was significantly reduced, suggesting that vaccination prevents or strongly reduces virus replication in the lower respiratory tract. Despite this marked difference in virus replication in the lungs, we did not observe reduction in viral shedding from the nose in either the prime-only or prime-boost regimen. Interestingly, viral RNA in nose swabs from vaccinated animals was also detected by Yu et al10, whereas Gao et al9 challenged only the lower respiratory tract with SARS-CoV-2 and did not assess nasal shedding.
Our primary goal for a vaccine against SARS-CoV-2 is to prevent disease, and we did not observe pneumonia or viral antigen in the lungs of vaccinated animals. Based on the data presented here, it is possible that a single or double dose of ChAdOx1 nCoV-19 will not prevent infection nor transmission of SARS-CoV-2. However, it could significantly reduce illness. Animals in this study were challenged with a high dose of virus via multiple routes as a stringent test of the protective efficacy of the vaccine and absence of enhanced disease upon infection. This does not reflect a realistic human exposure regarding route and dose. Future studies will determine whether changing the route of vaccination to expose mucosal surfaces will induce mucosal immunity, which may result in reduced nasal shedding and onward transmission. It should be noted that detection of sgRNA in nasal swabs was low with lower levels also detected in intestinal tissues. No viral antigen could be detected by immunohistochemistry, and it is not yet clear whether virus is replicating in the nasal mucosa of vaccinated animals.
Several preclinical studies of vaccines against SARS-CoV resulted in immunopathology after vaccination and subsequent challenge, with more severe disease in vaccinated animals than in controls11–13. Immune-enhanced disease could be observed as early as 2 days post challenge in mice14,15 and 7 days post challenge in NHPs13. Immune-enhanced disease was associated with a Th2 response in mice14 and diffuse alveolar damage and cellular infiltrates in NHPs13. In this study, there was no evidence of immune-enhanced disease in vaccinated animals. The immune response was not skewed towards a Th2 response in mice or NHPs, there was no increase in clinical signs or virus replication throughout the study in vaccinated NHPs compared to controls and no markers of disease enhancement in pulmonary tissue of NHPs such as an influx of cells or diffuse alveolar damage were observed. Clinical scoring of vaccinated animals was lower than clinical scoring in control animals, again strongly suggesting absence of immune-enhanced disease.
Results from ongoing clinical trials will be the most informative in determining whether ChAdOx1 nCoV-19 will be an appropriate vaccine candidate, but the results presented in the current study are encouraging. The data presented here informed the start of the phase I clinical trial with ChAdOx1 nCoV-19 on April 23, 2020. As of July 1st, 2020, more than 8000 volunteers have participated in the randomised controlled clinical trials. This study is thus an important step towards the development of a safe and efficacious SARS-CoV-2 vaccine.
Methods
Ethics Statement
Mice - Mice were used in accordance with the UK Animals (Scientific Procedures) Act under project license number P9804B4F1 granted by the UK Home Office. Animals were group housed in IVCs under SPF conditions, with constant temperature (20–24°C) and humidity (45 to 65%) with lighting on a fixed light/dark cycle (12-hours/12-hours)
NHPs - Animal experiment approval was provided by the Institutional Animal Care and Use Committee (IACUC) at Rocky Mountain Laboratories. Animal experiments were executed in an Association for Assessment and Accreditation of Laboratory Animal Care (AALAC)-approved facility by certified staff, following the basic principles and guidelines in the NIH Guide for the Care and Use of Laboratory Animals, the Animal Welfare Act, United States Department of Agriculture and the United States Public Health Service Policy on Humane Care and Use of Laboratory Animals. Rhesus macaques were housed in individual primate cages allowing social interactions, in a climate-controlled room with a fixed light/dark cycle (12-hours/12-hours) and monitored a minimum of twice daily. Commercial monkey chow, treats, and fruit were provided by trained personnel. Water was available ad libitum. Environmental enrichment consisted of a variety of human interaction, commercial toys, videos, and music. The Institutional Biosafety Committee (IBC) approved work with infectious SARS-CoV-2 virus strains under BSL3 conditions. All sample inactivation was performed according to IBC approved standard operating procedures for removal of specimens from high containment.
Generation of vaccine ChAdOx1 nCoV-19
The spike protein of SARS-CoV-2 (GenBank accession number YP_009724390.1), the surface glycoprotein responsible for receptor binding and fusion/entry into the host cell, was codon optimised for expression in human cell lines and synthesised with the tissue plasminogen activator (tPA) leader sequence at the 5’ end by GeneArt Gene Synthesis (Thermo Fisher Scientific). The sequence, encoding SARS-CoV-2 amino acids 2–1273 and tPA leader, was cloned into a shuttle plasmid using InFusion cloning (Clontech). The shuttle plasmid encodes a modified human cytomegalovirus major immediate early promoter (IE CMV) with tetracycline operator (TetO) sites, poly adenylation signal from bovine growth hormone (BGH), between Gateway® recombination cloning sites. ChAdOx1 nCoV-19 was prepared using Gateway® recombination technology (Thermo Fisher Scientific) between the shuttle plasmid described and the ChAdOx1 destination DNA BAC vector described in5 resulting in the insertion of the SARS-CoV-2 expression cassette at the E1 locus. The ChAdOx1 adenovirus genome was excised from the BAC using unique PmeI sites flanking the adenovirus genome sequence. The virus was rescued and propagated in T-Rex 293 HEK cells (Invitrogen) which repress antigen expression during virus propagation. Purification was by CsCl gradient ultracentrifugation. Virus titers were determined by hexon immunostaining assay and viral particles calculated based on spectrophotometry16,17.
Study design animal experiments
Mice – Female BALB/cOlaHsd (BALB/c) (Envigo) and outbred Crl:CD1(ICR) (CD1) (Charles River) mice of at least 6 weeks of age, were immunized IM in the musculus tibialis with 6x109 VP of ChAdOx1 nCoV-19 unless otherwise stated.
NHPs - 18 adult rhesus macaques (17M, 1F) between 2–4 years old were randomly divided into three groups of six animals. Animal group size was based on initial model development. Group 1 was vaccinated with ChAdOx1 nCoV-19 at −28 DPI, group 2 was vaccinated with ChAdOx1 nCoV-19 at −56 and −28 DPI, group 3 was vaccinated with ChAdOx1 GFP at −56 and −28 DPI (1 animal) or at −28 DPI (5 animals). All vaccinations were done with 2.5 x 1010 VP/animal diluted in sterile PBS. Blood samples were obtained before vaccination and 14 days thereafter. Animals were challenged with SARS-CoV-2 strain nCoV-WA1–2020 (MN985325.1) diluted in sterile DMEM on 0 DPI; with administration of 4 mL intratracheally, 1 mL intranasally, 1 mL orally and 0.5 mL ocularly of 4 x 105 TCID50/mL virus suspension. Animals were scored daily by the same person who was blinded to study group allocations using a standardized scoring sheet. Scoring was based on the following criteria: general appearance, appearance skin and coat, discharge, respiration, feces and urine appearance, appetite, and activity. The scoring sheet can be found in7. Clinical exams were performed on −28, −14, 0, 1, 3, and 5 and 7 DPI. Nasal swabs and blood were collected at all exam dates. BAL was performed on 3, 5, and 7 DPI by insertion of an endotracheal tube and bronchoscope into the trachea, then past the 3rd bifurcation, and subsequent installation of 10 mL of sterile saline. Manual suction was applied to retrieve the BAL sample. Serum biochemistry (albumin, AST, ALT, GGT, BUN, creatinine) was analyzed using the Vetscan VS2 Chemistry Analyzer and VetScan 2 Preventative Care 12 discs (Abaxis) (Tables S1–3). Necropsy was performed on 7 DPI and the following tissues were collected: cervical lymph node, mediastinal lymph node, conjunctiva, nasal mucosa, oropharynx, tonsil, trachea, all six lung lobes, right and left bronchus, heart, liver, spleen, kidney, stomach, duodenum, jejunum, ileum, cecum, colon, urinary bladder.
Cells and virus
SARS-CoV-2 strain nCoV-WA1–2020 (MN985325.1) was provided by CDC, Atlanta, USA. Virus propagation was performed in VeroE6 cells in DMEM supplemented with 2% fetal bovine serum, 1 mM L-glutamine, 50 U/ml penicillin and 50 μg/ml streptomycin. The used virus stock was 100% identical to the initial deposited genbank sequence (MN985325.1) and no contaminants were detected. VeroE6 cells were maintained in DMEM supplemented with 10% fetal bovine serum, 1 mM L-glutamine, 50 U/ml penicillin and 50 μg/ml streptomycin. VeroE6 cells were provided by Dr. Ralph Baric and were not authenticated in-house; mycoplasma testing is performed at regular intervals and no mycoplasma has been detected.
Virus isolation from tissue
Tissue sections were weighed and homogenized in 1 mL of DMEM. 250 µl of homogenate was added to VeroE6 cells in a 24 well plate in duplicate. After 1 hour at 37°C and 5% CO2, cells were washed with PBS and 500 µl of DMEM containing 2% FBS was added. Cells were incubated at 37°C and 5% CO2. CPE was read 6 days later.
Virus neutralization assay SARS-CoV-2
Sera were heat-inactivated (30 min, 56 °C), two-fold serial dilutions were prepared in 2% DMEM and 100 TCID50 of SARS-CoV-2 was added. After 1hr incubation at 37 °C and 5% CO2, virus:serum mixture was added to VeroE6 cells and incubated at 37°C and 5% CO2. At 5 dpi, cytopathic effect was scored. The virus neutralization titer was expressed as the reciprocal value of the highest dilution of the serum which still inhibited virus replication.
Virus neutralization assay ChAdOx1
Chimpanzee adenovirus ChAdOx1-specific neutralizing antibody titers were assessed using a secreted placental alkaline phosphatase (SEAP) quantitation assay as described6. Briefly, GripTite MSR 293 cells (Invitrogen, catalog no. R795–07) were infected with the serial diluted serum in phenol red–free DMEM (Life Technologies, catalog no. 31053028) and the ChAdOx1-SEAP reporter virus in a 1:1 mixture for 1 hour before replacing with phenol red–free 10% FBS DMEM for 24 hours. For each sample, SEAP concentration was assessed in 50 μl aliquots of culture supernatant, with CPSD as an indicator substrate (Tropix Phospha-Light Chemiluminescent Assay Kit, Life Technologies, catalog no. T1017). Luminescence intensity was measured using a Varioskan Flash luminometer (Thermo Fisher Scientific). Serum dilution neutralization titers were measured by linear interpolation of adjacent values (to 50% inhibition) to determine the serum dilution required to reduce SEAP concentration by 50% compared to wells with virus alone.
RNA extraction and quantitative reverse-transcription polymerase chain reaction
Tissues (up to 30 mg) were homogenized in RLT buffer and RNA was extracted using the RNeasy kit (Qiagen) according to the manufacturer’s instructions. RNA was extracted from BAL fluid and nasal swabs using the QiaAmp Viral RNA kit (Qiagen) according to the manufacturer’s instructions. Viral gRNA18 and sgRNA19 specific assays were used for the detection of viral RNA. Five μl RNA was tested with the Rotor-GeneTM probe kit (Qiagen) according to instructions of the manufacturer. Dilutions of SARS-CoV-2 standards with known genome copies were run in parallel.
Enzyme-linked immunosorbent assay for mouse sera
MaxiSorp plates (Nunc) were coated with S1 (monomeric, AA 1–674) or S2 (monomeric, AA 685–1211 The Native Antigen Company; 50 ng/well) or with prefusion stabilized SARS-CoV-2 spike protein (250 ng/well) in PBS for overnight adsorption at 4°C. Plates were washed in PBS/Tween (0.05% v/v) and wells blocked using casein (ThermoFisher Scientific) for 1hr at RT. Serially diluted mouse serum samples were added and incubated overnight at 4°C or 37°C for 2hr for specific IgM detection. Plates were washed and Alkaline Phosphatase-conjugated goat anti-mouse IgG (Sigma) or IgM (Abcam) was added to all wells for 1hr at RT or 2hr at 37°C, respectivelly. After washing pNPP substrate (Sigma) was added. Optical density (OD) values for each well were measured at 405 nm. Endpoint titers were calculated as follows: the log10 OD against log10 sample dilution was plotted and a regression analysis of the linear part of this curve allowed calculation of the endpoint titer with an OD of three times the background. The same calculation was used for diluting the sera to the same amounts of total IgG for further testing on different IgG subclasses with anti-mouse IgG subclass-specific antibodies (Abcam). The results of the IgG subclass ELISA are presented using OD values.
Enzyme-linked immunosorbent assay for NHP sera
Prefusion stabilized SARS-CoV-2 spike protein with a T4 fibritin trimerization motif20 was obtained from the Vaccine Research Centre, Bethesda, USA. Maxisorp plates (Nunc) were coated overnight at 4°C with 100 or 250 ng/well spike protein in PBS for IgG and IgM detection, respectively. Plates were blocked with 100 µl of casein in PBS (Thermo Fisher) for 1hr at RT. Serum serially diluted 2x in casein in PBS was incubated at RT for 1hr or 37°C for 2hr. Antibodies were detected using affinity-purified polyclonal antibody peroxidase-labeled goat-anti-monkey IgG (Seracare, 074–11-021) in casein and TMB 2-component peroxidase substrate (Seracare, 5120–0047), developed for 5–10 min, and reaction was stopped using stop solution (Seracare, 5150–0021) and read at 450 nm or anti-monkey IgM – AP labelled after adding the pNPP substrate and measuring OD values at 405 nm. All wells were washed 4x with PBST 0.1% tween in between steps. Threshold for positivity was set at 3x OD value of negative control (serum obtained from non-human primates prior to start of the experiment) or 0.2, whichever one was higher, or OD of three times the background for the calculation of the IgM endpoint values.
ELISpot assay and ICS analysis
Single cell suspension of murine splenocytes were prepared by passing cells through 70μM cell strainers and ACK lysis prior to resuspension in complete media. Rhesus macaque PBMCs were isolated from ethylene diamine tetraaceticacid (EDTA) whole blood using LeucosepTM tubes (Greiner Bio-one International GmbH) and Histopaque®−1077 density gradient cell separation medium (Sigma-Aldrich) according to the manufacturers’ instructions.
Mice - For analysis of IFN-γ production by ELISpot, cells were stimulated with pools of S1 or S2 peptides (final concentration of 2μg/ml) on IPVH-membrane plates (Millipore) coated with 5μg/ml anti-mouse IFN-γ (AN18). After 18–20 hours of stimulation, IFN-γ spot forming cells (SFC) were detected by staining membranes with anti-mouse IFN-γ biotin (1μg/ml) (R46A2) followed by streptavidin-Alkaline Phosphatase (1μg/ml) and development with AP conjugate substrate kit (BioRad, UK).
For analysis of intracellular cytokine production, cells were stimulated at 37°C for 6 hours with 2μg/ml S1 or S2 pools of peptide, media or cell stimulation cocktail (containing PMA-Ionomycin, Biolegend), together with 1μg/ml Golgi-plug (BD) with the addition of 2μl/ml CD107a-Alexa647. Cell supernatant was collected and frozen at –20̊C for subsequent analysis by MesoScale Discovery (MSD) assay (see below). Following surface staining with CD4-BUV496, CD8-PerCPCy5.5, CD62L-BV711, CD127-BV650 and live-dead Aqua, cells were fixed with 4% paraformaldehyde and stained intracellularly with TNF-α-A488, IL-2-PECy7, IL-4-BV605, IL-10-PE and IFN-γ-e450 diluted in Perm-Wash buffer (BD).
Sample acquisition was performed on a Fortessa (BD) and data analyzed in FlowJo V10 (TreeStar). An acquisition threshold was set at a minimum of 5000 events in the live CD3+ gate. Antigen specific T cells were identified by gating on LIVE/DEAD negative, doublet negative (FSC-H vs FSC-A), size (FSC-H vs SSC), CD3+, CD4+ or CD8+ cells and cytokine positive (Gating strategy detailed in Supplementary Figure 1). Cytokine positive responses are presented after subtraction of the background response detected in the corresponding unstimulated sample (media containing CD107a and Golgi-plug) of each individual spleen sample. To determine whether there was a statistically significant differences in the level of cytokines produced by CD4+ or CD8+ T cells, data in each graph was analysed with a 2-way ANOVA (repeated measure) and post-hoc Tukey’s multiple comparison test.
NHPs - IFN-γ ELISpot assay of PBMCs was performed using the ImmunoSpot® Human IFN- γ Single-Color Enzymatic ELISpot Assay Kit according to the manufacturer’s protocol (Cellular Technology Limited). PBMCs were plated at a concentration of 100,000 cells per well and were stimulated with four contiguous peptide pools spanning the length of the SARS-CoV-2 spike protein sequence at a concentration of 2 µg/mL per peptide (Mimotopes). ELISpot plates were subjected to overnight formalin inactivation prior to removal from BSL4 for reading. Analysis was performed using the CTL ImmunoSpot® Analyzer and ImmunoSpot® Software (Cellular Technology Limited). Spot forming units (SFU) per 1.0x106 PBMCs were summed across the 4 peptide pools for each animal.
Measurement of cytokines and chemokines
Mouse samples were assayed using MSD Technology V-PLEX Mouse Cytokine 29-Plex kit according to the manufacturer’s instructions. Non-human primate samples were inactivated with γ-radiation (2 MRad) according to standard operating procedures and assayed on a Bio-Plex 200 instrument (Bio-Rad) using the Non-Human Primate Cytokine MILLIPLEX map 23-plex kit (Millipore) according to the manufacturer’s instructions. LLOD was used for all undetectable and extrapolated values. Only data for cytokines consistently above the lower limit of quantification were included in further analyses.
Log10 Fold Change (Log10FC) for mouse samples was calculated as follows:
Log10FC = Log10((Stimulated (pg/ml) + 1) / (Unstimulated (pg/ml) + 1))
Fold change for NHP samples was calculated as follows:
FC = Concentration (pg/mL) on DX (1, 3, 5, or 7)/Concentration (pg/mL) on D0
Histology and immunohistochemistry
Necropsies and tissue sampling were performed according to IBC-approved protocols. Lungs were perfused with 10% formalin and processed for histologic review. Harvested tissues were fixed for eight days in 10% neutral-buffered formalin, embedded in paraffin, processed using a VIP-6 Tissue Tek (Sakura Finetek, USA) tissue processor, and embedded in Ultraffin paraffin polymer (Cancer Diagnostics, Durham, NC). Samples were sectioned at 5 µm, and resulting slides were stained with hematoxylin and eosin. Specific anti-CoV immunoreactivity was detected using an in-house SARS-CoV-2 nucleocapsid protein rabbit antibody (Genscript) at a 1:1000 dilution. The IHC assay was carried out on a Discovery ULTRA automated staining instrument (Roche Tissue Diagnostics) with a Discovery ChromoMap DAB (Ventana Medical Systems) kit. All tissue slides were evaluated by a board-certified veterinary anatomic pathologist blinded to study group allocations. 18 sections, taken from 6 different lung lobes are evaluated for each animal; a representative lesion from each group was selected for the figure.
Statistical analyses
Two-tailed Mann-Whitney’s rank or Wilcoxon tests were conducted to compare differences between groups using Graphpad Prism version 8.3.0. A Bonferroni correction was used to control for type I error rate where required.
Extended Data
Extended Data Figure 1. Antigen specific responses following ChAdOx1 nCov19 vaccination.
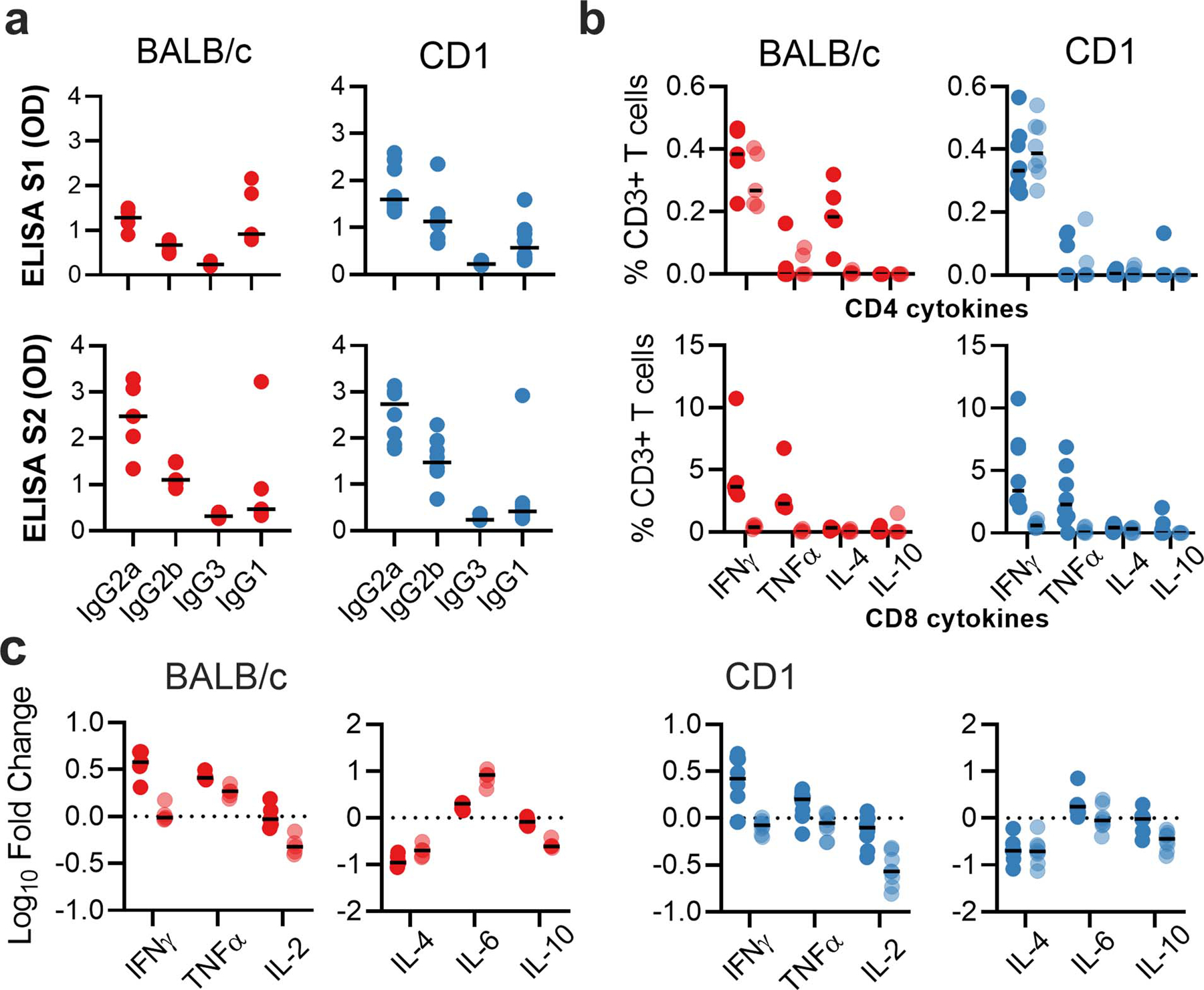
a. IgG subclass antibodies detected against S1 or S2 protein in sera of BALB/c or CD1 mice. b. Frequency of cytokine positive CD3+ T cells following stimulation of splenocytes with S1 pool (dark) or S2 pool (transparent) peptides in BALB/c (red) and CD1 (blue) mice. c. Log10 fold change in cytokine levels in supernatant from S1 (dark) and S2 (transparent) stimulated splenocytes when compared to corresponding unstimulated splenocyte sample for BALB/c and CD1 mice. n=5 (BALB/c) and 8 (CD1) animals examined over 1 independent experiment for all figure panels.
Extended Data Figure 2. Spike-specific serum IgM.
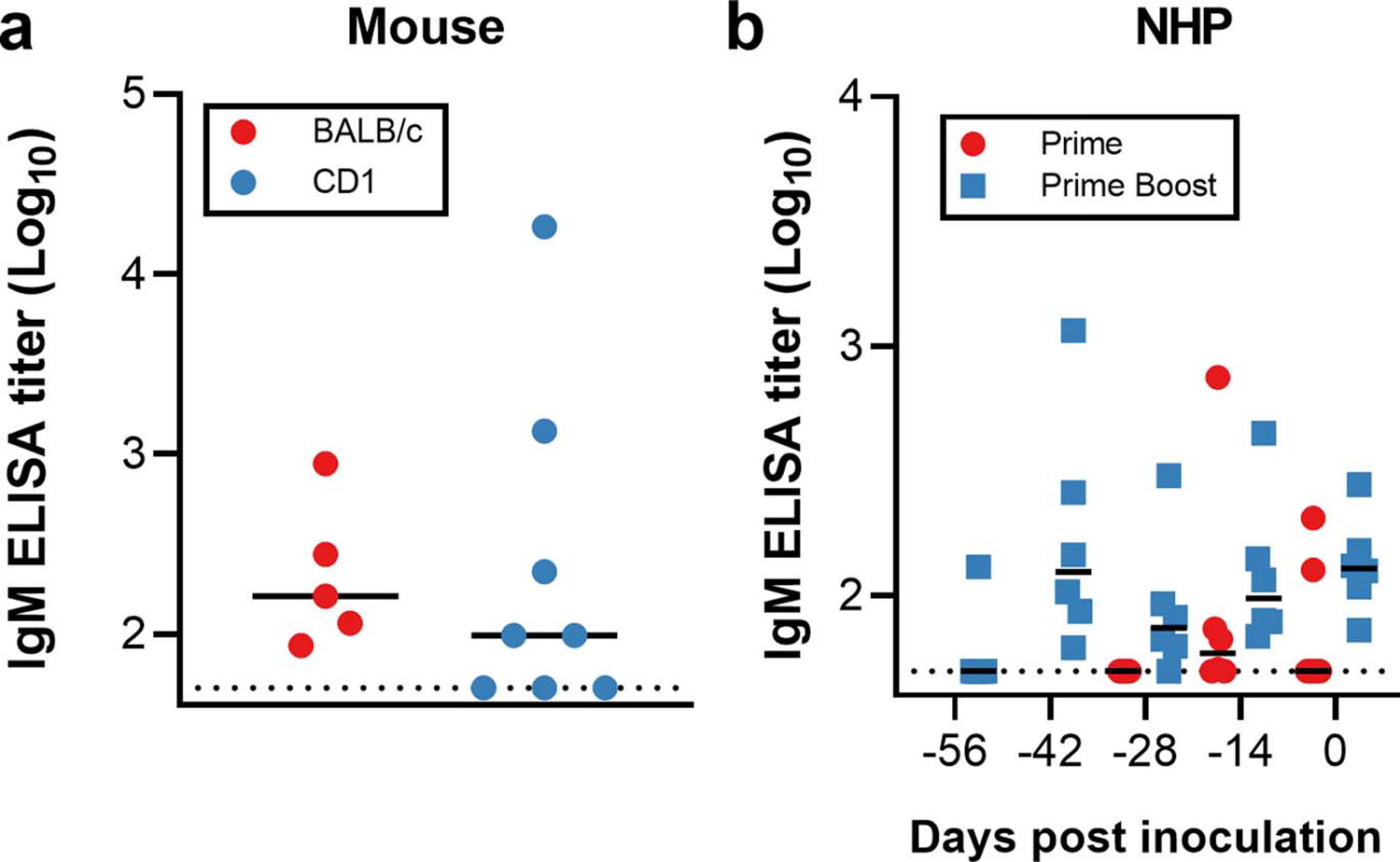
a. Spike-specific serum IgM in mice 14 days post vaccination. n=5 (BALB/c) and 8 (CD1) animals examined over 1 independent experiment. b. Spike-specific serum IgM in NHPs upon prime-boost or prime-only vaccination. n=6 animals per group examined over 2 independent experiments.
Extended Data Figure 3.
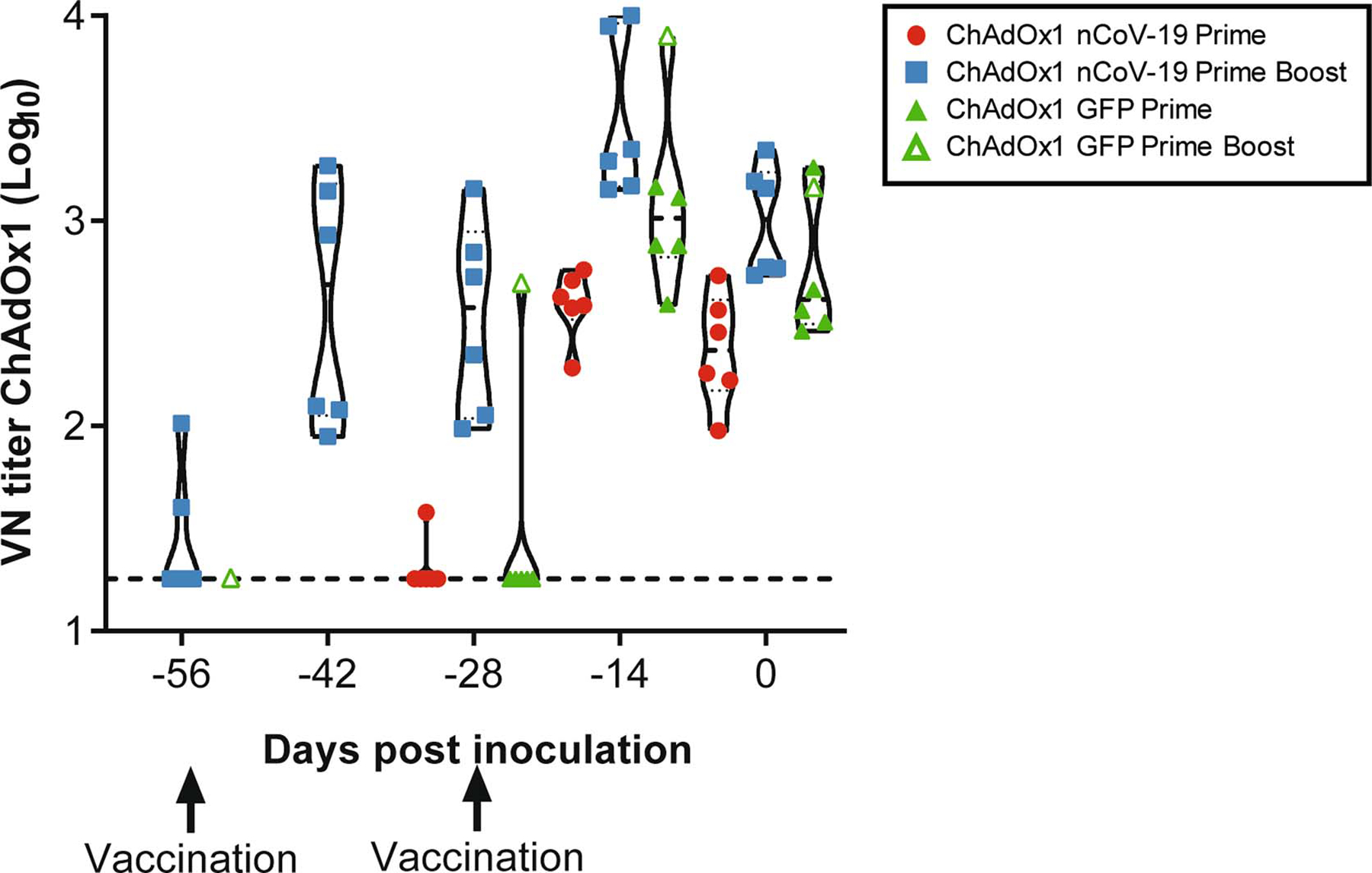
ChAdOx1 neutralizing antibodies in serum of vaccinated NHPs. Control animal with prime-boost regimen is highlighted with open triangle symbol. VN = virus neutralizing. n=6 animals per group examined over 2 independent experiments.
Extended Data Figure 4. Serum cytokines in rhesus macaques challenged with SARS-CoV-2.

Fold increase in cytokines in serum compared to pre-challenge values. ** = p-value<0.01; Line = median. Statistical significance determined via two-tailed Mann-Whitney test. P-values: IFN-γ = 0.0087; IL-10 = 0.0043; IL-13 = 0.0043 and 0.0065. n=6 animals per group examined over 2 independent experiments.
Extended Data Figure 5.
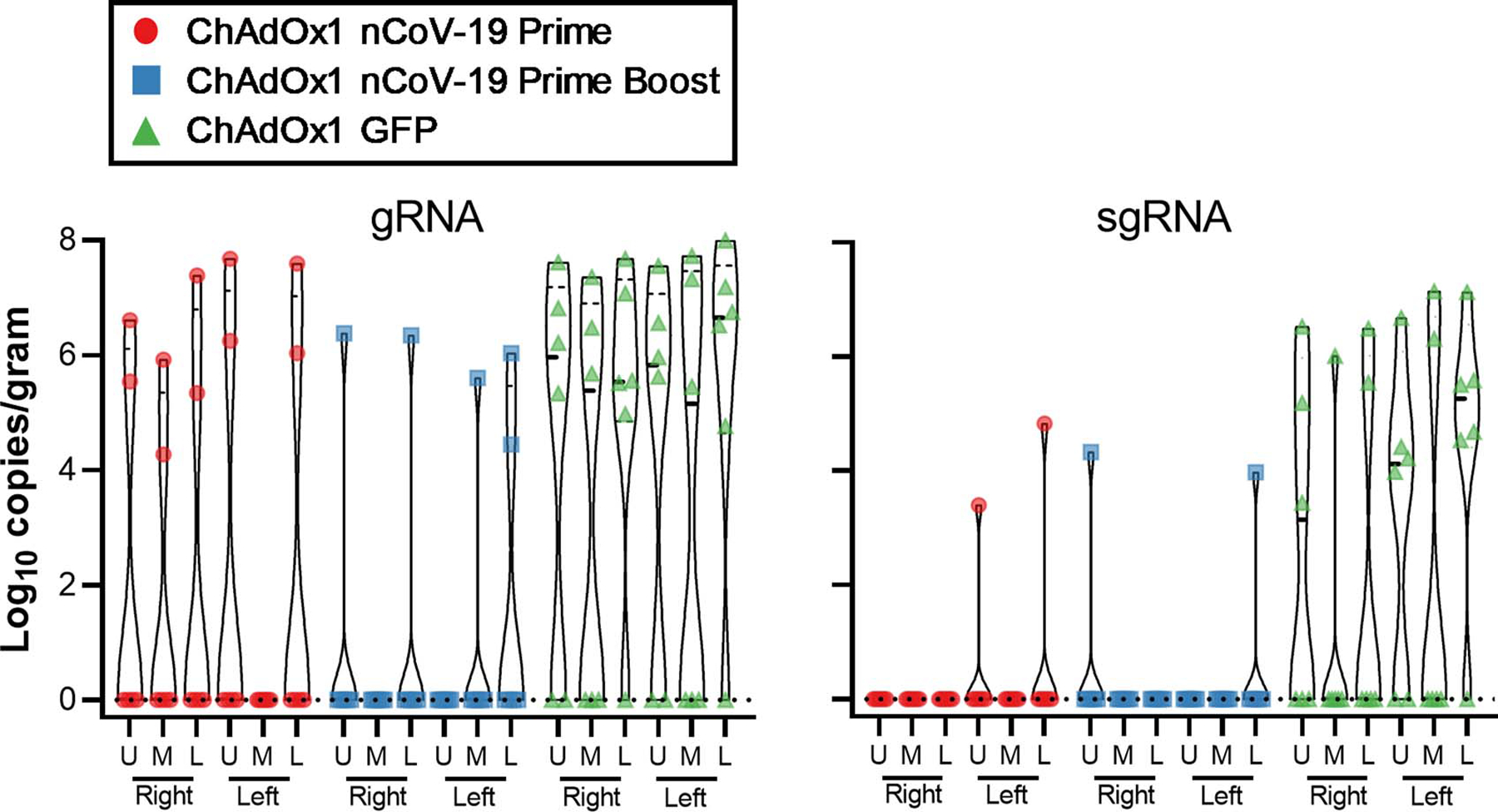
Viral load in lung tissue of rhesus macaques challenged with SARS-CoV-2 at 7 DPI.
Extended Data Figure 6. Viral load in tissues of rhesus macaques challenged with SARS-CoV-2 at 7 DPI.
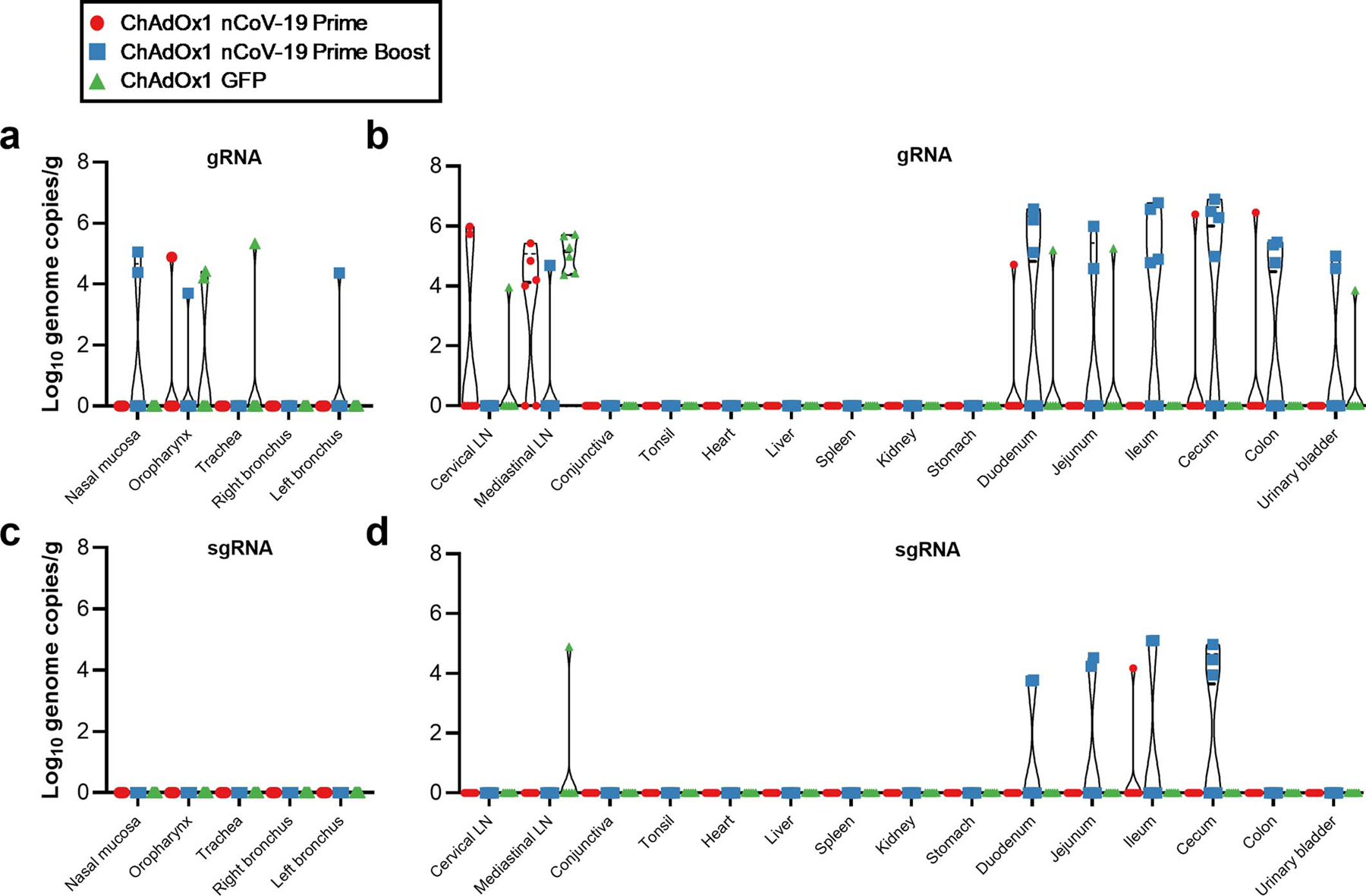
a. Viral gRNA in respiratory tissues excluding lung tissue. b. Viral gRNA in non-respiratory tissues. c. Viral sgRNA in respiratory tissues excluding lung tissue. d. Viral sgRNA in non-respiratory tissues.
Extended Data Table 1 |.
Virus isolation from nasal swabs
| ChAdOx1 nCoV-19 Prime | ChAdOx1 nCoV-19 Prime-boost | ChAdOx1 GFP | |
|---|---|---|---|
| 1 DPI | 4/6 | 2/6 | 4/6 |
| 3 DPI | 2/6 | 0/6 | 1/6 |
| 5 DPI | 0/6 | 0/6 | 0/6 |
| 7 DPI | 0/6 | 0/6 | 0/6 |
Virus was isolated from nasal swabs of rhesus macaques after challenge with SARS-CoV-2.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Olubukola Abiona, Brandon Bailes, Aaron Carmody, Kizzmekia Corbett, Kathleen Cordova, Jayne Faris, Heinz Feldmann, Susan Gerber, Barney Graham, Elaine Haddock, Ryan Kissinger, Michael Jones, Mary Marsh, Kay Menk, Anita Mora, Stephanie Seifert, Les Shupert, Brian Smith, Natalie Thornburg, Amanda Weidow, Marissa Woods, and Kwe Claude Yinda for their contributions to this study. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) (1ZIAAI001179–01) and the Department of Health and Social Care using UK Aid funding managed by the NIHR.
Footnotes
Competing interests
SCG is a board member of Vaccitech and named as an inventor on a patent covering use of ChAdOx1-vectored vaccines and a patent application covering a SARS-CoV-2 (nCoV-19) vaccine (UK Patent Application No. 2003670.3). Teresa Lambe is named as an inventor on a patent application covering a SARS-CoV-2 (nCoV-19) vaccine (UK Patent Application No. 2003670.3). The University of Oxford and Vaccitech, having joint rights in the vaccine, entered into a partnership with AstraZeneca in April 2020 for further development, large-scale manufacture and global supply. Equitable access to the vaccine is a key component of the partnership. Neither Oxford University nor Vaccitech will receive any royalties during the pandemic period or from any sales of the vaccine in developing countries. The remaining authors declare no competing interests.
Data availability statement
Data have been deposited in Figshare21.
References
- 1.Zhu N et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 382, 727–733, doi: 10.1056/NEJMoa2001017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269, doi: 10.1038/s41586-020-2008-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Coronavirus disease (COVID-19) Situation Report 113, <https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200512-covid-19-sitrep-113.pdf?sfvrsn=feac3b6d_2> (2020).
- 4.Lurie N, Saville M, Hatchett R & Halton J Developing Covid-19 Vaccines at Pandemic Speed. N Engl J Med, doi: 10.1056/NEJMp2005630 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Dicks MD et al. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PLoS One 7, e40385, doi: 10.1371/journal.pone.0040385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Doremalen N et al. A single dose of ChAdOx1 MERS provides protective immunity in rhesus macaques. Science Advances, doi: 10.1126/sciadv.aba8399 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munster VJ et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature, doi: 10.1038/s41586-020-2324-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiang G et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science, doi: 10.1126/science.abc1932 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science, doi: 10.1126/science.abc6284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weingartl H et al. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol 78, 12672–12676, doi: 10.1128/JVI.78.22.12672-12676.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolles M et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol 85, 12201–12215, doi: 10.1128/JVI.06048-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight 4, doi: 10.1172/jci.insight.123158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tseng CT et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One 7, e35421, doi: 10.1371/journal.pone.0035421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasui F et al. Prior immunization with severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS-CoV. J Immunol 181, 6337–6348, doi: 10.4049/jimmunol.181.9.6337 (2008). [DOI] [PubMed] [Google Scholar]
References Methods
- 15.Yasui F et al. Prior immunization with severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS-CoV. J Immunol 181, 6337–6348, doi: 10.4049/jimmunol.181.9.6337 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Bewig B & Schmidt WE Accelerated titering of adenoviruses. Biotechniques 28, 870–873, doi: 10.2144/00285bm08 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Maizel JV Jr., White DO & Scharff MD The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology 36, 115–125, doi: 10.1016/0042-6822(68)90121-9 (1968). [DOI] [PubMed] [Google Scholar]
- 18.Corman VM et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25, doi: 10.2807/1560-7917.ES.2020.25.3.2000045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfel R et al. Virological assessment of hospitalized patients with COVID-2019. Nature, doi: 10.1038/s41586-020-2196-x (2020). [DOI] [PubMed] [Google Scholar]
- 20.Wrapp D et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263, doi: 10.1126/science.abb2507 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.<https://figshare.com/articles/dataset/Figshare_document_xlsx/12290696> (2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been deposited in Figshare21.


