Abstract
Background
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in late 2019 and spread globally, prompting an international effort to accelerate development of a vaccine. The candidate vaccine mRNA-1273 encodes the stabilized prefusion SARS-CoV-2 spike protein.
Methods
We conducted a phase 1, dose-escalation, open-label trial including 45 healthy adults, 18 to 55 years of age, who received two vaccinations, 28 days apart, with mRNA-1273 in a dose of 25 μg, 100 μg, or 250 μg. There were 15 participants in each dose group.
Results
After the first vaccination, antibody responses were higher with higher dose (day 29 enzyme-linked immunosorbent assay anti–S-2P antibody geometric mean titer [GMT], 40,227 in the 25-μg group, 109,209 in the 100-μg group, and 213,526 in the 250-μg group). After the second vaccination, the titers increased (day 57 GMT, 299,751, 782,719, and 1,192,154, respectively). After the second vaccination, serum-neutralizing activity was detected by two methods in all participants evaluated, with values generally similar to those in the upper half of the distribution of a panel of control convalescent serum specimens. Solicited adverse events that occurred in more than half the participants included fatigue, chills, headache, myalgia, and pain at the injection site. Systemic adverse events were more common after the second vaccination, particularly with the highest dose, and three participants (21%) in the 250-μg dose group reported one or more severe adverse events.
Conclusions
The mRNA-1273 vaccine induced anti–SARS-CoV-2 immune responses in all participants, and no trial-limiting safety concerns were identified. These findings support further development of this vaccine. (Funded by the National Institute of Allergy and Infectious Diseases and others; mRNA-1273 ClinicalTrials.gov number, NCT04283461).
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in December 2019 and spread globally, causing a pandemic of respiratory illness designated coronavirus disease 2019 (Covid-19).1 The urgent need for vaccines prompted an international response, with more than 120 candidate SARS-CoV-2 vaccines in development within the first 5 months of 2020.2 The candidate vaccine mRNA-1273 is a lipid nanoparticle–encapsulated, nucleoside-modified messenger RNA (mRNA)–based vaccine that encodes the SARS-CoV-2 spike (S) glycoprotein stabilized in its prefusion conformation. The S glycoprotein mediates host cell attachment and is required for viral entry3; it is the primary vaccine target for many candidate SARS-CoV-2 vaccines.4-7
We conducted a first-in-human phase 1 clinical trial in healthy adults to evaluate the safety and immunogenicity of mRNA-1273. Here we report interim results of the trial.
Methods
Trial Design and Participants
We conducted a phase 1, dose-escalation, open-label clinical trial designed to determine the safety, reactogenicity, and immunogenicity of mRNA-1273. Eligible participants were healthy adults 18 to 55 years of age who received two injections of trial vaccine 28 days apart at a dose of 25 μg, 100 μg, or 250 μg. On the basis of the results obtained in patients at these dose levels, additional groups were added to the protocol; those results will be reported in a subsequent publication. Participants were not screened for SARS-CoV-2 infection by serology or polymerase chain reaction before enrollment. The trial was conducted at the Kaiser Permanente Washington Health Research Institute in Seattle and at the Emory University School of Medicine in Atlanta. The protocol, available with the full text of this article at NEJM.org, permitted interim analyses to inform decisions regarding vaccine strategy and public health; this interim analysis reports findings through day 57. Full details of the trial design, conduct, oversight, and analyses can be found in the protocol and statistical analysis plan (available at NEJM.org).
The trial was reviewed and approved by the Advarra institutional review board, which functioned as a single board and was overseen by an independent safety monitoring committee. All participants provided written informed consent before enrollment. The trial was conducted under an Investigational New Drug application submitted to the Food and Drug Administration. The vaccine was codeveloped by researchers at the National Institute of Allergy and Infectious Diseases (NIAID, the trial sponsor) and at Moderna (Cambridge, MA). Moderna was involved in discussions of the trial design, provided the vaccine candidate, and, as part of the writing group, contributed to drafting the manuscript. The Emmes Company, as a subcontractor to the NIAID, served as the statistical and data coordinating center, developed the statistical analysis plan, and performed the analyses. The manuscript was written entirely by the authors, with the first author as the overall lead author, the fourth author as the lead NIAID author, and the last two authors as senior authors (details are provided in the Supplementary Appendix, available at NEJM.org). The authors had full access to the data reports, which were prepared from the raw data by the statistical and data coordinating center, and vouch for the completeness and accuracy of the data and for the fidelity of the trial to the protocol.
Vaccine
The mRNA-1273 vaccine candidate, manufactured by Moderna, encodes the S-2P antigen, consisting of the SARS-CoV-2 glycoprotein with a transmembrane anchor and an intact S1–S2 cleavage site. S-2P is stabilized in its prefusion conformation by two consecutive proline substitutions at amino acid positions 986 and 987, at the top of the central helix in the S2 subunit.8 The lipid nanoparticle capsule composed of four lipids was formulated in a fixed ratio of mRNA and lipid. The mRNA-1273 vaccine was provided as a sterile liquid for injection at a concentration of 0.5 mg per milliliter. Normal saline was used as a diluent to prepare the doses administered.
Trial Procedures
The vaccine was administered as a 0.5-ml injection in the deltoid muscle on days 1 and 29; follow-up visits were scheduled for 7 and 14 days after each vaccination and on days 57, 119, 209, and 394. The dose-escalation plan specified enrollment of four sentinel participants in the 25-μg group, followed by four sentinel participants in the 100-μg group, followed by full enrollment of those two dose groups. If no halting rules were met after all participants in those two dose groups completed day 8, four sentinel participants in the 250-μg group were enrolled, followed by the remainder of that dose group.
Participants recorded local and systemic reactions, using a memory aid, for 7 days after each vaccination. Participants were not instructed to routinely use acetaminophen or other analgesics or antipyretics before or after the vaccinations but were asked to record any new medications taken. Adverse events were graded according to a standard toxicity grading scale (Table S1 in the Supplementary Appendix).9
Assessment of SARS-CoV-2 Binding Antibody and Neutralizing Responses
Binding antibody responses against S-2P and the isolated receptor-binding domain, located in the S1 subunit, were assessed by enzyme-linked immunosorbent assay (ELISA). Vaccine-induced neutralizing activity was assessed by a pseudotyped lentivirus reporter single-round-of-infection neutralization assay (PsVNA) and by live wild-type SARS-CoV-2 plaque-reduction neutralization testing (PRNT) assay. ELISA and PsVNA were performed on specimens collected from all participants on days 1, 15, 29, 36, 43, and 57. Because of the time-intensive nature of the PRNT assay, for this report of the interim analysis, results were available only for the day 1 and day 43 time points in the 25-μg and 100-μg dose groups.
For comparison of the participants’ immune responses with those induced by SARS-CoV-2 infection, 41 convalescent serum specimens were also tested. The assays were performed at the NIAID Vaccine Research Center (ELISA and PsVNA) and the Vanderbilt University Medical Center (PRNT).
Assessment of SARS-CoV-2 T-Cell Responses
T-cell responses against the spike protein were assessed by an intracellular cytokine–staining assay, performed on specimens collected at days 1, 29, and 43. For this report of the interim analysis, results were available only for the 25-μg and 100-μg dose groups. These assays were performed at the NIAID Vaccine Research Center. (See the Supplementary Appendix for details of all assay methods and for characteristics of the convalescent serum specimens.)
Statistical Analysis
Results of immunogenicity testing of the 45 enrolled participants excluded findings for day 36, day 43, and day 57 for 3 participants who did not receive the second vaccination and for time points at which specimens were not collected (in the 100-μg group: 1 participant at day 43 and day 57; in the 250-μg group: 1 participant at day 29 and 1 at day 57). Confidence intervals of the geometric means were calculated with the Student’s t distribution on log-transformed data. Seroconversion as measured by ELISA was defined as an increase by a factor of 4 or more in antibody titer over baseline.
Results
Trial Population
The 45 enrolled participants received their first vaccination between March 16 and April 14, 2020 (Fig. S1). Three participants did not receive the second vaccination, including one in the 25-μg group who had urticaria on both legs, with onset 5 days after the first vaccination, and two (one in the 25-μg group and one in the 250-μg group) who missed the second vaccination window owing to isolation for suspected Covid-19 while the test results, ultimately negative, were pending. All continued to attend scheduled trial visits. The demographic characteristics of participants at enrollment are provided in Table 1.
Table 1. Characteristics of the Participants in the mRNA-1273 Trial at Enrollment.*.
| Characteristic | 25-μg Group (N=15) |
100-μg Group (N=15) |
250-μg Group (N=15) |
Overall (N=45) |
|---|---|---|---|---|
| Sex — no. (%) | ||||
| Male | 9 (60) | 7 (47) | 6 (40) | 22 (49) |
| Female | 6 (40) | 8 (53) | 9 (60) | 23 (51) |
| Age — yr | 36.7±7.9 | 31.3±8.7 | 31.0±8.0 | 33.0±8.5 |
| Race or ethnic group — no. (%)† | ||||
| American Indian or Alaska Native | 0 | 1 (7) | 0 | 1 (2) |
| Asian | 0 | 0 | 1 (7) | 1 (2) |
| Black | 0 | 2 (13) | 0 | 2 (4) |
| White | 15 (100) | 11 (73) | 14 (93) | 40 (89) |
| Unknown | 0 | 1 (7) | 0 | 1 (2) |
| Hispanic or Latino — no. (%) | 1 (7) | 3 (20) | 2 (13)‡ | 6 (13) |
| Body-mass index§ | 24.6±3.4 | 26.7±2.6 | 24.7±3.1 | 25.3±3.2 |
Plus–minus values are means ±SD.
Race or ethnic group was reported by the participants.
One participant did not report ethnic group.
The body-mass index is the weight in kilograms divided by the square of the height in meters. This calculation was based on the weight and height measured at the time of screening.
Vaccine Safety
No serious adverse events were noted, and no prespecified trial halting rules were met. As noted above, one participant in the 25-μg group was withdrawn because of an unsolicited adverse event, transient urticaria, judged to be related to the first vaccination.
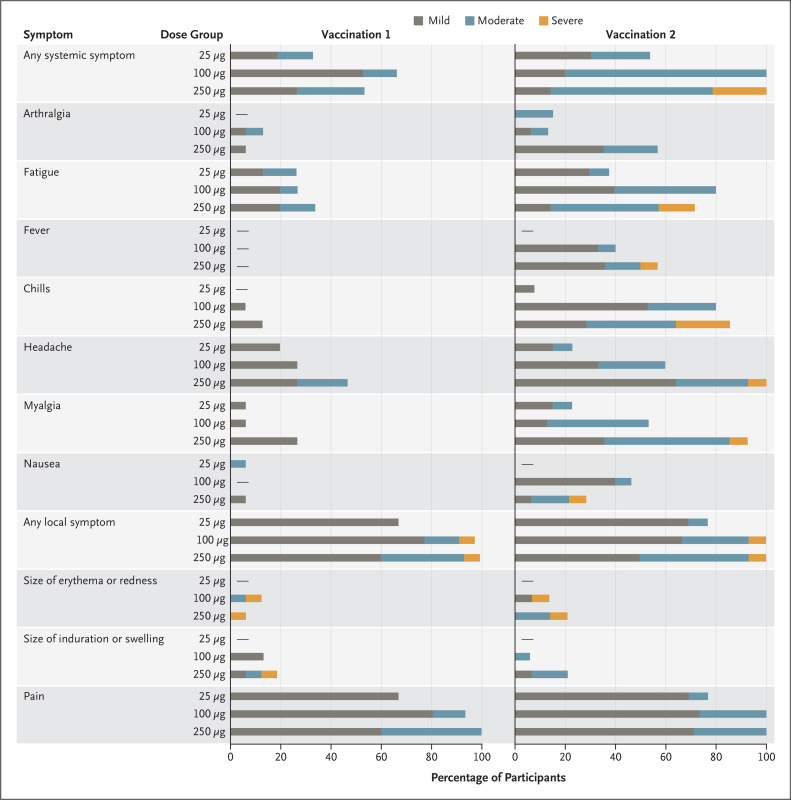
After the first vaccination, solicited systemic adverse events were reported by 5 participants (33%) in the 25-μg group, 10 (67%) in the 100-μg group, and 8 (53%) in the 250-μg group; all were mild or moderate in severity (Figure 1 and Table S2). Solicited systemic adverse events were more common after the second vaccination and occurred in 7 of 13 participants (54%) in the 25-μg group, all 15 in the 100-μg group, and all 14 in the 250-μg group, with 3 of those participants (21%) reporting one or more severe events.
Figure 1. Systemic and Local Adverse Events.
The severity of solicited adverse events was graded as mild, moderate, or severe (see Table S1).
None of the participants had fever after the first vaccination. After the second vaccination, no participants in the 25-μg group, 6 (40%) in the 100-μg group, and 8 (57%) in the 250-μg group reported fever; one of the events (maximum temperature, 39.6°C) in the 250-μg group was graded severe. (Additional details regarding adverse events for that participant are provided in the Supplementary Appendix.)
Local adverse events, when present, were nearly all mild or moderate, and pain at the injection site was common. Across both vaccinations, solicited systemic and local adverse events that occurred in more than half the participants included fatigue, chills, headache, myalgia, and pain at the injection site. Evaluation of safety clinical laboratory values of grade 2 or higher and unsolicited adverse events revealed no patterns of concern (Supplementary Appendix and Table S3).
SARS-CoV-2 Binding Antibody Responses
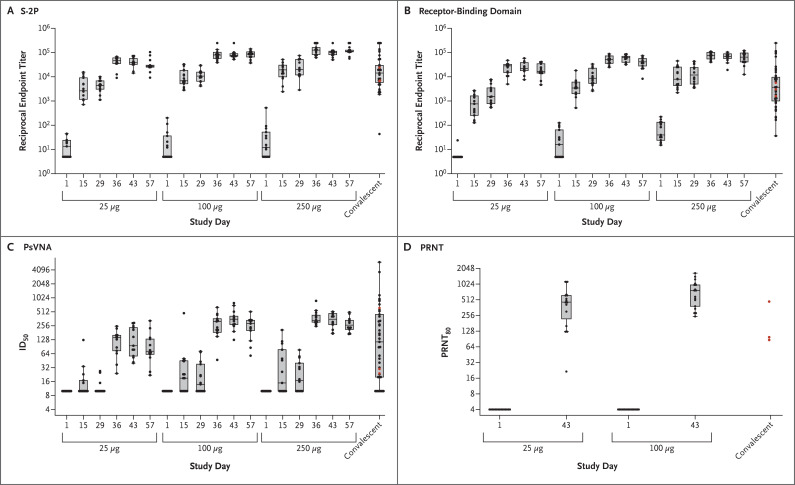
Binding antibody IgG geometric mean titers (GMTs) to S-2P increased rapidly after the first vaccination, with seroconversion in all participants by day 15 (Table 2 and Figure 2A). Dose-dependent responses to the first and second vaccinations were evident. Receptor-binding domain–specific antibody responses were similar in pattern and magnitude (Figure 2B). For both assays, the median magnitude of antibody responses after the first vaccination in the 100-μg and 250-μg dose groups was similar to the median magnitude in convalescent serum specimens, and in all dose groups the median magnitude after the second vaccination was in the upper quartile of values in the convalescent serum specimens. The S-2P ELISA GMTs at day 57 (299,751 [95% confidence interval {CI}, 206,071 to 436,020] in the 25-μg group, 782,719 [95% CI, 619,310 to 989,244] in the 100-μg group, and 1,192,154 [95% CI, 924,878 to 1,536,669] in the 250-μg group) exceeded that in the convalescent serum specimens (142,140 [95% CI, 81,543 to 247,768]).
Table 2. Geometric Mean Humoral Immunogenicity Assay Responses to mRNA-1273 in Participants and in Convalescent Serum Specimens.*.
| Time Point | 25-μg Group | 100-μg Group | 250-μg Group | Convalescent Serum | ||||
|---|---|---|---|---|---|---|---|---|
| no. | GMT (95% CI) | no. | GMT (95% CI) | no. | GMT (95% CI) | no. | GMT (95% CI) | |
| ELISA anti–S-2P | 38 | 142,140 (81,543–247,768) |
||||||
| Day 1 | 15 | 116 (72–187) |
15 | 131 (65–266) |
15 | 178 (81–392) |
||
| Day 15† | 15 | 32,261 (18,723–55,587) |
15 | 86,291 (56,403–132,016) |
15 | 163,449 (102,155–261,520) |
||
| Day 29 | 15 | 40,227 (29,094–55,621) |
15 | 109,209 (79,050–150,874) |
14 | 213,526 (128,832–353,896) |
||
| Day 36 | 13 | 391,018 (267,402–571,780) |
15 | 781,399 (606,247–1,007,156) |
14 | 1,261,975 (973,972–1,635,140) |
||
| Day 43 | 13 | 379,764 (281,597–512,152) |
14 | 811,119 (656,336–1,002,404) |
14 | 994,629 (806,189–1,227,115) |
||
| Day 57 | 13 | 299,751 (206,071–436,020) |
14 | 782,719 (619,310–989,244) |
13 | 1,192,154 (924,878–1,536,669) |
||
| ELISA anti–receptor-binding domain | 38 | 37,857 (19,528–73,391) |
||||||
| Day 1 | 15 | 55 (44–70) |
15 | 166 (82–337) |
15 | 576 (349–949) |
||
| Day 15† | 15 | 6567 (3651–11,812) |
15 | 34,073 (21,688–53,531) |
15 | 87,480 (51,868–147,544) |
||
| Day 29 | 15 | 18,149 (11,091–29699) |
15 | 93,231 (59,895–145,123) |
14 | 120,088 (71,013–203,077) |
||
| Day 36 | 13 | 208,652 (142,803–304,864) |
15 | 499,539 (400,950–622,369) |
14 | 720,907 (591,860–878,090) |
||
| Day 43 | 13 | 233,264 (164,756–330,259) |
14 | 558,905 (462,907–674,810) |
14 | 644,395 (495,808–837,510) |
||
| Day 57 | 13 | 183,652 (122,763–274,741) |
14 | 371,271 (266,721–516,804) |
13 | 582,259 (404,019–839,134) |
||
| GMR (95% CI) | GMR (95% CI) | GMR (95% CI) | GMR (95% CI) | |||||
| PsVNA ID50‡ | 38 | 109.2 (59.6–199.9) |
||||||
| Day 1 | 15 | 10 | 15 | 10 | 15 | 10 | ||
| Day 15† | 15 | 14.5 (9.8–21.4) |
15 | 23.7 (13.3–42.3) |
15 | 26.1 (14.1–48.3) |
||
| Day 29 | 15 | 11.7 (9.7–14.1) |
15 | 18.2 (12.1–27.4) |
14 | 20.7 (13.3–32.2) |
||
| Day 36 | 13 | 105.8 (69.8–160.4) |
15 | 256.3 (182.0–361.1) |
14 | 373.5 (308.6–452.2) |
||
| Day 43 | 13 | 112.3 (71.2–177.1) |
14 | 343.8 (261.2–452.7) |
14 | 332.2 (266.3–414.5) |
||
| Day 57 | 13 | 80.7 (51.0–127.6) |
14 | 231.8 (163.2–329.3) |
14 | 270.2 (221.0–330.3) |
||
| Live virus PRNT80 | 3 | 158.3 (15.1–1663.0) |
||||||
| Day 1§ | 15 | 4 | 15 | 4 | NA | |||
| Day 43 | 13 | 339.7 (184.0–627.1) |
14 | 654.3 (460.1–930.5) |
NA | |||
ELISA denotes enzyme-linked immunosorbent assay, GMT geometric mean titer, GMR geometric mean response, ID50 50% inhibitory dilution, NA not available, PRNT80 plaque-reduction neutralization testing assay that shows reduction in SARS-CoV-2 infectivity by 80% or more, and PsVNA pseudotyped lentivirus reporter neutralization assay.
Seroconversion occurred in all participants at day 15.
Samples that did not neutralize at the 50% level are expressed as less than 20 and plotted at half that dilution (i.e., 10).
All day 1 specimens exhibited less than 80% inhibitory activity at the lowest dilution tested (1:8) and were assigned a titer of 4.
Figure 2. SARS-CoV-2 Antibody and Neutralization Responses.
Shown are geometric mean reciprocal end-point enzyme-linked immunosorbent assay (ELISA) IgG titers to S-2P (Panel A) and receptor-binding domain (Panel B), PsVNA ID50 responses (Panel C), and live virus PRNT80 responses (Panel D). In Panel A and Panel B, boxes and horizontal bars denote interquartile range (IQR) and median area under the curve (AUC), respectively. Whisker endpoints are equal to the maximum and minimum values below or above the median ±1.5 times the IQR. The convalescent serum panel includes specimens from 41 participants; red dots indicate the 3 specimens that were also tested in the PRNT assay. The other 38 specimens were used to calculate summary statistics for the box plot in the convalescent serum panel. In Panel C, boxes and horizontal bars denote IQR and median ID50, respectively. Whisker end points are equal to the maximum and minimum values below or above the median ±1.5 times the IQR. In the convalescent serum panel, red dots indicate the 3 specimens that were also tested in the PRNT assay. The other 38 specimens were used to calculate summary statistics for the box plot in the convalescent panel. In Panel D, boxes and horizontal bars denote IQR and median PRNT80, respectively. Whisker end points are equal to the maximum and minimum values below or above the median ±1.5 times the IQR. The three convalescent serum specimens were also tested in ELISA and PsVNA assays. Because of the time-intensive nature of the PRNT assay, for this preliminary report, PRNT results were available only for the 25-μg and 100-μg dose groups.
SARS-CoV-2 Neutralization Responses
No participant had detectable PsVNA responses before vaccination. After the first vaccination, PsVNA responses were detected in less than half the participants, and a dose effect was seen (50% inhibitory dilution [ID50]: Figure 2C, Fig. S8, and Table 2; 80% inhibitory dilution [ID80]: Fig. S2 and Table S6). However, after the second vaccination, PsVNA responses were identified in serum samples from all participants. The lowest responses were in the 25-μg dose group, with a geometric mean ID50 of 112.3 (95% CI, 71.2 to 177.1) at day 43; the higher responses in the 100-μg and 250-μg groups were similar in magnitude (geometric mean ID50, 343.8 [95% CI, 261.2 to 452.7] and 332.2 [95% CI, 266.3 to 414.5], respectively, at day 43). These responses were similar to values in the upper half of the distribution of values for convalescent serum specimens.
Before vaccination, no participant had detectable 80% live-virus neutralization at the highest serum concentration tested (1:8 dilution) in the PRNT assay. At day 43, wild-type virus–neutralizing activity capable of reducing SARS-CoV-2 infectivity by 80% or more (PRNT80) was detected in all participants, with geometric mean PRNT80 responses of 339.7 (95% CI, 184.0 to 627.1) in the 25-μg group and 654.3 (95% CI, 460.1 to 930.5) in the 100-μg group (Figure 2D). Neutralizing PRNT80 average responses were generally at or above the values of the three convalescent serum specimens tested in this assay. Good agreement was noted within and between the values from binding assays for S-2P and receptor-binding domain and neutralizing activity measured by PsVNA and PRNT (Figs. S3 through S7), which provides orthogonal support for each assay in characterizing the humoral response induced by mRNA-1273.
SARS-CoV-2 T-Cell Responses
The 25-μg and 100-μg doses elicited CD4 T-cell responses (Figs. S9 and S10) that on stimulation by S-specific peptide pools were strongly biased toward expression of Th1 cytokines (tumor necrosis factor α > interleukin 2 > interferon γ), with minimal type 2 helper T-cell (Th2) cytokine expression (interleukin 4 and interleukin 13). CD8 T-cell responses to S-2P were detected at low levels after the second vaccination in the 100-μg dose group (Fig. S11).
Discussion
We report interim findings from this phase 1 clinical trial of the mRNA-1273 SARS-CoV-2 vaccine encoding a stabilized prefusion spike trimer, S-2P. Experience with the mRNA platform for other candidate vaccines and rapid manufacturing allowed the deployment of a first-in-human clinical vaccine candidate in record time. Product development processes that normally require years10 were finished in about 2 months. Vaccine development was initiated after the SARS-CoV-2 genome was posted on January 10, 2020; manufacture and delivery of clinical trials material was completed within 45 days, and the first trial participants were vaccinated on March 16, 2020, just 66 days after the genomic sequence of the virus was posted. The accelerated timeline generated key interim data necessary to launch advanced large-scale clinical trials within 6 months after initial awareness of a new pandemic threat.
The two-dose vaccine series was generally without serious toxicity; systemic adverse events after the first vaccination, when reported, were all graded mild or moderate. Greater reactogenicity followed the second vaccination, particularly in the 250-μg group. Across the three dose groups, local injection-site reactions were primarily mild. This descriptive safety profile is similar to that described in a report of two trials of avian influenza mRNA vaccines (influenza A/H10N8 and influenza A/H7N9) that were manufactured by Moderna with the use of an earlier lipid nanoparticle capsule formulation11 and is consistent with an interim report of a phase 1–2 evaluation of a Covid-19 mRNA vaccine encoding the S receptor-binding domain.6 Those studies showed that solicited systemic adverse events tended to be more frequent and more severe with higher doses and after the second vaccination.
The mRNA-1273 vaccine was immunogenic, inducing robust binding antibody responses to both full-length S-2P and receptor-binding domain in all participants after the first vaccination in a time- and dose-dependent fashion. Commensurately high neutralizing antibody responses were also elicited in a dose-dependent fashion. Seroconversion was rapid for binding antibodies, occurring within 2 weeks after the first vaccination, but pseudovirus neutralizing activity was low before the second vaccination, which supports the need for a two-dose vaccination schedule. It is important to note that both binding and neutralizing antibody titers induced by the two-dose schedule were similar to those found in convalescent serum specimens. However, interpretation of the significance of those comparisons must account for the variability in Covid-19 convalescent antibody titers according to factors such as patient age, disease severity, and time since disease onset and for the number of samples in the panel.12,13
Though correlates of protection from SARS-CoV-2 infection have not yet been determined, measurement of serum neutralizing activity has been shown to be a mechanistic correlate of protection for other respiratory viruses, such as influenza14 and respiratory syncytial virus,15 and is generally accepted as a functional biomarker of the in vivo humoral response.16 In rhesus macaques given DNA vaccine candidates expressing different forms of the SARS-CoV-2 spike protein, post-vaccination neutralizing antibody titers were correlated with protection against SARS-CoV-2 challenge.17 Humoral and cell-mediated immune responses have been associated with vaccine-induced protection against challenge18 or subsequent rechallenge after SARS-CoV-2 infection in a rhesus macaque model.19 We found strong correlations between the binding and neutralization assays and between the live virus and pseudovirus neutralization assays. The latter finding suggests that the pseudovirus neutralization assay, performed under biosafety level 2 containment, may, when validated, serve as a relevant surrogate for live virus neutralization, which requires biosafety level 3 containment. In humans, phase 3 efficacy trials will allow assessment of the correlation of vaccine-induced immune responses with clinical protection.
In this interim report of follow-up of participants through day 57, we were not able to assess the durability of the immune responses; however, participants will be followed for 1 year after the second vaccination with scheduled blood collections throughout that period to characterize the humoral and cellular immunologic responses. This longitudinal assessment is relevant given that natural history studies suggest that SARS-CoV and MERS-CoV (Middle East respiratory syndrome coronavirus) infections, particularly mild illnesses, may not generate long-lived antibody responses.20-22
The rapid and robust immunogenicity profile of the mRNA-1273 vaccine most likely results from an innovative structure-based vaccine antigen design,23 coupled with a potent lipid-nanoparticle delivery system, and the use of modified nucleotides that avoid early intracellular activation of interferon-associated genes. These features of the mRNA composition and formulation have been associated with prolonged protein expression, induction of antigen-specific T-follicular helper cells, and activation of germinal center B cells.24 Stabilizing coronavirus spike proteins by substituting two prolines at the top of heptad repeat 1 prevents structural rearrangements of the fusion (S2) subunit. This has enabled the determination of atomic-level structure for the prefusion conformation of spike from both endemic and pandemic strains, including HKU1,25 SARS-CoV,26 and MERS-CoV.27 Moreover, S-2P conformational stability translates into greater immunogenicity,27-29 based on preservation of neutralization-sensitive epitopes at the apex of the prefusion molecule, as shown for respiratory syncytial virus F glycoprotein,30 and improved protein expression,27 which is particularly advantageous for gene-based antigen delivery. Thus, presentation of the naturally folded prefusion conformation of the S glycoprotein to the immune system from an mRNA template enables efficient within-host antigen production and promotes both high-quality and high-magnitude antibody responses to SARS-CoV-2.
Previous experience with veterinary coronavirus vaccines and animal models of SARS-CoV and MERS-CoV infection have raised safety concerns about the potential for vaccine-associated enhanced respiratory disease. These events were associated either with macrophage-tropic coronaviruses susceptible to antibody-dependent enhancement of replication or with vaccine antigens that induced antibodies with poor neutralizing activity and Th2-biased responses.31 Reducing the risk of vaccine-associated enhanced respiratory disease or antibody-dependent enhancement of replication involves induction of high-quality functional antibody responses and Th1-biased T-cell responses. Studies of mRNA-1273 in mice show that the structurally defined spike antigen induces robust neutralizing activity and that the gene-based delivery promotes Th1-biased responses, including CD8 T cells that protect against virus replication in lung and nose without evidence of immunopathology.32 It is important to note that mRNA-1273 also induces Th1-biased CD4 T-cell responses in humans. Additional testing in animals and ongoing T-cell analysis of clinical specimens will continue to define the safety profile of mRNA-1273.
These safety and immunogenicity findings support advancement of the mRNA-1273 vaccine to later-stage clinical trials. Of the three doses evaluated, the 100-μg dose elicits high neutralization responses and Th1-skewed CD4 T cell responses, coupled with a reactogenicity profile that is more favorable than that of the higher dose. A phase 2 trial of mRNA-1273 in 600 healthy adults, evaluating doses of 50 μg and 100 μg, is ongoing (ClinicalTrials.gov number, NCT04405076). A large phase 3 efficacy trial, expected to evaluate a 100-μg dose, is anticipated to begin during the summer of 2020.
Acknowledgments
We thank the members of the mRNA-1273 Study Team (see the Supplementary Appendix) for their many and invaluable contributions, the members of the safety monitoring committee (Stanley Perlman, M.D., Ph.D. [chair], University of Iowa; Gregory Gray, M.D., M.P.H., Duke University; and Kawsar Talaat, M.D., Johns Hopkins Bloomberg School of Public Health) for their oversight; Huihui Mu and Michael Farzan for providing the ACE2-overexpressing 293 cells; and Dominic Esposito, Ph.D., Director of the Protein Expression Laboratory at the Frederick National Laboratory for Cancer Research, for providing the S-2P protein for the immunologic assays. We also thank the participants themselves for their altruism and their dedication to this trial. The Emory University study team thanks the Georgia Research Alliance and Children’s Healthcare of Atlanta for their support. The Kaiser Washington study team thanks Howard Crabtree, R.Ph., and Sheena Mangicap, Seattle Pharmacy Relief, for their many years of support for vaccine trials.
Protocol
Supplementary Appendix
Disclosure Forms
Data Sharing Statement
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does any mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Moderna provided mRNA-1273 for use in this trial but did not provide any financial support. Employees of Moderna collaborated on protocol development, significantly contributed to the Investigational New Drug (IND) application, and participated in weekly protocol team calls. The National Institute of Allergy and Infectious Diseases (NIAID) ultimately made all decisions regarding trial design and implementation.
This article was published on July 14, 2020, at NEJM.org.
A data sharing statement provided by the authors is available with the full text of this article at NEJM.org.
Footnotes
Supported by the NIAID, National Institutes of Health (NIH), Bethesda, under award numbers UM1AI148373 (Kaiser Washington), UM1AI148576 (Emory University), UM1AI148684 (Emory University), UM1Al148684-01S1 (Vanderbilt University Medical Center), and HHSN272201500002C (Emmes); by the National Center for Advancing Translational Sciences, NIH, under award number UL1 TR002243 (Vanderbilt University Medical Center); and by the Dolly Parton COVID-19 Research Fund (Vanderbilt University Medical Center). Funding for the manufacture of mRNA-1273 phase 1 material was provided by the Coalition for Epidemic Preparedness Innovation (CEPI).
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Helmy YA, Fawzy M, Elaswad A, Sobieh A, Kenney SP, Shehata AA. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J Clin Med 2020;9:1225-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Draft landscape of COVID-19 candidate vaccines. July 14, 2020. (https://www.who.int/who-documents-detail/draft-landscape-of-covid-19-candidate-vaccines).
- 3.Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012;4:1011-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callaway E. The race for coronavirus vaccines: a graphical guide. Nature 2020;580:576-577. [DOI] [PubMed] [Google Scholar]
- 5.Zhu F-C, Li Y-H, Guan X-H, et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020;395:1845-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulligan MJ, Lyke KE, Kitchin N, et al. Phase 1/2 study to describe the safety and immunogenicity of a COVID-19 RNA vaccine candidate (BNT162b1) in adults 18 to 55 years of age: interim report. July 1, 2020. (https://www.medrxiv.org/content/10.1101/2020.06.30.20142570v1). preprint.
- 7.Smith TRF, Patel A, Ramos S, et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun 2020;11:2601-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367:1260-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Food and Drug Administration. Toxicity grading scale for healthy adult and adolescent volunteers enrolled in preventive vaccine clinical trials: guidance for industry. September 2007. (https://www.fda.gov/regulatory-information/search-fda-guidance-documents/toxicity-grading-scale-healthy-adult-and-adolescent-volunteers-enrolled-preventive-vaccine-clinical). [DOI] [PubMed]
- 10.Pronker ES, Weenen TC, Commandeur H, Claassen EHJHM, Osterhaus ADME. Risk in vaccine research and development quantified. PLoS One 2013;8(3):e57755-e57755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldman RA, Fuhr R, Smolenov I, et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine 2019;37:3326-3334. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Guo X, Xin Q, et al. Neutralizing antibodies responses to SARS-CoV-2 in COVID-19 inpatients and convalescent patients. Clin Infect Dis 2020. June 4 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu F, Wang A, Liu M, et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. April 20, 2020. (https://www.medrxiv.org/content/10.1101/2020.03.30.20047365v2). preprint.
- 14.Verschoor CP, Singh P, Russell ML, et al. Microneutralization assay titres correlate with protection against seasonal influenza H1N1 and H3N2 in children. PLoS One 2015;10(6):e0131531-e0131531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulkarni PS, Hurwitz JL, Simões EAF, Piedra PA. Establishing correlates of protection for vaccine development: considerations for the respiratory syncytial virus vaccine field. Viral Immunol 2018;31:195-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerging Infect Dis 2020;26:1478-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J, Tostanoski LH, Peter L, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020. May 20 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Doremalen N, Lambe T, Spencer A, et al. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. May 13, 2020. (https://www.biorxiv.org/content/10.1101/2020.05.13.093195v1). preprint. [DOI] [PubMed]
- 19.Chandrashekar A, Liu J, Martinot AJ, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 2020. May 20 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choe PG, Perera RAPM, Park WB, et al. MERS-CoV antibody responses 1 year after symptom onset, South Korea, 2015. Emerg Infect Dis 2017;23:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao W-C, Liu W, Zhang P-H, Zhang F, Richardus JH. Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med 2007;357:1162-1163. [DOI] [PubMed] [Google Scholar]
- 22.Wu L-P, Wang N-C, Chang Y-H, et al. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis 2007;13:1562-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham BS, Gilman MSA, McLellan JS. Structure-based vaccine antigen design. Annu Rev Med 2019;70:91-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardi N, Hogan MJ, Naradikian MS, et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J Exp Med 2018;215:1571-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchdoerfer RN, Cottrell CA, Wang N, et al. Pre-fusion structure of a human coronavirus spike protein. Nature 2016;531:118-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirchdoerfer RN, Wang N, Pallesen J, et al. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep 2018;8:15701-15701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pallesen J, Wang N, Corbett KS, et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A 2017;114:E7348-E7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrapp D, De Vlieger D, Corbett KS, et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell 2020;181(5):1004.e15-1015.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang N, Rosen O, Wang L, et al. Structural definition of a neutralization-sensitive epitope on the MERS-CoV S1-NTD. Cell Rep 2019;28(13):3395.e6-3405.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crank MC, Ruckwardt TJ, Chen M, et al. A proof of concept for structure-based vaccine design targeting RSV in humans. Science 2019;365:505-509. [DOI] [PubMed] [Google Scholar]
- 31.Graham BS. Rapid COVID-19 vaccine development. Science 2020;368:945-946. [DOI] [PubMed] [Google Scholar]
- 32.Corbett KS, Edwards D, Leist SR, et al. SARS-CoV-2 mRNA vaccine development enabled by prototype pathogen preparedness. June 11, 2020. (https://www.biorxiv.org/content/10.1101/2020.06.11.145920v1). preprint. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.