Abstract
Recent studies have reported protective efficacy of both natural immunity1 and vaccine-induced immunity2–7 against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) challenge in rhesus macaques. However, the importance of humoral and cellular immunity for protection against SARS-CoV-2 infection remains to be determined. Here we show that adoptive transfer of purified IgG from convalescent macaques protects naïve recipient rhesus macaques against SARS-CoV-2 challenge in a dose dependent fashion. Depletion of CD8+ T cells in convalescent animals partially abrogated the protective efficacy of natural immunity against SARS-CoV-2 re-challenge, suggesting the importance of cellular immunity in the context of waning or subprotective antibody titers. These data demonstrate that relatively low antibody titers are sufficient for protection against SARS-CoV-2 in rhesus macaques, and that cellular immune responses may also contribute to protection if antibody responses are suboptimal. We also show that higher antibody titers are required for therapy of SARS-CoV-2 infection in macaques. These findings have important implications for the development of SARS-CoV-2 vaccines and immune-based therapeutics.
The protective efficacy of several SARS-CoV-2 vaccine candidates2–7 and the protective efficacy of natural immunity against re-exposure to SARS-CoV-21 have recently been reported in rhesus macaques. Both neutralizing antibody (NAb) titers as well as Fc functional antibody responses correlated with protection4,5, but direct data demonstrating the immunologic determinants of protection have been lacking. In particular, the relative importance of humoral immunity, cellular immunity, and innate immunity in protecting against SARS-CoV-2 has not yet been determined. Such knowledge would be important in the development of vaccines, antibody-based therapeutics, and public health strategies.
IgG Adoptive Transfer Protection Study
We previously reported that 9 rhesus macaques infected with SARS-CoV-2 by the intranasal and intratracheal routes were protected against SARS-CoV-2 re-challenge1. Following re-challenge, these animals developed increased log10 neutralizing antibody (NAb) titers of 2.56–3.38 (median 3.07) using a luciferase-based pseudovirus neutralization assay1,4,8 (Extended Data Fig. 1). To develop a high titer IgG stock for adoptive transfer studies, we purified IgG from plasma from each of these 9 animals and prepared individual IgG stocks at concentrations of 10 mg/ml. These stocks had similar NAb titers as the original plasma samples (Extended Data Fig. 1). We then pooled these individual IgG stocks to create a combined IgG stock with a log10 NAb titer of 3.20 (NAb titer of 1,581).
We assessed the protective efficacy of this purified IgG stock in an adoptive transfer study in rhesus macaques. 12 Indian-origin male and female rhesus macaques (Extended Data Table 1) received an intravenous infusion with 250 mg/kg (Group I; N=3), 25 mg/kg (Group II; N=3), or 2.5 mg/kg (Group III; N=3) of the SARS-CoV-2 IgG stock or sham IgG (Group IV; N=3) on day −3. On day 0, all animals were challenged with 1.0×105 TCID50 SARS-CoV-2 by the intranasal and intratracheal routes, as we have previously described5. Following infusion, serum antibody titers were observed in the recipient animals as expected. On day 0, log10 NAb titers were 2.71–2.76 in Group I (NAb titers 511–571), 1.62–1.87 in Group II (NAb titers 42–49), and <1.30–1.36 in Group III (NAb titers <20–23) (Extended Data Fig. 2a). Binding ELISA titers to spike (S) and the receptor binding domain (RBD) similarly showed a titration of antibody titers in the recipient animals on day 0 (Extended Data Fig. 2b, c). Following SARS-CoV-2 challenge, NAb and ELISA titers gradually decreased in Group I and in a subset of animals in Group II over the 14 days of follow-up. In contrast, NAb and ELISA titers increased sharply in Groups III and IV by day 10–14 following challenge, presumably reflecting the autologous antibody response to productive infection1.
We profiled S-, RBD-, and nucleocapsid (N)-specific antibody responses in the recipient animals by systems serology9,10, as we previously observed diverse Fc functional antibody responses in SARS-CoV-2 infected animals1. IgG subclasses, Fc-receptors (FcR2A-1, FcR2A-2, FcR3A), antibody-dependent neutrophil phagocytosis (ADNP), antibody-dependent complement deposition (ADCD), antibody-dependent cellular phagocytosis (ADCP), and antibody-dependent natural killer cell activation (ADNKA) responses were assessed. Consistent with the NAb and ELISA data, the antibody Fc effector profiles showed higher responses in Group I and marginal responses in Group III (Extended Data Fig. 3). Moreover, a principal component analysis (PCA) of the antibody features showed that animals in Group I separated from the other groups.
To assess the protective efficacy of the adoptively transferred IgG, we assessed viral loads in BAL and NS by RT-PCR for subgenomic RNA (sgRNA), which is believed to measure replicating virus1,11. All sham controls in Group IV were infected and had a median peak of 5.18 (range 3.82–5.45) log10 sgRNA copies/ml in BAL (Fig. 1a) and a median peak of 5.72 (range 5.12–6.62) log10 sgRNA copies/ml in NS (Fig. 1b), consistent with our previous experience with this challenge stock1,4,5. As expected, viral replication was observed for 7–10 days in BAL and for 10–14 days in NS. All animals that received 2.5 mg/kg IgG in Group III were also infected but generally showed shorter duration of viral replication of 3–10 days in BAL and 7–10 days in NS. In contrast, 1 of 3 animals that received 25 mg/kg IgG in Group II and all animals that received 250 mg/kg IgG in Group I were protected with no detectable virus in BAL or NS at any timepoint following challenge (limit of quantitation 1.69 log10 sgRNA copies/ml or copies/swab). These data demonstrate that purified IgG from convalescent macaques, in the absence of cellular and innate immunity, can effectively protect naïve recipient macaques against SARS-CoV-2 challenge in both the upper and lower respiratory tracts in a dose-dependent fashion.
Figure 1. Viral loads following prophylactic adoptive transfer of IgG prior to SARS-CoV-2 challenge.
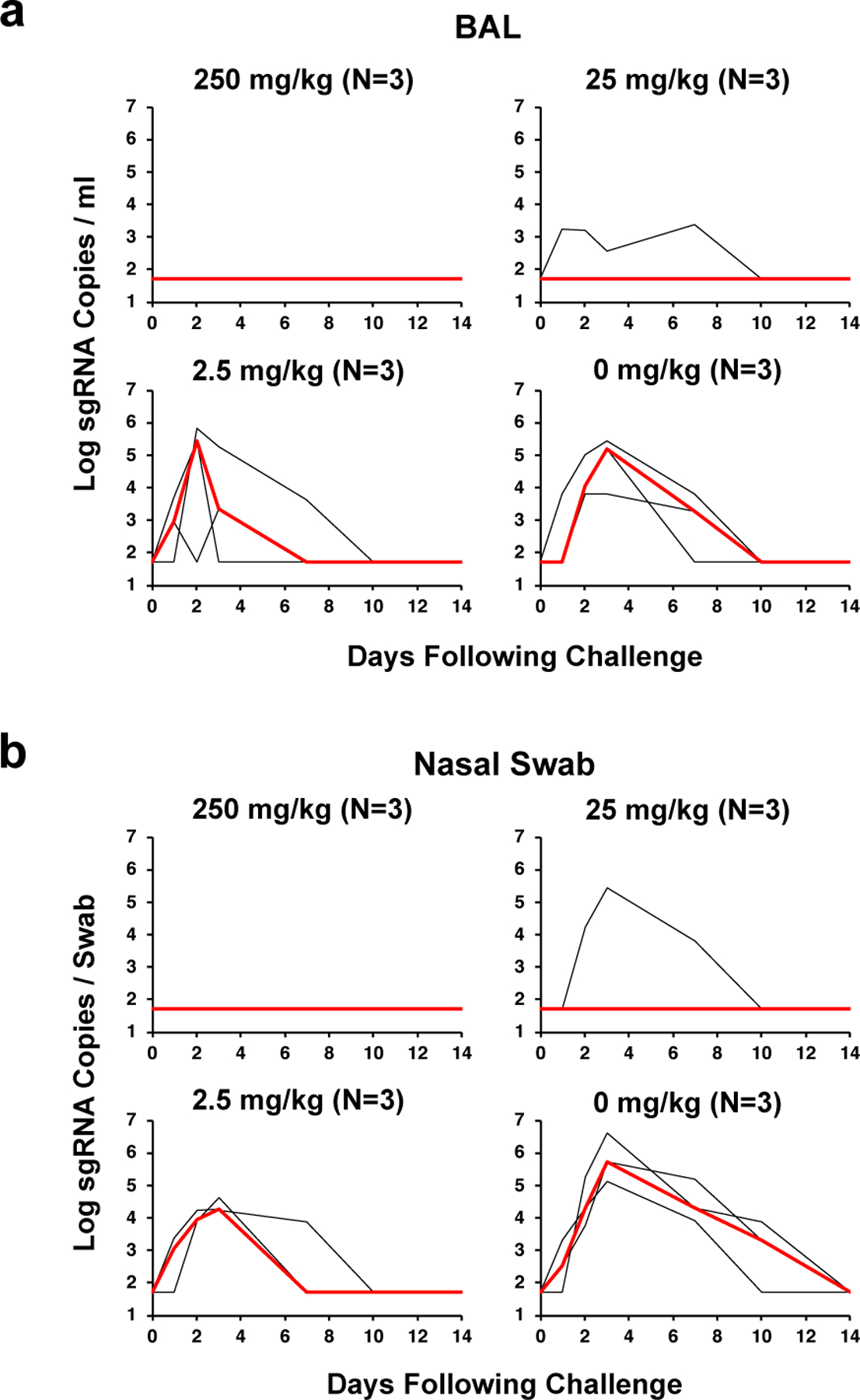
Rhesus macaques were challenged by the intranasal and intratracheal routes with 105 TCID50 SARS-CoV-2. (a) Log10 sgRNA copies/ml (limit of quantification 50 copies/ml) were assessed in bronchoalveolar lavage (BAL) following challenge. (b) Log10 sgRNA copies/swab (limit of quantification 50 copies/swab) were assessed in nasal swabs (NS) following challenge. Days following challenge is shown on the x-axis. Red lines reflect median values. The number of animals is denoted in each panel and the median line overlaps with data lines.
We employed logistic regression models to determine the antibody titer thresholds for protection against SARS-CoV-2 in this experiment. These regression models indicated that pseudovirus NAb titers of approximately 50 (Fig. 2a, d), RBD ELISA titers of approximately 100 (Fig. 2b, e), and S ELISA titers of approximately 400 (Fig. 2c, f) were required for protection. More precise estimates of NAb titers required for protection will require evaluation of titers between those achieved in Group II (NAb titers approximately 50) and those achieved in Group I (NAb titers approximately 500). Correlation analysis showed that NAb titers correlated with protective efficacy (P=0.013 for BAL, P=0.023 for NS, two-sided Spearman rank-correlation tests; Fig. 2) and that NAb titers effectively differentiated fully protected vs. non-protected animals (P=0.0082, two-sided Mann-Whitney test; Fig. 2). Other antibody features also correlated with protective efficacy, including IgG1, ADCD, ADNKA, and ADNP (P=0.00032, P=0.00097, P=0.0048, and P=0.0074, respectively, two-sided Spearman rank-correlation tests; Extended Data Fig. 4). Future studies could assess if there are different correlates of protection in the lower and upper respiratory tracts.
Figure 2. Regression analysis of antibody threshold for protection.

Differences in (a) pseudovirus NAb titers, (b) RBD ELISA titer, and (c) S (FLED) ELISA titer between protected and not protected animals. Animals were considered to be protected if they had no detectable virus in bronchoalveolar lavage (BAL, n = 5 protected animals, left), nasal swab (NS, n = 5 protected animals, middle) or in both (n = 4 protected animals, right). The threshold titers are indicated as horizontal lines and were obtained by logistic regression for scenarios where the groups were not completely separable using the corresponding titer (95% predicted probability of protection, solid lines), or represent the mean between the highest and lowest titers of not protected and protected animals, respectively (dotted lines). In the box plots, whiskers indicate range, and boxes indicates 25th to 75th percentile. Relationship between (d) pseudovirus NAb titers, (e) RBD ELISA titer, and (f) S (FLED) ELISA titer and peak viral loads in BAL (left) and NS (right). Red lines indicate fitted exponential curves.
IgG Adoptive Transfer Therapy Study
Given the clinical interest in the therapeutic use of convalescent plasma for SARS-CoV-2 infection in humans12,13, we next assessed the therapeutic efficacy of purified IgG in infected rhesus macaques. 6 Indian-origin rhesus macaques were infected with 1.0×105 TCID50 SARS-CoV-2 by the intranasal and intratracheal routes on day 0 as described above, and then received an intravenous infusion of 250 mg/kg (Group I; N=2), 25 mg/kg (Group II; N=2), or 0 mg/kg (Sham; Group III; N=2) of the SARS-CoV-2 IgG stock on day 1. Following infusion, serum antibody titers were similar to those observed in the prior experiment (NAb titers approximately 500 in Group I and 50 in Group II) (data not shown).
The sham controls in Group III had a median peak of 5.22 log10 sgRNA copies/ml in BAL and 5.62 log10 sgRNA copies/swab in NS with 7–10 days of virus replication (Fig. 3), similar to the previous experiment. Animals in Group II that received 25 mg/kg IgG had a median peak of 4.58 log10 sgRNA copies/ml in BAL and 5.74 log10 sgRNA copies/swab in NS. In contrast, animals in Group I that received 250 mg/kg IgG had a median peak of 3.38 log10 sgRNA copies/ml in BAL and 4.36 log10 sgRNA copies/swab in NS with 3–7 days of virus replication. These data suggest a 1.84 log reduction of peak sgRNA copies/ml in BAL and a 1.26 log reduction of peak sgRNA copies/swab in NS with the 250 mg/kg IgG dose, but minimal or no efficacy with the 25 mg/kg IgG dose.
Figure 3. Viral loads following therapeutic adoptive transfer of IgG after SARS-CoV-2 challenge.
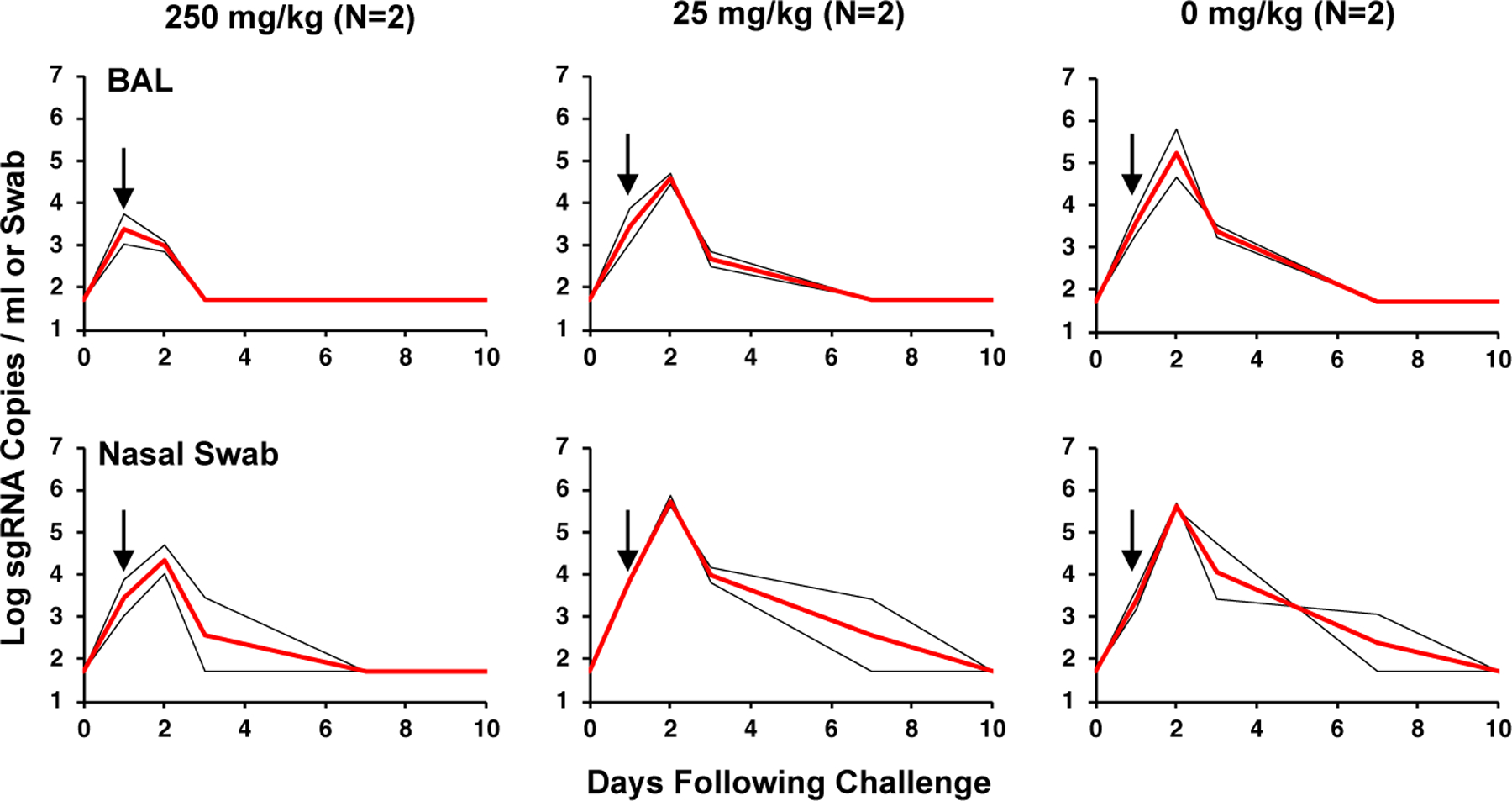
Rhesus macaques were challenged by the intranasal and intratracheal routes with 105 TCID50 SARS-CoV-2 on day 0 and then received intravenous infusion of 250 mg/kg, 25 mg/kg, or 0 mg/kg purified SARS-CoV-2 IgG on day 1. Log10 sgRNA copies/ml (limit of quantification 50 copies/ml) were assessed in bronchoalveolar lavage (BAL) and log10 sgRNA copies/swab (limit of quantification 50 copies/swab) were assessed in nasal swabs (NS) following challenge. Days following challenge is shown on the x-axis. Red lines reflect median values. Arrows represent the day of IgG infusion. The number of animals is denoted in each panel and the median line overlaps with data lines.
CD8 Depletion Study
To evaluate the potential role of cellular immune responses in protection against SARS-CoV-2, we CD8 depleted14 convalescent rhesus macaques prior to re-challenge1. 13 rhesus macaques were infected with 1.0×105 TCID50 SARS-CoV-2 by the intranasal and intratracheal routes as described above, and all animals demonstrated robust viral replication in BAL and NS as expected (data not shown). At week 4 following infection, the median log10 NAb titer of these convalescent animals was 2.30, which was lower than the median log10 NAb titer of 3.07 in the re-challenged animals in the prior study (P=0.0001, two-sided Mann-Whitney test; Extended Data Fig. 5). NAb titers in these convalescent animals declined to a median log10 NAb titer of 1.91 by week 7 (P=0.0003, two-sided Mann-Whitney test, comparing week 4 and week 7 titers; Extended Data Fig. 5).
To evaluate the role of CD8+ T cells cells in contributing to protective efficacy against re-challenge, macaques received a single intravenous infusion of 50 mg/kg of the anti-CD8α CDR-grafted rhesus IgG1 antibody MT807R1 (MassBiologics, Mattapan, MA) (N=5), the anti-CD8β rhesus IgG1 antibody CD8b255R1 (MassBiologics, Mattapan, MA) (N=3), or a sham control antibody (N=5) at week 7 after primary infection, reflecting day −3 relative to re-challenge. The anti-CD8α monoclonal antibody depletes both CD8+ T cells and NK cells in rhesus macaques, whereas the anti-CD8β monoclonal antibody depletes only CD8+ T cells14. Following infusion, all animals that received the anti-CD8α or anti-CD8β antibodies showed complete or near-complete CD8 depletion in peripheral blood, including total CD8+ cells, CD8+CD3+ cells, and CD8+CD3- cells (Extended Data Fig. 6). On day 0, all animals were re-challenged with 1.0×105 TCID50 SARS-CoV-2 by the intranasal and intratracheal routes. Following re-challenge, S-specific IFN-γ+ CD8+ T cell responses were undetectable in the CD8-depleted animals, as expected, but increased in the undepleted sham controls (P=0.0157, two-sided Mann-Whitney test; Extended Data Fig. 7). In contrast, S-specific IFN-γ+ CD4+ T cell responses were similar in both groups (Extended Data Fig. 8). NAb responses also increased in both groups following re-challenge (Extended Data Fig. 9), consistent with our previous findings1.
Following re-challenge, animals that received the sham control antibody demonstrated no virus in BAL (Fig. 4a) and only transient virus in 1 of 5 animals in NS (Fig. 4b) indicative of natural immunity, similar to our previous observations1. In contrast, in the CD8α depleted animals, 1 of 5 animals showed breakthrough virus in BAL (Fig. 4a), but 5 of 5 animals exhibited breakthrough virus in NS (Fig. 4b) following re-challenge. Similarly, in the CD8β depleted animals, 1 of 3 animals showed breakthrough virus in BAL (Fig. 4a), but 3 of 3 animals exhibited breakthrough virus in NS (Fig. 4b) following re-challenge. Peak viral loads in NS were higher in the CD8 depleted animals compared with the sham controls (P=0.0085, two-sided Mann-Whitney test; Fig. 4c) but lower than in naïve animals following primary challenge (P=0.0242, two-sided Mann-Whitney test; Fig. 4c). These data suggest that cellular immunity, including CD8+ T cells, can contribute to protection of convalescent animals against SARS-CoV-2 re-challenge in the setting of waning and subprotective antibody titers.
Figure 4. Viral loads following CD8 depletion and SARS-CoV-2 re-challenge.
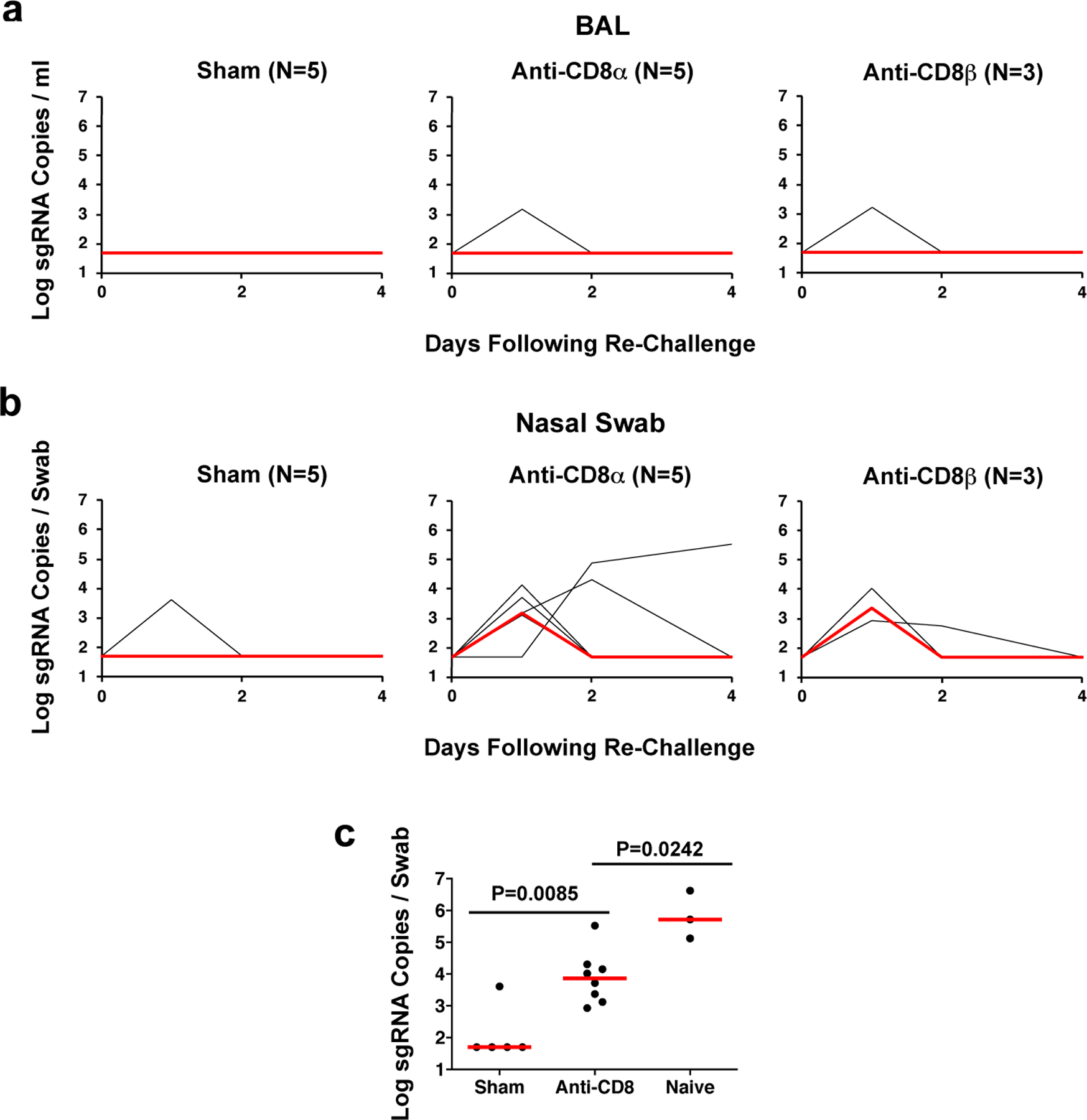
Rhesus macaques were infected with SARS-CoV-2 and received 50 mg/kg anti-CD8α mAb, anti-CD8β mAb, or sham mAb at week 7, reflecting day −3 relative to re-challenge. On day 0, all animals were re-challenged with 105 TCID50 SARS-CoV-2. (a) Log10 sgRNA copies/ml (limit of quantification 50 copies/ml) were assessed in bronchoalveolar lavage (BAL) following re-challenge. (b) Log10 sgRNA copies/swab (limit of quantification 50 copies/swab) were assessed in nasal swabs (NS) following re-challenge. Days following challenge is shown on the x-axis. (c) Comparison of peak log sgRNA in NS following re-challenge in sham and anti-CD8 groups. Peak log sgRNA in NS from naïve animals following primary challenge from the adoptive transfer study are shown for comparison. Red lines reflect median values. The number of animals is denoted in each panel (Sham n=5, Anti-CD8 n=8, Naïve n=3 independent animals). P values reflects two-sided Mann-Whitney tests.
Discussion
Our data demonstrate that adoptive transfer of purified polyclonal IgG from convalescent macaques robustly protected naïve recipient rhesus macaques against SARS-CoV-2 challenge in a dose dependent fashion. These data indicate that relatively low titers of antibodies are sufficient for protection in both the upper and lower respiratory tracts. CD8 depletion studies also showed that cellular immunity can contribute to protection against SARS-CoV-2 re-challenge in convalescent macaques with waning antibody titers. Taken together, these data demonstrate important proof-of-concept findings that define key immunologic determinants for protection against SARS-CoV-2 in rhesus macaques.
These findings extend our DNA and Ad26 vaccine studies in rhesus macaques in which we observed that NAb titers and other Fc functional antibody responses correlated with protective efficacy4,5. Our data also extend recent studies that have shown that potent RBD-specific monoclonal antibodies can protect against SARS-CoV-2 challenge in macaques15,16. In the present study, we show that polyclonal antibodies from convalescent macaques, in the absence of cellular and innate immune responses, protect when administered at an appropriate titer. In particular, NAb titers of approximately 500 fully protected and titers of approximately 50 partially protected macaques. These titers are comparable to those observed in convalescent macaques and are readily achievable by vaccination in humans. These data demonstrate that relatively low NAb titers are sufficient to protect against SARS-CoV-2 in rhesus macaques.
We also observed that protection of convalescent animals against SARS-CoV-2 re-challenge was partially abrogated by CD8 depletion prior to re-challenge. NAb titers declined in convalescent animals from week 4 to week 7, with over half of the animals exhibiting NAb titers <100 by week 7. CD8 depletion in these animals resulted in loss of protection in the upper respiratory tract against SARS-CoV-2 re-challenge, suggesting that CD8+ T cells are likely critical for virologic control if NAb titers are suboptimal or subprotective. Future studies could also be designed to evaluate the potential protective efficacy of cellular immune responses in the absence of antibody responses.
Our findings have important implications for vaccines and immune-based therapeutics. Our data suggest the importance of SARS-CoV-2 vaccines to induce potent and durable humoral as well as cellular immune responses. We speculate that NAb titers above a certain threshold are sufficient for protection but that CD8+ T cells may also contribute to protection when NAb titers decline. It is possible, however, that the specific NAb threshold for protection identified in the present study (IC50 ∼50) may be model-specific and may be dependent on technical details, such as donor plasma characteristics, challenge virus stock infectivity and inoculum dose, and recipient macaque specifics. Further adoptive transfer studies should be performed with IgG purified from plasma from vaccinated macaques or humans.
The present data also demonstrate the therapeutic efficacy of convalescent plasma for treatment of SARS-CoV-2 infection, which is currently being explored in clinical trials12,13. However, our data should be interpreted cautiously, because only high serum NAb titers in recipient animals (approximately 500) showed therapeutic efficacy in this model, whereas lower serum NAb titers in recipient animals (approximately 50) did not have detectable efficacy. Such high serum NAb titers in recipient macaques likely exceed typical serum NAb titers achieved in human recipients of convalescent human plasma4,17, and thus implications for the clinical use of convalescent plasma for treatment of SARS-CoV-2 infection remain limited. Monoclonal antibodies achieve substantially higher neutralization titers than convalescent plasma.
Our data provide a key proof-of-concept that antibodies can protect against SARS-CoV-2 challenge in a dose dependent fashion. If antibodies are near or below the threshold titer required for protection, then cellular immune responses may be important for rapid virologic control. Collectively, these data provide key insights into the immunologic determinants and correlates of protection against SARS-CoV-2 and have important implications for the development of vaccines, immune-based therapeutics, and public health strategies.
Methods
Animals and study design.
31 outbred Indian-origin male and female rhesus macaques (Macaca mulatta), 2–5 years old, were randomly allocated to groups. All animals were housed at Bioqual, Inc. (Rockville, MD). In the prophylactic adoptive transfer study (N=12), animals received an intravenous (IV) infusion of 250 mg/kg, 25 mg/kg, 2.5 mg/kg purified SARS-CoV-2 IgG or sham IgG (N=3/group) on day −3. On day 0, all animals were challenged with 1.0×105 TCID50 (1.2×108 RNA copies, 1.1×104 PFU) SARS-CoV-2, which was derived from USA-WA1/2020 (NR-52281; BEI Resources) and fully sequenced revealing no mutations1. In the therapeutic adoptive transfer study (N=6), animals were challenged with 1.0×105 TCID50 on day 0 and then received an IV infusion of 250 mg/kg, 25 mg/kg, or 0 mg/kg purified SARS-CoV-2 IgG (N=2/group) on day 1. Virus was administered as 1 ml by the intranasal (IN) route (0.5 ml in each nare) and 1 ml by the intratracheal (IT) route. In the CD8 depletion study (N=13), animals were infected with SARS-CoV-2 as above and then received an IV infusion of 50 mg/kg of the anti-CD8α CDR-grafted rhesus IgG1 antibody MT807R1 (MassBiologics, Mattapan, MA) (N=5), the anti-CD8β rhesus IgG1 antibody CD8b255R1 (MassBiologics, Mattapan, MA) (N=3), or a sham antibody (N=5) at week 7, reflecting day −3 relative to re-challenge. On day 0, all animals were re-challenged IN+IT with SARS-CoV-2 as above. All immunologic and virologic assays were performed blinded. All animal studies were conducted in compliance with all relevant local, state, and federal regulations and were approved by the Bioqual Institutional Animal Care and Use Committee (IACUC).
IgG purification.
Polyclonal IgG was purified from rhesus macaque plasma using the NAb™ Protein G Spin Columns (Thermo Scientific). Plasm was incubated for 1 h with Protein G columns pre-conditioned with Protein G IgG Binding Buffer (Thermo Scientific) to capture maximum IgG. The columns were then washed six times, and IgG was eluted with IgG Elution Buffer (Thermo Scientific). The eluted IgG was buffer exchanged with 1x DPBS and analyzed by SDS-PAGE and spectrophotometry.
Subgenomic mRNA assay.
SARS-CoV-2 E gene subgenomic mRNA (sgRNA or sgmRNA) was assessed by RT-PCR using primers and probes as previously described1,4,11. Briefly, to generate a standard curve, the SARS-CoV-2 E gene sgRNA was cloned into a pcDNA3.1 expression plasmid; this insert was transcribed using an AmpliCap-Max T7 High Yield Message Maker Kit (Cellscript) to obtain RNA for standards. Prior to RT-PCR, samples collected from challenged animals or standards were reverse-transcribed using Superscript III VILO (Invitrogen) according to the manufacturer’s instructions. A Taqman custom gene expression assay (ThermoFisher Scientific) was designed using the sequences targeting the E gene sgRNA11. Reactions were carried out on a QuantStudio 6 and 7 Flex Real-Time PCR System (Applied Biosystems) according to the manufacturer’s specifications. Standard curves were used to calculate sgRNA in copies per ml or per swab; the quantitative assay sensitivity was 50 copies per ml or per swab.
Enzyme-linked immunosorbent assay (ELISA).
Binding antibodies were assessed by ELISA essentially as described1,4. Briefly, 96-well plates were coated with 1 µg/ml SARS-CoV-2 spike (S) or receptor binding domain (RBD) protein in 1X DPBS and incubated at 4°C overnight. After incubation, plates were washed once with wash buffer (0.05% Tween 20 in 1 X DPBS) and blocked with 350 µL Casein block/well for 2–3 h at room temperature. After incubation, block solution was discarded and plates were blotted dry. Serial dilutions of heat-inactivated serum diluted in casein block were added to wells and plates were incubated for 1 h at room temperature, prior to three further washes and a 1 h incubation with a 1:1000 dilution of anti-macaque IgG HRP (NIH NHP Reagent Program) at room temperature in the dark. Plates were then washed three times, and 100 µL of SeraCare KPL TMB SureBlue Start solution was added to each well; plate development was halted by the addition of 100 µL SeraCare KPL TMB Stop solution per well. The absorbance at 450nm was recorded using a VersaMax or Omega microplate reader. ELISA endpoint titers were defined as the highest reciprocal serum dilution that yielded an absorbance > 0.2. Log10 endpoint titers are reported.
Pseudovirus neutralization assay.
The SARS-CoV-2 pseudoviruses expressing a luciferase reporter gene were generated in an approach similar to as described previously1,4,8. Briefly, the packaging construct psPAX2 (AIDS Resource and Reagent Program), luciferase reporter plasmid pLenti-CMV Puro-Luc (Addgene), and spike protein expressing pcDNA3.1-SARS-CoV-2 SΔCT were co-transfected into HEK293T cells with calcium phosphate. The supernatants containing the pseudotype viruses were collected 48 h post-transfection; pseudotype viruses were purified by filtration with 0.45 µm filter. To determine the neutralization activity of the antisera from vaccinated animals, HEK293T-hACE2 cells were seeded in 96-well tissue culture plates at a density of 1.75 × 104 cells/well overnight. Two-fold serial dilutions of heat inactivated serum samples were prepared and mixed with 50 µL of pseudovirus. The mixture was incubated at 37oC for 1 h before adding to HEK293T-hACE2 cells. After 48 h, cells were lysed in Steady-Glo Luciferase Assay (Promega) according to the manufacturer’s instructions. SARS-CoV-2 neutralization titers were defined as the sample dilution at which a 50% reduction in RLU was observed relative to the average of the virus control wells.
Intracellular cytokine staining (ICS) assay.
106 PBMCs/well were re-suspended in 100 µL of R10 media supplemented with CD49d monoclonal antibody (1 µg/mL). Each sample was assessed with mock (100 µL of R10 plus 0.5% DMSO; background control), peptide pools (2 µg/mL), or 10 pg/mL phorbol myristate acetate (PMA) and 1 µg/mL ionomycin (Sigma-Aldrich) (100µL; positive control) and incubated at 37°C for 1 h. After incubation, 0.25 µL of GolgiStop and 0.25 µL of GolgiPlug in 50 µL of R10 was added to each well and incubated at 37°C for 8 h and then held at 4°C overnight. The next day, the cells were washed twice with DPBS, stained with Near IR live/dead dye for 10 mins and then stained with predetermined titers of mAbs against CD279 (clone EH12.1, BB700), CD38 (clone OKT10, PE), CD28 (clone 28.2, PE CY5), CD4 (clone L200, BV510), CD45 (clone D058–1283, BUV615), CD95 (clone DX2, BUV737), CD8 (clone SK1, BUV805), for 30 min. Cells were then washed twice with 2% FBS/DPBS buffer and incubated for 15 min with 200µL of BD CytoFix/CytoPerm Fixation/Permeabilization solution. Cells were washed twice with 1X Perm Wash buffer (BD Perm/WashTM Buffer 10X in the CytoFix/CytoPerm Fixation/ Permeabilization kit diluted with MilliQ water and passed through 0.22µm filter) and stained with intracellularly with mAbs against Ki67 (clone B56, FITC), CD69 (clone TP1.55.3, ECD), IL10 (clone JES3–9D7, PE CY7), IL13 (clone JES10–5A2, BV421), TNF-α (clone Mab11, BV650), IL4 (clone MP4–25D2, BV711), IFN-γ (clone B27; BUV395), IL2 (clone MQ1–17H12, APC), CD3 (clone SP34.2, Alexa 700), for 30 min. Cells were washed twice with 1X Perm Wash buffer and fixed with 250µL of freshly prepared 1.5% formaldehyde. Fixed cells were transferred to 96-well round bottom plate and analyzed by BD FACSymphonyTM system.
Luminex isotype assay.
To determine relative concentrations of antigen-specific antibody isotypes and Fc receptor binding activity, a Luminex isotype assay was performed10. Protein antigens included prefusion stabilized spike ectodomain (S), receptor binding domain (RBD), nucleocapsid (N), and S1+S2 (Aalto Bio Reagents; Sino Biological). Antigens were covalently coupled to different Luminex microplex carboxylated bead regions (Luminex Corporation) using NHS-ester linkages by utilizing EDC and NHS (Thermo Scientific) according to manufacturer recommendation. Antigen-coupled beads were added to 384-well plates and incubated with diluted samples (1:100 for isotypes, 1:250 for FcR binding) for 2 h to form immune complexes. The plates were then washed. Mouse-anti-NHP secondary detectors were added (40μL at 0.65μg/mL) for each antibody isotype (IgG1, IgG2, IgG3, IgG4, Nonhuman Primate Reagent Resource) and allowed to bind for an hour. Following a wash, tertiary anti-mouse-IgG-PE detector antibodies were added (45 μL at 0.5μg/mL). FcR binding was measured in a similar procedure by using recombinant NHP FcRs (FcR2A-1, FcR2A-2, FcR3A, Duke Protein Production Facility) conjugated to PE as secondary detectors. Flow cytometry was performed using an iQue (Intellicyt).
Functional antibody assays.
To quantify and compare antibody functionality of plasma samples, bead-based assays were used to measure antibody-dependent cellular phagocytosis (ADCP), antibody-dependent neutrophil phagocytosis (ADNP) and antibody-dependent complement deposition (ADCD)9. Biotinylated antigens S, RBD, and N were coupled to fluorescent streptavidin beads (Thermo Fisher) and incubated with diluted plasma (ADCP at 1:100, ADNP at 1:50, and ADCD at 1:10) to allow antibody binding to occur. For ADCP, cultured THP-1s were incubated with immune complexes for 16 h at 37°C, during which phagocytosis occurred. For ADNP, primary neutrophils were isolated from whole blood using ammonium-chloride-potassium (ACK) lysis buffer. After phagocytosis of immune complexes for 1 h at 37°C, neutrophils were stained with an anti-CD66b Pacific Blue detection antibody (Biolegend) prior to flow cytometry. For ADCD, lyophilized guinea pig complement (Cedarlane) was reconstituted according to manufacturer’s instructions and diluted in gelatin veronal buffer with calcium and magnesium (Boston BioProducts). After allowing for complement deposition, C3 bound to immune complexes was detected with FITC-Conjugated Goat IgG Fraction to Guinea Pig Complement C3 (MP Biomedicals). For quantification of antibody-dependent NK cell activation, plasma samples diluted at 1:25 were incubated for 2 h at 37°C in ELISA plates coated with 0.3μg of antigen per well. Human NK cells were isolated the evening before using RosetteSep (STEMCELL Technologies) from healthy buffy coat donors and incubated overnight in 1ng/mL human recombinant Interleukin 15 (STEMCELL Technologies). NK cells were incubated with immune complexes for 5 h at 37°C, after which they were stained with CD107a PE-Cy5 (BD), Golgi stop (BD) and Brefeldin A (BFA, Sigma-Aldrich). Post incubation, cells were fixed (Perm A, Life Tech) and stained using anti-CD16 APC-Cy7 (BD), anti-CD56 PE-Cy7 (BD) and anti-CD3 Pacific Blue (BD). Intracellular staining using anti-MIP-1β PE (BD) was performed after permeabilizing the NK cells using Perm B (Thermo Fisher). Flow cytometry acquisition of all assays was performed using an iQue (Intellicyt) and an S-LAB robot (PAA). For ADCP, phagocytosis events were gated on bead-positive cells. For ADNP, neutrophils were identified by gating on CD3 negative, CD14 negative, CD66b positive cells. Neutrophil phagocytosis was identified by gating on bead-positive cells. A phagocytosis score for ADCP and ADNP was calculated as (percentage of bead-positive cells) x (MFI of bead-positive cells) divided by 10,000. ADCD quantification was reported as MFI of FITC-anti-C3. NK cells were identified by gating on CD3 negative, CD16 positive and CD56 positive cells. Data were reported as the percentage of cells positive for CD107a and MIP-1β.
Statistical analyses.
Comparisons of virologic and immunologic data was performed using GraphPad Prism 8.4.2 (GraphPad Software). Comparison of data between groups was performed using two-sided Mann-Whitney tests. P-values of less than 0.05 were considered significant. For regression analyses, pseudovirus NAb titers, ELISA titers, IgG subclass titers, FcR binding levels, ADCD, and the maximum/peak viral loads were log10 transformed. To determine a threshold in antibody titers predicting protection, we classified protected and not protected animals with logistic regression models fitted using the R function ‘glm’. The thresholds required for protection were then determined as the titers for which the models predict a 95% probability of being protected. When the titers were higher for all protected animals than for the not protected animals, the two groups were completely separable, and more data would be required to obtain reliable estimates for the titer threshold corresponding to a 95% probability of protection. In these cases, the mean value of the highest titer of the not protected animals and the lowest titer of the protected animals was reported. To relate the antibody features to the peak viral loads, we fitted exponential curves, y = a*exp(b*x) + log10(50), with log10 antibody titer x and log10 peak viral load y using the R function ‘nls’. This curve has the asymptote log10(50) = 1.7 which corresponds to the limit of quantification of the assay at 50 copies/ml (BAL) or copies/swab (NS). A principal component analysis (PCA) was constructed using the R package ‘ropls’ to compare multivariate profiles. Correlations were assessed using Spearman rank correlations, and p-values were corrected for multiple testing using the Benjamini-Hochberg procedure. The analyses were performed in R version 3.6.1.
Extended Data
Extended Data Figure 1. Pseudovirus NAb titers in plasma and purified IgG of donor macaques.

9 rhesus macaques were infected and re-challenged with SARS-CoV-2. IgG was purified from plasma and formulated at 10 mg/ml.
Extended Data Figure 2. Antibody titers following prophylactic adoptive transfer of IgG prior to SARS-CoV-2 challenge.
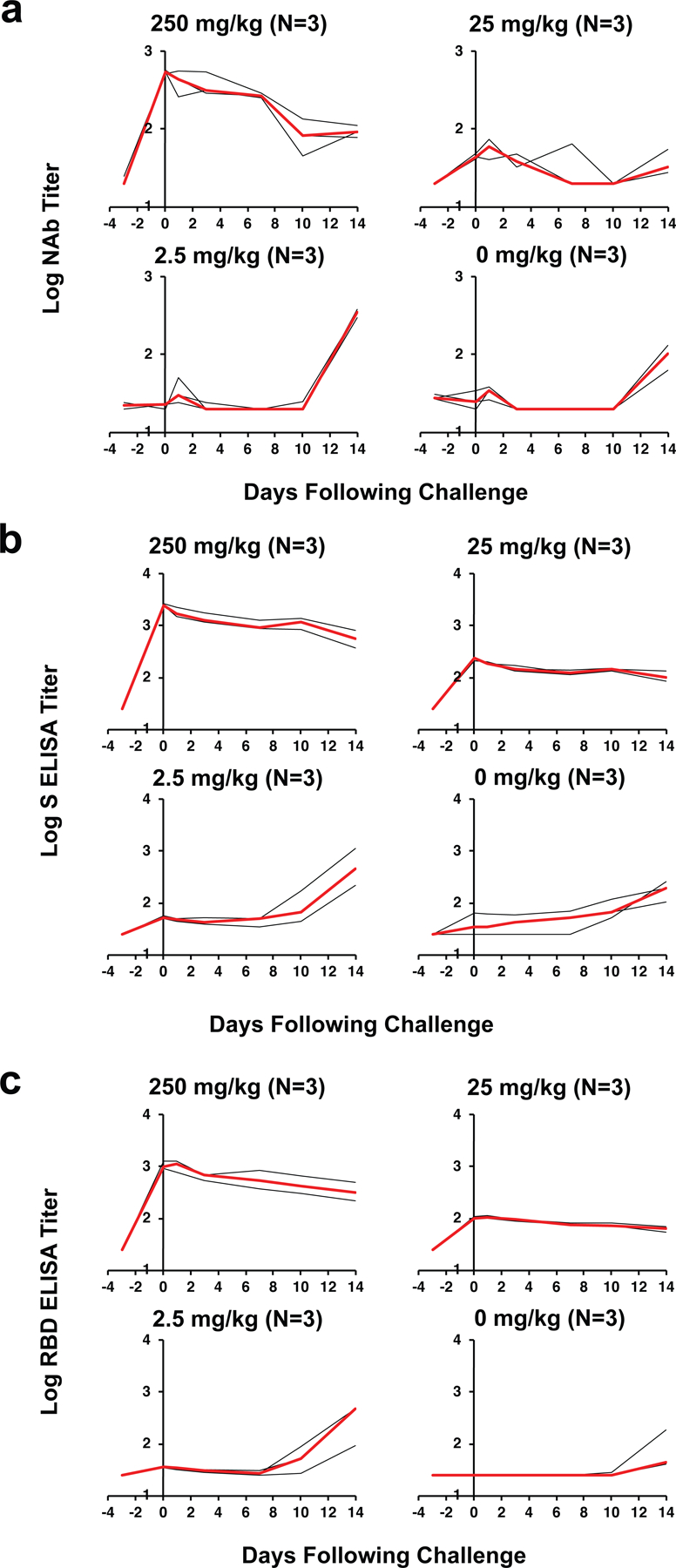
(a) Log10 pseudovirus neutralizing antibody (NAb) titers, (b) Log10 S-specific ELISA titers, and (c) Log10 RBD-specific ELISA titers in rhesus macaques following intravenous infusion of 250 mg/kg, 25 mg/kg, 2.5 mg/kg of purified SARS-CoV-2 IgG or sham IgG on day −3. On day 0, all animals were challenged with 105 TCID50 SARS-CoV-2. Days following challenge is shown on the x-axis. Red lines reflect median responses. The number of animals is denoted in each panel and the median line overlaps with data lines.
Extended Data Figure 3. Systems serology following adoptive transfer of IgG in rhesus macaques.
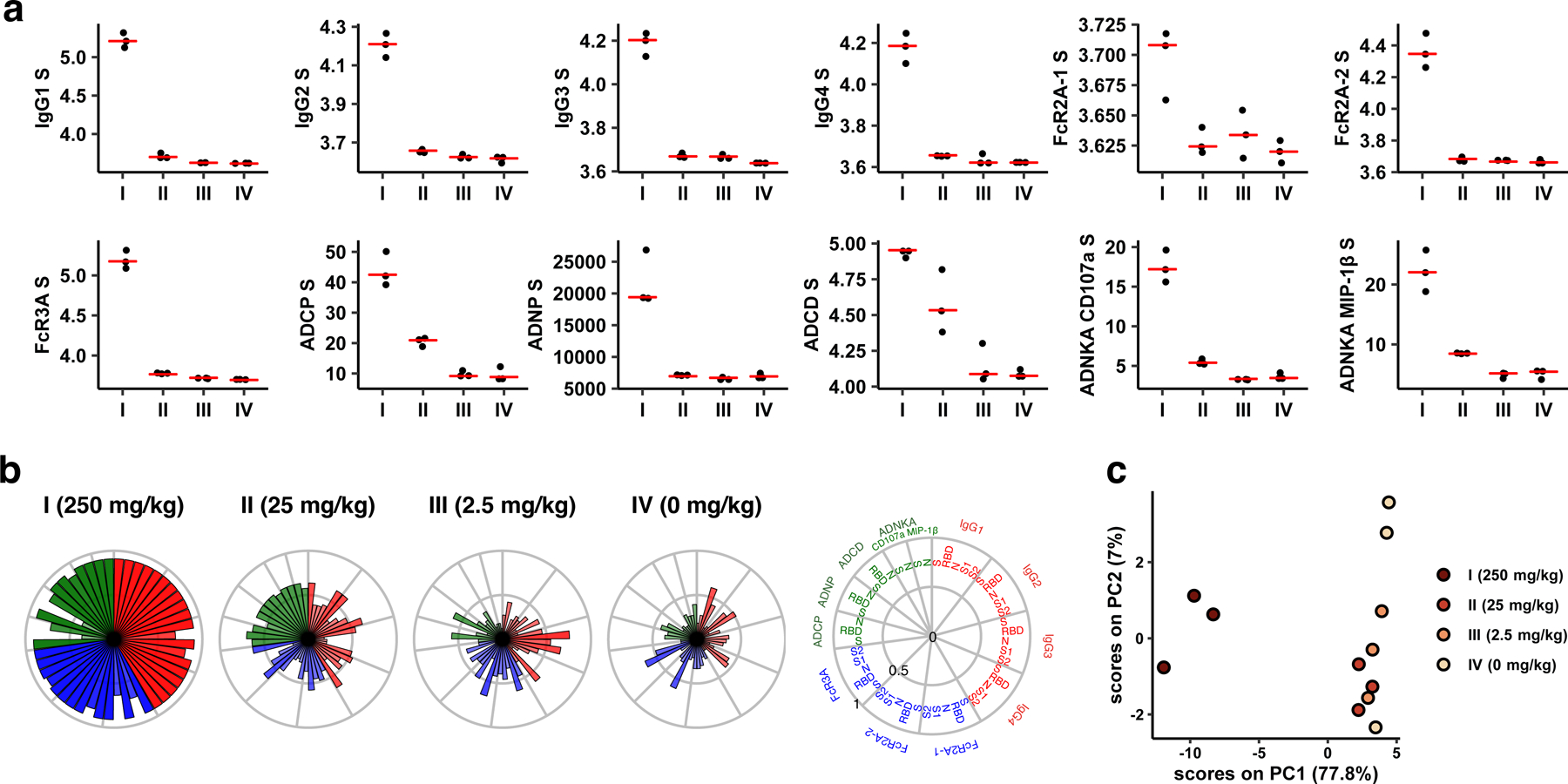
(a) S-specific IgG subclass, FcR, and antibody-dependent cellular phagocytosis (ADCP), antibody-dependent neutrophil phagocytosis (ADNP), antibody-dependent complement deposition (ADCD) and antibody-dependent NK cell activation (ADNKA) responses. (b) Radar plot showing the mean percentile of the antibody levels across passive transfer groups. The size and color intensity of the wedges indicate the mean percentile of the feature for the corresponding group (antibody subclass, red; FcR binding, blue; effector function, green). (c) Principal component analysis (PCA) plot showing the multivariate antibody profiles across groups. Each dot represents one animal (n=12 independent animals) and the color of the dot indicates the group. Red lines reflect median responses.
Extended Data Figure 4. Systems serology immune correlates.
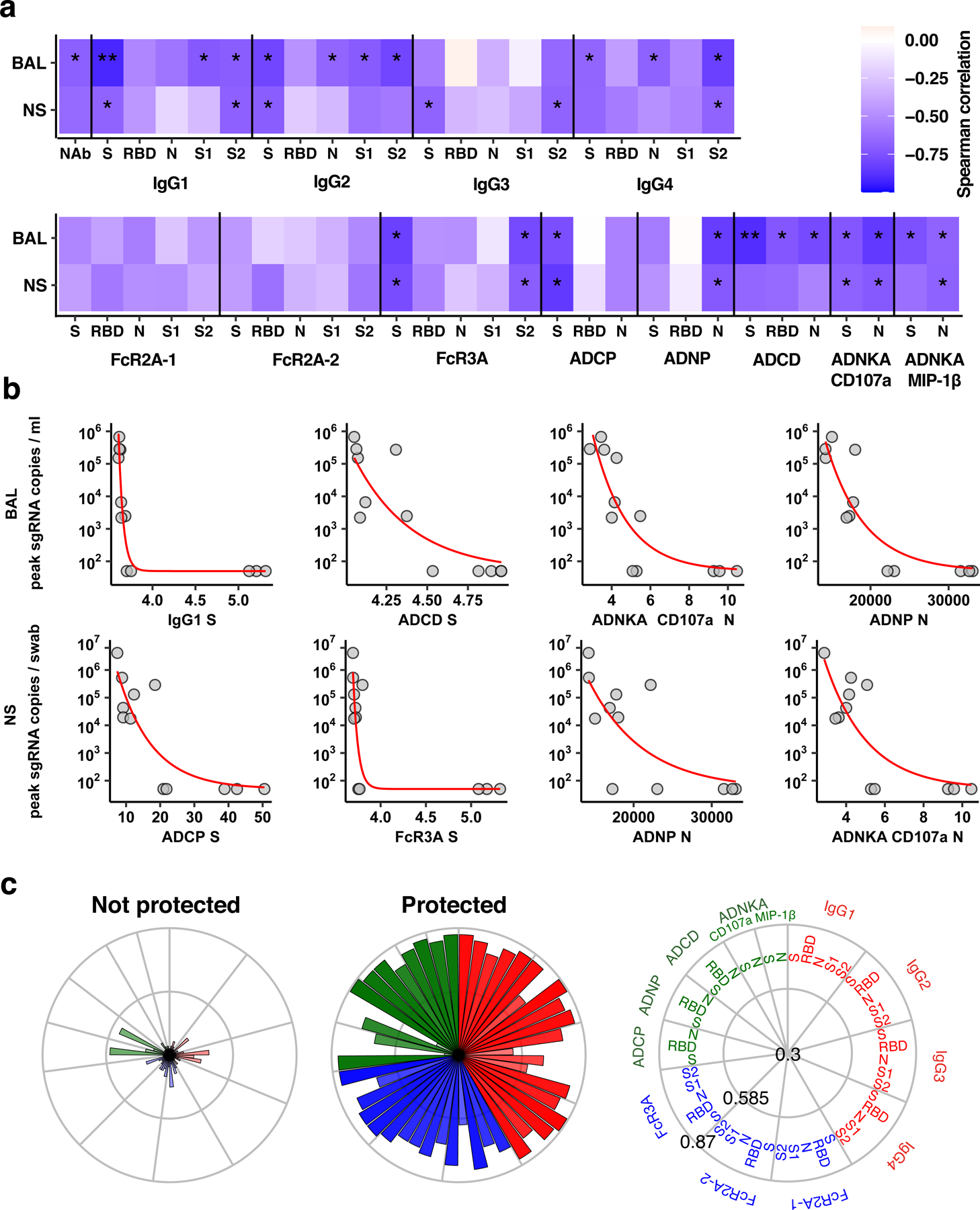
(a) The heat map shows the Spearman correlations between antibody features and peak sgRNA in BAL and NS (*q-value < 0.05, **q-value < 0.01 with q-value obtained by Benjamini-Hochberg correction for multiple testing). (b) Relations between the top four antibody features (n=12 independent animals) that are highest correlated to peak viral loads in BAL (upper row) and NS (bottom row). Red lines indicate fitted exponential curves. (c) The radar plots depict the mean percentile of each antibody feature for protected and non-protected animals. The size and color intensity of the wedges indicate the mean percentile of the feature for the corresponding group (antibody subclass, red; FcR binding, blue; effector function, green).
Extended Data Figure 5. Decline of NAb titers in convalescent rhesus macaques following SARS-CoV-2 infection.

Log10 pseudovirus NAb titers in the 9 re-challenged macaques used as donors in the adoptive transfer study and in the 10 macaques used in the CD8 depletion study at week 4 and week 7 following SARS-CoV-2 infection. Red lines reflect median values. P values reflect two-sided Mann-Whitney tests.
Extended Data Figure 6. CD8+ cells following CD8 depletion and SARS-CoV-2 re-challenge.
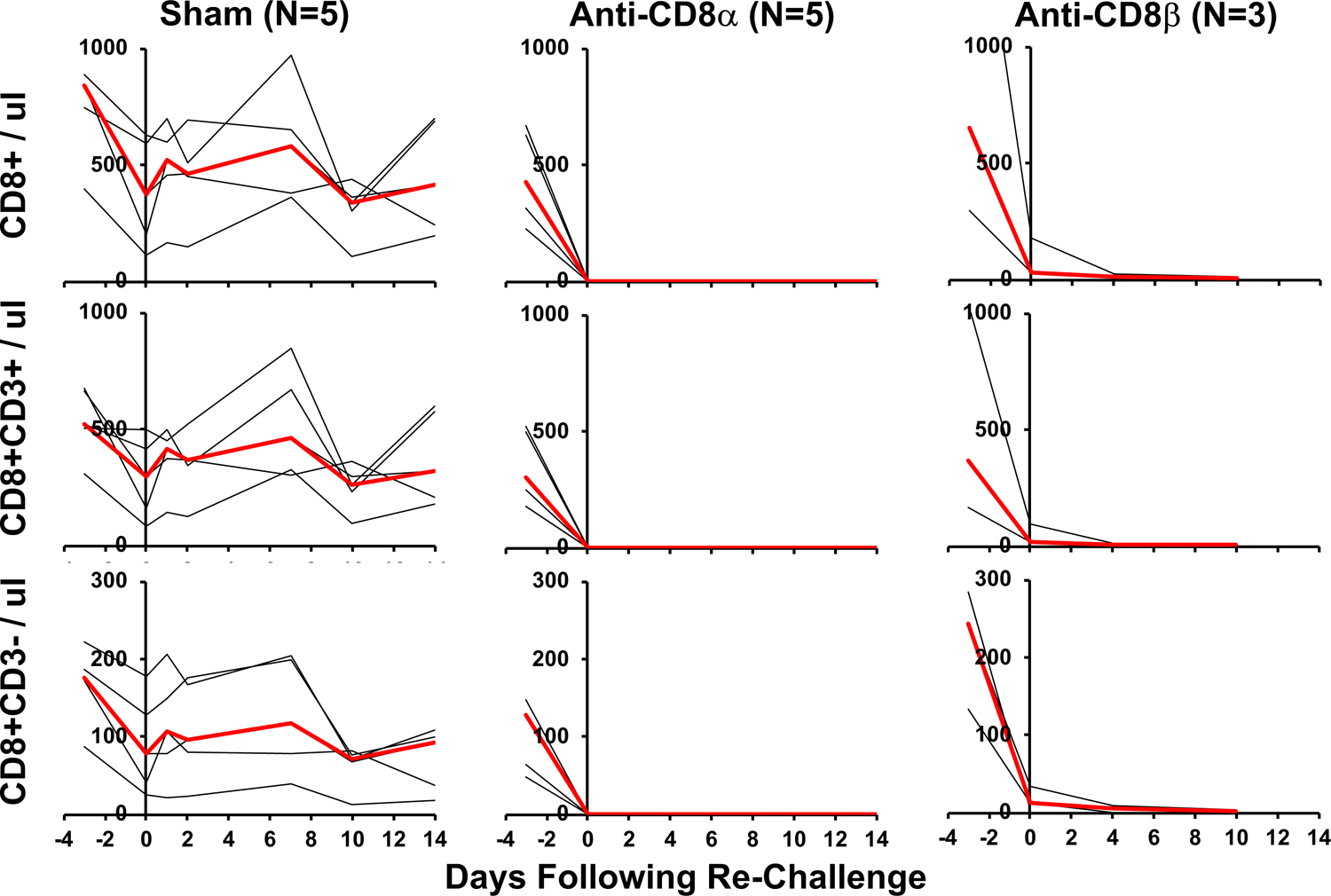
Rhesus macaques were infected with SARS-CoV-2 and received 50 mg/kg anti-CD8α mAb, anti-CD8β mAb, or sham mAb at week 7, reflecting day −3 relative to re-challenge. On day 0, all animals were re-challenged with 105 TCID50 SARS-CoV-2. Total CD8+ cells, CD8+CD3+ cells, and CD8+CD3- cells per ul peripheral blood are shown. Days following challenge is shown on the x-axis. Red lines reflect median values.
Extended Data Figure 7. CD8+ T cell responses following CD8 depletion and SARS-CoV-2 re-challenge.
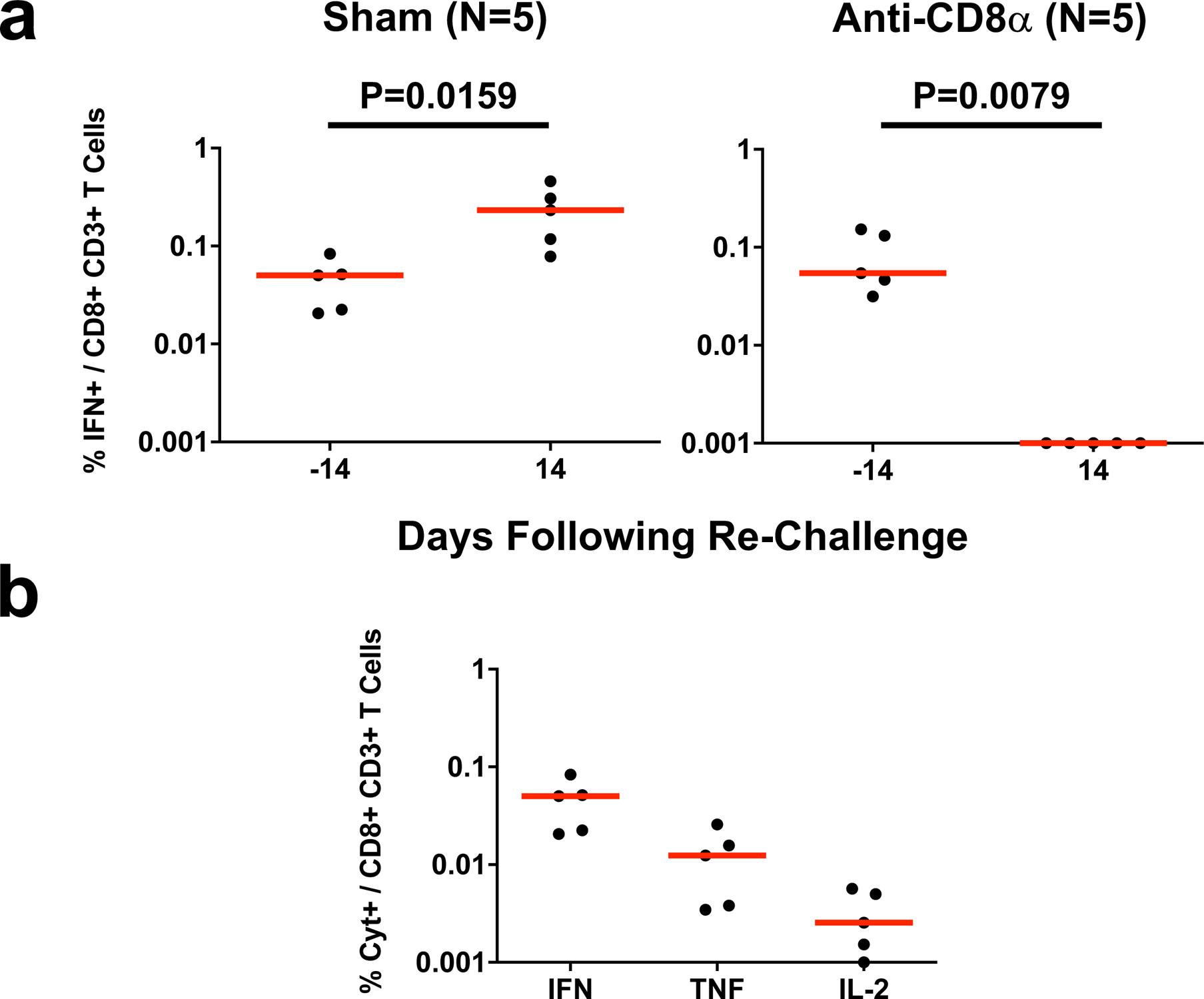
(a) IFN-γ S-specific CD8+ T cell responses assessed by intracellular cytokine staining in non-depleted (Sham) and CD8 depleted (anti-CD8α) rhesus macaques prior to and following SARS-CoV-2 re-challenge. Days following re-challenge is shown on the x-axis. Red lines reflect median values. P values reflect two-sided Mann-Whitney tests. (b) IFN-γ, TNF-α, and IL-2 S-specific CD8+ T cell responses in non-depleted (Sham) animals prior to re-challenge (n=5 independent animals). Red lines reflect median responses.
Extended Data Figure 8. CD4+ T cell responses following CD8 depletion and SARS-CoV-2 re-challenge.
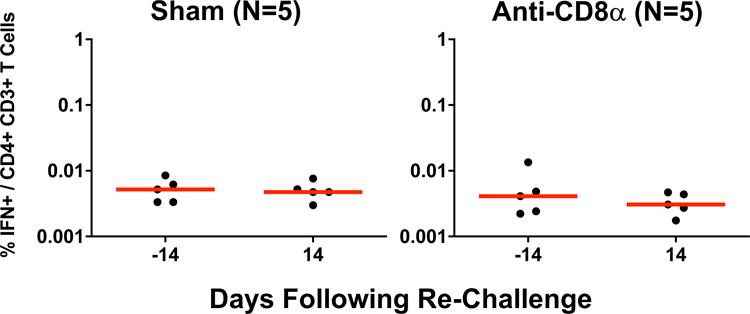
IFN-γ S-specific CD4+ T cell responses assessed by intracellular cytokine staining in non-depleted (Sham) and CD8 depleted (anti-CD8α) rhesus macaques prior to and following SARS-CoV-2 re-challenge. Days following re-challenge is shown on the x-axis. Red lines reflect median values.
Extended Data Figure 9. NAb responses following CD8 depletion and SARS-CoV-2 re-challenge.
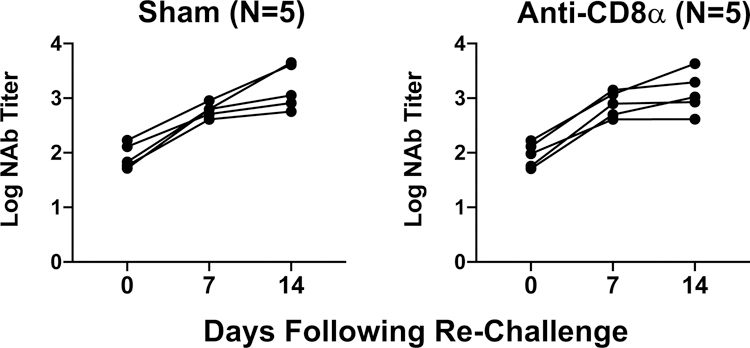
Log10 pseudovirus NAb titers in non-depleted (Sham) and CD8 depleted (anti-CD8α) rhesus macaques prior to and following SARS-CoV-2 re-challenge. Days following re-challenge is shown on the x-axis.
Extended Data Table 1.
Characteristics of rhesus macaques.
| Animal ID | Sex | Age (years) | Weight (kg) |
|---|---|---|---|
| Group I: 250 mg/kg | |||
| 18C279 | Female | 2 | 3.29 |
| 18C300 | Female | 2 | 2.5 |
| 18C289 | Male | 2 | 2.31 |
| Group II: 25 mg/kg | |||
| 18C221 | Female | 2 | 2.96 |
| 18C200 | Male | 2 | 2.83 |
| 18C277 | Male | 2 | 2.92 |
| Group III: 2.5 mg/kg | |||
| 18C297 | Female | 2 | 2.29 |
| 18C278 | Male | 2 | 2.76 |
| 18C284 | Male | 2 | 2.83 |
| Group IV: Sham | |||
| 18C290 | Female | 2 | 3.28 |
| 18C139 | Male | 2 | 2.85 |
| 18C287 | Male | 2 | 2.91 |
Supplementary Material
Acknowledgements
We thank Jason Velasco, Deandre Beuno-Wilkerson, Emily Hoffman, Felix Nampanya, Marinela Kirilova, Nicole Kordana, Zijin Lin, Lori Maxfield, Michelle Lifton, Erica Borducchi, Morgana Silva, and Alyssa Richardson for generous advice, assistance, and reagents. We acknowledge support from the Ragon Institute of MGH, MIT, and Harvard, Mark and Lisa Schwartz Foundation, Massachusetts Consortium on Pathogen Readiness (MassCPR), the Bill & Melinda Gates Foundation (INV-006131), and the National Institutes of Health (OD024917, AI129797, AI124377, AI128751, AI126603, CA260476).
Footnotes
Competing Interests
The authors declare no financial conflicts of interest. D.H.B. is a co-inventor on provisional SARS-CoV-2 vaccine patents (62/969,008; 62/994,630).
Data Availability Statement
All data are available in the manuscript and the supplementary material.
References
- 1.Chandrashekar A et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 369, 812–817, doi: 10.1126/science.abc4776 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell 182, 713–721 e719, doi: 10.1016/j.cell.2020.06.008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao Q et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 369, 77–81, doi: 10.1126/science.abc1932 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu J et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369, 806–811, doi: 10.1126/science.abc6284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercado NB et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature, doi: 10.1038/s41586-020-2607-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbett KS et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N Engl J Med, doi: 10.1056/NEJMoa2024671 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Doremalen N et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature, doi: 10.1038/s41586-020-2608-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang ZY et al. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 428, 561–564, doi: 10.1038/nature02463 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung AW et al. Dissecting Polyclonal Vaccine-Induced Humoral Immunity against HIV Using Systems Serology. Cell 163, 988–998, doi: 10.1016/j.cell.2015.10.027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown EP et al. Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J Immunol Methods 443, 33–44, doi: 10.1016/j.jim.2017.01.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfel R et al. Virological assessment of hospitalized patients with COVID-2019. Nature, doi: 10.1038/s41586-020-2196-x (2020). [DOI] [PubMed] [Google Scholar]
- 12.Joyner MJ et al. Effect of Convalescent Plasma on Mortality among Hospitalized Patients with COVID-19: Initial Three-Month Experience. medRxiv, doi: 10.1101/2020.08.12.20169359 (2020). [DOI] [Google Scholar]
- 13.Casadevall A, Joyner MJ & Pirofski LA SARS-CoV-2 viral load and antibody responses: the case for convalescent plasma therapy. J Clin Invest 130, 5112–5114, doi: 10.1172/JCI139760 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitz JE et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283, 857–860 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Zost SJ et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449, doi: 10.1038/s41586-020-2548-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi R et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124, doi: 10.1038/s41586-020-2381-y (2020). [DOI] [PubMed] [Google Scholar]
- 17.Robbiani DF et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442, doi: 10.1038/s41586-020-2456-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript and the supplementary material.


