Abstract
Blood myeloid cells are known to be dysregulated in coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2. It is unknown whether the innate myeloid response differs with disease severity and whether markers of innate immunity discriminate high-risk patients. Thus, we performed high-dimensional flow cytometry and single-cell RNA sequencing of COVID-19 patient peripheral blood cells and detected disappearance of non-classical CD14LowCD16High monocytes, accumulation of HLA-DRLow classical monocytes (Human Leukocyte Antigen - DR isotype), and release of massive amounts of calprotectin (S100A8/S100A9) in severe cases. Immature CD10LowCD101−CXCR4+/− neutrophils with an immunosuppressive profile accumulated in the blood and lungs, suggesting emergency myelopoiesis. Finally, we show that calprotectin plasma level and a routine flow cytometry assay detecting decreased frequencies of non-classical monocytes could discriminate patients who develop a severe form of COVID-19, suggesting a predictive value that deserves prospective evaluation.
Keywords: COVID-19, calprotectin, emergency myelopoiesis, monocyte subsets, neutrophils, S100A8, S100A9, SARS-CoV-2, type I interferon
Graphical Abstract
An analysis of patients with severe COVID-19 reveals immature neutrophil and non-classical monocyte pools, with levels of the protein calprotectin linked to disease severity.
Introduction
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which infects the lungs, leading to fever, cough, and dyspnea (Guan and Zhong, 2020). Most patients presenting with mild disease develop an efficient immune response (Thevarajan et al., 2020), but some go on to develop acute respiratory distress syndrome, leading to admission to the intensive care unit (ICU), often culminating in multi-organ dysfunction and death (Wang et al., 2020).
In addition to cell-autonomous effects of the viral infection, a dysregulated immune response participates in the sudden deterioration of COVID-19 patients, ultimately overwhelming infected and non-infected tissues (Vabret et al., 2020). This overt inflammatory response centers around a cytokine storm (Chen et al., 2020a) with elevated blood concentrations of interleukin-6 (IL-6). Accordingly, therapeutic agents targeting the IL-6/IL-6R-gp130 axis can alleviate the inflammatory response (Michot et al., 2020) and ameliorate immune dysregulation (Giamarellos-Bourboulis et al., 2020), emphasizing the clinical significance of this cytokine. Marked lymphocytopenia is also associated with COVID-19 severity (Chen et al., 2020a); however, the primary source of the cytokine storm and of the mechanisms behind lymphocytopenia remains elusive (Li et al., 2020).
A growing body of evidence points to dysregulation of innate immune cells of the granulomonocytic lineage during viral infections of the lungs. A variety of human viruses infect monocytes and macrophages to spread through the tissues (Al-Qahtani et al., 2017; Desforges et al., 2007; Nottet et al., 1996; Smith et al., 2004; Yilla et al., 2005). SARS-CoV-2 mRNA is detectable in lung monocytes/macrophages of severe COVID-19 patients (Bost et al., 2020), but its ability to enter these cells in the peripheral blood and activate them directly remains unclear. Also, tissue damage induced by SARS-CoV-2 infection may lead to release of pathogen- and damage-associated molecular patterns that, in turn, induce activation and recruitment of inflammatory cytokine- and chemokine-producing innate immune cells in an amplifying loop (Liao et al., 2020).
It remains unclear to what extent immune patterns associated with COVID-19 pathophysiology are causative and exacerbate the disease and/or could be used for accurate patient stratification. Here, using high-dimensional single-cell approaches, including single-cell RNA sequencing, mass cytometry, and 25-parameter spectral flow cytometry, we show that patients who develop severe disease exhibit a massive release of S100A8/S100A9 calprotectin accompanied by changes in monocyte and neutrophil subsets. We further discover that this pathological immune system reorganization is initiated by onset of emergency myelopoiesis, which release immature myeloid cells with an immunosuppressive phenotype into the peripheral blood and lungs. Our study integrates frequencies of non-classical monocytes and immature neutrophils with calprotectin plasma levels as robust biomarkers of COVID-19 severity and suggests potential therapeutic strategies targeting calprotectin to alleviate severe COVID-19.
Results
Introduction to the Patient Cohort
This non-interventional study enrolled 158 patients (Table S1), including 86 and 72 subjects referred to the hospital with various flu-like symptoms who were diagnosed or not with COVID-19 based on positive and negative RT-PCR of pharyngeal swabs, respectively. Patients were stratified according to disease severity. Mild disease (n = 27) was defined as having no or limited clinical symptoms and not requiring computed tomography (CT) scanning or hospitalization. Moderate disease (n = 16) was defined as being symptomatic, with dyspnea and radiological findings of pneumonia upon thoracic CT scan, requiring hospitalization with a maximum of 9 L/min of oxygen. Severe disease (n = 43) was defined as respiratory distress requiring admission into the ICU. Mild and moderate cases were mixed in the discovery part of the study and considered separately to explore the ability of a routine flow assay to discriminate patients who require hospitalization.
Circulating Innate Immune Cells in Patients with Mild and Severe COVID-19 Exhibit Distinct Phenotypes
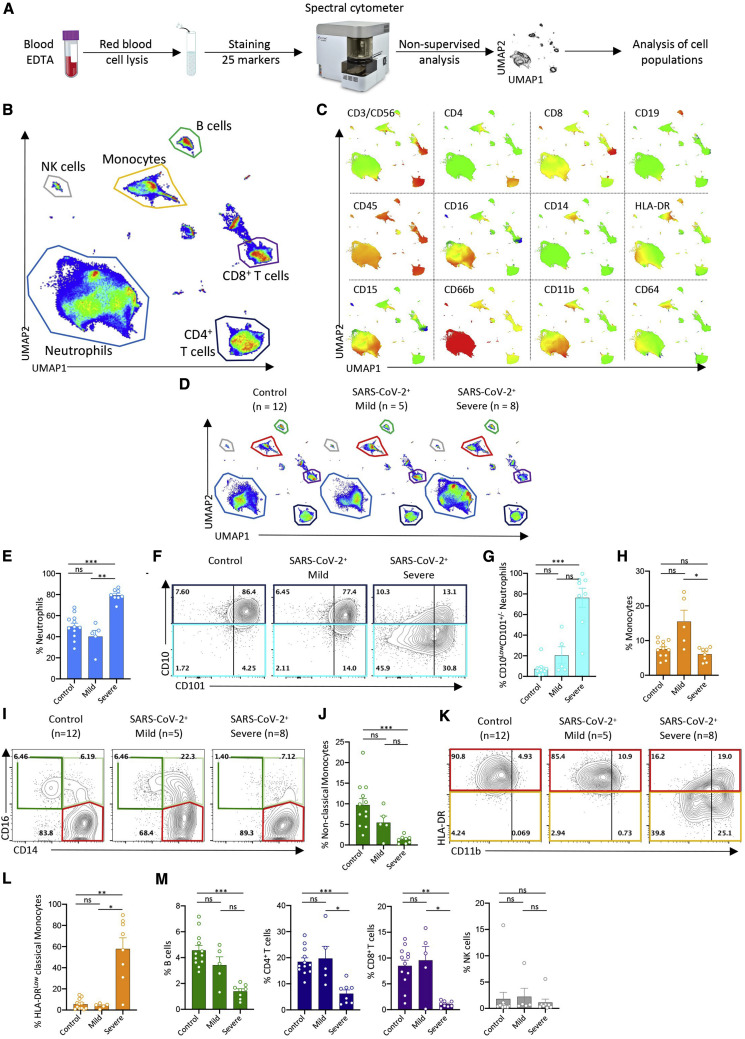
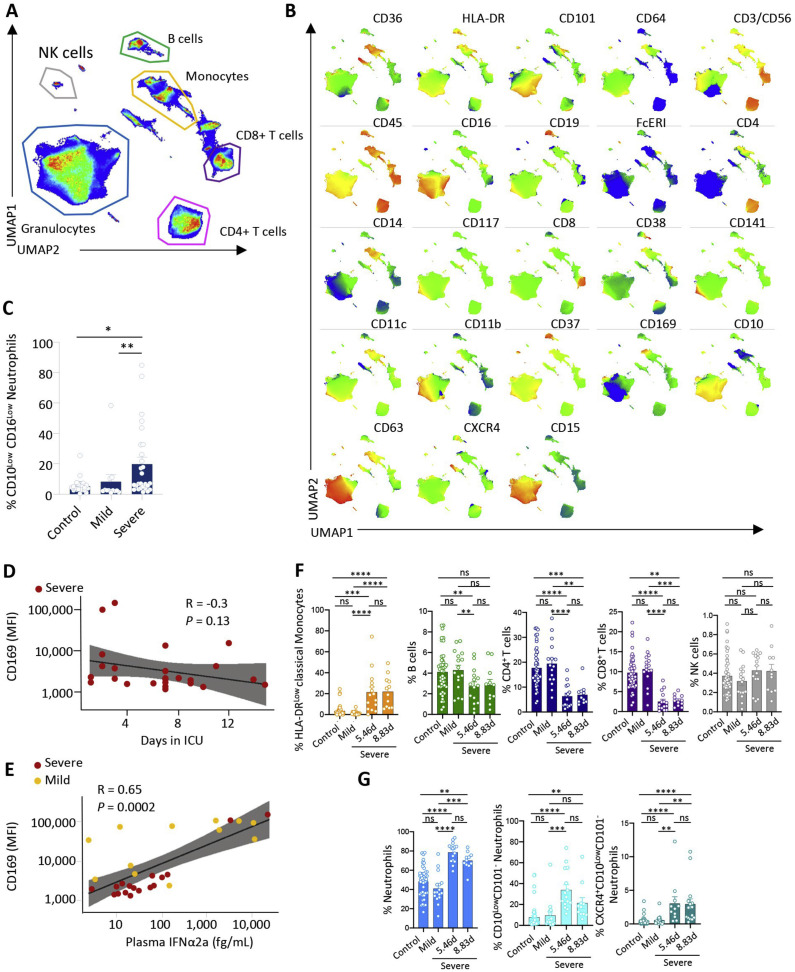
To explore changes in circulating immune cell phenotype induced by SARS-CoV-2 infection, we first collected peripheral blood samples from a discovery cohort of 13 patients positive for SARS-CoV-2 (hereafter called COVID-19 patients) by RT-PCR and 12 patients suffering from flu-like symptoms but negative for SARS-CoV-2. The former group included 5 patients with mild disease and 8 patients with severe COVID-19 (Table S2). After red blood cell lysis, we labeled peripheral blood cells with a panel of 25 antibodies recognizing immune cell surface markers (Key Resources Table) and analyzed them by spectral flow cytometry (Figures 1 A, S1 A, and S1B). By pooling the data from the 25 control and COVID-19 patients and subjecting them to dimensionality reduction using the non-supervised Uniform Manifold Approximation and Projection (UMAP) for dimension reduction algorithm (Becht et al., 2018), we identified populations of CD4+ T cells, CD8+ T cells, CD19+ B cells, CD14High monocytes, and CD15+CD66b+ neutrophils (Figures 1B and 1C). We also identified HLA-DRHighCD11b+ and CD16High monocytes as well as neutrophils expressing CD11b, CD15, CD16, and CD64 (Figures 1B and 1C). Analysis and visualization, using UMAP dimensionality reduction to the cell surface marker expression datasets from control and mild and severe COVID-19 groups suggested differences in the repartition of cell populations (Figure 1D).
Figure 1.
Spectral Flow Analysis of Peripheral Blood Cells in a Learning Cohort of Controls and COVID-19 Patients
(A) Peripheral blood sample collection pipeline.
(B) Non-supervised UMAP analysis of data from 25 patients (controls, 12; mild, 5; critical, 8).
(C) Cell surface marker expression in the UMAP analysis shown in (B).
(D) Non-supervised UMAP analysis of patient blood samples in the control, mild, and severe groups.
(E) Percentage of neutrophils among total cells in each individual sample in the indicated patient groups.
(F) Partition of neutrophil subsets, based on CD101 and CD10 expression, in each patient group (data pooled per group).
(G) Percentage of CD10LowCD101+/− neutrophils among total neutrophils as in (E).
(H) Percentage of monocytes among total cells as in (E).
(I) Partition of monocyte subsets in each individual sample in patient groups, based on CD14 and CD16 expression (left panels) or CD11b and HLA-DR expression (right panels).
(J) Fractions of non-classical monocytes among total monocytes as in (E).
(K) CD11b and HLA-DR expression on classical monocytes in each patient group (data pooled per group).
(L) Percentage of HLA-DRlow classical monocytes among classical monocytes as in (E).
(M) Percentage of B, CD4+ T, CD8+ T, and NK cells among total cells as in (E).
Kruskal-Wallis test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, non-significant.
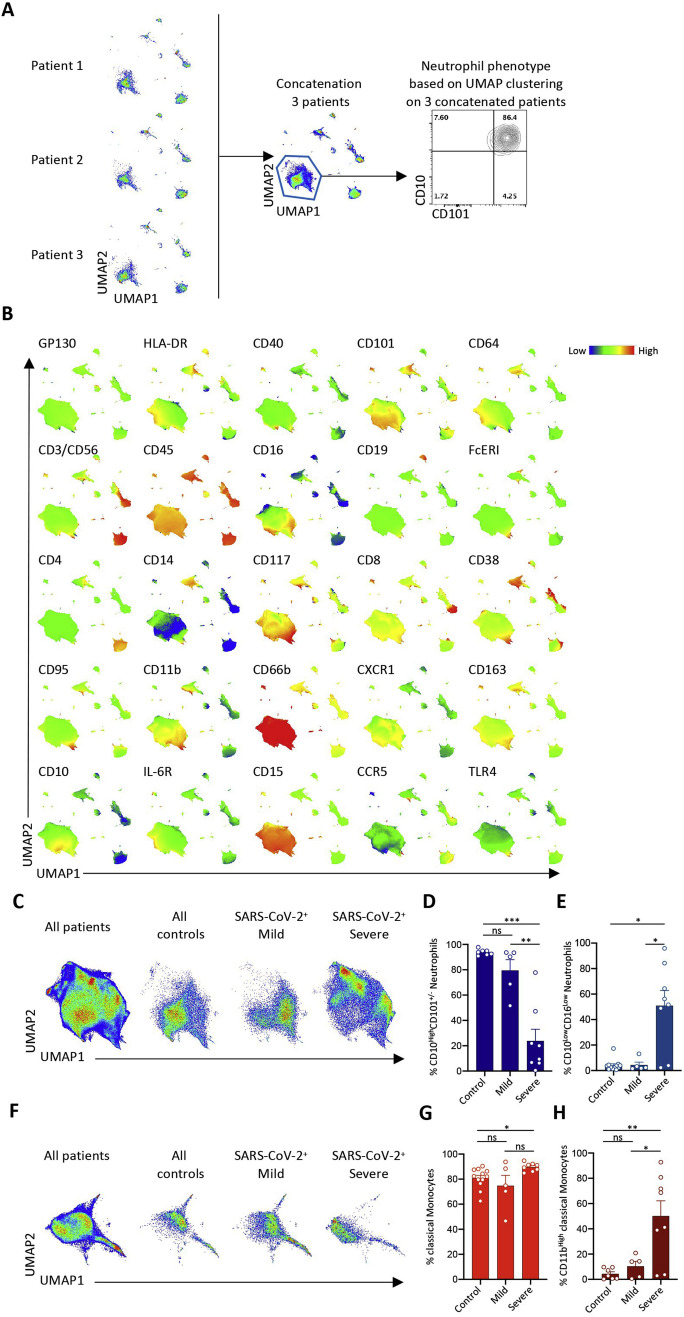
Figure S1.
Spectral Flow Analysis of Peripheral Blood Cells in a Learning Cohort of Controls and COVID-19 Patients, Related to Figure 1 and Table S2
A. Representative example of data pooling of individual UMAP profiles obtained from 3 patients of the same group. Here, neutrophil subsets are identified based on CD10 and CD101 expression across cells from patients 1, 2 and 3, allowing the analysis of cell subset repartition within the group; B. Cell surface marker expression identifying cell populations on UMAP analysis generated by data pooling from all the tested samples; C. UMAP profile from pooled data on neutrophils in all patients or indicated group of patients; D-E. Percentage of CD10High (D) or CD16Low neutrophils (E) among total neutrophils in all individuals of indicated groups; F. UMAP profile from pooled data on monocytes in all patients or indicated group of patients; G-H. Percentage of CD14+ (G) or CD11bHigh (H) monocytes among total monocytes in all individuals of indicated groups; Kruskal-Wallis test, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ns, non-significant.
Patients with severe disease exhibited an expansion in the proportion of circulating neutrophils within the peripheral blood cell population (Figure 1E) that was associated with an increase in their absolute numbers (Table S2), as already reported (Huang et al., 2020). Focusing on neutrophil subsets, we noticed a slight increase in the fraction of CD10LowCD101+ neutrophils in patients with mild COVID-19 (Figure 1F), whereas the fraction of CD10LowCD101− neutrophils was remarkably amplified in patients with severe disease, suggesting an accumulation of immature subsets of neutrophils (Ng et al., 2019) in the peripheral blood of these patients (Figures 1G and S1C–S1E).
In patients with severe disease, the absolute numbers of circulating monocytes (Table S2) and the proportion of total monocytes among peripheral blood leucocytes (Figures 1H and S1F) were similar to controls, but we noticed changes in monocyte subset repartition. The fraction of CD14HighCD16High intermediate monocytes was significantly greater in patients with mild COVID-19 (16.95% ± 6.75%) than in the control (5.84% ±1.02%) or severe (6.77% ± 1.10%) groups, whereas the non-classical CD14LowCD16High monocyte fraction was lower in patients with severe COVID-19 (1.31% ± 0.35%) than in the mild (5.46% ± 1.57%) or control groups (6.68% ± 1.14%) (Figures 1I and 1J). Within the CD14HighCD16Low classical monocyte subset (Figure S1G), we detected higher frequencies of CD11bHigh monocytes with increased disease severity (Figure S1H), whereas the intensity of HLA-DR expression was lower across the CD11b+ and CD11b− monocyte populations of patients with severe COVID-19 (Figures 1K and 1L).
Changes in myeloid cell repartition observed in patients with severe disease were associated with lower frequencies of B cells compared with controls (p < 0.001) and of CD4+ (p < 0.001) and CD8+ T cells (p < 0.01) relative to controls and patients with mild disease, whereas CD56+ natural killer (NK) cell frequencies remained comparable across all groups (Figure 1M).
Altogether, these data suggested that SARS-CoV-2-induced changes in the relative abundance of monocyte and neutrophil subsets within the peripheral blood cell population, with loss of non-classical CD14LowCD16High monocytes, reduced the expression of HLA-DR on classical monocytes and a drop in CD101 and CD10 expression on neutrophils, characterizing severe cases.
Serial Single-Cell Analysis of Mild versus Severe Patient Blood Cells Identifies Dynamic Changes in Monocyte Subsets
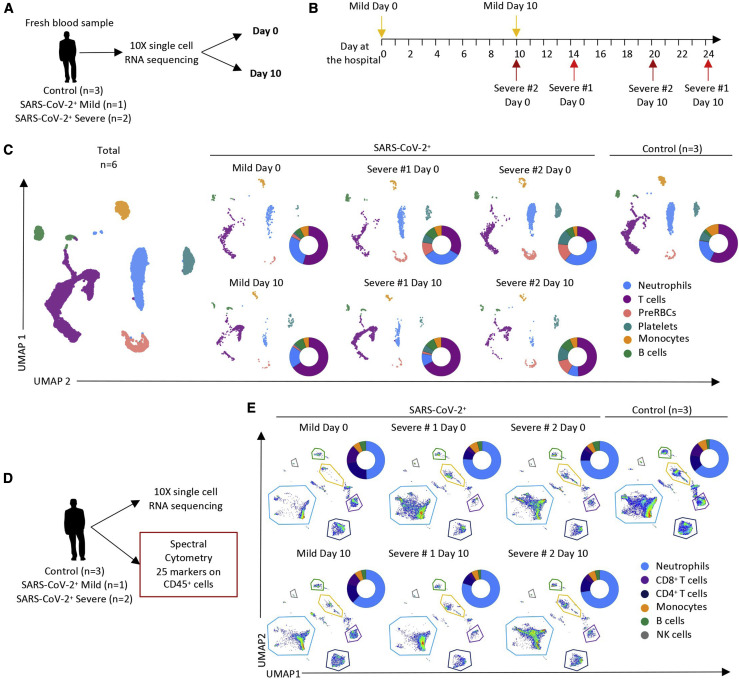
As a second step in our discovery process, we collected peripheral blood samples from three control patients with flu-like symptoms who tested negative for SARS-CoV-2 and three SARS2-CoV-2-positive patients, one outpatient with mild disease and two patients with severe disease admitted to the ICU (Figures 2A and 2B; Table S3). Using the 10X Chromium droplet-based platform, these samples were subjected to single-cell RNA sequencing (scRNA-seq) immediately after collection and red blood cell lysis, without additional sorting or freezing, to preserve fragile cell populations, mainly neutrophils. Unsupervised clustering based on gene expression identified B and T cells as well as neutrophils, monocytes, erythroid cells, and platelets (Figures 2C, S2 A, and S2B). Samples analyzed by scRNA-seq were analyzed simultaneously by spectral flow cytometry for comparison (Figure 2D). UMAP analysis of spectral flow cytometry data suggested lower proportions of CD4+ and CD8+ T cells, whereas the neutrophil fraction was greater in patients with severe disease compared with controls and with the unique patient with mild disease (Figures 2E and S2C). The three SARS2-CoV-2-infected patients were sampled again 10 days later to monitor progression of the immune response in relation to clinical status (Figures 2A–2E).
Figure 2.
scRNA-Seq of Peripheral Blood Cells in SARS-CoV-Negative and SARS-CoV-Positive Patients
(A) Two blood samples were collected 10 days apart from 3 COVID-19 patients. Blood was also collected once from 3 outpatient controls whose SARS-CoV-2 RT-PCR was negative. Individual cell mRNAs were sequenced using Chromium 10X technology.
(B) Timeline of sample collection in the three patients (further details in Table S4).
(C) UMAP analysis of the 9 sequenced samples showing repartition of the indicated cell populations. Patient samples were analyzed individually at days 0 and 10; for control patient individual analyses, see Figure S2.
(D) Spectral flow cytometry analysis of surface marker expression performed on the same samples.
(E) UMAP analysis of cell populations detected by spectral flow cytometry data in each patient at day 0 and at day 10 and in controls (for individual analyses, see Figure S2).
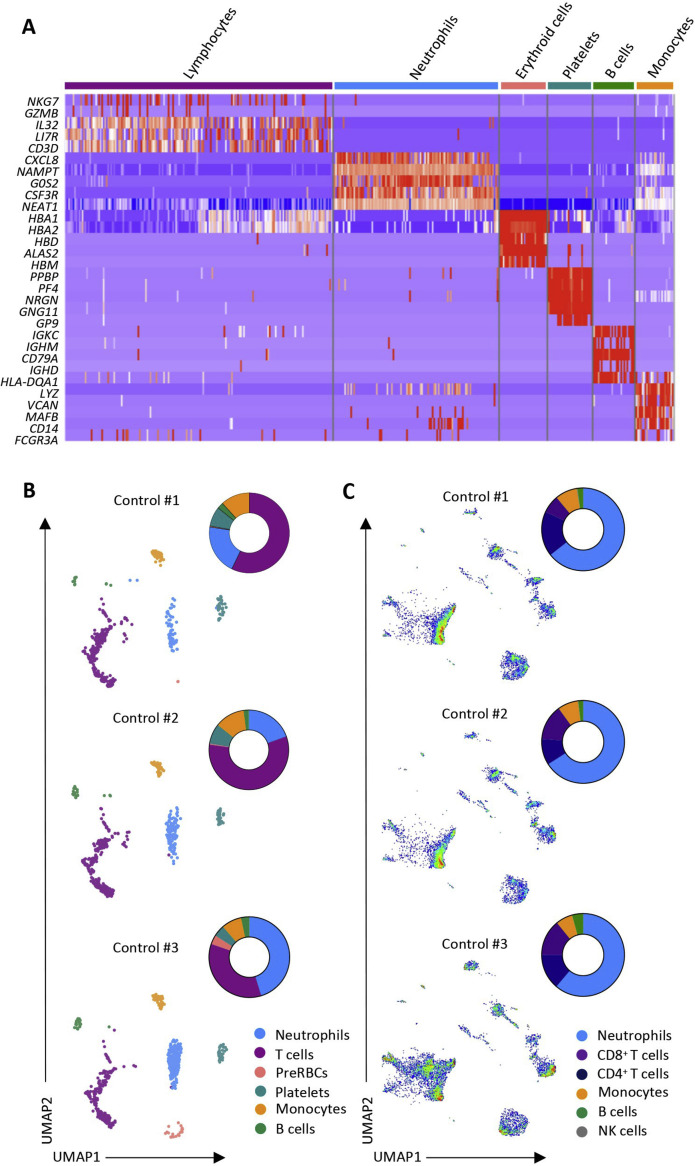
Figure S2.
Single-Cell Analysis of Peripheral Blood Cells, Related to Figure 2 and Tables S3 and S4
A. Heatmap of gene expression used to identify cell populations in scRNaseq experiments; B-C. Individual UMAP analysis of each control sample analyzed by scRNaseq (B) and spectral flow cytometry (C).
UMAP visualization of monocytes analyzed by scRNA-seq identified three clusters (Figure 3 A) that may correspond to well-defined monocyte subsets (Guilliams et al., 2018). Cells of cluster 1 expressed CD14, ITGAM (encoding CD11b), and KLF4 while poorly expressing FCGR3A (encoding CD16), suggesting classical monocytes. Cells of cluster 3, which expressed high levels of FCGR3A and low levels of CD14, may correspond to non-classical monocytes, and cluster 2, in which cells expressed CD14 and FCGR3A, evoked intermediate monocytes (Figure 3A). Differentially expressed genes (DEGs) and pathway analyses delineated a type I interferon signature in monocytes of patients with mild COVID-19 (Figures 3B, S3 A, and S3B; Table S4). This signature was less pronounced in the two severe COVID-19 samples, contrasting with the elevated expression of genes involved in production of reactive oxygen species (ROS) and nitric oxygen species (NOS) (Figures S3A and S3B).
Figure 3.
Single-Cell Analysis of Monocytes by scRNA-Seq, Spectral Flow Cytometry, and Mass Cytometry
(A) UMAP profile of monocytes in the samples described in Figure 2A and violin plots of gene expression in three statistically defined clusters.
(B) Heatmap of differentially expressed genes (DEGs; logFC ± 0.25; false discovery rate (FDR) < 0.05) in total monocytes; columns labeled “0” identify DEGs generated by comparing each patient sample at day 0 with the pool of the three controls and the two other patient samples at day 0. Columns labeled “10” identify the expression of these genes in each patient sample at day 10 compared with day 0. Genes are shown in Table S4.
(C–E) Spectral flow analysis of pooled controls and each individual patient sample at day 0 and day 10 of monocyte subset partition in samples analyzed by scRNA-seq (C), CD11b and CD141 expression among classical monocytes (D), and CD169 and HLA-DR expression among classical monocytes (E).
(F–I) Mass cytometry analysis of monocyte subsets in 4 patients within each group (pooled data) (F), non-classical monocyte fraction among total monocytes in each individual sample within the 3 groups (G), p65/NF-κB expression in HLA-DRlow classical monocyte subset as in (F) (H), and fraction of p65/NF-κBhighHLA-DRlow classical monocytes among classical monocytes as in (G) (I). Kruskal-Wallis test, ∗p < 0.05.
Figure S3.
Monocyte Analysis by scRNA-Seq, Spectral Flow Cytometry, and Mass Cytometry, Related to Figure 3 and Tables S3, S4, and S5
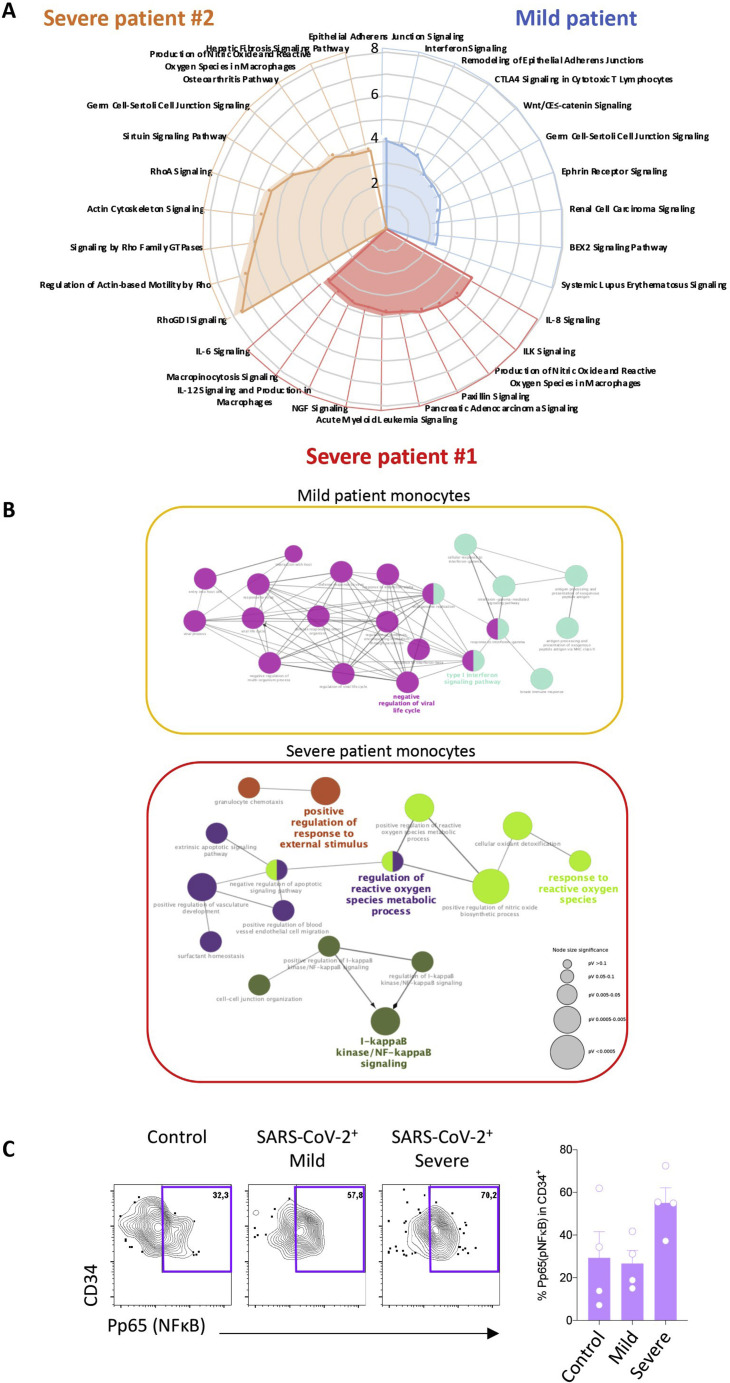
A. Pathway analysis generated by comparing DEGs in monocytes of each SARS-CoV-2 positive patient to the same population in the three control patients considered together using IPA software (mild patient in blue, severe #1 in red, severe # 2 in orange); B. The same DEGs were used to perform a gene ontology network analysis using clueGO software, considering the two severe patients together; C. Combined (left panel) and individual (right panel) mass cytometry analysis of p65/NF-κB expression in circulating CD34+ cells in each group. Kruskal-Wallis test, non-significant.
A non-supervised UMAP analysis of the data collected by spectral flow cytometry of the same samples detected variations in monocyte subset repartition among patients; compared with controls and the patient with mild disease, patient 1 with severe disease showed a lower fraction of CD14LowCD16High non-classical monocytes at day 0, whereas the other patient with severe disease showed a high level of this monocyte fraction (Figure 3C). Additionally, the two patients with severe disease showed markedly higher levels of classical CD14HighCD16Low monocytes, expressing more CD141 (THBD) at their surface (Figure 3D), in accordance with the scRNA-seq analysis (Table S4).
In the patient with mild disease, one of the most highly expressed genes in classical monocytes was the interferon-stimulated gene (Sevelsted et al., 2015) SIGLEC-1, consistent with the high level of expression of CD169, the corresponding protein, at the surface of classical monocytes at day 0 (Figure 3E). Ten days later, SIGLEC-1 gene expression was downregulated, and CD169 expression was undetectable at the surface of HLA-DRHigh classical monocytes (Figures 3B and 3E). The two patients with severe disease exhibited low expression of HLA-DR protein on monocyte surfaces at day 0, without significant change at day 10 (Figure 3E).
Validating these discovery experiments, we performed mass cytometry analysis of an independent cohort of 12 patients (four in each group; control, mild, and severe) (Table S5), which showed a lower fraction of CD14LowCD16High non-classical monocytes in patients with severe compared with mild disease (Figures 3F and 3G). In accordance with pathway analysis of scRNA-seq data highlighting nuclear factor κB (NF-κB) activation as a prominent feature in monocytes of patients with severe disease (Figures 3B and S3B), we observed significantly higher levels of the phosphorylated transcription factor RelA/p65 (P-p65), a critical effector of the canonical NF-κB pathway, in HLA-DRLowCD14High classical monocytes from patients with severe disease compared with controls (Figures 3H and 3I). We also measured P-p65 expression in circulating CD34+ cells, identifying increased expression in severe disease (Figure S3C).
Serial Single-Cell Analysis of Blood Cells from Patients with Mild versus Severe Disease Identifies Changes in Neutrophil Subsets
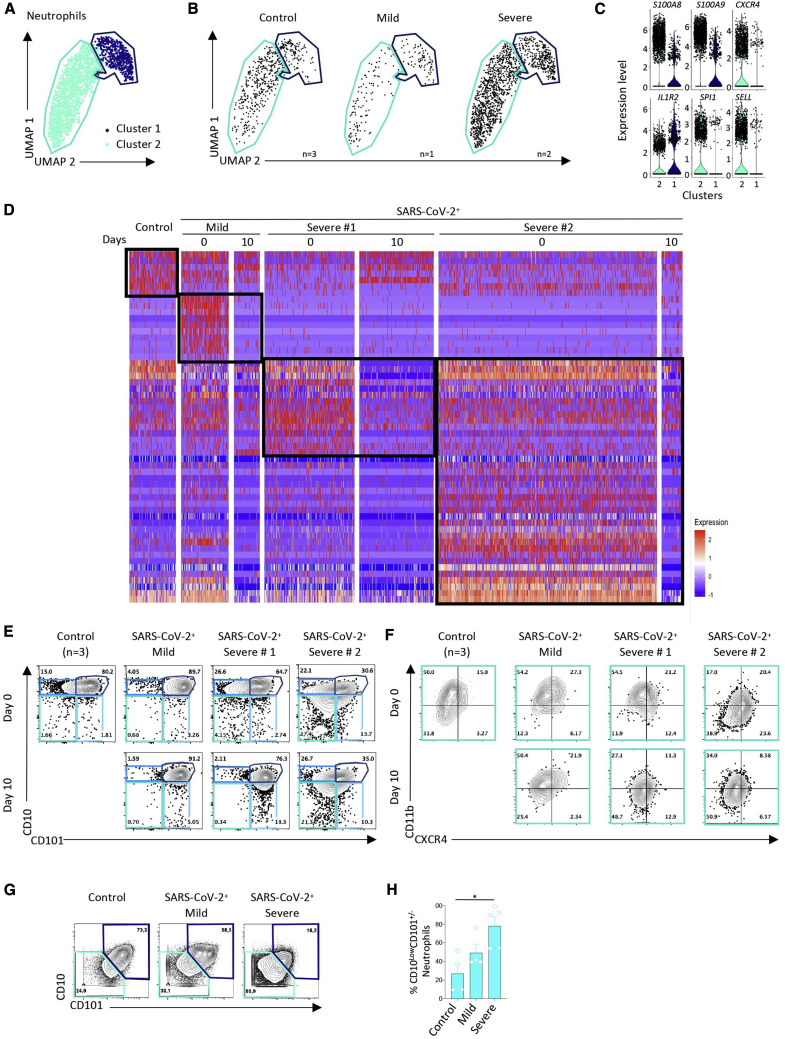
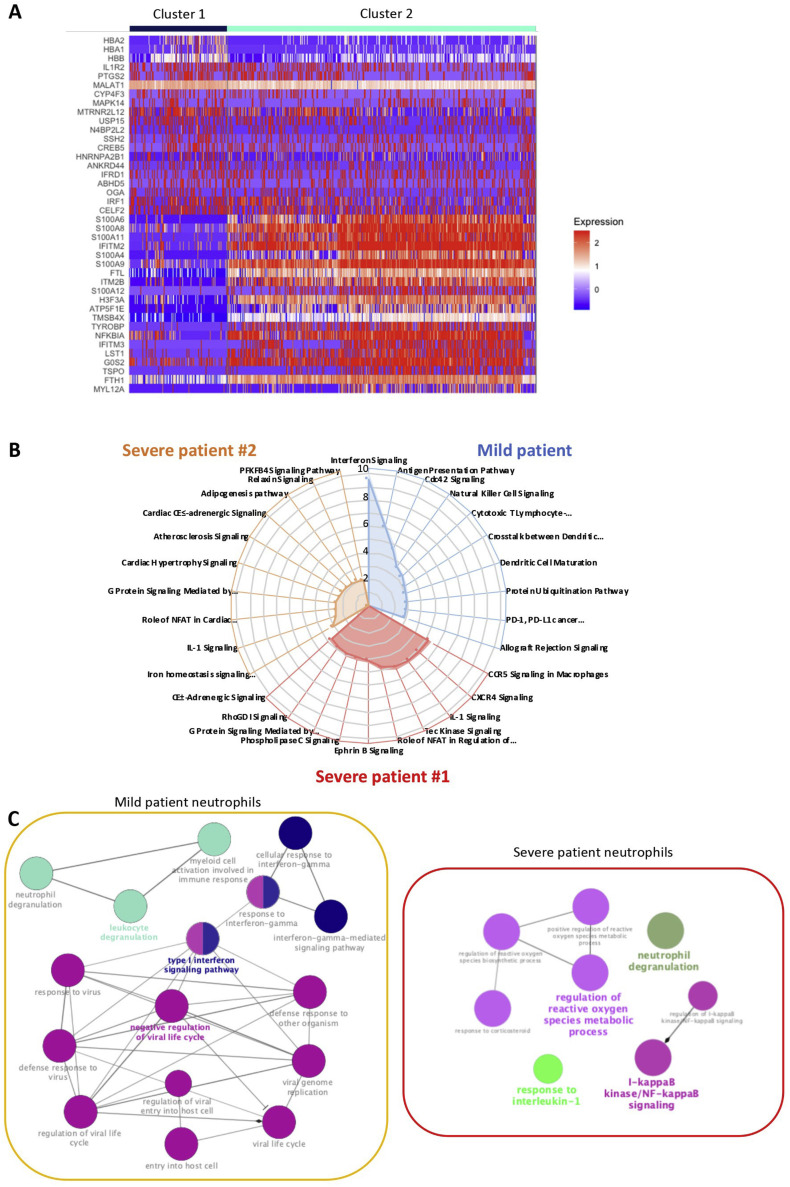
UMAP analysis of neutrophils identified two clusters (Figure 4 A). We observed an increase of cluster 2 cells in patients with severe COVID-19 (Figure 4B). Cluster 1 expressed the IL1R2 gene, whereas cluster 2 also expressed high levels of S100A8 and S100A9, CXCR4, SELL, and SPI1 (Figures 4C and S4 A). DEGs and pathway analyses in neutrophils of patients with mild disease informed about a type I interferon response at day 0 that was lost by day 10 (Figures 4D, S4B, and S4C). This signature was absent in controls and also in the two samples collected from patients with severe disease at later time points (Figure 4D), demonstrating high expression of genes involved in production of ROS, the inducible NOS pathway, IL-1 signaling, and NF-κB activation pathways (Figures S4B and S4C).
Figure 4.
Single-Cell Analysis of Neutrophils by scRNA-Seq, Spectral Flow Cytometry, and Mass Cytometry
(A) UMAP profile of neutrophils in the 9 samples analyzed as described in Figure 2A.
(B) UMAP profile of neutrophils within the 3 controls and the mild and the two severe cases with the cluster gates overlaid.
(C) Violin plots of expression of the indicated genes in two statistically defined neutrophil clusters.
(D) Heatmap of DEGs in total neutrophils generated as described in Figure 3B.
(E and F) Spectral flow analysis of neutrophil subsets in pooled controls and each individual patient sample at day 0 and day 10, based on CD10 and CD101 expression (E) and CXCR4 and CD11b expression among CD10LowCD101− neutrophils (F) in the indicated samples (pooled controls).
(G and H) Mass cytometry analysis of neutrophil subsets in 4 patients within each group (pooled data) as in Figures 3F–3I, based on CD10 and CD101 expression (G) and the fraction of CD10LowCD101– neutrophils among total neutrophils in each sample within the 3 groups (H). Kruskal-Wallis test, ∗p < 0.05.
Figure S4.
Neutrophil Analysis by scRNA-Seq, Spectral Flow Cytometry, and Mass Cytometry, Related to Figure 4 and Tables S3, S4, and S5
A. Heatmap of the top 20 DEGs defining two neutrophil clusters. B. Pathway analysis generated by comparing DEGs in neutrophils of each SARS-CoV-2 patient to the same population in the three control patients considered together using IPA software (mild patient in blue, severe #1 in red, severe # 2 in orange); C. The same DEGs identified in neutrophils were used to perform a gene ontology network analysis using clueGO software, considering the two severe patients together.
Analysis of the data collected by spectral flow cytometry of the same samples distinguished CD10+CD101+ mature neutrophils from CD10LowCD101− immature neutrophils. At day 0, the two patients with severe disease had more circulating CD10LowCD101− immature neutrophils compared with controls or the patient with mild disease (Figure 4E). Neutrophils from patient 1 with severe disease had increased expression of CD101 on their surfaces at day 10, whereas neutrophils from patient 2 with severe disease retained their immature phenotype at day 10. Focusing on expression of a pre-neutrophil hallmark, CXCR4, on the surface of CD10LowCD101− immature neutrophils (Ng et al., 2019), we observed an increase in the proportion of neutrophils with a CD10LowCD101−CXCR4+ phenotype, which, presumably, are pre-neutrophils (Figure 4F). Mass cytometry analysis of an independent cohort of 12 patients (four controls, four patients with mild COVID-19, and four patients with severe COVID-19; Table S5) again suggested a higher fraction of CD10LowCD101− immature neutrophils in patients with severe disease compared with control patients (Figures 4G and 4H).
Altogether, the results of these exploratory scRNA-seq experiments identified a transient type I interferon response in cells of a patient with mild disease and the presence of phenotypically immature subsets of monocytes and neutrophils in two patients with severe disease, which was further confirmed by mass cytometry.
Calprotectin Plasma Levels Distinguish Patients with Mild from Patients with Severe COVID-19
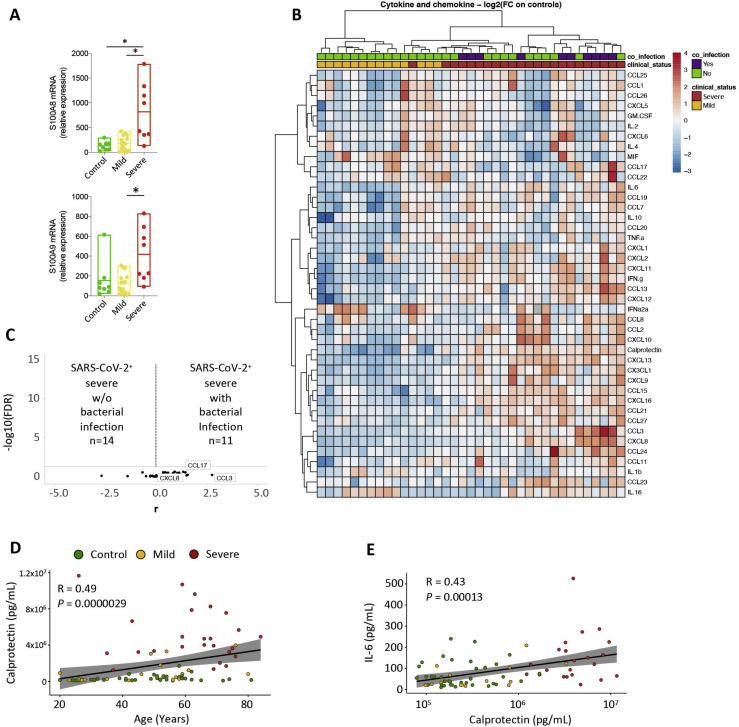
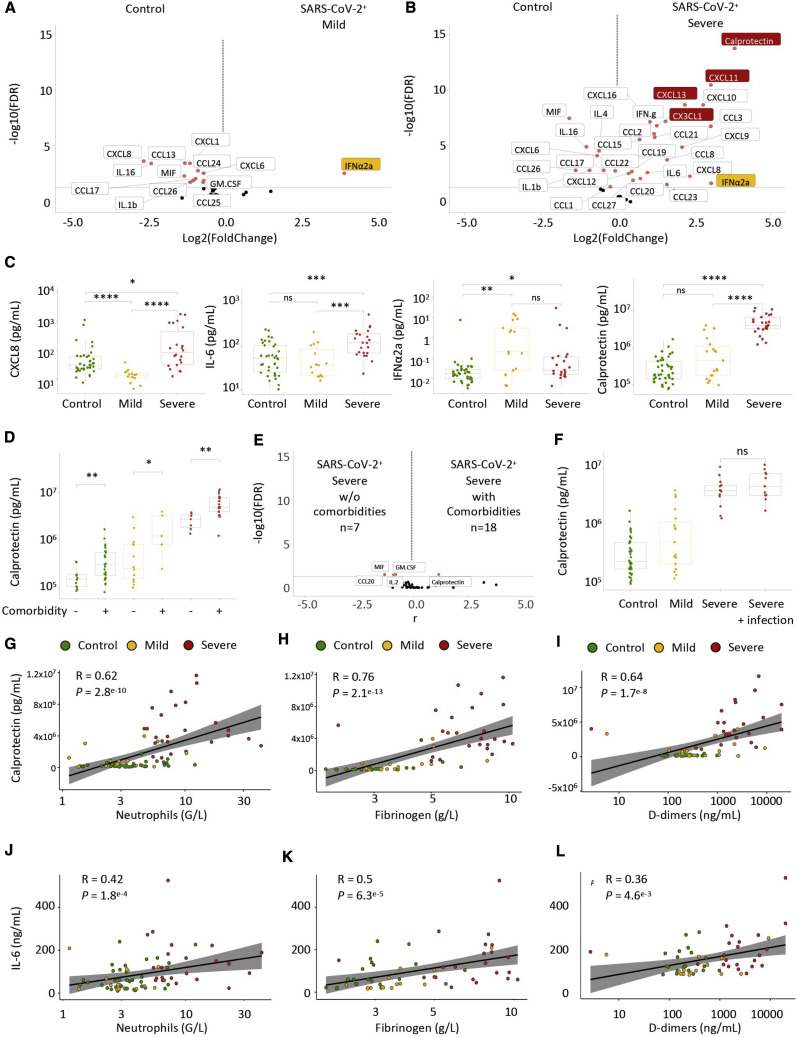
S100A8 and S100A9 alarmins, representing ∼45% of the cytoplasmic proteins in neutrophils, are released under inflammatory conditions and form a stable heterodimer known as “calprotectin” (Wang et al., 2018). In accordance with preliminary results generated by scRNA-seq (Figure S4A), qRT-PCR analysis detected higher expression of the S100A8 and S100A9 genes in peripheral blood nucleated cells of patients with severe COVID-19 (n = 8) compared with controls (n = 8) and patients with mild disease (n = 16) (Figure S5 A; Table S5). This led us to measure the plasma level of calprotectin, together with type I interferon (IFNα) and 40 other cytokines and chemokines, in samples from a cohort of 84 patients (Table S6). As seen in Figure 5 A, patients with mild disease showed significantly less CXCL8 (Figures 5C and S5B) and significantly more type I IFNα (Figures 5A and 5C) compared with controls. Patients with severe disease exhibited dramatically higher calprotectin levels compared with patients with mild COVID-19 or controls, without a further increase in IFNα plasma levels above mild disease levels (Figures 5B, 5C, and S5C). Calprotectin was the most significantly increased circulating biomarker in patients with severe disease, accompanied by a rise in 23 other tested chemokines and cytokines, including CXCL-8, CXCL-12, and IL-6 (Figures 5B, 5C, and S5B).
Figure S5.
Calprotectin Is the Most Abundant Immune Mediator/Immune Protein Detected in the Plasma of Patients with Severe COVID-19, Related to Figure 5 and Tables S5 and S6
A. RT-qPCR analysis of S100A8 and S100A9 gene expression in the three groups of patients, using HPRT as a control gene; B. Heatmap of cytokines, chemokines, IFNα2a and calprotectin plasma levels in 37 COVID-19 patients compared to 40 controls. SARS-CoV-2-positive patients included 14 mild and 23 severe patients. Associated bacterial infection at sample collection is indicated in purple. The heatmap shows z score-normalized concentrations, each column represents one patient and each row one protein; the color gradient from blue to red indicates increasing concentrations. Rows and columns are clustered using correlation distance and average linkage; C. Volcano-plot representation of cytokine levels in severe SARS-CoV-2 patients with (n = 11) or without (n = 14) bacterial infection at the time of sample collection; D. Spearman correlation between calprotectin concentration and age showing control patients in green, mild in orange and severe in red; E. Spearman correlation between IL-6 and calprotectin concentrations (color code as in D).
Figure 5.
Calprotectin Is the Most Abundant Immune Mediator Detected in the Plasma of Patients with Severe COVID-19
Shown are plasma levels of calprotectin (S100A8/S100A9), interferon alpha (IFNα2a), and 40 cytokines and chemokines in blood samples collected from 84 patients (controls, 40; mild disease, 18; moderate or severe disease, 25).
(A) Volcano plot of cytokine levels in patients with mild COVID-19 compared with controls; IFNα2a is shown in orange.
(B) Volcano plot of cytokine levels in patients with severe COVID-19 compared with control patients; IFNα2a is shown in orange. Calprotectin, CXCL11, CXCL13, and CX3CL1, shown in red, are most significantly associated with the severe forms.
(C) Circulating levels of CXCL8, IFNα2a, calprotectin, and IL-6 in individual samples in each group.
(D) Effect of comorbidities (Table S6) on calprotectin plasma levels in each group.
(E) Volcano plot of cytokine levels in patients with severe disease with and without comorbidities.
(F) Effect of bacterial infection on calprotectin plasma levels in each group.
(G–I) Spearman correlations between calprotectin plasma levels and neutrophil count (G), fibrinogen (H), and D dimers (I).
(J–L) Spearman correlations between IL-6 plasma levels and neutrophil count (J), fibrinogen (K), and D dimers (L).
Wilcoxon rank-sum test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Age and comorbidities (including obesity, diabetes mellitus, cardiovascular and respiratory diseases, and cancer) are predictors of severe COVID-19 disease (Richardson et al., 2020). We found that plasma calprotectin levels were significantly higher in control patients with comorbidities as well as in mild or severe COVID-19 patients with comorbidities (Figure 5D). Nevertheless, the increase in calprotectin in patients with severe COVID-19 far exceeds correlations associated with comorbidities. None of the other measured circulating proteins were significantly higher in patients with comorbidities (Figure 5E). Bacterial infections can occur in patients with severe COVID-19 (Chen et al., 2020c; Llitjos et al., 2020) and were present in some of our patients with severe disease but did not significantly modify the profile of released proteins (Figures S5B and S5C), including calprotectin (Figure 5F). No correlation between calprotectin and age was observed in any group of patients (Figure S5D). Calprotectin concentration correlated with neutrophil counts (Figure 5G), fibrinogen plasma levels (Figure 5H), and D dimers (Figure 5I), the latter being fibrin degradation products reflecting a hypercoagulability state. When modeling calprotectin plasma levels using multivariable linear regression to take into account potential confounding factors (age, sex, and comorbidities) and the correlation with neutrophil count, fibrinogen, and D dimers, these associations were still statistically significant (neutrophils, p = 1.154e−04; fibrinogen, p = 5.688e−05; D dimers, p = 2.099e−03). We also uncovered a weak correlation between IL-6 plasma concentration and levels of calprotectin (Figure S5E), blood neutrophil count (Figure 5J), fibrinogen (Figure 5K), and D dimers (Figure 5L), which disappeared after adjusted multilinear regression. Finally, a logistic regression including age, sex, and comorbidities together with biological parameters identified plasma levels of calprotectin, CX3CL1, CXCL11, and CXCL13 as the parameters that best discriminate controls/patients with mild COVID-19 from patients with severe disease.
These results indicate that high plasma levels of calprotectin are seen in patients with severe COVID-19 but not in those with mild disease. Importantly, this increase is independent of confounding factors for prognosis, such as advanced age, comorbidities, or concurrent bacterial infection, which have only minor effects on plasma calprotectin levels.
Spectral Flow Analyses Validate a Contrasted Innate Immune Cell Signature in Mild versus Severe COVID-19
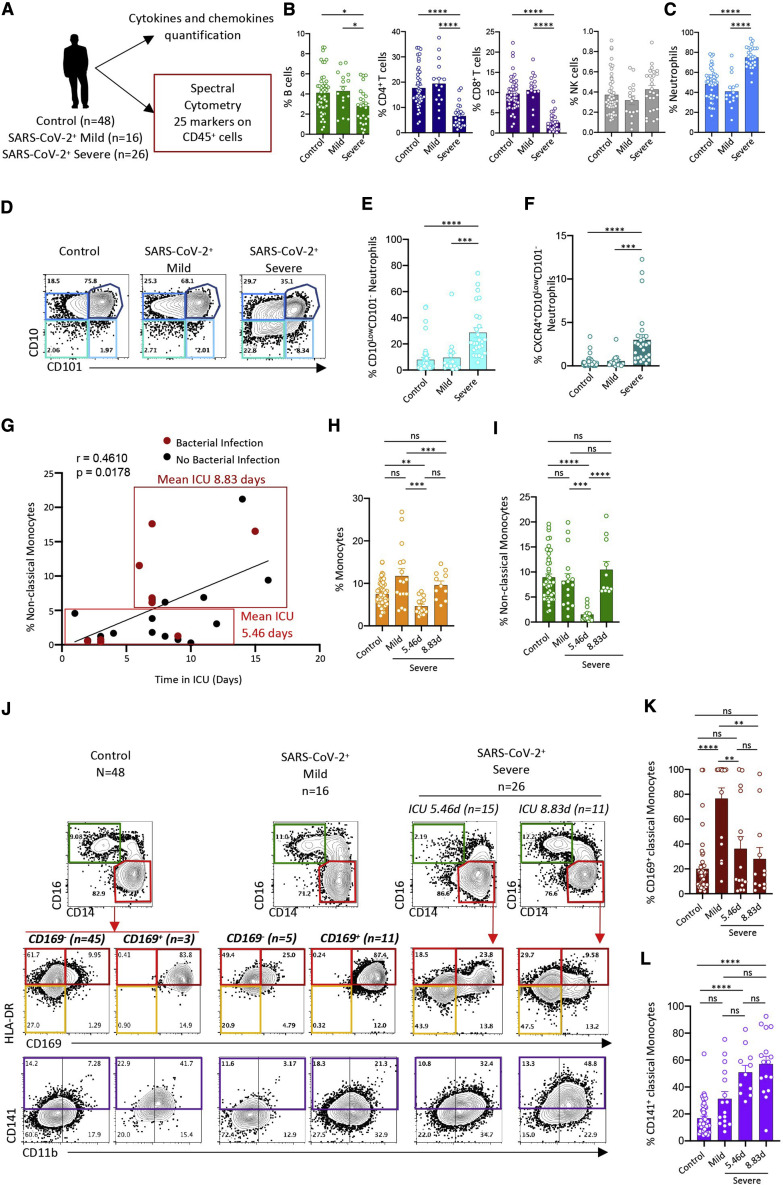
The hypothesis from the scRNA-seq-based identification of CD37, CD63 (LAMP3), CD169 (SIGLEC-1), and CD184 (CXCR4) biomarkers of blood cell subsets, whose relative proportions differ in patients with mild and severe COVID-19, prompted us to add antibodies targeting these proteins to the spectral flow cytometry panel. We applied this new panel to samples from an independent validation cohort of 90 patients. This cohort included 48 control patients and 42 COVID-19-positive patients, of whom 16 had mild disease and 26 had severe disease (Figure 6 A; Table S6). Non-supervised analysis and UMAP visualization identified the main cell populations in the three categories of patients combined (Figures S6 A and S6B). Analyzing patients individually confirmed the significant decrease in B cell, CD4+ T cell, and CD8+ T cell fractions in severe patients compared with the control and mild disease groups (Figure 6B), which may be a consequence of the increased neutrophil fraction (Figure 6C) and absolute numbers (Table S6). More specifically, within neutrophils, we observed a shift in CD10LowCD101− neutrophils (Figures 6D and 6E) and the subset of CD10LowCD101− neutrophils that express CXCR4 (CD10LowCD101−CXCR4+ cells) (Figure 6F) we observed previously in patients with severe disease. Finally, the fraction of CD10LowCD16Low neutrophils was also higher in patients with severe disease (Figure S6C), suggesting accumulation of immature neutrophils in the blood of patients with severe COVID-19.
Figure 6.
Validation of the Severe COVID-19 Innate Immune Signature
(A) Spectral flow pipeline.
(B) Fraction of B cells, CD4+ T cells, CD8+ T cells, and NK cells among total cells in individual samples (circles) in each group.
(C) Fraction of neutrophils in individual samples in each group.
(D) Neutrophil subsets identified by CD101 and CD10 expression in each group (pooled data).
(E) Fraction of CD10LowCD101− neutrophils among total neutrophils in individual samples within each group.
(F) Fraction of CXCR4+ neutrophils among CD10LowCD101– neutrophils as in (E).
(G) Spearman correlation of time spent in the ICU and non-classical monocyte fraction among total monocytes. Patients with a bacterial infection are shown as red dots. Mean time spent in the ICU was 5.46 days for patients with a low (≤5%) and 8.83 days in those with a high (>5%) CD14lowCD16High monocyte fraction.
(H) Fraction of classical monocytes among white blood cells in each individual sample in each group.
(I) Fraction of non-classical monocytes among total monocytes as in (H).
(J) Monocyte subset partition in each group (pooled data), with the severe group split in two groups based on mean time spent in the ICU. Top panels: monocyte subsets identified by CD14 and CD16 expression. Bottom panels: HLA-DR and CD169 or CD11b and CD141 expression in the classical monocyte subset.
(K) Fraction of monocytes expressing CD169 among classical monocytes as in (H).
(L) Fraction of monocytes expressing CD141 among classical monocytes as in (H).
Kruskal-Wallis test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Figure S6.
Validation of the Innate Immune Signature of Severe COVID-19, Related to Figure 6 and Table S6
A-B Non-supervised UMAP representation generated by pooling data from all the patient samples; cell identification (A) surface marker expression (B); C. Bar plots representing the percentage of CD10LowCD16Low neutrophils among all neutrophils in individual patients from each group in the validation cohort (n = 90). D. Spearman correlation between CD169 (SIGLEC-1) mean fluorescence intensity (MFI) and days spent by severe patients in ICU. E. Spearman correlation between CD169 (SIGLEC-1) mean fluorescence intensity (MFI) and plasma IFNα concentration; yellow, mild COVID-19 patients; red, severe COVID-19 patients. F.G. Bar plots representing the percentage of HLA-DRLow classical monocytes, B cells, CD4+ and CD8+ T cells and NK cells (F) and neutrophils among CD45+ cells, CD10LowCD101- neutrophils among all neutrophils and CD10LowCXCR4+ neutrophils among CD10LowCD101- neutrophils (G) in individual patients from each group in the validation cohort (n = 90). Kruskal-Wallis test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, non-significant.
scRNA-seq analyses of monocyte subsets indicated differential changes in the distribution of non-classical CD14LowCD16High monocyte fractions in the two patients with severe disease (Figure 3C). Because samples were collected from patients at various time points after admission to the ICU, we wanted to find out whether the duration of ICU stay affects monocyte subset distribution. In the 26 patients of this cohort with severe disease (Table S6), we observed a significant correlation between the time spent in the ICU and the fraction of the non-classical monocyte subset, irrespective of the presence or absence of concurrent bacterial infection (Figure 6G). The mean time spent in the ICU was 5.46 days for patients with fewer than 5% non-classical monocytes compared with 8.83 days for those with 5% and more non-classical monocytes (Figures 6H and 6I).
Then we examined other monocyte subsets. In the majority of patients with mild disease, we observed a fraction of classical monocytes that express CD169, which was decreased in patients with severe disease (Figures 6J, 6K, and S6D). CD169 expression correlated with IFNα plasma levels (Figure S6E). Independent of the time spent in the ICU, patients with severe disease also showed a larger fraction of classical monocytes expressing high levels of CD141 compared with controls (Figures 6J and 6L) and of monocytes expressing low levels of HLA-DR compared with controls and patients with mild disease (Figure S6F). Finally, the time spent in the ICU did not significantly affect repartition of lymphocyte populations or neutrophil subsets (Figures S6F and S6G).
Thus, patients with severe COVID-19 exhibited a transient decrease in non-classical monocyte frequencies, a stable decrease in HLA-DRLowCD141+ classical monocytes, and a major increase in CD10LowCD101−CXCR4+/− immature neutrophils.
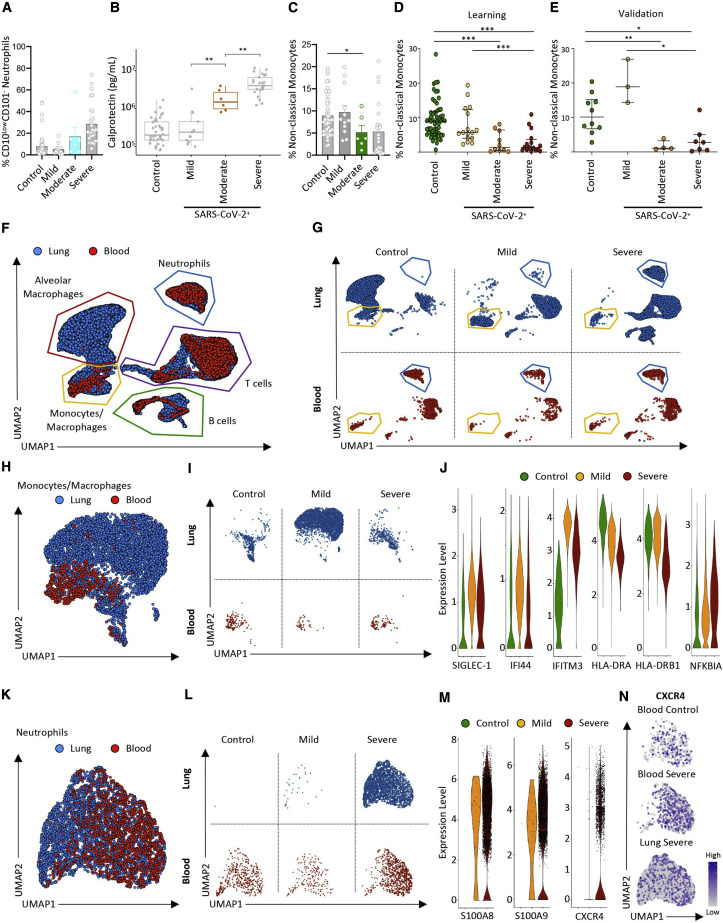
A High Calprotectin Level and Loss of Non-classical Monocytes Correlate with COVID-19 Severity
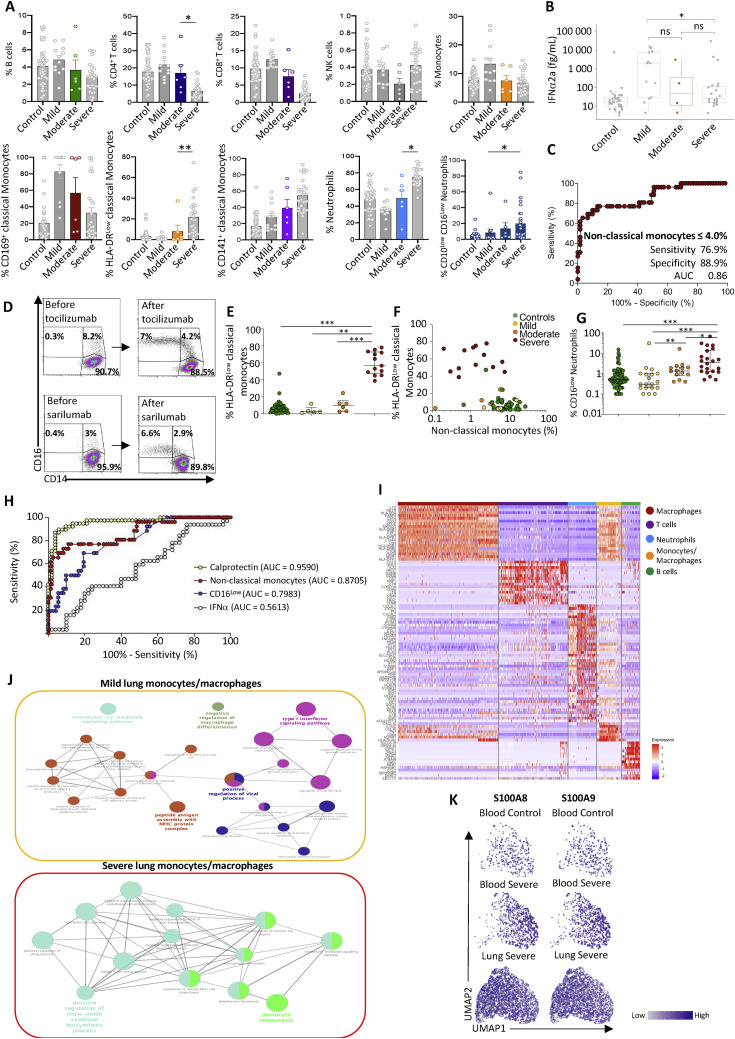
We next investigated whether changes in circulating myeloid cell phenotypes could be used to discriminate patients who develop severe COVID-19. Within our previous cohort, we separated patients with mild (n = 12) from patients with moderate (n = 6) and severe (n = 27) disease using clinical criteria. Patients classified as having “moderate” disease demonstrated intermediate changes between those of outpatients with mild disease and patients with severe disease in the ICU (Figure S7 A). The fraction of CD10LowCD101− neutrophils in patients with moderate disease was intermediate but not significantly different from any group (Figure 7 A). However, the amount of calprotectin measured in patients with moderate COVID-19 was significantly higher than in outpatients with mild disease but still significantly lower than in patients with severe COVID-19 (Figure 7B). In comparison, IFNα levels were not significantly different between patients with moderate and mild disease or patients with severe disease (Figure S7B). The difference in non-classical monocyte fraction was significant between patients with mild and moderate disease, dropping to levels comparable with patients with severe disease (Figure 7C).
Figure S7.
Changes in Innate Immune Cell Phenotype Are Detected in Patients with Moderate COVID-19, Related to Figure 7 and Tables S6 and S7
A. Bar plots representing the percentage of B cells, CD4+ T cells, CD8+ T cells, NK cells, total monocytes, CD169+, HLA-DRLow and CD141+ classical monocyte subsets, total neutrophils among CD45+ cells, and CD10LowCD101- and CD10LowCD16Low neutrophil subset among all neutrophils in individual patients from each group, with the moderate category (6 patients) highlighted. B. Plasma concentration of IFNα in moderate COVID-19 patients compared to the three other groups. C. ROC analysis showing performance of a diagnostic test using percentage of non-classical monocytes among total monocytes to distinguish controls and mild COVID patients from moderate and severe COVID patients; D. Monocyte subset analysis in the peripheral blood of 2 severe patients, before (left panels) and after (right panels) treatment with the indicated anti-IL-6 antibodies; E. Percentage of HLA-DRlow classical monocytes among classical monocytes in a cohort of 22 patients and 17 controls grouped into 4 clinical categories; F. Correlation between the percentage of HLA-DRlow classical monocytes and non-classical monocytes; G. Percentage of CD16low neutrophils among neutrophils in control and COVID-19 patients of the learning cohort described in Figure 7. H. ROC curves evaluating the discriminating significance of calprotectin plasma level (yellow), nonclassical monocyte fraction (red), CD16low circulating neutrophils (blue) and IFNα2a plasma level (white) between controls/mild patients and moderate/severe patients. AUC, Area Under the Curve; Mann Whitney test; I. Heatmap of blood and bronchoalveolar lavage fluid scRNaseq cells integrated defining the 5 regions of cell populations; J. Pathway analysis (Cytoscape and ClueGo) of DEGs expressed at a higher level in bronchoalveolar monocytes/macrophages from mild versus severe patients. K. UMAP analysis of neutrophils with S100A8 (left panel) and S100A9 (right panel) gene expression level projection (low expression = gray dots; high expression = dark blue dots). Kruskal-Wallis test, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Figure 7.
Low-Dimensional Flow Analysis of Non-classical Monocyte Subsets in COVID-19
(A) Fraction of CD10LcowCD101− neutrophils among total neutrophils in individual samples within each group, separating moderate and severe COVID-19.
(B) Calprotectin plasma levels in patients with moderate disease (orange dots) compared with patients of the three other groups.
(C) Fraction of non-classical monocytes among total monocytes in patients with moderate disease (green bar plot) compared with patients of the three other three groups.
(D and E) Fraction of non-classical monocytes among total monocytes in a learning cohort of 98 patients (controls, n = 56; mild, n = 16; moderate, n = 10; severe, n = 16) (D) and a validation cohort of 24 patients (controls, n = 10; mild n = 3; moderate, n = 4; severe, n = 7) (E). Mann-Whitney test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(F–M) Integration of scRNA-seq data from blood and lung (BALF) cells of COVID-19 patients. Blood samples are described in Tables S3 and S4. Lung data are from Liao et al. (2020) (controls, n = 3; mild, n = 3; severe, n = 6).
(F) UMAP analysis of integrated scRNA-seq from blood and lung samples.
(G) UMAP analysis of blood and lung samples in each patient category.
(H) UMAP analysis of integrated scRNA-seq data of monocytes/macrophages from blood and lungs.
(I) UMAP analysis of blood and lung monocytes/macrophages in each patient group.
(J) Violin plot of gene expression in lung monocytes/macrophages of control patients and patients with mild and severe disease.
(K) UMAP analysis of integrated scRNA-seq data of neutrophils from blood and lung samples.
(L) UMAP analysis of blood and lung neutrophils in each patient group.
(M) Violin plot of gene expression in lung neutrophils of control patients and patients with mild and severe COVID-19.
(N) UMAP analysis of neutrophils with CXCR4 gene expression level projection (low expression, gray dots; high expression, dark blue dots).
Thus, we hypothesized that the decreased non-classical monocyte fraction could be used as a fast and simple diagnostic test to distinguish moderate from mild COVID-19, especially in patients in whom clinical symptoms may be substantially overlapping. This would facilitate rapid and accurate identification of patients currently classified as having mild disease at the cusp of potentially progressing to more severe disease. We therefore employed a low-dimensional flow cytometry approach that measures the fraction of classical (CD14HighCD16Low), intermediate (CD14HighCD16High), and non-classical (CD14LowCD16High) monocyte subsets among total peripheral blood monocytes and applied it initially to a learning cohort of 98 patients, consisting of 16 patients with mild disease, 10 with moderate disease, and 16 with severe COVID-19, along with 56 controls (Table S7). All hospitalized patients were sampled within 10 days of admission to limit the potential effect of time spent in the ICU (see Figure 6G); the mean time spent in the ICU was 5.5 days at the point of sampling. The cohort also included 56 controls. Patients with mild disease showed a fraction of non-classical monocytes similar to that observed in controls. In contrast, moderate patients showed lower levels of non-classical monocytes, as observed in patients with severe disease (Figure 7D). To measure the global performance of this test, we used a receiver operating characteristic (ROC) curve (Hajian-Tilaki, 2013). The point of the ROC curve corresponding to the best sensitivity/specificity compromise indicated that a non-classical monocyte fraction below 4% separated patients with moderate or severe COVID-19 from those with mild or no disease with 76.9% sensitivity (95% bootstrap confidence interval [BCI] [61.5%, 92.3%]) and 89% specificity (95% BCI [80.6%, 95.8%]) (Figure S7C).
We then applied these analyses to blood samples from an independent validation cohort of 24 hospitalized patients from a different clinical center (10 controls, 3 patients with mild disease, 4 with moderate disease, and 7 with severe COVID-19) (Table S7). A non-classical monocyte fraction below 4% of total circulating monocytes, as defined in the learning cohort, also segregated those with mild disease from those with moderate and severe disease with high sensitivity (81.8%, 95% BCI [72.7%, 100%]) and specificity (92.3%, 95% BCI [83.3%, 100%]) (Figure 7E). These results confirmed the specificity and sensitivity of our assay to discriminate patients with different COVID-19 severity.
Further confirming these observations, serial sampling of two patients with severe disease who responded to anti-IL-6R antibodies documented that their clinical recovery was associated with reappearance of non-classical monocytes in the blood (Figure S7D). One patient who was initially referred with limited symptoms (atypical thoracic pain) and was SARS-Cov-2 PCR negative unexpectedly exhibited a low fraction of non-classical monocytes (3.4%) accompanied by 10% HLA-DRLow classical monocytes. The following day, pulmonary symptoms appeared and the patient was hospitalized, requiring oxygen therapy, and a lung CT scan revealed characteristic COVID-19-associated injury. Such cases suggest that loss of the non-classical monocyte fraction could be a strong indicator of existing or impending severe COVID-19.
Additional informative parameters could be added to this flow assay to increase its specificity to identify transition to severe COVID-19, including decreased expression of HLA-DR on the surface of classical monocytes (Figure S7E), which is associated with a decrease in the non-classical monocyte fraction below 4% (Figure S7F), and an increase in the fraction of CD16Low neutrophils (Figure S7G). Comparison of ROC curves indicated that calprotectin plasma level and monocyte or neutrophil subset analyses distinguished mild COVID-19 in outpatients from moderate or severe disease in hospitalized patients whereas IFNα2a plasma levels did not (Figure S7H).
Together with calprotectin plasma levels, flow identification of a decrease in the non-classical monocyte fraction below 4% of total monocytes could provide improved resolution for clinical observations when categorizing patients at the border of mild and moderate/severe COVID-19. This would potentially identify individuals at greatest risk of rapid decline and highlight the need for pro-active management/intervention and intensive monitoring. This assay could be reinforced by analysis of HLA-DRLow classical monocyte and CD16Low neutrophil fractions.
Integration of Lung and Blood scRNA-Seq Reveals Abnormal Myeloid Cell Populations Discriminating Severe and Mild COVID-19
The lungs are major organs affected in patients with severe COVID-19. To better understand how the distinctive cell signatures found in the blood of patients with severe COVID-19 affect immune cell compartments in the lungs, particularly the presence of immature neutrophils and HLA-DRLow monocytes, we integrated our dataset using the Seurat V3 pipeline (Stuart et al., 2019) with the published scRNA-seq dataset of cells from 12 bronchoalveolar lavage fluid samples (BALFs) of controls (n = 3) and patients with mild (n = 3) and severe (n = 6) COVID-19 (Liao et al., 2020; Database: GSE145926). This analysis provided an unbiased global map of immune cells in the blood and BALF of controls and patients with mild and severe COVID-19. Using dimensional reduction, we identified 5 regions based on DEGs across pooled data from all samples (Figures 7F and S7I), including T cells (characterized by expression the NKG7, CD8A, CST7, GZMB, and GZMA genes), B cells (IGLV3-19, IGHV4-34, IGHG1, IGHA1, and JCHAIN), neutrophils (G0S2, RSAD2, IL1R2, and IL1RN), alveolar macrophages (APOE, MSR1, MARCO, and FBP1), and monocytes/macrophages (FN1, CXCL10, CD68, and NUPR1). Validating this approach, the alveolar macrophage region was mainly present in BALF of control patients but decreased dramatically in patients with mild and severe COVID-19,and only one cell from our blood scRNA-seq matched in this region (Figures 7F and 7G). We also observed changes in the monocyte/macrophage region of BALF from patients with mild or severe disease versus controls and dramatic neutrophil accumulation in patients with severe disease (Figure 7G).
Monocytes/macrophages were increased in BALF of the mild compared with the control and severe groups (Figures 7H and 7I), and these cells were characterized by expression of the interferon-stimulated genes (ISGs) (SIGLEC-1, IFI44, and IFITM3) (Figure 7J), with pathway analyses indicating upregulation of the viral replication and type I IFN signaling pathways. In contrast, the NOS biosynthetic process and monocyte chemotaxis were upregulated in BALF monocytes/macrophages of patients with severe disease (Figure S7J), which, similar to blood monocytes, expressed lower levels of HLA-DRA and HLA-DRB1 and higher levels of NFKBIA mRNA compared with controls or patients with mild COVID-19 (Figure 7J). Finally, neutrophils were present at high frequencies in BALF from patients with severe COVID-19 but not in BALF from controls or patients with mild disease (Figures 7K and 7L). UMAP integration of samples from patients with severe disease samples indicated that BALF neutrophils, similar to blood neutrophils (Figure 7L), were characterized by high expression of S100A8, S100A9, as well as CXCR4, indicating an immature state (Figures 7M, 7N, and S7K).
Integration of blood and BALF myeloid cells identified, in patients with severe COVID-19, loss of HLA-DRA and HLA-DRB1 and high NFKBIA expression in monocytes/macrophages (not including alveolar macrophages) together with accumulation of neutrophils expressing high levels of S100A8/A9 and CXCR4.
Discussion
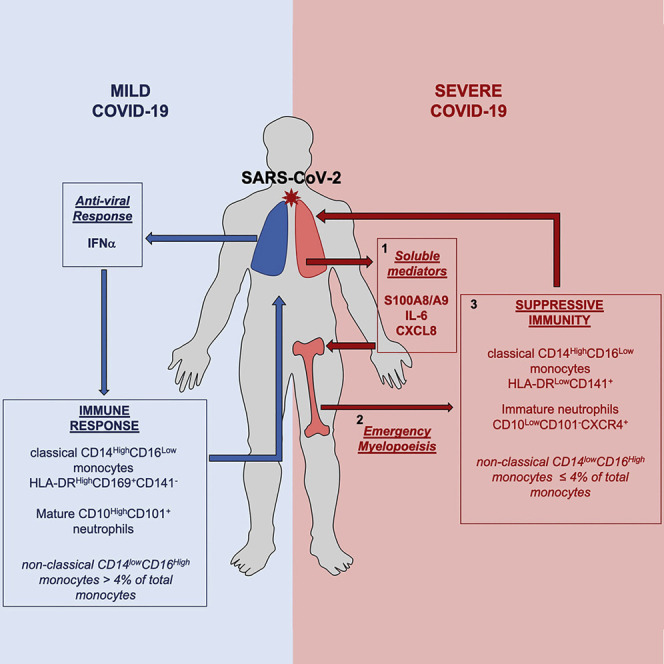
This study presents evidence that patients who develop severe COVID-19 exhibit high levels of calprotectin and inflammatory cytokines and chemokines, correlating with emergency myelopoiesis generating ROS- and NOS-expressing immunosuppressive myeloid cells (HLA-DRLow monocytes and immature subsets of neutrophils).
The first line of defense in virus-infected patients typically involves a protective innate response incorporating transient and strong production of type I IFNs. By inducing expression of ISGs, type I IFNs inhibit virus replication and promote an effective innate and adaptive immune response (Thevarajan et al., 2020; Totura and Baric, 2012). This antiviral response may be impaired in COVID-19 patients who suddenly develop clinically life-threatening disease (Hadjadj et al., 2020). Severe COVID-19 frequently develops in the context of advanced age and comorbidities, which provide a degree of underlying systemic chronic inflammation (Furman et al., 2019). Such inflammation could disrupt the timing of the type I IFN response relative to the kinetics of virus replication (Terán-Cabanillas and Hernández, 2017), which has been shown to be critical in mouse models of coronavirus infection (Channappanavar et al., 2019). An imbalance between type I IFN and inflammatory responses could also be favored by the highly efficient replication of SARS-CoV-2 in human tissues (Chu et al., 2020) and by the IFN-neutralizing effects of structural and non-structural viral components shared between SARS-CoV-2 and other virulent human coronaviruses (Chen et al., 2014; Yang et al., 2015).
Patients with severe COVID-19 exhibit abnormal partition of circulating monocytes and of neutrophils expressing the S100A8 (calgranulin A/myeloid-related protein 8) and S100A9 (calgranulin B/myeloid-related protein 14) alarmin genes. Importantly, accumulation of neutrophils expressing high levels of the S100A8/A9 genes was also observed in the BALF of these patients. The release of massive amounts of calprotectin, the heterodimer formed by S100A8 and S100A9 proteins, is a striking event associated with severe COVID-19. This heterodimer promotes cell migration and boosts NADPH (nicotinamide adenine dinucleotide phosphate) oxidase activity. Calprotectin is a TLR4 and RAGE (receptor for advanced glycation end products) ligand that, upstream of tumor necrosis factor alpha (TNF-α) (Vogl et al., 2018) and CXCL8 (Simard et al., 2014) synthesis and secretion, promotes NF-κB activation (Riva et al., 2012) and secretion of multiple inflammatory proteins, such as IL-6 (Wang et al., 2018). Thus, we propose that calprotectin may account for and possibly trigger the cytokine release syndrome that characterizes severe COVID-19. Its production may be amplified by tissue damage, generating a harmful hyperinflammation loop (Kuipers et al., 2013) that precludes these peptides from exerting more protective functions (Austermann et al., 2014; Freise et al., 2019; Ulas et al., 2017; Vogl et al., 2018). Chronic inflammation from comorbidities may synergize with SARS-CoV-2 infection to induce systemic release of calprotectin, which translates into upregulation of NF-κB and loss of HLA-DR on classical monocytes and the presence of immature neutrophils, converging to a state of chronic inflammation-induced immunosuppression. Abnormal neutrophils have been observed previously in patients with severe COVID-19 (Wilk et al., 2020). However, the authors concluded that these neutrophils transdifferentiate from B cells. We have no supporting results suggesting that this could be the case.
Under healthy conditions, roughly 85% of total circulating monocytes are CD14HighCD16LowHLA-DRHigh cells that are rapidly recruited to inflamed tissues (Guilliams et al., 2018). As in other severe illnesses (Lukaszewicz et al., 2009), expression of HLA-DR on CD14High circulating monocytes is low in severe COVID-19, which correlates with, and could be mediated by, IL-6 overproduction (Giamarellos-Bourboulis et al., 2020). A more specific feature of COVID-19 is the low fraction of CD14LowCD16High non-classical monocytes. This fraction commonly increases in patients with sepsis and inflammatory diseases, including viral infections (Kratofil et al., 2017). The decrease in non-classical monocyte fraction could involve the ability of calprotectin to hasten trans-endothelial migration of leucocytes (Fassl et al., 2015), unless these cells strongly adhere to the endothelium or conversion of classical into non-classical monocytes is stuck (Hanna et al., 2011; Hofer et al., 2015; Selimoglu-Buet et al., 2018). Whatever the mechanism, the lower-than-normal frequencies of non-classical monocytes (Thevarajan et al., 2020; Hadjadj et al., 2020) suggest a SARS-CoV-2-characteristic effect that is not observed in other viral infections. Most importantly, this decrease generates a highly characteristic biological signature of COVID-19’s aggressive form with the potential to be measured easily using standard diagnostic flow cytometry and provide information regarding the real-time immunological severity of the infection.
The burst of calprotectin detected in COVID-19 patients may trigger NF-κB-driven emergency myelopoiesis, generating immature and dysplastic cells (Basiorka et al., 2016; Chen et al., 2013). Given the considerable hematopoietic potential of the lungs (Lefrançais et al., 2017), the burst of calprotectin could also promote the contribution of lung megakaryocytes to disease pathogenesis in these organs. Whatever the mechanism, the immature and mature cells released into the peripheral blood by emergency myelopoiesis may be endowed with immunosuppressive functions, suggesting that myeloid-derived suppressive cells (MDSCs), as detected in cancer, inflammation, and other diseases (Veglia et al., 2018), might be important in COVID-19. In addition to HLA-DRLow monocytes, whose phenotype is that of monocytic MDSCs (M-MDSCs), CD10LowCD101−CXCR1+ immature cells are reminiscent of granulocytic MDSCs (G-MDSCs) (Aarts et al., 2019; Mastio et al., 2019; Veglia et al., 2018). Thus, neutrophil precursors, such as the pre-neutrophil (preNeu) population, which is CXCR4 positive (Evrard et al., 2018), may be released prematurely into the blood from the bone marrow and infiltrate the lung tissue in patients with severe disease. Emergence of these populations could be a predictor of the switch to severe disease. Further research will be required to determine their specific role in disease development.
We observed that severe COVID-19 is specifically associated with (1) a burst of circulating calprotectin that precedes cytokine release syndrome, (2) low levels of non-classical monocytes in the peripheral blood, and (3) emergency myelopoiesis, which releases immature and dysplastic myeloid cells with an immune-suppressive phenotype. Monitoring calprotectin plasma levels and non-classical monocyte in the blood of patients could be implemented routinely in the lab to discriminate patients with early immunological signs consistent with developing more severe disease, as suggested recently (Chen et al., 2020b). Finally, in addition to the network of potential drug targets depicted recently by analysis of SARS-CoV-2 interactions (Gordon et al., 2020), our work provides a further rationale for testing several clinical strategies, including blocking emergency myelopoiesis with lenzilumab (NCT04351152), a recombinant anti-human granulocyte-macrophage colony-stimulating factor (GM-CSF) antibody (Patnaik et al., 2020); testing the oral quinoline-3-carboxamide tasquinimod (Fizazi et al., 2017) and related molecules, such as ABR-215757 (paquinimod), which block binding of S100A9 to TLR4 and RAGE (Kraakman et al., 2017; Raquil et al., 2008); and preclinical anti-CD33 monoclonal antibodies (Walter, 2018), which may prevent interaction of S100A9 with myeloid progenitors (Eksioglu et al., 2017).
Limitations of Study
These analyses provide snapshots of the differences in innate immune cell phenotype and calprotectin plasma levels between outpatients with mild disease at the time of sampling, having no or limited clinical symptoms and not requiring a CT scan or hospitalization, and patients with moderate to severe disease whose clinical situation requires hospitalization and, in most cases, oxygen supply. Although all statistical analyses indicate that these biomarkers efficiently discriminate these two clinical situations and may help with urgent patient triage, a serial analysis is now required to evaluate how these biomarkers can predict the switch from mild to moderate or severe COVID-19 and inform on the mechanisms involved in this switch.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD195/CCR5 | BD | Cat# 560635 |
| CD10 | eBiosciences | Cat# 15-0106-42 |
| CD101 | BioLegend | Cat# 331010 |
| CD117 (ckit) | BD | Cat# 746848 |
| CD11b | BD | Cat# 740965 |
| CD11c | BD | Cat# 563130 |
| CD14 | BioLegend | Cat# 301830 |
| CD141 | BioLegend | Cat# 740604 |
| CD15 | BioLegend | Cat# 323022 |
| CD16 | BD | Cat# 564653 |
| CD163 | BD | Cat# 562670 |
| CD169 | BioLegend | Cat# 346004 |
| CD19 | BD | Cat# 612916 |
| CD3 | BD | Cat# 566575 |
| CD36 | BioLegend | Cat# 336208 |
| CD37 | BioLegend | Cat# 356304 |
| CD38 | BD | Cat# 562665 |
| CD4 | BD | Cat# 612887 |
| CD40 | BD | Cat# 565179 |
| CD45 | BD | Cat# 563792 |
| CD56 | BD | Cat# 566400 |
| CD63 | BioLegend | Cat# 353010 |
| CD64 | R&D | Cat# FAB12571N-100UG |
| CD66b | Beckman Coulter | Cat# IM0531U |
| CD8 | BD | Cat# 563919 |
| CD95 | BD | Cat# 740589 |
| CD181/CXCR1 | BioLegend | Cat# 320608 |
| CD184/CXCR4 | BioLegend | Cat# 306526 |
| FcεRI | BD | Cat# 749338 |
| CD130/GP130 | BioLegend | Cat# 362005 |
| HLA-DR | BD | Cat# 561358 |
| CD126/IL-6Rc | BioLegend | Cat# 352810 |
| CD284/TLR4 | BD | Cat# 564404 |
| CD41 | Fluidigm | Pro# 3089004B |
| CD45 | Fluidigm | Pro# 3141009B |
| CD19 | Fluidigm | Pro# 3142001B |
| CD45RA | Fluidigm | Pro# 3143006C |
| CD42 | Fluidigm | Pro# 3144020B |
| CD4 | Fluidigm | Pro# 3145001B |
| CD8 | Fluidigm | Pro# 3146001B |
| CD303 | Fluidigm | Pro# 3147009B |
| CD16 | Fluidigm | Pro# 3148004B |
| CD34 | Fluidigm | Pro# 3149013B |
| CD123/IL-3R | Fluidigm | Pro# 3151001B |
| CD66b | Fluidigm | Pro# 3152011B |
| CD192/CCR2 | Fluidigm | Pro# 3153023B |
| CD3 | Fluidigm | Pro# 3154003B |
| CD36 | Fluidigm | Pro# 3155012B |
| CD10 | Fluidigm | Pro# 3156001B |
| CD101 | Fluidigm | Pro# 3158020C |
| CD11c | Fluidigm | Pro# 3159001B |
| CD14 | Fluidigm | Pro# 3160006B |
| CD90 | Fluidigm | Pro# 3161009B |
| CD33 | Fluidigm | Pro# 3163023B |
| CD95/Fas | Fluidigm | Pro# 3164008B |
| CD163 | Fluidigm | Pro# 3165017B |
| Pp65/PNFκB | Fluidigm | Pro# 3166006A |
| CD38 | Fluidigm | Pro# 3167001B |
| CD71 | Fluidigm | Pro# 3168014B |
| CD40 | BioLegend | Cat# 334302 |
| CX3CR1 | Fluidigm | Pro# 3172017B |
| Granzyme B | Fluidigm | Pro# 3173006C |
| HLA-DR | Fluidigm | Pro# 3174001B |
| CD56 | Fluidigm | Pro# 3176008B |
| CD11b | Fluidigm | Pro# 3209003B |
| Biological Samples | ||
| Human blood samples | APHP-Paris | Tables S1–S3, S4, and S5–S7 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cytodelics | Cytodelics | hC001-1000 |
| Cytofix/Cytoperm | BD | 554714 |
| BSA | GE Healthcare Hyclone | SH30574.02 |
| EDTA | SIGMA | 3690 |
| PBS | ThermoFisher | 20012068 |
| RNase OUT Recombinant Ribonuclease Inhibitor, 40U/mL | Invitrogen | 10777019 |
| Chromium Single Cell 3′ Solution v3 | 10x Genomics | PN-1000075 |
| RNeasy Mini Kit | QIAGEN | 74104 |
| reverse transcribed with SuperScript IV VILO Master Mix with ezDNase Enzyme | Invitrogen | 11766050 |
| Power SYBR Green | Applied Biosystems | 4368577 |
| Foxp3/Transcription Factor Staining Buffer kit | eBiosciences | 00-5523-00 |
| Maxpar Cell Acquisition Solution | Fluidigm | 107950 |
| EQ Beads | Fluidigm | 201078 |
| Bio-Plex Pro™ Human Chemokine Panel 40-plex Assay | Bio-rad | 171AK99MR2 |
| R-plex Human Calprotectin Antibody Set | Meso Scale Discovery | F21YB-3 |
| ultra-sensitive assay S-plex Human IFNa2a kit | Meso Scale Discovery | K151P3S-1 |
| 20-Plex Pd barcoding kit | Fluidigm | 201060 |
| Deposited Data | ||
| COVID Blood cells | ArrayExpress Archive of Functional Genomics Data | E-MTAB-9221 |
| COVID BALF cells (Liao et al., 2020) | GEO | GSE145926 |
| Oligonucleotides | ||
| GUS foward | GAAAATATGTGGTTGGAGAGCTCATT | N/A |
| GUS reverse | CCGAGTGAAGATCCCCTTTTTA | N/A |
| HPRT foward | GGACAGGACTGAACGTCTTGC | N/A |
| HPRT reverse | CTTGAGCACACAGAGGGCTACA | N/A |
| S100A8 foward | CAACACTGATGGTGCAGTTAACTTC | N/A |
| S100A8 reverse | CTGCCACGCCCATCTTTATC | N/A |
| S100A9 foward | CTGAGCTTCGAGGAGTTCATCA | N/A |
| S100A9 reverse | CGTCACCCTCGTGCATCTTC | N/A |
| Software and Algorithms | ||
| Bio-Plex Manager 6.1 Software | Bio-Rad | https://www.bio-rad.com/fr-fr/product/bio-plex-manager-software-standard-edition?ID=5846e84e-03a7-4599-a8ae-7ba5dd2c7684 |
| CyTOF Software v.6.7.1014 | Fluidigm | https://www.fluidigm.com/binaries/content/documents/fluidigm/resources/cytof-software-6.7-rl-400314/cytof-software-6.7-rl-400314/fluidigm%3Afile |
| FlowJo v6.05 | FlowJo | https://www.flowjo.com/ |
| Cytoscape | Shannon et al., 2003 | https://cytoscape.org/ |
| GraphPad Prism 7 | GraphPad Software | https://www.graphpad.com/ |
| Ingenuity Pathway Analysis | Krämer et al., 2014 | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/content-exploration-and-databases/qiagen-ipa/ |
| STAR aligner | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Seurat v3 | Stuart et al., 2019 | https://satijalab.org/seurat/install.html |
Resource Availability
Lead Contact
Further information and request for resources and reagents should be directed to and will be fulfilled by the lead contact: Florent_Ginhoux@immunol.a-star.edu.sg (F.Gi.).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All blood scRNA-seq data used in this study can be accessed by ArrayExpress Archive of Functional Genomics Data under the accession number E-MTAB-9221. Integrated BALF scRNA-seq data can be accessed in GEO under the accession number GSE145926.
Experimental Model and Subject Details
Patients
This non-interventional study was approved by institutional review boards of Cochin-Port Royal (Paris, France) and Gustave Roussy (Villejuif, France) hospitals and the ethical committee of Cochin-Port Royal Hospital (CLEP Decision N°: AAA-2020-08023), and conformed to the principles outlined in the Declaration of Helsinki. Controls (n = 72) were symptomatic patients who were seen at Hôtel-Dieu or Gustave Roussy COVID-19 screening unit and were negative for SARS-CoV-2 RT-PCR on pharyngeal swab. Mild COVID-19 patients (n = 27) were defined by having limited clinical symptoms (fever, cough, diarrhea, myalgia, anosmia/ageusia) that did not require CT-scan or hospitalization. Moderate cases (n = 16) were defined as symptomatic patients with dyspnea and radiological findings of pneumonia on thoracic CT scan, requiring hospitalization and a maximum of 9 L/min of oxygen. In the larger part of this study, mild and moderate cases were analyzed together and grouped under “mild category.” Severe patients (n = 43) were those hospitalized in the ICU with respiratory distress requiring 10L/min of oxygen or more, without or with endotracheal intubation and mechanical ventilation.
Method Details
Sampling
Whole human peripheral blood was collected into sterile vacutainer tubes containing EDTA or heparin. Except for single cell RNA sequencing, tubes were centrifuged at 300 g for 5 min at room temperature and plasma was collected. Whole blood was mixed at a 1:1 ratio with Whole Blood Cell Stabilizer (Cytodelics), incubated at room temperature for 10 min and transferred to −80°C freezer to await analysis. These samples were secondarily thawed in a water bath set to +37°C. Cells were fixed at a ratio 1:1 with Fixation Buffer (Cytodelics, ratio 1:1) and incubated for 10 min at room temperature. Red blood cells were lysed by addition of 2 mL of Lysis Buffer (Cytodelics, ratio 1:4) at room temperature for 10 min. White blood cells were washed with 2 mL of Wash Buffer (Cytodelics, ratio 1:5).
Spectral flow Cytometry
Cells were resuspended in 100 μL extra-cellular antibody cocktail and incubated at room temperature for 15 min. For intra-cellular labeling, a step of permeabilization was performed using 200 μL of BD Cytofix/Cytoperm Kit (BD); cells were then incubated for 40 min at +4°C, washed in Perm Buffer (BD) and resuspended in intra-cellular antibody cocktail. After incubation, cells were washed in Flow Cytometry Buffer (1% BSA, 0.5% Na-Azide and 0.5M EDTA in PBS) and resuspended to proceed to the acquisition. All antibodies are listed in the Key Resources Table. Samples were acquired on CyTEK Aurora flow cytometer (Cytek Biosciences). Fcs files were exported and analyzed using FlowJo software.
3′ scRNaseq analysis of human blood cells
To fully capture peripheral blood cell heterogeneity, we analyzed fresh samples without cell sorting or freezing and without Ficoll enrichment, minimizing time of incubation and processing. Sample preparation was done at room temperature. After red cell lysis, single-cell suspensions were loaded onto a Chromium Single Cell Chip (10x Genomics) according to the manufacturer’s instructions for co-encapsulation with barcoded Gel Beads at a target capture rate of ∼7000 individual cells per sample. To analyze neutrophils, we added RNase inhibitor (RNase OUT Recombinant Ribonuclease Inhibitor Invitrogen, 40U/mL) into the loading buffer. Captured mRNAs were barcoded during cDNA synthesis using the Chromium Single Cell 3′ Solution v3 (10x Genomics) according to the manufacturer’s instructions. Of note, we increased the PCR cycles by two during cDNA amplification. All samples (at Day 0 and Day 10) were processed simultaneously with the Chromium Controller (10x Genomics) and the resulting libraries were prepared in parallel in a single batch. We pooled all of the libraries for sequencing in a single SP Illumina flow cell. All of the libraries were sequenced with an 8-base index read, a 28-base Read1 containing cell-identifying barcodes and unique molecular identifiers (UMIs), and a 91-base Read2 containing transcript sequences on an Illumina NovaSeq 6000. Reads were aligned to the hg19 genome and were used for subsequent analysis.
Analysis of scRNaseq and integration of dataset from bronchoalveolar lavage fluid of COVID-19 patients
Using the package Seurat V3 (Stuart et al., 2019), we normalized and scaled scRNA sequencing data. We next applied a principle component analysis to the scRNA sequencing results yielding a number of significant PCs (Using Jackstraw plot analysis). In addition, the standard deviation differences from one PC to another was taken into account as described by the Seurat V3 manual (Stuart et al., 2019). To generate UMAP plots, min_distance was set as 0.3 and n_neighbors was set to 30. By dimensionality reduction, distinct clusters were identified and described by performing the FindClusters feature. The resolution of this feature was reduced to 0.3 to identify main cellular population only. Following this, differential genes were identified by performing the FindAllMarkers function and selecting genes that were differentially expressed (logFC > / = +/− 0.25 and FDR < 0.05). This approach identified a number of well characterized blood cell populations. Clustering and analysis of specific cell populations were performed in a similar manner to as previously stated. Cells were clustered and separated based on well described markers (CD14/CD16 as describing monocyte populations).
The bronchoalveolar dataset was downloaded from the NIH GEO database (Liao et al., dataset GSE145926) and integrated with our own blood scRNaseq data using the Seurat V3 anchoring method (Stuart et al., 2019). Briefly, the datasets were normalized independently and the highly variable genes were identified for each dataset using the Seurat pipeline. A corrected data matrix with both datasets was then generated using the Seurat v3 anchoring procedure to allow for joint analysis. The matrix was scaled and a Principal Component Analysis (PCA) was performed using the Seurat v3 pipeline. A UMAP was performed on the 30 first Principal Components (PCs) (Becht et al., 2018). These principle components and subsequent clustering and analysis of scRNA sequencing data was performed as previously described.
Comparisons between patient samples were performed by a variation of the FindMarkers function that compared the differentially expressed genes from different samples, patient groups, and organs. Cutoff values were determined as previously described.
RT-qPCR analysis
Total RNA was extracted with RNeasy Mini Kit (QIAGEN) and reverse transcribed with SuperScript IV VILO Master Mix with ezDNase Enzyme (Invitrogen). Real-time quantitative polymerase chain reaction (RT-qPCR) was performed using Power SYBR Green PCR Master Mix in a BioRad CFX96 thermocycler using the standard SyBR Green detection protocol as outlined by the manufacturer (Applied Biosystems). Briefly, 12 ng of total cDNA, 50nM (each) primers and 1 × SyBR Green mixture were used in a total volume of 20 μL. Human primer sequences are the following: GUS (F: GAAAATATGTGGTTGGAGAGCTCATT; R: CCGAGTGAAGATCCCCT TTTTA); HPRT (F: GGACAGGACTGAACGTCTTGC; R: CTTGAGCACACAGAGGGCTACA); S100A8 (F: CAACACTG ATGGTGCAGTTAACTTC; R: CTGCCACGCCCATCTTTATC); S100A9 (F: CTGAGCTTCGAGG AGTTCATCA; R: CGTCACCCTCGTGCATCTTC).
Cytokine and chemokine measurements
Plasma samples (Table S6) were centrifuged for 15 min at 1,000 g, diluted 1:4, then monitored using the Bio-Plex Pro™ Human Chemokine Panel 40-plex Assay (Bio-rad, ref: 171AK99MR2) according to the manufacturer’s instructions. 40-plex cytokines and chemokines provided are: CCL1, CCL11, CCL13, CCL15, CCL17, CCL19, CCL2, CCL20, CCL21, CCL22, CCL23, CCL24, CCL25, CCL26, CCL27, CCL3, CCL7, CCL8, CX3CL1, CXCL1, CXCL10, CXCL11, CXCL12, CXCL13, CXCL16, CXCL2, CXCL5, CXCL6, CXCL8, CXCL9, GM-CSF, IFNα, IL-10, IL-16, IL-1b, IL-2, IL-4, IL-6, MIF, TNFa. Acquisitions and analyses were performed on a Bio-Plex 200 system (Bio-rad) and a Bio-Plex Manager 6.1 Software (Bio-rad), respectively. Soluble Calprotectin (diluted 1:100) and IFNα2a were analyzed using a R-plex Human Calprotectin Antibody Set (Meso Scale Discovery, ref: F21YB-3) and the ultra-sensitive assay S-plex Human IFNα2a kit (Meso Scale Discovery, ref: K151P3S-1), respectively, following manufacturer’s instructions. Acquisitions and analyses of soluble Calprotectin and IFNα were performed on a MESO QuickPlex SQ120 reader and the MSD’s Discovery Workbench 4.0. Each plasma sample was assayed twice with the average value taken as the final result. Data representation was performed with software R v3.3.3 using tidyverse, dplyr, ggplot2, ggpubr, pheatmap, corrplot or Hmisc packages.
Mass Cytometry
Cells were barcoded using the 20-Plex Pd barcoding kit (Fluidigm). Briefly, they were washed in Barcode Perm Buffer, resuspended in 800 μL of Barcode Perm Buffer and 100 μL of each barcode were transferred to the appropriate sample. Cell suspensions were incubated for 30 min at room temperature, washed twice with Cell Staining Buffer (Fluidigm) and pooled, suspended in 100 μL filtered antibody cocktail, and incubated for 30 min at +4°C. All antibodies used are listed in Key Resources Table. After staining, cells were washed with Cell Staining Buffer and permeabilized with 200 μL of Fix/Perm from Foxp3/Transcription Factor Staining Buffer kit (eBiosciences), 40 min at +4°C. After incubation, cells were washed in Perm Buffer from Foxp3/Transcription Factor Staining Buffer kit (eBiosciences), resuspended in 100 μL filtered antibody cocktail, incubated for 30 min at +4°C, washed in Cell Staining Buffer and resuspended in 50 μL of CytoFix/Perm for 5 min at room temperature. Then, 400 μL of PBS containing 1.6% PFA + Iridium (1:4000) were added for 35 min at room temperature. Finally, cells were washed in Cell Staining Buffer, resuspended in 50 μL and stored at +4°C until acquisition. Cells were counted, washed and resuspended in Maxpar Cell Acquisition Solution at 0.5x 106 / mL and mixed with 10% EQ Beads immediately before acquisition on Helios mass cytometer using noise reduction, event length limits of 10-150 pushes. An average of 500,000 events were acquired per sample at a flow rate of 0.03mL/min. Mass cytometry standard files were normalized to a global standard determined for each log of EQ beads using CyTOF Software v.6.7.1014 (Fluidigm). Fcs files were exported and analyzed using FlowJo software. UMAP was performed with n_neighbors of 15 and a min_distance of 0.2. Clusters were identified by the detection of commonly used cell markers (T cells expressing CD4 or CD8, neutrophils expressing CD15, and monocytes expressing CD14 and or CD16).
Routine multiparameter flow analysis test
Whole-blood samples (200μL) were labeled with anti-CD14-PC7 (clone RMO52); CD16-PB (clone 3G8); CD2-FITC (clone 39C1.5); CD56-PC5.5 (clone N901); CD24-PE (clone ALB9); CD45-KO (clone J33) and HLA-DR-APC (clone Immu-357) antibodies, all purchased from Beckman-Coulter. Red blood cells were lysed with 1 mL VersalyseTM (Beckman Coulter) before sample analysis with a Navios Cytometer (Beckman Coulter) as described (Tarfi et al., 2019). Monocytes were selected as CD45High/SSCInt cells among living cells and singlets before excluding T cells as CD2+/SSCLow, NK cells as CD56+/SSCLow/Int, B cells as CD24+/SSCLow, immature and mature granulocytes as CD24+/SSCInt/High, CD16Bright residual granulocytes, and remaining CD14−CD16− cells corresponding mainly to basophils and NK cells not previously excluded. Monocyte subsets were detected on a CD45/SCC dot plot, using a CD14/CD16 scattergram that separates CD14HighCD16Low (classical), CD14High/CD16High (intermediate) and CD14LowCD16High (non-classical) subsets. Finally, the proportion of monocytes HLA-DRLow was evaluated on a HLA-DR/CD14 scattergram.
Quantification and Statistical Analysis
Data analysis
Calculations and statistical tests were performed using R v3.3.3. Unless stated, p values are two-sided with 95% confidence intervals for the reported statistic of interest. Individual data points representing the measurement from one patient are systematically calculated from the corresponding distribution. Wilcoxon rank-sum test was applied to assess differences in concentration between two different groups. When indicated, the false discovery rate (FDR, p > 0.05) was controlled using the Benjamini–Hochberg procedure. Spearman correlations were computed using Hmisc R package and cytokine results were shown using R package Pheatmap. Soluble factor fold ratios were calculated as log2 transformation of values of mild and severe patients versus median value of all control patients, and were converted to z scores. Hierarchical clustering of the patients based on the z score of 42 soluble factors was performed using euclidean distance and ward.D clustering. Gene ontology networks were made by subjecting the DEGs from previous scRNA sequencing analysis to the Cytoscape addon ClueGO. The selected DEGs were specific to those with increased expression by monocytes and neutrophils from mild or severe SARS-CoV-2 positive patients. Biological Process gene ontologies selected had an FDR < 0.05. Other statistical analyses were performed using GraphPad Prism 7.
A generalized linear model was also used to analyze interactions between biological parameters. First, neutrophil count, calprotectin, fibrinogen, IL-6 and D-dimers were normalized using log transformation. Then, calprotectin plasma level was modeled using multivariable linear regression adjusted for the other parameters, and their interaction with the groups. Similar approach was performed to model IL-6. Backward selection was applied to obtain a parsimonious model.
To identify the most discriminant markers, we used a logistic regression adjusted for the scaled log2-transformed markers. Parameters were penalized using the least absolute shrinkage and selection operator (lasso) to limit overfitting due to the high number of markers. The regularization parameter was selected from 10-folds cross-validation using the glmnet R package (Friedman et al., 2010). The final AUC estimate was corrected for optimism using the Harrell’s method (Harrell et al., 1996), and its confidence interval was computed using the two-stage approach proposed by Noma and colleagues (Noma et al., 2020) with 2000 bootstrap samples for each stage. In this analysis, we included age, sex and comorbidities together with biological parameters. Given the absence of validation cohort, AUC was corrected to limit overfitting bias. This correction indicated an AUC at 99.7% (95% confidence interval [98.8%; 100.0%]). The final score corresponds to the following equation:
With , , , and , the values of the calprotectin, CX3CL1, CXCL11 and CXCL13 which are log2 transformed and scaled according to the value training cohort: , , , and .
Acknowledgments
Our teams are supported by grants from Ligue Nationale Contre le Cancer (Equipes labellisées), Agence Nationale de la Recherche (IHU PRISM), Institut National du Cancer (SIRIC CARPEM and SOCRATE), Fondation ARC, Fondation pour la Recherche Médicale, and private donations (Dassault Systems, Agnes B, Izipizi, Ralph Lauren, and Malakoff Humanis) as well as Fondation Gustave Roussy (including the Paediatric Campaign). F.G. is supported by Singapore Immunology Network (SIgN) core funding and Singapore National Research Foundation Senior Investigatorship (NRFI) NRF2016NRF-NRFI001-02 and is a European Molecular Biology Organization (EMBO) YIP awardee. L.G.N. is supported by SIgN core funding. We thank Emmanuelle Gallois, Jean-François Méritet, and Flore Rosenberg for SARS-CoV-2 PCR tests and the following individuals for technical support: Jean-Daniel Chiche and Jean-Paul Mira (Medical Intensive Care Unit, Cochin Hospital); Maela Francillette, Betty Leite, and Valérie Camara-Clayette (AMMICa platforms, CRB, Gustave Roussy); Audrey Naimo (AMMICa platforms, Gustave Roussy); Karine Bailly (CYBIO platform, Institut Cochin); and Jérôme Duchemin, Catherine Gicquel, Amandine Houvert, Georges Jourdi, Françoise Levavasseur, Laurence Marnet, Bruno Montout, Loetitia Rhino, Isabelle Souville, and Christine Scamps. We also thank Julien Hadoux and Jean-Marie Michot for providing samples, Dorothée Selimoglu-Buet and Xavier Mariette for helpful scientific discussions, and Dr. Lucy Robinson (Insight Editing London) for editing the manuscript.
Author Contributions
Conceptualization, A. Silvin, N.C., F. Ginhoux, M.F., and E. Solary; Methodology, L.G.N., I.A., Z.Z., L.Z., and B.G.; Investigation, G.D., A.-G.G., A.D., C.A., C.H., O.K., N.D., P.R., C.C., C.D., C.F., D.C., D. Bredel, S.M., M.A., A.-S.L., V.S., F. Griscelli, and P.B.; Formal Analysis, N.C., G.D., A.A., D. Borderie, M.D., and D.D.; Funding Acquisition, A.M., F.A., L.Z., F. Ginhoux, M.F., and E. Solary; Resources, L.D., E. Sourdeau, N.M., T.-A.S., D.C., L.M., A. Stoclin, C.W., F. Pommeret, F.B., A.M., and F. Pène; Data Curation, L.D., C.A., C.H., and M.F.; Validation, M.F., and E. Solary; Visualization, A. Silvin, G.D., A.-G.G., A.D., F. Ginhoux, M.F., and E. Solary; Writing – Original Draft, E. Solary; Writing – Review & Editing, A. Silvin, F. Ginhoux, M.F., and E. Solary; Supervision, F. Ginhoux, M.F., and E. Solary.
Declaration of Interests
A. Silvin, N.C., M.F., E. Solary, and F. Ginhoux are inventors of patent EP 20305624.7, “Methods for detecting and treating COVID patients requiring intensive care,” submitted on June 9, 2020 under Gustave Roussy.
Published: August 5, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.08.002.
Supplemental Information
Detailed Results of the scRNA-Seq Experiments, Including Metadata, Cell Population Numbers, Cell Population DEGs, Monocyte DEGs, and Neutrophil DEGs, Related to Figures 2, 3, 4, and S2, S3, and S4
References
- Aarts C.E.M., Hiemstra I.H., Béguin E.P., Hoogendijk A.J., Bouchmal S., van Houdt M., Tool A.T.J., Mul E., Jansen M.H., Janssen H. Activated neutrophils exert myeloid-derived suppressor cell activity damaging T cells beyond repair. Blood Adv. 2019;3:3562–3574. doi: 10.1182/bloodadvances.2019031609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Qahtani A.A., Lyroni K., Aznaourova M., Tseliou M., Al-Anazi M.R., Al-Ahdal M.N., Alkahtani S., Sourvinos G., Tsatsanis C. Middle east respiratory syndrome corona virus spike glycoprotein suppresses macrophage responses via DPP4-mediated induction of IRAK-M and PPARγ. Oncotarget. 2017;8:9053–9066. doi: 10.18632/oncotarget.14754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austermann J., Friesenhagen J., Fassl S.K., Petersen B., Ortkras T., Burgmann J., Barczyk-Kahlert K., Faist E., Zedler S., Pirr S. Alarmins MRP8 and MRP14 induce stress tolerance in phagocytes under sterile inflammatory conditions. Cell Rep. 2014;9:2112–2123. doi: 10.1016/j.celrep.2014.11.020. [DOI] [PubMed] [Google Scholar]
- Basiorka A.A., McGraw K.L., Eksioglu E.A., Chen X., Johnson J., Zhang L., Zhang Q., Irvine B.A., Cluzeau T., Sallman D.A. The NLRP3 inflammasome functions as a driver of the myelodysplastic syndrome phenotype. Blood. 2016;128:2960–2975. doi: 10.1182/blood-2016-07-730556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H. Host-Viral Infection Maps Reveal Signatures of Severe COVID-19 Patients. Cell. 2020;181:1475–1488.e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Jr., Meyerholz D.K., Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;129:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Eksioglu E.A., Zhou J., Zhang L., Djeu J., Fortenbery N., Epling-Burnette P., Van Bijnen S., Dolstra H., Cannon J. Induction of myelodysplasia by myeloid-derived suppressor cells. J. Clin. Invest. 2013;123:4595–4611. doi: 10.1172/JCI67580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Bracht J.R., Goldman A.D., Dolzhenko E., Clay D.M., Swart E.C., Perlman D.H., Doak T.G., Stuart A., Amemiya C.T. The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell. 2014;158:1187–1198. doi: 10.1016/j.cell.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Long X., Xu Q., Tan J., Wang G., Cao Y., Wei J., Luo H., Zhu H., Huang L. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0492-x. Published online July 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan J.F., Wang Y., Yuen T.T., Chai Y., Hou Y., Shuai H., Yang D., Hu B., Huang X. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa410. Published online April 9, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desforges M., Miletti T.C., Gagnon M., Talbot P.J. Activation of human monocytes after infection by human coronavirus 229E. Virus Res. 2007;130:228–240. doi: 10.1016/j.virusres.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eksioglu E.A., Chen X., Heider K.H., Rueter B., McGraw K.L., Basiorka A.A., Wei M., Burnette A., Cheng P., Lancet J. Novel therapeutic approach to improve hematopoiesis in low risk MDS by targeting MDSCs with the Fc-engineered CD33 antibody BI 836858. Leukemia. 2017;31:2172–2180. doi: 10.1038/leu.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard M., Kwok I.W.H., Chong S.Z., Teng K.W.W., Becht E., Chen J., Sieow J.L., Penny H.L., Ching G.C., Devi S. Developmental Analysis of Bone Marrow Neutrophils Reveals Populations Specialized in Expansion, Trafficking, and Effector Functions. Immunity. 2018;48:364–379.e8. doi: 10.1016/j.immuni.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Fassl S.K., Austermann J., Papantonopoulou O., Riemenschneider M., Xue J., Bertheloot D., Freise N., Spiekermann C., Witten A., Viemann D. Transcriptome assessment reveals a dominant role for TLR4 in the activation of human monocytes by the alarmin MRP8. J. Immunol. 2015;194:575–583. doi: 10.4049/jimmunol.1401085. [DOI] [PubMed] [Google Scholar]
- Fizazi K., Ulys A., Sengeløv L., Moe M., Ladoire S., Thiery-Vuillemin A., Flechon A., Guida A., Bellmunt J., Climent M.A. A randomized, double-blind, placebo-controlled phase II study of maintenance therapy with tasquinimod in patients with metastatic castration-resistant prostate cancer responsive to or stabilized during first-line docetaxel chemotherapy. Ann. Oncol. 2017;28:2741–2746. doi: 10.1093/annonc/mdx487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freise N., Burghard A., Ortkras T., Daber N., Imam Chasan A., Jauch S.L., Fehler O., Hillebrand J., Schakaki M., Rojas J. Signaling mechanisms inducing hyporesponsiveness of phagocytes during systemic inflammation. Blood. 2019;134:134–146. doi: 10.1182/blood.2019000320. [DOI] [PubMed] [Google Scholar]
- Friedman J., Hastie T., Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Furman D., Campisi J., Verdin E., Carrera-Bastos P., Targ S., Franceschi C., Ferrucci L., Gilroy D.W., Fasano A., Miller G.W. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019;25:1822–1832. doi: 10.1038/s41591-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Zhong N.S. Clinical Characteristics of Covid-19 in China. Reply. N. Engl. J. Med. 2020;382:1861–1862. doi: 10.1056/NEJMc2005203. [DOI] [PubMed] [Google Scholar]
- Guilliams M., Mildner A., Yona S. Developmental and Functional Heterogeneity of Monocytes. Immunity. 2018;49:595–613. doi: 10.1016/j.immuni.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P. Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients. medRxiv. 2020 doi: 10.1101/2020.04.19.20068015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajian-Tilaki K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Caspian J. Intern. Med. 2013;4:627–635. [PMC free article] [PubMed] [Google Scholar]
- Hanna R.N., Carlin L.M., Hubbeling H.G., Nackiewicz D., Green A.M., Punt J.A., Geissmann F., Hedrick C.C. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat. Immunol. 2011;12:778–785. doi: 10.1038/ni.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell F.E., Jr., Lee K.L., Mark D.B. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Hofer T.P., Zawada A.M., Frankenberger M., Skokann K., Satzl A.A., Gesierich W., Schuberth M., Levin J., Danek A., Rotter B. slan-defined subsets of CD16-positive monocytes: impact of granulomatous inflammation and M-CSF receptor mutation. Blood. 2015;126:2601–2610. doi: 10.1182/blood-2015-06-651331. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman M.J., Lee M.K., Al-Sharea A., Dragoljevic D., Barrett T.J., Montenont E., Basu D., Heywood S., Kammoun H.L., Flynn M. Neutrophil-derived S100 calcium-binding proteins A8/A9 promote reticulated thrombocytosis and atherogenesis in diabetes. J. Clin. Invest. 2017;127:2133–2147. doi: 10.1172/JCI92450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratofil R.M., Kubes P., Deniset J.F. Monocyte Conversion During Inflammation and Injury. Arterioscler. Thromb. Vasc. Biol. 2017;37:35–42. doi: 10.1161/ATVBAHA.116.308198. [DOI] [PubMed] [Google Scholar]
- Kuipers M.T., Vogl T., Aslami H., Jongsma G., van den Berg E., Vlaar A.P., Roelofs J.J., Juffermans N.P., Schultz M.J., van der Poll T. High levels of S100A8/A9 proteins aggravate ventilator-induced lung injury via TLR4 signaling. PLoS ONE. 2013;8:e68694. doi: 10.1371/journal.pone.0068694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançais E., Ortiz-Muñoz G., Caudrillier A., Mallavia B., Liu F., Sayah D.M., Thornton E.E., Headley M.B., David T., Coughlin S.R. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature. 2017;544:105–109. doi: 10.1038/nature21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu L., Zhang D., Xu J., Dai H., Tang N., Su X., Cao B. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet. 2020;395:1517–1520. doi: 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Llitjos J.F., Leclerc M., Chochois C., Monsallier J.M., Ramakers M., Auvray M., Merouani K. High incidence of venous thromboembolic events in anticoagulated severe COVID-19 patients. J. Thromb. Haemost. 2020;18:1743–1746. doi: 10.1111/jth.14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A.C., Grienay M., Resche-Rigon M., Pirracchio R., Faivre V., Boval B., Payen D. Monocytic HLA-DR expression in intensive care patients: interest for prognosis and secondary infection prediction. Crit. Care Med. 2009;37:2746–2752. doi: 10.1097/CCM.0b013e3181ab858a. [DOI] [PubMed] [Google Scholar]
- Mastio J., Condamine T., Dominguez G., Kossenkov A.V., Donthireddy L., Veglia F., Lin C., Wang F., Fu S., Zhou J. Identification of monocyte-like precursors of granulocytes in cancer as a mechanism for accumulation of PMN-MDSCs. J. Exp. Med. 2019;216:2150–2169. doi: 10.1084/jem.20181952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot J.M., Albiges L., Chaput N., Saada V., Pommeret F., Griscelli F., Balleyguier C., Besse B., Marabelle A., Netzer F. Tocilizumab, an anti-IL6 receptor antibody, to treat Covid-19-related respiratory failure: a case report. Ann. Oncol. 2020;31:961–964. doi: 10.1016/j.annonc.2020.03.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L.G., Ostuni R., Hidalgo A. Heterogeneity of neutrophils. Nat. Rev. Immunol. 2019;19:255–265. doi: 10.1038/s41577-019-0141-8. [DOI] [PubMed] [Google Scholar]
- Noma H., Nagashima K., Furukawa T.A. Permutation inference methods for multivariate meta-analysis. Biometrics. 2020;76:337–347. doi: 10.1111/biom.13134. [DOI] [PubMed] [Google Scholar]
- Nottet H.S., Persidsky Y., Sasseville V.G., Nukuna A.N., Bock P., Zhai Q.H., Sharer L.R., McComb R.D., Swindells S., Soderland C., Gendelman H.E. Mechanisms for the transendothelial migration of HIV-1-infected monocytes into brain. J. Immunol. 1996;156:1284–1295. [PubMed] [Google Scholar]
- Patnaik M.M., Sallman D.A., Mangaonkar A., Heuer R., Hirvela J., Zblewski D., Al-Kali A., Binder M., Balasis M.E., Newman H. Phase 1 study of lenzilumab, a recombinant anti-human GM-CSF antibody, for chronic myelomonocytic leukemia (CMML) Blood. 2020 doi: 10.1182/blood.2019004352. blood.2019004352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raquil M.A., Anceriz N., Rouleau P., Tessier P.A. Blockade of antimicrobial proteins S100A8 and S100A9 inhibits phagocyte migration to the alveoli in streptococcal pneumonia. J. Immunol. 2008;180:3366–3374. doi: 10.4049/jimmunol.180.5.3366. [DOI] [PubMed] [Google Scholar]
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., Northwell COVID-19 Research Consortium Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva M., Källberg E., Björk P., Hancz D., Vogl T., Roth J., Ivars F., Leanderson T. Induction of nuclear factor-κB responses by the S100A9 protein is Toll-like receptor-4-dependent. Immunology. 2012;137:172–182. doi: 10.1111/j.1365-2567.2012.03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selimoglu-Buet D., Rivière J., Ghamlouch H., Bencheikh L., Lacout C., Morabito M., Diop M., Meurice G., Breckler M., Chauveau A. A miR-150/TET3 pathway regulates the generation of mouse and human non-classical monocyte subset. Nat. Commun. 2018;9:5455. doi: 10.1038/s41467-018-07801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevelsted A., Stokholm J., Bønnelykke K., Bisgaard H. Cesarean section and chronic immune disorders. Pediatrics. 2015;135:e92–e98. doi: 10.1542/peds.2014-0596. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard J.C., Noël C., Tessier P.A., Girard D. Human S100A9 potentiates IL-8 production in response to GM-CSF or fMLP via activation of a different set of transcription factors in neutrophils. FEBS Lett. 2014;588:2141–2146. doi: 10.1016/j.febslet.2014.04.027. [DOI] [PubMed] [Google Scholar]
- Smith M.S., Bentz G.L., Alexander J.S., Yurochko A.D. Human cytomegalovirus induces monocyte differentiation and migration as a strategy for dissemination and persistence. J. Virol. 2004;78:4444–4453. doi: 10.1128/JVI.78.9.4444-4453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarfi S., Badaoui B., Freynet N., Morabito M., Lafosse J., Toma A., Etienne G., Micol J.B., Sloma I., Fenaux P., Groupe Francophone des Myelodysplasies (GFM) Disappearance of slan-positive non-classical monocytes for diagnosis of chronic myelomonocytic leukemia with associated inflammatory state. Haematologica. 2019 doi: 10.3324/haematol.2019.219782. haematol.2019.219782. [DOI] [PubMed] [Google Scholar]
- Terán-Cabanillas E., Hernández J. Role of Leptin and SOCS3 in Inhibiting the Type I Interferon Response During Obesity. Inflammation. 2017;40:58–67. doi: 10.1007/s10753-016-0452-x. [DOI] [PubMed] [Google Scholar]
- Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulas T., Pirr S., Fehlhaber B., Bickes M.S., Loof T.G., Vogl T., Mellinger L., Heinemann A.S., Burgmann J., Schöning J. S100-alarmin-induced innate immune programming protects newborn infants from sepsis. Nat. Immunol. 2017;18:622–632. doi: 10.1038/ni.3745. [DOI] [PubMed] [Google Scholar]
- Vabret N., Samstein R., Fernandez N., Merad M., Sinai Immunology Review Project. Trainees. Faculty Advancing scientific knowledge in times of pandemics. Nat. Rev. Immunol. 2020;20:338. doi: 10.1038/s41577-020-0319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veglia F., Perego M., Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat. Immunol. 2018;19:108–119. doi: 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl T., Stratis A., Wixler V., Völler T., Thurainayagam S., Jorch S.K., Zenker S., Dreiling A., Chakraborty D., Fröhling M. Autoinhibitory regulation of S100A8/S100A9 alarmin activity locally restricts sterile inflammation. J. Clin. Invest. 2018;128:1852–1866. doi: 10.1172/JCI89867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter R.B. Investigational CD33-targeted therapeutics for acute myeloid leukemia. Expert Opin. Investig. Drugs. 2018;27:339–348. doi: 10.1080/13543784.2018.1452911. [DOI] [PubMed] [Google Scholar]
- Wang S., Song R., Wang Z., Jing Z., Wang S., Ma J. S100A8/A9 in Inflammation. Front. Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Du Z., Zhu F., Cao Z., An Y., Gao Y., Jiang B. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet. 2020;395:e52. doi: 10.1016/S0140-6736(20)30558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ye F., Zhu N., Wang W., Deng Y., Zhao Z., Tan W. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 2015;5:17554. doi: 10.1038/srep17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilla M., Harcourt B.H., Hickman C.J., McGrew M., Tamin A., Goldsmith C.S., Bellini W.J., Anderson L.J. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res. 2005;107:93–101. doi: 10.1016/j.virusres.2004.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed Results of the scRNA-Seq Experiments, Including Metadata, Cell Population Numbers, Cell Population DEGs, Monocyte DEGs, and Neutrophil DEGs, Related to Figures 2, 3, 4, and S2, S3, and S4
Data Availability Statement
All blood scRNA-seq data used in this study can be accessed by ArrayExpress Archive of Functional Genomics Data under the accession number E-MTAB-9221. Integrated BALF scRNA-seq data can be accessed in GEO under the accession number GSE145926.