Abstract
A key goal to controlling coronavirus disease 2019 (COVID-19) is developing an effective vaccine. Development of a vaccine requires knowledge of what constitutes a protective immune response and also features that might be pathogenic. Protective and pathogenic aspects of the response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are not well understood, partly because the virus has infected humans for only 6 months. However, insight into coronavirus immunity can be informed by previous studies of immune responses to non-human coronaviruses, common cold coronaviruses, and SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV). Here, we review the literature describing these responses and discuss their relevance to the SARS-CoV-2 immune response.
While the immune response to SARS-CoV-2 is not yet well understood, insights may be gained from studies of other coronavirus infections. Here, Sariol and Perlman review the literature on animal and human coronavirus infections and discuss the critical outstanding questions for understanding SARS-CoV-2 vaccination and protective immunity.
Introduction
Coronavirus disease 2019 (COVID-19), caused by a novel coronavirus, SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), is the cause of a pandemic that has infected over 15,000,000 people worldwide with a mortality of ∼4% to date (World Health Organization, 2020a). Two previously identified coronaviruses, SARS-CoV and MERS-CoV (Middle East respiratory syndrome coronavirus), caused severe pneumonia but, unlike SARS-CoV-2, exhibited only limited person-to-person spread, resulting in dramatically lower numbers of confirmed cases (∼8,100 and 2,500, respectively). Because COVID-19 has been associated with huge mortality and economic loss, efforts are underway to rapidly develop a vaccine, which will result in a safer and more expedient path to herd immunity. After vaccination, the goal will be not only protecting the vaccinated individual but also decreasing transmission by minimizing the number of susceptible individuals. Vaccine development is highly dependent on understanding the immune response to SARS-CoV-2, especially those components that are protective. However, the immune response to coronaviruses is not well understood, and specific aspects that are protective versus pathogenic are not well defined. While some aspects of SARS-CoV-2 immunity appear to be novel, much of the immune response parallels that observed in humans, domestic and companion non-human animals naturally infected with coronaviruses, and experimentally infected laboratory animals. In this review, we will focus on studies that described innate and adaptive immune responses in the setting of these non-SARS-CoV-2 infections, focusing on those studies that potentially provide insight into COVID-19 immunity and vaccine development in humans.
Coronavirus Biology
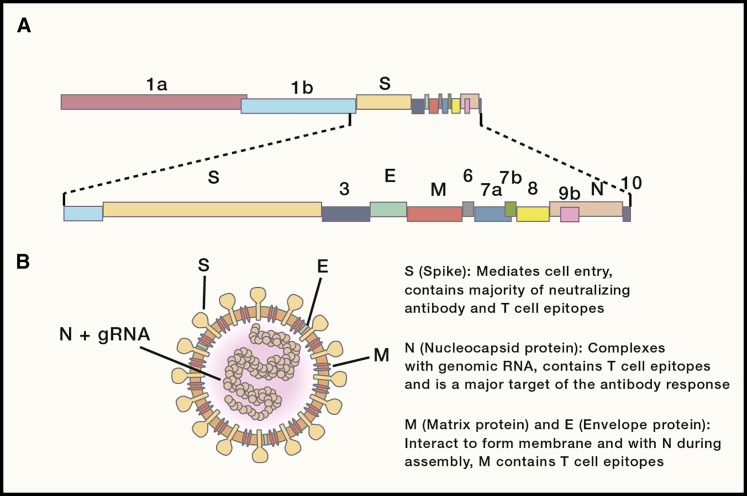
Coronaviridae is a family of large (31 kb) single-stranded positive-sense RNA viruses that consist of viruses from four genera (alpha, beta, gamma, and delta coronavirus). SARS-CoV, MERS-CoV, and SARS-CoV-2 are all betacoronaviruses. Genomic coronavirus RNA is translated into a long polyprotein that contains proteins involved in RNA replication (Figure 1 A). Structural proteins, which encompass the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, and accessory proteins believed to be involved in immunoevasion, are translated from a nested set of subgenomic RNAs that have the same 5′ and 3′ ends (Figure 1B). A coronavirus protein, nonstructural protein 14 (nsp14), has proofreading capabilities and is critical for maintaining and is responsible for the increased replication fidelity of coronaviruses. This is especially important given the size of the coronavirus genome. Coronaviruses have an estimated error rate of 10−6 to 10−7 errors per nucleotide, which is much lower than that of smaller RNA viruses (error rates of 10−3-10−5) (Smith et al., 2014).
Figure 1.
Genomic Organization and Virion Structure
(A) Schematic of the 30-kb SARS-CoV-2 genome. The first two-thirds of CoV genomes encode a polyprotein that is cleaved into constituent nonstructural proteins involved in replication and immune evasion, while the remaining one-third encodes the four main structural proteins (S, E, M, and N), along with accessory proteins.
(B) Schematic representation of a CoV virion. gRNA, genomic RNA.
Animal Coronaviruses
Coronaviruses are known to cause a wide variety of mild and severe diseases in domestic and companion animals, including livestock such as chickens, pigs, and cattle, as well as companion animals such as cats and dogs (Table 1 ). Because these coronaviruses have significant economic and psychological importance to humans, correlates of immunity have been investigated to guide development of protective vaccines against these pathogens.
Table 1.
Summary of Discussed Coronaviruses
| Virus | Genus | Host | Tropism | Available vaccine? |
|---|---|---|---|---|
| IBV | Gammacoronavirus | chicken | respiratory, kidney, reproductive tract | LAVs against several heterologous strains (Cavanagh, 2003) |
| TGEV | Alphacoronavirus | pig | enteric | LAV, PRCV as natural vaccine (Saif et al., 2019) |
| PRCV | Alphacoronavirus | pig | respiratory | no |
| BCoV | Betacoronavirus | cattle | respiratory, enteric | enteric disease only; inactivated virus, LAV (Saif, 2010) |
| FCoV/FIPV | Alphacoronavirus | cat | enteric (FCoV) | Temperature-sensitive LAV (Fehr et al., 1997; Gerber et al., 1990) |
| systemic (FIP) | ||||
| MHV | Betacoronavirus | mouse | strain dependent (enteric, hepatic, respiratory, CNS) | no |
| HCoV-229E | Alphacoronavirus | human | respiratory | no |
| HCoV-NL63 | Alphacoronavirus | human | respiratory | no |
| HCoV-OC43 | Betacoronavirus | human | respiratory | no |
| HCoV-HKU1 | Betacoronavirus | human | respiratory | no |
| SARS-CoV | Betacoronavirus | human | respiratory | no; multiple phase 1 trials (Lin et al., 2007; Martin et al., 2008) |
| MERS-CoV | Betacoronavirus | human | respiratory | no; three recently concluded phase 1 trials (Folegatti et al., 2020; Koch et al., 2020; Modjarrad et al., 2019) |
| SARS-CoV-2 | Betacoronavirus | human | respiratory | no; several ongoing trials (World Health Organization, 2020c) |
IBV, infectious bronchitis virus; TGEV, transmissible gastroenteritis virus; PRCV, porcine respiratory coronavirus; BCoV, bovine coronavirus; FCoV, feline coronavirus; FIPV, feline infectious peritonitis virus; MHV, mouse hepatitis virus; HCoV, human coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; CNS, central nervous system; LAV, live attenuated vaccine.
Infectious bronchitis virus (IBV), a chicken coronavirus, causes bronchitis and kidney and reproductive tract disease (Cavanagh, 2007). A number of vaccines, particularly live virus attenuated by passage in chicken eggs, generate neutralizing antibodies and have been successfully used for decades to protect flocks against IBV. Caveats are that protection provided by many of these vaccines is short-lived, with protective immunity waning after about 9 weeks (Cavanagh, 2007), and these vaccines afford poor protection from heterologous strains of IBV (Jackwood, 2012). The strength of cross-protection between variants is predicted by differences in the S1 subunit of the S protein, the site of most neutralizing antibody epitopes (Cavanagh et al., 1997). Of note for live attenuated vaccine strategies, high rates of recombination and frequent exposure of chickens to multiple vaccine and field IBV strains can contribute to the generation of novel virus strains and reversion to virulence (Jackwood and Lee, 2017; Jia et al., 1995; Zhang et al., 2010). S1 and the N protein contain CD8 T cell epitopes, which are known to confer protection, though little is known about the longevity of the T cell response to IBV or their relevance in vaccine-mediated immunity (Collisson et al., 2000).
Transmissible gastroenteritis virus (TGEV) is an enteropathic and highly contagious coronavirus that causes mild disease in adult animals but is nearly always fatal in piglets under 2 weeks of age, causing significant economic burden (Saif et al., 2019). As such, a substantive portion of vaccine and immunity research has focused on the generation of immunoglobulin A (IgA) responses in sows, which confer passive lactogenic immunity to suckling pigs via colostrum or milk (Chattha et al., 2015; Saif et al., 1972). T cell responses are correlated with the generation of lactogenic immunity in immunized sows, highlighting the importance of a cellular response (Antón et al., 1995; Park et al., 1998). While various live attenuated vaccines have been used to immunize sows, these vaccines do not induce as strong an IgA response as infection with virulent TGEV and are thus less protective to newborn piglets (Saif et al., 2019). Interestingly, the decline of TGEV incidence worldwide was correlated with the emergence of the closely related porcine respiratory coronavirus (PRCV) in 1983 (Schwegmann-Wessels and Herrler, 2006). PRCV, which generally causes subclinical to mild respiratory infections, is an S protein deletion mutant of TGEV. This roughly 200-amino-acid deletion in the N-terminal region of the S protein results in a shift of tropism from the enteric and respiratory tracts to only the respiratory tract (Sánchez et al., 1992). Because this virus provides protection against TGEV infection, including in newborn piglets, it represents a natural TGEV vaccine (Brim et al., 1995; Wesley and Woods, 1996). Interesting and perhaps relevant for COVID-19 immunity, IgA-mediated mucosal immunity following PRCV infection wanes over time, requiring reinfection or reimmunization prior to farrowing to produce protective lactogenic immunity (Callebaut et al., 1990; Wesley, 2002).
Similar features have also been observed in cattle infected with bovine coronavirus (BCoV), which causes respiratory and enteric disease. Reinfection is commonly observed in animals with measurable antibody titers, with concomitant virus shedding from the respiratory tract. Of note, disease was mitigated in duration in animals with high IgA titers prior to infection (Cho et al., 2001; Heckert et al., 1990). This recurrent theme of waning immunity and need for periodic boosts following initial infection or vaccination is relevant to our understanding of human respiratory coronaviruses and has implications for vaccine strategies against these viruses, especially SARS-CoV-2, in humans.
Feline infectious peritonitis virus (FIPV), a highly lethal feline coronavirus (FCoV), is thought to arise from mutations in the S protein during persistent infection with otherwise mild enteric strains of FCoV, resulting in a shift in tropism from intestinal epithelial cells to macrophages that allows systemic spread of the virus (Rottier et al., 2005). FIPV results in lymphopenia and severe serositis and is uniformly fatal. Antibody-dependent enhancement (ADE), a phenomenon in which virus-specific antibodies potentiate infection via Fc or complement-receptor-mediated uptake of infectious virus into myeloid cells, has been observed after vaccination against FIPV. This development of ADE reflects the macrophage tropism of FIPV and complicates vaccine development (Vennema et al., 1990; Weiss and Scott, 1981). ADE in the context of FIPV vaccination raises concerns about the same phenomenon occurring after SARS-CoV-2 vaccination, but ADE has not been described in any other coronavirus infection in vivo.
In addition, the most extensively studied coronavirus is mouse hepatitis virus (MHV), the prototypical laboratory coronavirus, which causes a wide variety of respiratory, enteric, neurological, and hepatic disease in susceptible rodents (Barthold and Smith, 1984). Some strains, such as the neurotropic JHMV or the neurotropic and hepatotropic MHV-A59, cause immune-mediated demyelination as a consequence of viral clearance (Bergmann et al., 2006; Wang et al., 1990). The immune responses to these neurotropic coronaviruses are among the most-studied aspects of coronavirus immunology, as these central nervous system (CNS) infections require an exquisite balance between immune activation to clear virus and suppression to prevent immunopathology. A crucial aspect of the immune response to these viruses is type I interferon (IFN), as mice lacking the type I IFN receptor (IFNAR) rapidly succumb to strains and doses of MHV that would ordinarily be sublethal or even nonpathogenic (Ireland et al., 2008; Khanolkar et al., 2009; Roth-Cross et al., 2008). Type I IFN is produced both by macrophages or microglia and plasmacytoid dendritic cells (pDCs) in these infections, and signals through LysM+ macrophages and CD11c+ dendritic cells (DCs) for protection (Cervantes-Barragán et al., 2009; Roth-Cross et al., 2008).
Virus-specific CD4+ and CD8+ T cell responses are necessary to clear CNS infection with JHMV (Savarin et al., 2008; Williamson and Stohlman, 1990); however, these same virus-specific T cell responses are pathogenic and mediate myelin destruction (Anghelina et al., 2006; Castro and Perlman, 1995; Wu et al., 2000). The role of type II IFN in T cells in protection and pathogenesis differs between CD4+ and CD8+ T cells, as IFN-γ produced by CD4+ T cells is protective against demyelination, whereas that produced by CD8+ T cells, while critical for viral clearance, contributes to demyelination (Bergmann et al., 2004; Pewe and Perlman, 2002; Pewe et al., 2002). Adding further to the complexity of the role of T cells in balancing viral clearance and immunopathogenesis, some subsets of these T cells, including T regulatory (Treg) cells and interleukin-10 (IL-10)-producing CD8+ T cells, are necessary for protection against excessive immune responses in the CNS (Anghelina et al., 2009; Cervantes-Barragán et al., 2012; Trandem et al., 2011). Interestingly, virus-specific Treg cells targeting the same immunodominant epitope as effector T cells are particularly critical for suppressing immunopathology (Zhao et al., 2011a, 2014a). Of particular relevance to SARS-CoV-2 and other highly pathogenic human coronaviruses, T cells are essential to prevent cytokine storm in MHV-A59 via tempering the innate immune response in both a Treg and non-Treg cell-dependent manner (Kim et al., 2007).
In addition to T cells, a virus-specific antibody response is required to prevent recrudescence after initial virus clearance, and passively transferred antibodies against MHV are protective against subsequent infection (Lin et al., 1999; Matthews et al., 2001; Ramakrishna et al., 2003). Neutralizing antibodies are also thought to play a key role in preventing CD8+ T cell escape, a feature of some persistent MHV infections (Butler et al., 2007; Chua et al., 2004; Dandekar et al., 2003). An immunopathogenic role for infiltrating monocytes and macrophages, which are found at high numbers in demyelinating lesions, has also been implicated (Templeton et al., 2008). The role for these myeloid cells in demyelination is further supported by the finding that in Rag1 −/− mice, expression of a macrophage chemoattractant (CCL2) encoded by recombinant virus is sufficient to mediate demyelination (Kim and Perlman, 2005).
Together, these studies of coronavirus infections of domestic and companion animals and experimentally infected animals illustrate the waning nature of the immune response, the requirement for both T cell and antibody responses for protection, and the fine balance between protective and pathogenic immune responses, which may all be relevant for understanding SARS-CoV-2 immunity.
Human Common Cold Coronaviruses
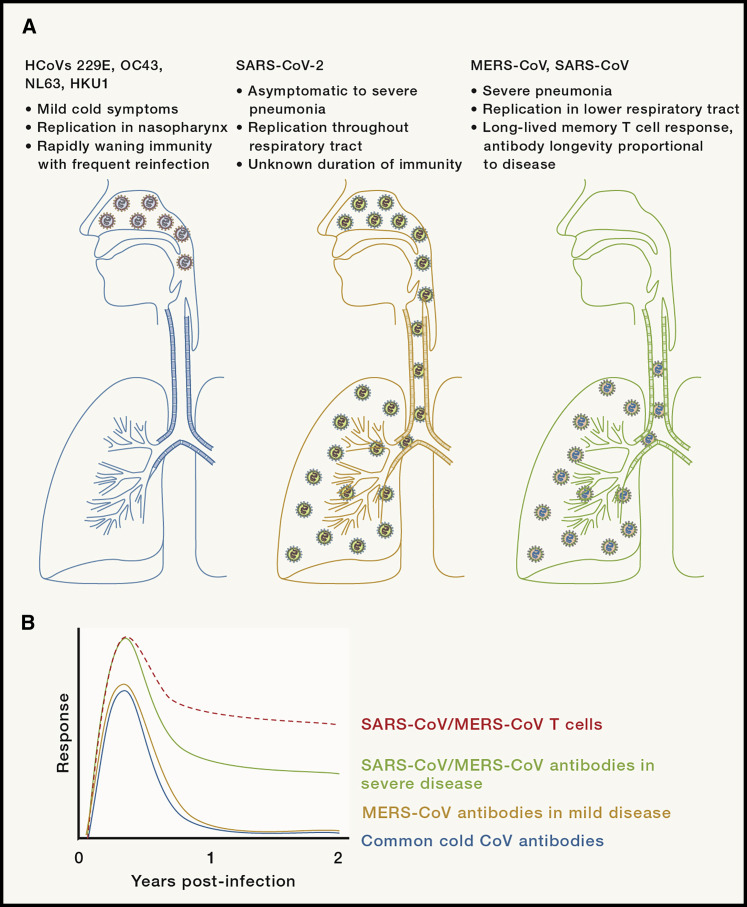
The first human coronaviruses were isolated in the 1960s from nasopharyngeal samples of individuals experiencing common colds (McIntosh et al., 1967; Tyrrell and Bynoe, 1965). These viruses were found to be morphologically similar to IBV and ultimately classified as coronaviruses. Four strains of these common cold coronaviruses are known to circulate globally, HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1, the latter two of which were discovered following heightened attention on human coronaviruses in the aftermath of the SARS epidemic (Fouchier et al., 2004; van der Hoek et al., 2004; Woo et al., 2005). Together, these coronaviruses are thought to cause ∼15% of common colds, and it has been estimated that ≥90% of adults have serum antibodies against these four viruses (Gorse et al., 2010; Perlman and McIntosh, 2019). Because these coronaviruses are the closest to SARS-CoV-2 in transmissibility and ability to replicate in the nasopharyngeal tract, albeit without the same predilection for severe lower respiratory tract disease, studies of the immune responses to these viruses may be of particular relevance to the current pandemic (Figure 2 A; Dijkman et al., 2013; Sungnak et al., 2020).
Figure 2.
Human Coronavirus Tropism and Longevity of Immune Responses
Schematic depicting sites of replication of human coronaviruses (A) and schematic of longevity of immune responses to common cold coronaviruses, SARS-CoV, and MERS-CoV (B) (not drawn to scale). Data not available for antibody longevity in patients following mild disease caused by SARS-CoV.
The majority of our understanding of immunity against these viruses comes from experimental infections of volunteers with HCoV-229E or HCoV-OC43, as well as longitudinal serological surveys monitoring respiratory infections. These volunteer studies demonstrated that reinfection with a homologous virus can occur even when measurable titers of neutralizing antibodies are present in serum prior to infection, though there was an inverse correlation between antibody titer and likelihood of symptomatic infection (Bradburne et al., 1967). Similar to studies of animal coronaviruses, antibody titers in volunteers had waned substantially 1 year after initial infection and many could be reinfected and shed virus, though these secondary infections did not cause clinical symptoms (Figure 2B; Callow et al., 1990). Large serological surveys of natural infections with HCoV-229E and HCoV-OC43 have revealed similar trends of reinfection and patterns of waning and rising antibody titers, with estimates of anywhere between 30% and 80% of infections with HCoV-229E or HCoV-OC43 representing reinfections based on pre-infection antibody titers (Hendley et al., 1972; Monto and Lim, 1974; Schmidt et al., 1986). It should be noted that in all of these studies, serum antibody titers are measured. In other experimental respiratory virus infections, levels of mucosal antibody appear to be more relevant for establishing the likelihood of reinfection (Habibi et al., 2015; Singleton et al., 2003). Measurements of mucosal antibodies in COVID-19 patients will be critical for fully determining the likelihood of developing clinical disease after primary infection or vaccination. Reinforcing this notion of frequent reinfections with community-acquired respiratory coronaviruses, a recent longitudinal survey of 196 individuals using qRT-PCR-based detection of viral genetic material found that over an 18-month period, 12 of these individuals became reinfected with the same coronavirus at least once, with a mean of 37 weeks between positive tests (Shaman and Galanti, 2020). In terms of therapeutic interventions, intranasal administration of recombinant IFN-α prior to infection protected against experimental infection, resulting in reduced viral loads and diminished incidence and severity of symptoms (Higgins et al., 1983). Early IFN-α treatment is thus likely to induce an antiviral state independent of the presence of optimal antivirus antibody and T cell responses.
SARS-CoV
Beginning in Guangdong Province in China in November 2002, SARS-CoV, the causative agent of SARS, infected an estimated 8,098 people with a near 10% mortality rate until being declared contained by the World Health Organization (WHO) in 2003 (Peiris et al., 2004). This epidemic caused substantial global concern and sparked interest in coronavirus research. Relative to the common cold coronaviruses and SARS-CoV-2, SARS-CoV was significantly less transmissible and, despite worldwide dissemination, predominantly spread within healthcare settings and among households, with no evidence of asymptomatic transmission (Peiris et al., 2003a).
SARS-CoV, like SARS-CoV-2, caused severe pulmonary pathology, manifested by edema, hyaline membrane formation, infiltration of inflammatory cells (including lymphocytes and macrophages in the alveoli and interstitium), and epithelial denudation (Lee et al., 2003; Nicholls et al., 2003). This pulmonary disease progressed to acute respiratory distress syndrome (ARDS), particularly in older individuals and those with underlying conditions (Lew et al., 2003). Vasculitis, lymphopenia, spleen and lymph node atrophy, and virus presence in several tissues, including the blood, brain, spleen, and other tissues, were observed. Similar features are shared with COVID-19 (Ding et al., 2003; Gu and Korteweg, 2007; Puelles et al., 2020). While viral load began to decline around day 10 post-onset, clinical disease worsened in many patients around this time, suggesting potential immunopathology, rather than excessive viral replication, as the cause of clinical progression (Peiris et al., 2003b). SARS-CoV, like SARS-CoV-2, uses angiotensin-converting enzyme 2 (ACE2) as its receptor for entry into the cell and primarily infects airway and alveolar epithelial cells (Figure 2A; He et al., 2006; Zhou et al., 2020). Some studies suggested that the use of ACE2 as the viral receptor contributed directly to the virulence and pathology of SARS-CoV, as ACE2, a regulator of renin-angiotensin signaling, is downregulated upon entry and this inhibition can lead to enhanced lung pathology (Imai et al., 2005; Kuba et al., 2005). Downregulation of ACE2 has been observed in mice infected with influenza A virus (IAV), which results in elevated serum angiotensin II compared to wild-type mice (Yang et al., 2014; Zou et al., 2014). In IAV-infected mice and patients, elevated serum angiotensin II is correlated with disease severity. Further, Ace2 −/− mice develop more severe IAV-mediated disease than do their wild-type counterparts. These results support the notion that downregulation of ACE2 may contribute to SARS-CoV pathogenesis.
Studies of the immune response to SARS-CoV have consisted of both direct study of human SARS patients and animal models of SARS, including macaques, marmosets, and ferrets, as well as smaller animals, such as hamsters and particularly mice (Gretebeck and Subbarao, 2015). While mice are susceptible to infection with SARS-CoV, young mice develop no illness, and aged mice develop mild clinical disease (Roberts et al., 2005). In order to address this limitation, transgenic mice expressing the human ACE2 (hACE2) gene were developed; however, though they develop pulmonary disease, they also develop a lethal encephalitis (McCray et al., 2007; Tseng et al., 2007). A different approach instead adapted SARS-CoV to mice by serially passaging the virus in mouse lungs. MA15, the first of these mouse-adapted SARS-CoV strains, caused severe pulmonary disease in young mice (Roberts et al., 2007). This virus had six coding mutations relative to the original virus, four of which were in located in ORF1 nonstructural proteins, one in the receptor-binding domain of the S protein, and one in the M protein. Subsequent experiments determined that the substitution in the receptor-binding domain of the S protein and, to a lesser extent, a second one in nsp9 contributed to the enhanced disease in mice relative to unadapted virus (Frieman et al., 2012).
Innate Immune Responses
The cytokine response to SARS-CoV was frequently characterized by high-level production of pro-inflammatory chemokines and cytokines (e.g., CCL2, CCL3, CCL5, and CXCL10) and IL-6, tumor necrosis factor (TNF), and IL-8 production, all of which were further upregulated in patients with more severe disease (He et al., 2006; Jiang et al., 2005; Zhang et al., 2004). Similar findings were also observed in SARS animal models and in vitro, both in human airway epithelial (HAE) cells and in human monocyte-derived macrophages and DCs after infection (Cheung et al., 2005; Law et al., 2005; Yen et al., 2006). Interestingly, while macrophages and DCs can be infected by SARS-CoV and produce cytokines following infection, replication is abortive in these cells. Of note, infected monocytes/macrophages and monocyte-derived DCs do not produce type I IFN, suggesting that SARS-CoV immune evasion strategies are effective in these cells (Cheung et al., 2005; Law et al., 2005; Yilla et al., 2005).
As is the case of other viruses, such as measles virus, influenza virus, dengue virus, and Ebola virus (García-Sastre, 2017), coronaviruses, including SARS-CoV, have developed numerous mechanisms to counter the type I IFN response, both via evasion and direct antagonism of IFN signaling. Evasion of sensors of viral double-stranded RNA (dsRNA), including Mda5, RIG-I, and mitochondrial antiviral signaling protein (MAVS), is mediated by an array of mechanisms, including 2′-O-methylation of the 5′ viral mRNA cap by nsp16 (Menachery et al., 2014; Züst et al., 2011), as well as endoribonuclease degradation of viral RNAs and selective RNA packaging mediated by nsp15 (Athmer et al., 2018; Deng et al., 2017; Kindler et al., 2017). Other coronavirus proteins mediate inhibition of pattern recognition receptors or IFN production and signaling pathway molecules, reflecting an extensive array of mechanisms of immune evasion (Fehr et al., 2016; Frieman et al., 2009; Hu et al., 2017; Zhao et al., 2012b). Despite these immune evasion strategies, it has been shown that both primary human and mouse plasmacytoid DCs are capable of inducing a type I IFN response after SARS-CoV infection in a manner dependent on TLR7, which detects single-stranded RNA (Cervantes-Barragan et al., 2007; Channappanavar et al., 2016).
The type I IFN response in SARS patients was observed to be dysregulated in patients that experienced adverse outcomes and severe disease, with one report finding that IFN responses persisted significantly longer than in those patients who went on to recover, and were accompanied by the lack of a protective antivirus neutralizing antibody response (Cameron et al., 2007). Other reports did not describe this persistent IFN expression and instead found a poor IFN response relative to other respiratory viruses, a pattern that seems to be reflected in patients infected with SARS-CoV-2 (Blanco-Melo et al., 2020; Reghunathan et al., 2005). A pathogenic role for dysregulated IFN signaling is reflected in mouse studies of SARS-CoV. While pre-infection or early treatment with recombinant IFN-β or poly(I:C) to induce a type I IFN response resulted in complete protection from lethal disease, administration of IFN-β to mice at the peak of SARS-CoV replication led to delayed viral clearance and enhanced lethality rather than protection (Channappanavar et al., 2016, 2019; Zhao et al., 2012a). This delayed IFN-enhanced disease was characterized by T cell apoptosis and elevated inflammatory monocyte and macrophage accumulation in the lungs, with production of inflammatory cytokines such as IL-6, CCL2, and TNF. Antibody-mediated depletion of these inflammatory monocytes and macrophages was fully protective against lethality, suggesting that these cells were responsible for significant immunopathology. Together, these data suggest a role for dysregulated IFN signaling in the immunopathogenesis of SARS-CoV and other coronaviruses.
Adaptive Immune Responses
The antibody response to SARS-CoV is characterized by seroconversion as early as 4 days and generally ∼10–16 days post-onset, with titers peaking ∼15–20 days post-infection (Hsueh et al., 2004; Lee et al., 2006; Wu et al., 2007). Several neutralizing antibody epitopes, predominantly against the S1 and S2 subunits of the S protein, and particularly the receptor binding domain (RBD) in the S1 subunit, have been identified (Buchholz et al., 2004; Zhong et al., 2005). While antibodies against other structural proteins have been observed, these are largely non-neutralizing (Akerström et al., 2006; Qiu et al., 2005). A positive correlation between N- and S-protein-specific serum antibody titers and recovery from SARS-CoV was observed, and passive transfer of neutralizing antibodies was found to prevent replication in mouse models of SARS (Bisht et al., 2004; Subbarao et al., 2004; Zhang et al., 2006). However, these antibody responses have been found to lack longevity. While serum antibody titers remain high for the first 2 years after infection, by 3 years post-infection, only 55% of patients tested had detectable IgG responses to SARS-CoV proteins, and by 6 years post-infection, no detectable memory B cell responses remained in the periphery (Tang et al., 2011; Wu et al., 2007). These data suggest that antibody responses to SARS-CoV, like those to common cold coronaviruses, wane significantly with time (Figure 2B). However, a recent study suggests that as late as 13 years post-infection, low levels of IgG against whole SARS-CoV could be detected some survivors, suggesting variability in the longevity of the antibody response (Guo et al., 2020). Further, a meta-analysis convalescent plasma therapy during the SARS epidemic demonstrated some efficacy, highlighting the importance of the humoral response to SARS-CoV (Mair-Jenkins et al., 2015).
In contrast to the poor longevity of the antibody response to SARS-CoV, the memory T cell response has been reported to have significant longevity in patients (Figure 2B), with CD4 and CD8 T cell responses in the blood identified by ELISpot at 4 and 6 years after infection in 70%–100% of patients, including those with no identifiable memory B cell responses in the blood (Fan et al., 2009; Oh et al., 2011; Tang et al., 2011). These T cells were frequently polyfunctional, expressing IFN-γ, TNF, and other cytokines. Others have identified positive CD8 T cell responses as late as 11 years post-infection (Ng et al., 2016b). The duration of this memory T cell response was positively correlated, at least in part, with disease severity (Tang et al., 2011). A number of T cell epitopes have been identified, with most found in the S, N, and M proteins (Liu et al., 2017). While the longevity and specificity of the T cell response in human studies have been well studied, the kinetics of the T cell response during acute infection and the correlates of protection are less well understood. Lymphopenia was a commonly observed phenotype in SARS patients during acute disease, and T cell activation was also found to be suppressed, particularly in patients with severe disease (Cameron et al., 2008; He et al., 2005; Yu et al., 2003). Additionally, increased T helper 2 (Th2) cytokine expression correlate with poor outcomes in patients, a finding also supported in mouse studies of SARS-CoV (Li et al., 2008; Page et al., 2012).
Because of the general lack of human data regarding T cell kinetics and function during acute coronavirus infections, much of our understanding of their role comes from studies of mice. Zhao et al. found that suboptimal T cell responses result from an impairment of respiratory DC migration from the lungs to the lymph nodes (Zhao et al., 2009). This inhibition of DC migration was mediated, at least in part, by inhibitory alveolar macrophages, as depletion of these cells prior to infection resulted in enhanced T cell responses, viral clearance, and survival in mice. Aging is thought to play a significant role in this process, as expression of prostaglandin D2 (PGD2) and an upstream phospholipase (PLA2G2D) increase with age and are strongly correlated with this migration defect and suboptimal T cell response. Inhibition or depletion of these factors reverse these age-related impairments (Vijay et al., 2015; Zhao et al., 2011b). PGD2 signaling through its receptor on DCs may contribute to the exacerbated disease and enhanced mortality observed with age in the SARS epidemic. Infected HAE cells have also been observed to produce cytokines, particularly IL-6 and IL-8, that inhibit priming of naive T cells by DCs (Yoshikawa et al., 2009).
Further, T cells alone are sufficient to partly control SARS-CoV in mice, as adoptive transfer of activated T cells into Rag1 −/− mice lacking T cell responses ameliorated disease and conferred significant protection and viral clearance. Depletion of CD4 T cells during SARS-CoV infection resulted in impaired viral clearance and reduced neutralizing antibody titers (Chen et al., 2010; Zhao et al., 2010). Immunization with vaccinia encoding CD4 or CD8 T cell epitopes or DCs coated with these peptides prior to infection with SARS-CoV resulted in significant protection from SARS-CoV lethality (Channappanavar et al., 2014; Zhao et al., 2010). Intranasal immunization of mice with Venezuelan equine encephalitis replicon particles (VRPs) encoding an N-protein-specific CD4 T cell epitope resulted in the generation of an airway memory T cell response that conferred complete protection from lethality and reduced pulmonary edema upon SARS-CoV infection and resulted in enhanced DC and CD8 T cell responses in an IFN-γ-dependent manner, highlighting the role of type II IFN in the lungs (Zhao et al., 2016). However, others have observed enhanced immunopathology and eosinophilic infiltrates in the lungs in mice immunized peripherally with VRP-N and challenged with SARS-CoV, demonstrating the importance of the route of immunization in protection (Deming et al., 2006). Together, these data highlight the important roles that both the humoral and cellular response to SARS-CoV have in controlling disease and guide vaccine studies.
MERS-CoV
MERS-CoV was first identified in 2012 and to date has resulted in ∼2,500 confirmed infections, with a mortality rate of ∼35% (World Health Organization, 2020b; Zaki et al., 2012). Like SARS-CoV, this virus is significantly less transmissible than the common cold coronaviruses or SARS-CoV-2, with all cases occurring on the Arabian Peninsula or travelers from this region with local transmission. The most notable outbreak occurred in South Korea, in which the index case had recently returned from the Arabian Peninsula (Cho et al., 2016). Primary infections are believed to occur via camel-to-human transmission, as the virus circulates widely in dromedary camels throughout Asia and Africa (Chu et al., 2018; Kiambi et al., 2018; Zheng et al., 2019). Secondary infections largely occur in healthcare and household settings (Drosten et al., 2014; Memish et al., 2020). In severe cases, MERS manifests clinically as pneumonia than can rapidly progress to ARDS and multiorgan failure (Arabi et al., 2017). While only two autopsies have been performed, diffuse alveolar damage with hyaline membrane formation in the lungs and alveolar edema was observed (Alsaad et al., 2018; Ng et al., 2016a). MERS-CoV predominantly infects cells of the respiratory tract, using dipeptidyl peptidase 4 (DPP4) as its receptor for cell entry, which is highly expressed on airway epithelial cells and some hematopoietic cells (Figure 2A; Raj et al., 2013).
As in SARS, study of the human immune response is limited. This has led to the use of MERS-CoV-susceptible animal models, such as macaques and marmosets, and the development of mouse models that express humanized DPP4, as well as mouse-adapted MERS-CoV, to facilitate study of the immune response to the virus (Cockrell et al., 2016; Falzarano et al., 2014; Li et al., 2017; de Wit et al., 2013; Zhao et al., 2014b).
Innate Immune Responses
The innate immune response in severe MERS is characterized by elevated levels of pro-inflammatory cytokines in the blood, particularly IL-6 and CXCL10, along with IL-8, CCL5, and IFN-α (Kim et al., 2016; Min et al., 2016). These elevated cytokines correlated with elevated numbers of neutrophils and macrophages and lymphopenia in peripheral blood mononuclear cell (PBMC) samples, suggesting that they contribute to immunopathology. Unlike SARS-CoV, which only abortively infected macrophages and DCs, MERS-CoV replicates in human monocyte-derived macrophages and DCs, causing elevated production of these cytokines relative to SARS-CoV in these cells (Chu et al., 2014; Tynell et al., 2016; Zhou et al., 2014). Infection of HAE cells also resulted in production of pro-inflammatory cytokines, as well as downregulation of major histocompatibility complex (MHC) class I and II molecules, in part via epigenetic modulation of MHC class I components (Josset et al., 2013; Menachery et al., 2018).
MERS infection of myeloid cells or HAEs did not result in type I IFN production, again reflecting the ability of coronaviruses to evade viral RNA sensing and antagonize the IFN response. Similar to SARS-CoV, however, infection of human pDCs resulted in abortive infection with expression of both type I and type III IFNs in a TLR7-mediated manner (Scheuplein et al., 2015). In experimentally infected macaques, early treatment with IFN-α (beginning 8 h post-infection) and ribavirin resulted in reduced pathology and viral load (Falzarano et al., 2013). Similar to experiments with SARS-CoV, treatment of mice with recombinant IFN-β was protective when provided prior to the peak of viral replication (1 day post-infection), whereas delaying treatment until the peak of viral replication (2–4 days post-infection) resulted in elevated viral load, an increase in neutrophils and inflammatory monocytes and macrophages in the lungs, and enhanced lethality (Channappanavar et al., 2019). Clinical trials of recombinant IFNs in combination with ribavirin, a nucleoside inhibitor and antiviral, have demonstrated no changes in 28- and 90-day survival rates or kinetics of viral clearance (Arabi et al., 2020a; Omrani et al., 2014), though clinical trials of IFN in combination with antiviral protease inhibitors (lopinavir and ritonavir) are ongoing (Arabi et al., 2020b). Taken together, these data reflect a role for dysregulation of the innate immune response in the pathogenesis of MERS-CoV similar to that observed with SARS-CoV.
Adaptive Immune Responses
While most patients showed seroconversion ∼2–3 weeks post-onset of disease, absent or delayed antibody responses were strongly associated with severe or fatal disease (Corman et al., 2016; Park et al., 2015). Neutralizing antibodies identified in human patients, as well as convalescent patient sera, showed both prophylactic and therapeutic protection in mice (Corti et al., 2015; Zhao et al., 2017). However, while these antibodies had a protective effect in mice, MERS-CoV clearance was demonstrated in mice lacking B cells, showing that antibodies are not necessary to clear acute infection (Zhao et al., 2014b). Because MERS-CoV frequently crosses from camels to infect humans, antibodies that allow neutralization of diverse camel strains of MERS-CoV, as well as antibodies or combinations of antibodies that prevent antibody escape, have been identified (Tai et al., 2016; Tang et al., 2014; Wang et al., 2018). While MERS-CoV-specific antibody responses persist for at least 2 years in patients who recovered from severe disease, responses are not detected or transient in patients with subclinical or mild disease, waning to low or undetectable levels by 2 years post-infection (Figure 2B; Drosten et al., 2014; Zhao et al., 2017).
The T cell response to MERS-CoV has been partially characterized. Despite lymphopenia in many patients, MERS-specific CD8 T cell response were observed in patients with severe disease, whereas CD4 T cell responses did not develop until convalescence (Shin et al., 2019). It has been shown that activated T cells can be directly infected ex vivo with MERS-CoV, resulting in activation of apoptosis pathways, which could, along with the downregulation of MHC molecules in airway epithelial cells and dysregulated cytokine response, contribute to the lymphopenia observed in many patients (Chu et al., 2016). Memory T cell responses in MERS survivors were polyfunctional, expressing both IFN-γ and TNF, consistent with greater protective ability. These responses could be detected in all patients as late as 2 years post-infection, including in patients with no detectable antibody response, suggesting that at least some immune memory remains intact despite transient antibody responses (Zhao et al., 2017). Further, T cell responses have been demonstrated to play critical protective roles in MERS-CoV infections of mice, as animals lacking T cells were incapable of clearing virus, resulting in persistent infection (Zhao et al., 2014b). Intriguingly, immunization with a VRP encoding a SARS-CoV N protein CD4 T cell epitope resulted in some degree of cross-protection against MERS-CoV, resulting in reduced viral load (Zhao et al., 2016). This epitope is fairly well conserved between these two and related bat coronaviruses. It was observed that mice immunized with the MERS-CoV-specific epitope mediated some protection upon SARS-CoV infection, and the homologous epitope in a MERS-like bat coronavirus (HKU4) mediated protection against MERS-CoV challenge. Despite these protective roles for T cells in MERS-CoV infection, others have found that depletion of CD8 T cells in a sublethal mouse model of MERS-CoV results in diminished lung pathology and clinical disease without impacting the viral titers, suggesting that these cells may also play a role in immunopathogenesis (Coleman et al., 2016).
While there are no currently licensed vaccines for MERS-CoV, three vaccine candidates, an adenovirus-vectored vaccine, a modified vaccinia Ankara-vectored vaccine, and a DNA vaccine, each of which encodes the full-length S protein of MERS-CoV, have recently concluded phase 1 clinical trials and were demonstrated safe and capable of inducing neutralizing antibodies and virus-specific T cells responses in participants (Folegatti et al., 2020; Koch et al., 2020; Modjarrad et al., 2019). However, consistent with the results from the natural infection, immune responses, especially neutralizing antibody titers, had waned by 1 year after vaccination.
The Next Steps Forward
One can draw several conclusions relevant to our understanding of COVID-19 immunity from prior coronavirus studies. First, while less important for vaccine strategies, it is apparent from studies with MHV, as well as SARS-CoV and MERS-CoV, that an early type I IFN response is critical for protection from severe disease and preventing an exacerbated or aberrant pro-inflammatory cytokine response. However, coronaviruses engage in various immune evasion strategies that successfully prevent detection by pattern recognition receptors or inhibit IFN signaling pathways, resulting in either absent or delayed and dysregulated IFN responses that contribute to pathogenesis, rather than protection. Reflecting this, studies of SARS-CoV-2 have shown that the virus is sensitive to IFN pretreatment in vitro, but patients have impaired IFN responses with low levels of IFN production or signaling (Blanco-Melo et al., 2020; Hadjadj et al., 2020; Lokugamage et al., 2020). The cytokine and chemokine response in patients has also been found to be dysregulated in patients with impaired IFN responses, particularly those with severe disease, with many of the elevated pro-inflammatory cytokines and chemokines matching those seen in SARS and MERS, including CXCL10, CCL2, CCL3, IL-6, and TNF (Chen et al., 2020; Huang et al., 2020; Yang et al., 2020).
Second, studies of animal and human coronaviruses indicate that both the humoral and cellular adaptive immune responses are important mediators of protection, and a vaccine should robustly induce both. Neutralizing antibody responses are protective in coronavirus infections and are thus a primary target for vaccine strategies. However, as described above, antibody responses to coronaviruses can wane rapidly following infection or immunization, allowing for potential reinfection, particularly in mild or subclinical disease such as those caused by the common cold coronaviruses or mild MERS. Because SARS-CoV-2 infection often presents with asymptomatic or mild disease similar to the common cold viruses (Arashiro et al., 2020; Black et al., 2020), this is of particular concern, as it is possible that those cases will develop rapidly waning immunity relative to severe cases, potentially allowing for reinfection similar to the common cold coronaviruses. While seroconversion is observed in all COVID-19 patients 2–3 weeks post-symptom onset, early observations have suggested that antibody titers can wane significantly as early as 30–50 days post-symptom onset (Adams et al., 2020; Long et al., 2020; Robbiani et al., 2020). To date, while SARS-CoV-2 RNA has been detected after periods of negative testing, there is no available culture-based evidence confirming reinfection (Kirkcaldy et al., 2020). Together, these data suggest that vaccine strategies against SARS-CoV-2 could require boosting to maintain sufficient neutralizing antibody titers if the immune response is similar to that observed in mild disease. In contrast, based on our experience with MERS and SARS, it is probable that a more stable immune response will develop in patients with severe pneumonia.
Third, T cell responses are also sufficient for at least partial protection in many coronavirus infections and play a role in mitigating the exuberant innate immune responses involved in cytokine release syndromes. However, as in the case of SARS and MERS patients, COVID-19 patients develop lymphopenia, particularly those with severe disease, delaying the T cell response to the virus, though a virus-specific CD4 and CD8 T cell response is eventually mounted in most patients (Grifoni et al., 2020; Huang et al., 2020; Tan et al., 2020). T cell responses to SARS-CoV and MERS-CoV have also been observed to have enhanced durability relative to neutralizing antibody responses and thus are crucial for longevity of immunity induced by vaccination. However, T cells have also been observed to play immunopathogenic roles in some coronavirus infections, including Th2-skewed responses to SARS-CoV. Defining and generating a protective T cell response, rather than a pathogenic Th2-driven response, will also be important for vaccine efficacy.
Another consideration for vaccination strategies is the generation of mucosal immunity, as studies of both animal coronaviruses and SARS-CoV and MERS-CoV have highlighted the importance of IgA-driven immune responses and localization of T cells to the respiratory tract and lungs. Reflecting this, preclinical animal studies of SARS-CoV and MERS-CoV vaccine candidates have found enhanced protection correlated with intranasal immunization relative to parenteral routes (Jia et al., 2019; Kim et al., 2019; Zhao et al., 2016).
Finally, on a cautionary note for vaccination, the potential for enhanced disease, in the forms of ADE or vaccine-associated enhancement of respiratory disease (VAERD), must be considered. As mentioned above, ADE has not been observed in any human coronavirus or, for the most part, in non-human coronaviruses. Viruses associated with ADE show a preferential tropism for macrophages, unlike human respiratory coronaviruses. As SARS-CoV-2 primarily infects the respiratory tract and lungs, a markedly different tropism than the macrophage-tropic FIPV, ADE is unlikely in our estimation. ADE has also never been observed in SARS or MERS, and sera from rats immunized with the receptor-binding domain of the SARS-CoV-2 S protein does not enhance viral entry into Fcgamma receptor-expressing cells, further suggesting that this is unlikely for SARS-CoV-2 (Quinlan et al., 2020). VAERD, on the other hand, has some precedent in vaccination studies of animal models of SARS, including in non-human primates (Liu et al., 2019) and mice immunized against a human SARS-CoV isolate and challenged with a heterologous strain of the virus (Deming et al., 2006). These mouse studies also indicated that VAERD was most prominent in aged mice. While vaccines need to be carefully evaluated for evidence of VAERD, there is good reason to believe that proper vaccine and adjuvant formulation will minimize the risks of this problem yet still induce a protective immune response (Iwata-Yoshikawa et al., 2014). Critical will be to identify a vaccine strategy that elicits long-lasting immune responses.
Reaching these goals will require progress on several fronts. Much of our understanding of immune responses in the context of MERS and SARS resulted from studies of experimentally infected animals. Thus, the establishment of useful animal models of COVID-19 will be instrumental in understanding COVID-19 immunity. Taking cues from prior knowledge of SARS-CoV and MERS-CoV animal models, several of these models are currently being explored, including non-human primates, ferrets, hamsters, and mice. Each of these models presents unique advantages and disadvantages.
Infection of non-human primates, specifically macaques, results in mild clinical disease, virus replication in the respiratory tract, and pathological features such as pulmonary edema and hyaline membrane formation, features shared with COVID-19 (Munster et al., 2020; Rockx et al., 2020). As non-human primates are our closest evolutionary relatives, these animals may provide the most relevant data for human pathogenesis, immunity, and preclinical therapy and vaccine safety and efficacy (Chandrashekar et al., 2020; Gao et al., 2020; Williamson et al., 2020). However, the limited availability and expense associated with non-human primate studies limit their potential for widespread use. The use of smaller, more abundant, and readily available animal models is thus of importance as well.
While ferrets are often used in studies of respiratory disease and infection (Enkirch and von Messling, 2015), infection with SARS-CoV-2 results in relatively low viral titers and a lack of symptoms other than elevated body temperature (Kim et al., 2020; Shi et al., 2020). Ferrets will be useful for transmission studies, because infection of ferrets results in transmission of SARS-CoV-2 to co-housed naive ferrets. Similarly, infection of hamsters results in transmission to naive animals, both with direct and indirect contact. Despite elevated viral genomic material relative to ferrets, clinical symptoms are limited to relatively mild weight loss and moderate-to-severe lung pathology (Chan et al., 2020; Imai et al., 2020; Sia et al., 2020). These models will be best suited for study of SARS-CoV-2 transmission, particularly modeling asymptomatic or mild COVID-19 patient spread, as well as evaluation of vaccines and antiviral drugs.
Laboratory mice, due to their widespread availability, diverse array of reagents and tools for study, and past use as models for SARS-CoV and MERS-CoV, are of particular interest for studies of the immune response to SARS-CoV-2. While mice do not naturally support SARS-CoV-2 replication due to receptor incompatibility between the receptor-binding domain of the virus and mouse ACE2 (Zhou et al., 2020), several approaches are being used to render mice susceptible to SARS-CoV-2 infection, informed by similar mouse models of MERS and SARS. One such approach is the generation of transgenic mice expressing hACE2 under control of promoters that allow for expression in airway epithelial cells (Bao et al., 2020; Jiang et al., 2020). This approach allows for infection of the respiratory tract and the development of mild lung pathology following infection; however, hACE2 may also be expressed in additional tissues, including the brain. Infection of the brain results in a lethal encephalitis, similar to results observed when hACE2-transgenic mice were infected with SARS-CoV (McCray et al., 2007). While SARS-CoV-2 RNA has also been identified in the brains of COVID-19 patient autopsies (Puelles et al., 2020), the paucity of lung findings compared to the severe brain disease makes these mice not useful as models of the severe COVID-19 respiratory disease. Knockin (KI) mice, in which hACE2 replaces the mouse ACE2 gene rather than being randomly inserted into the genome, are also being developed. One such KI resulted in the expression of hACE2 in airways and in the development of pneumonia following infection; however, clinical disease was mild and restricted to aged mice (Sun et al., 2020b). Based on previous studies of hDPP4-KI mice, it is likely that passage through mouse lungs will be required for the generation of virulent SARS-CoV-2 that will provide a model for severe pneumonia (Cockrell et al., 2016; Li et al., 2017). An alternative method to generate hACE2 mice uses transduction of with an adenoviral vector encoding for hACE2, a method also used for the generation of a mouse model of MERS (Zhao et al., 2014b). Because vector is instilled intranasally, this results in expression of hACE2 exclusively in the respiratory tract of mice (Hassan et al., 2020; Sun et al., 2020a). Finally, another approach is to use reverse genetics to mutate residues in the receptor-binding domain, allowing for binding of the virus to mouse ACE2 and thereby facilitating viral entry into mouse cells. One such mouse-adapted virus, generated via targeted mutation without serial passage, is able to replicate in mice, though clinical disease was mild and observed primarily in aged mice (Dinnon et al., 2020). As in the case of hDPP4-KI mice, further passage of this virus through mouse lungs will likely result in more virulent virus. Thus, it is probable that some combination of mouse-adapted virus and wild-type or hACE2-expressing mice will produce the most robust mouse models for severe COVID-19, and these models will complement other models of mild disease.
Critical Questions for COVID-19 Immunity
Several outstanding questions are crucial to address to understand whether prior infection confers protection upon subsequent reinfection and for informed development of vaccines. First, determining whether a neutralizing antibody and/or SARS-CoV-2-specific T cell response is sufficient to prevent clinical disease and transmission is critical. If so, it will also be important to determine the magnitude of the responses required to provide protection in order to inform both social measures and vaccine strategies that can limit spread. Second, it will be essential to perform longitudinal studies to establish the longevity of these protective adaptive immune responses following natural infection or vaccination. Proper and detailed longitudinal studies will require substantial investment of resources by governments, industry sources, non-governmental agencies, and others. Third, identifying factors that contribute to the dysregulated immune response and immunopathology in patients with severe disease could inform early therapeutic options to limit disease severity. A critical part of these endeavors will require identification of biomarkers that identify patients predisposed to severe disease so that they can be managed aggressively to prevent poor outcomes. Finally, it will be essential, as vaccines are introduced into widespread use, to not only assess efficacy against severe disease and ability to minimize transmission but also identify vaccine-enhanced disease so that vaccination is safe and widely accepted by the public.
Acknowledgments
This work was supported in part by the NIH (grants PO1 AI060699 and RO1 AI129269).
Declaration of Interests
The authors declare no competing interests.
References
- Adams E.R., Ainsworth M., Anand R., Andersson M.I., Auckland K., Baillie J.K., Barnes E., Beer S., Bell J., Berry T., et al. Antibody testing for COVID-19: a report from the National COVID Scientific Advisory Panel. medRxiv. 2020 doi: 10.1101/2020.04.15.20066407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerström S., Tan Y.-J., Mirazimi A. Amino acids 15-28 in the ectodomain of SARS coronavirus 3a protein induces neutralizing antibodies. FEBS Lett. 2006;580:3799–3803. doi: 10.1016/j.febslet.2006.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsaad K.O., Hajeer A.H., Al Balwi M., Al Moaiqel M., Al Oudah N., Al Ajlan A., AlJohani S., Alsolamy S., Gmati G.E., Balkhy H., et al. Histopathology of Middle East respiratory syndrome coronovirus (MERS-CoV) infection - clinicopathological and ultrastructural study. Histopathology. 2018;72:516–524. doi: 10.1111/his.13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anghelina D., Pewe L., Perlman S. Pathogenic role for virus-specific CD4 T cells in mice with coronavirus-induced acute encephalitis. Am. J. Pathol. 2006;169:209–222. doi: 10.2353/ajpath.2006.051308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anghelina D., Zhao J., Trandem K., Perlman S. Role of regulatory T cells in coronavirus-induced acute encephalitis. Virology. 2009;385:358–367. doi: 10.1016/j.virol.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antón I.M., Suñé C., Meloen R.H., Borrás-Cuesta F., Enjuanes L. A transmissible gastroenteritis coronavirus nucleoprotein epitope elicits T helper cells that collaborate in the in vitro antibody synthesis to the three major structural viral proteins. Virology. 1995;212:746–751. doi: 10.1006/viro.1995.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y.M., Balkhy H.H., Hayden F.G., Bouchama A., Luke T., Baillie J.K., Al-Omari A., Hajeer A.H., Senga M., Denison M.R., et al. Middle East respiratory syndrome. N. Engl. J. Med. 2017;376:584–594. doi: 10.1056/NEJMsr1408795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y.M., Shalhoub S., Mandourah Y., Al-Hameed F., Al-Omari A., Al Qasim E., Jose J., Alraddadi B., Almotairi A., Al Khatib K., et al. Ribavirin and interferon therapy for critically ill patients with middle east respiratory syndrome: a multicenter observational study. Clin. Infect. Dis. 2020;70:1837–1844. doi: 10.1093/cid/ciz544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y.M., Asiri A.Y., Assiri A.M., Aziz Jokhdar H.A., Alothman A., Balkhy H.H., AlJohani S., Al Harbi S., Kojan S., Al Jeraisy M., et al. and the Saudi Critical Care Trials group Treatment of Middle East respiratory syndrome with a combination of lopinavir/ritonavir and interferon-β1b (MIRACLE trial): statistical analysis plan for a recursive two-stage group sequential randomized controlled trial. Trials. 2020;21:8. doi: 10.1186/s13063-019-3846-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arashiro T., Furukawa K., Nakamura A. COVID-19 in 2 persons with mild upper respiratory tract symptoms on a cruise ship, Japan. Emerg. Infect. Dis. 2020;26:1345–1348. doi: 10.3201/eid2606.200452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athmer J., Fehr A.R., Grunewald M.E., Qu W., Wheeler D.L., Graepel K.W., Channappanavar R., Sekine A., Aldabeeb D.S., Gale M., Jr., et al. Selective packaging in murine coronavirus promotes virulence by limiting type I interferon responses. MBio. 2018;9:9. doi: 10.1128/mBio.00272-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. Published online May 7, 2020. [DOI] [PubMed] [Google Scholar]
- Barthold S.W., Smith A.L. Mouse hepatitis virus strain—related patterns of tissue tropism in suckling mice. Arch. Virol. 1984;81:103–112. doi: 10.1007/BF01309300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C.C., Parra B., Hinton D.R., Ramakrishna C., Dowdell K.C., Stohlman S.A. Perforin and gamma interferon-mediated control of coronavirus central nervous system infection by CD8 T cells in the absence of CD4 T cells. J. Virol. 2004;78:1739–1750. doi: 10.1128/JVI.78.4.1739-1750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C.C., Lane T.E., Stohlman S.A. Coronavirus infection of the central nervous system: host-virus stand-off. Nat. Rev. Microbiol. 2006;4:121–132. doi: 10.1038/nrmicro1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. USA. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J.R.M., Bailey C., Przewrocka J., Dijkstra K.K., Swanton C. COVID-19: the case for health-care worker screening to prevent hospital transmission. Lancet. 2020;395:1418–1420. doi: 10.1016/S0140-6736(20)30917-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradburne A.F., Bynoe M.L., Tyrrell D.A. Effects of a “new” human respiratory virus in volunteers. BMJ. 1967;3:767–769. doi: 10.1136/bmj.3.5568.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brim T.A., VanCott J.L., Lunney J.K., Saif L.J. Cellular immune responses of pigs after primary inoculation with porcine respiratory coronavirus or transmissible gastroenteritis virus and challenge with transmissible gastroenteritis virus. Vet. Immunol. Immunopathol. 1995;48:35–54. doi: 10.1016/0165-2427(94)05416-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K., Collins P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. USA. 2004;101:9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler N.S., Dandekar A.A., Perlman S. Antiviral antibodies are necessary to prevent cytotoxic T-lymphocyte escape in mice infected with a coronavirus. J. Virol. 2007;81:13291–13298. doi: 10.1128/JVI.01580-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut P., Cox E., Pensaert M., Van Deun K. Induction of milk IgA antibodies by porcine respiratory coronavirus infection. Adv. Exp. Med. Biol. 1990;276:421–428. doi: 10.1007/978-1-4684-5823-7_58. [DOI] [PubMed] [Google Scholar]
- Callow K.A., Parry H.F., Sergeant M., Tyrrell D.A. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990;105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M., Muller M.P., Gold W.L., Richardson S.E., Poutanen S.M., et al. Canadian SARS Research Network Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., Bermejo-Martin J.F., Danesh A., Muller M.P., Kelvin D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro R.F., Perlman S. CD8+ T-cell epitopes within the surface glycoprotein of a neurotropic coronavirus and correlation with pathogenicity. J. Virol. 1995;69:8127–8131. doi: 10.1128/jvi.69.12.8127-8131.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathol. 2003;32:567–582. doi: 10.1080/03079450310001621198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Elus M.M., Cook J.K.A. Relationship between sequence variation in the S1 spike protein of infectious bronchitis virus and the extent of cross-protection in vivo. Avian Pathol. 1997;26:63–74. doi: 10.1080/03079459708419194. [DOI] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Züst R., Weber F., Spiegel M., Lang K.S., Akira S., Thiel V., Ludewig B. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragán L., Kalinke U., Züst R., König M., Reizis B., López-Macías C., Thiel V., Ludewig B. Type I IFN-mediated protection of macrophages and dendritic cells secures control of murine coronavirus infection. J. Immunol. 2009;182:1099–1106. doi: 10.4049/jimmunol.182.2.1099. [DOI] [PubMed] [Google Scholar]
- Cervantes-Barragán L., Firner S., Bechmann I., Waisman A., Lahl K., Sparwasser T., Thiel V., Ludewig B. Regulatory T cells selectively preserve immune privilege of self-antigens during viral central nervous system infection. J. Immunol. 2012;188:3678–3685. doi: 10.4049/jimmunol.1102422. [DOI] [PubMed] [Google Scholar]
- Chan J.F.-W., Yuan S., Zhang A.J., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., Fan Z., Li C., Liang R., Cao J., et al. Surgical mask partition reduces the risk of non-contact transmission in a golden Syrian hamster model for coronavirus disease 2019 (COVID-19) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa644. Published online May 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020 doi: 10.1126/science.abc4776. Published online May 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fett C., Zhao J., Meyerholz D.K., Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 2014;88:11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Jr., Meyerholz D.K., Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;129:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattha K.S., Roth J.A., Saif L.J. Strategies for design and application of enteric viral vaccines. Annu. Rev. Anim. Biosci. 2015;3:375–395. doi: 10.1146/annurev-animal-022114-111038. [DOI] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Lau Y.F., Lamirande E.W., Paddock C.D., Bartlett J.H., Zaki S.R., Subbarao K. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J. Virol. 2010;84:1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C.Y., Poon L.L.M., Ng I.H.Y., Luk W., Sia S.-F., Wu M.H.S., Chan K.-H., Yuen K.-Y., Gordon S., Guan Y., Peiris J.S. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K.-O., Hasoksuz M., Nielsen P.R., Chang K.-O., Lathrop S., Saif L.J. Cross-protection studies between respiratory and calf diarrhea and winter dysentery coronavirus strains in calves and RT-PCR and nested PCR for their detection. Arch. Virol. 2001;146:2401–2419. doi: 10.1007/s007050170011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.Y., Kang J.-M., Ha Y.E., Park G.E., Lee J.Y., Ko J.-H., Lee J.Y., Kim J.M., Kang C.-I., Jo I.J., et al. MERS-CoV outbreak following a single patient exposure in an emergency room in South Korea: an epidemiological outbreak study. Lancet. 2016;388:994–1001. doi: 10.1016/S0140-6736(16)30623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Hui K.P.Y., Perera R.A.P.M., Miguel E., Niemeyer D., Zhao J., Channappanavar R., Dudas G., Oladipo J.O., Traoré A., et al. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc. Natl. Acad. Sci. USA. 2018;115:3144–3149. doi: 10.1073/pnas.1718769115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Zhou J., Wong B.H.-Y., Li C., Cheng Z.-S., Lin X., Poon V.K.-M., Sun T., Lau C.C.-Y., Chan J.F.-W., et al. Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology. 2014;454-455:197–205. doi: 10.1016/j.virol.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Zhou J., Wong B.H.-Y., Li C., Chan J.F.-W., Cheng Z.-S., Yang D., Wang D., Lee A.C.-Y., Li C., et al. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J. Infect. Dis. 2016;213:904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua M.M., MacNamara K.C., San Mateo L., Shen H., Weiss S.R. Effects of an epitope-specific CD8+ T-cell response on murine coronavirus central nervous system disease: protection from virus replication and antigen spread and selection of epitope escape mutants. J. Virol. 2004;78:1150–1159. doi: 10.1128/JVI.78.3.1150-1159.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell A.S., Yount B.L., Scobey T., Jensen K., Douglas M., Beall A., Tang X.-C., Marasco W.A., Heise M.T., Baric R.S. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat. Microbiol. 2016;2:16226. doi: 10.1038/nmicrobiol.2016.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.M., Sisk J.M., Halasz G., Zhong J., Beck S.E., Matthews K.L., Venkataraman T., Rajagopalan S., Kyratsous C.A., Frieman M.B. CD8+ T cells and macrophages regulate pathogenesis in a mouse model of Middle East respiratory syndrome. J. Virol. 2016;91 doi: 10.1128/JVI.01825-16. e01825-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson E.W., Pei J., Dzielawa J., Seo S.H. Cytotoxic T lymphocytes are critical in the control of infectious bronchitis virus in poultry. Dev. Comp. Immunol. 2000;24:187–200. doi: 10.1016/s0145-305x(99)00072-5. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Albarrak A.M., Omrani A.S., Albarrak M.M., Farah M.E., Almasri M., Muth D., Sieberg A., Meyer B., Assiri A.M., et al. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin. Infect. Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Zhao J., Pedotti M., Simonelli L., Agnihothram S., Fett C., Fernandez-Rodriguez B., Foglierini M., Agatic G., Vanzetta F., et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc. Natl. Acad. Sci. USA. 2015;112:10473–10478. doi: 10.1073/pnas.1510199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar A.A., Jacobsen G., Waldschmidt T.J., Perlman S. Antibody-mediated protection against cytotoxic T-cell escape in coronavirus-induced demyelination. J. Virol. 2003;77:11867–11874. doi: 10.1128/JVI.77.22.11867-11874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming D., Sheahan T., Heise M., Yount B., Davis N., Sims A., Suthar M., Harkema J., Whitmore A., Pickles R., et al. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 2006;3:e525. doi: 10.1371/journal.pmed.0030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Hackbart M., Mettelman R.C., O’Brien A., Mielech A.M., Yi G., Kao C.C., Baker S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. USA. 2017;114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkman R., Jebbink M.F., Koekkoek S.M., Deijs M., Jónsdóttir H.R., Molenkamp R., Ieven M., Goossens H., Thiel V., van der Hoek L. Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J. Virol. 2013;87:6081–6090. doi: 10.1128/JVI.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Wang H., Shen H., Li Z., Geng J., Han H., Cai J., Li X., Kang W., Weng D., et al. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J. Pathol. 2003;200:282–289. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K.H., Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Hou Y.J., Adams L.E., et al. A mouse-adapted SARS-CoV-2 model for the evaluation of COVID-19 medical countermeasures. bioRxiv. 2020 doi: 10.1101/2020.05.06.081497. [DOI] [Google Scholar]
- Drosten C., Meyer B., Müller M.A., Corman V.M., Al-Masri M., Hossain R., Madani H., Sieberg A., Bosch B.J., Lattwein E., et al. Transmission of MERS-coronavirus in household contacts. N. Engl. J. Med. 2014;371:828–835. doi: 10.1056/NEJMoa1405858. [DOI] [PubMed] [Google Scholar]
- Enkirch T., von Messling V. Ferret models of viral pathogenesis. Virology. 2015;479-480:259–270. doi: 10.1016/j.virol.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., de Wit E., Rasmussen A.L., Feldmann F., Okumura A., Scott D.P., Brining D., Bushmaker T., Martellaro C., Baseler L., et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat. Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., de Wit E., Feldmann F., Rasmussen A.L., Okumura A., Peng X., Thomas M.J., van Doremalen N., Haddock E., Nagy L., et al. Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Pathog. 2014;10:e1004250. doi: 10.1371/journal.ppat.1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y.-Y., Huang Z.-T., Li L., Wu M.-H., Yu T., Koup R.A., Bailer R.T., Wu C.-Y. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch. Virol. 2009;154:1093–1099. doi: 10.1007/s00705-009-0409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Channappanavar R., Jankevicius G., Fett C., Zhao J., Athmer J., Meyerholz D.K., Ahel I., Perlman S. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. MBio. 2016;7 doi: 10.1128/mBio.01721-16. e017212-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr D., Holznagel E., Bolla S., Hauser B., Herrewegh A.A.P.M., Horzinek M.C., Lutz H. Placebo-controlled evaluation of a modified life virus vaccine against feline infectious peritonitis: safety and efficacy under field conditions. Vaccine. 1997;15:1101–1109. doi: 10.1016/S0264-410X(97)00006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folegatti P.M., Bittaye M., Flaxman A., Lopez F.R., Bellamy D., Kupke A., Mair C., Makinson R., Sheridan J., Rohde C., et al. Safety and immunogenicity of a candidate Middle East respiratory syndrome coronavirus viral-vectored vaccine: a dose-escalation, open-label, non-randomised, uncontrolled, phase 1 trial. Lancet Infect. Dis. 2020;20:816–826. doi: 10.1016/S1473-3099(20)30160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A.M., Hartwig N.G., Bestebroer T.M., Niemeyer B., de Jong J.C., Simon J.H., Osterhaus A.D.M.E. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. USA. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Agnihothram S., Page C., Donaldson E., Roberts A., Vogel L., Woodruff B., Scorpio D., Subbarao K., Baric R.S. Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J. Virol. 2012;86:884–897. doi: 10.1128/JVI.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020 doi: 10.1126/science.abc1932. Published online May 6, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sastre A. Ten strategies of interferon evasion by viruses. Cell Host Microbe. 2017;22:176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber J.D., Ingersoll J.D., Gast A.M., Christianson K.K., Selzer N.L., Landon R.M., Pfeiffer N.E., Sharpee R.L., Beckenhauer W.H. Protection against feline infectious peritonitis by intranasal inoculation of a temperature-sensitive FIPV vaccine. Vaccine. 1990;8:536–542. doi: 10.1016/0264-410X(90)90004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorse G.J., Patel G.B., Vitale J.N., O’Connor T.Z. Prevalence of antibodies to four human coronaviruses is lower in nasal secretions than in serum. Clin. Vaccine Immunol. 2010;17:1875–1880. doi: 10.1128/CVI.00278-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretebeck L.M., Subbarao K. Animal models for SARS and MERS coronaviruses. Curr. Opin. Virol. 2015;13:123–129. doi: 10.1016/j.coviro.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol. 2007;170:1136–1147. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Guo Z., Duan C., Chen Z., Wang G., Lu Y., Li M., Lu J. Long-term persistence of IgG antibodies in SARS-CoV infected healthcare workers. medRxiv. 2020 doi: 10.1101/2020.02.12.20021386. [DOI] [Google Scholar]
- Habibi M.S., Jozwik A., Makris S., Dunning J., Paras A., DeVincenzo J.P., de Haan C.A.M., Wrammert J., Openshaw P.J.M., Chiu C., Mechanisms of Severe Acute Influenza Consortium Investigators Impaired antibody-mediated protection and defective IgA B-cell memory in experimental infection of adults with respiratory syncytial virus. Am. J. Respir. Crit. Care Med. 2015;191:1040–1049. doi: 10.1164/rccm.201412-2256OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., et al. Impaired type I interferon activity and exacerbated inflammatory responses in severe COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.04.19.20068015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020 doi: 10.1016/j.cell.2020.06.011. Published online June 10, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Ding Y., Zhang Q., Che X., He Y., Shen H., Wang H., Li Z., Zhao L., Geng J., et al. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: relation to the acute lung injury and pathogenesis of SARS. J. Pathol. 2006;210:288–297. doi: 10.1002/path.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Zhao C., Dong Q., Zhuang H., Song S., Peng G., Dwyer D.E. Effects of severe acute respiratory syndrome (SARS) coronavirus infection on peripheral blood lymphocytes and their subsets. Int. J. Infect. Dis. 2005;9:323–330. doi: 10.1016/j.ijid.2004.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckert R.A., Saif L.J., Hoblet K.H., Agnes A.G. A longitudinal study of bovine coronavirus enteric and respiratory infections in dairy calves in two herds in Ohio. Vet. Microbiol. 1990;22:187–201. doi: 10.1016/0378-1135(90)90106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendley J.O., Fishburne H.B., Gwaltney J.M., Jr. Coronavirus infections in working adults. Eight-year study with 229 E and OC 43. Am. Rev. Respir. Dis. 1972;105:805–811. doi: 10.1164/arrd.1972.105.5.805. [DOI] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J.M., Wolthers K.C., Wertheim-van Dillen P.M.E., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins P.G., Phillpotts R.J., Scott G.M., Wallace J., Bernhardt L.L., Tyrrell D.A. Intranasal interferon as protection against experimental respiratory coronavirus infection in volunteers. Antimicrob. Agents Chemother. 1983;24:713–715. doi: 10.1128/aac.24.5.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh P.-R., Huang L.-M., Chen P.-J., Kao C.-L., Yang P.-C. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004;10:1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Li W., Gao T., Cui Y., Jin Y., Li P., Ma Q., Liu X., Cao C. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J. Virol. 2017;91 doi: 10.1128/JVI.02143-16. e02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland D.D.C., Stohlman S.A., Hinton D.R., Atkinson R., Bergmann C.C. Type I interferons are essential in controlling neurotropic coronavirus infection irrespective of functional CD8 T cells. J. Virol. 2008;82:300–310. doi: 10.1128/JVI.01794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Uda A., Suzuki T., Tsunetsugu-Yokota Y., Sato Y., Morikawa S., Tashiro M., Sata T., Hasegawa H., Nagata N. Effects of Toll-like receptor stimulation on eosinophilic infiltration in lungs of BALB/c mice immunized with UV-inactivated severe acute respiratory syndrome-related coronavirus vaccine. J. Virol. 2014;88:8597–8614. doi: 10.1128/JVI.00983-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W. Review of infectious bronchitis virus around the world. Avian Dis. 2012;56:634–641. doi: 10.1637/10227-043012-Review.1. [DOI] [PubMed] [Google Scholar]
- Jackwood M.W., Lee D.-H. Different evolutionary trajectories of vaccine-controlled and non-controlled avian infectious bronchitis viruses in commercial poultry. PLoS ONE. 2017;12:e0176709. doi: 10.1371/journal.pone.0176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W., Karaca K., Parrish C.R., Naqi S.A. A novel variant of avian infectious bronchitis virus resulting from recombination among three different strains. Arch. Virol. 1995;140:259–271. doi: 10.1007/BF01309861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W., Channappanavar R., Zhang C., Li M., Zhou H., Zhang S., Zhou P., Xu J., Shan S., Shi X., et al. Single intranasal immunization with chimpanzee adenovirus-based vaccine induces sustained and protective immunity against MERS-CoV infection. Emerg. Microbes Infect. 2019;8:760–772. doi: 10.1080/22221751.2019.1620083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R.-D., Liu M.-Q., Chen Y., Shan C., Zhou Y.-W., Shen X.-R., Li Q., Zhang L., Zhu Y., Si H.-R., et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58.e8. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Xu J., Zhou C., Wu Z., Zhong S., Liu J., Luo W., Chen T., Qin Q., Deng P. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am. J. Respir. Crit. Care Med. 2005;171:850–857. doi: 10.1164/rccm.200407-857OC. [DOI] [PubMed] [Google Scholar]
- Josset L., Menachery V.D., Gralinski L.E., Agnihothram S., Sova P., Carter V.S., Yount B.L., Graham R.L., Baric R.S., Katze M.G. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. MBio. 2013;4 doi: 10.1128/mBio.00165-13. e00165–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanolkar A., Hartwig S.M., Haag B.A., Meyerholz D.K., Epping L.L., Haring J.S., Varga S.M., Harty J.T. Protective and pathologic roles of the immune response to mouse hepatitis virus type 1: implications for severe acute respiratory syndrome. J. Virol. 2009;83:9258–9272. doi: 10.1128/JVI.00355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiambi S., Corman V.M., Sitawa R., Githinji J., Ngoci J., Ozomata A.S., Gardner E., von Dobschuetz S., Morzaria S., Kimutai J., et al. Detection of distinct MERS-coronavirus strains in dromedary camels from Kenya, 2017. Emerg. Microbes Infect. 2018;7:195. doi: 10.1038/s41426-018-0193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Perlman S. Viral expression of CCL2 is sufficient to induce demyelination in RAG1-/- mice infected with a neurotropic coronavirus. J. Virol. 2005;79:7113–7120. doi: 10.1128/JVI.79.11.7113-7120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.D., Zhao J., Auh S., Yang X., Du P., Tang H., Fu Y.-X. Adaptive immune cells temper initial innate responses. Nat. Med. 2007;13:1248–1252. doi: 10.1038/nm1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.S., Choe P.G., Park W.B., Oh H.S., Kim E.J., Nam E.Y., Na S.H., Kim M., Song K.-H., Bang J.H., et al. Clinical progression and cytokine profiles of Middle East respiratory syndrome coronavirus infection. J. Korean Med. Sci. 2016;31:1717–1725. doi: 10.3346/jkms.2016.31.11.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.H., Kim H.J., Chang J. Superior immune responses induced by intranasal immunization with recombinant adenovirus-based vaccine expressing full-length Spike protein of Middle East respiratory syndrome coronavirus. PLoS ONE. 2019;14:e0220196. doi: 10.1371/journal.pone.0220196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-I., Kim S.-G., Kim S.-M., Kim E.-H., Park S.-J., Yu K.-M., Chang J.-H., Kim E.J., Lee S., Casel M.A.B., et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E., Gil-Cruz C., Spanier J., Li Y., Wilhelm J., Rabouw H.H., Züst R., Hwang M., V’kovski P., Stalder H., et al. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13:e1006195. doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkcaldy R.D., King B.A., Brooks J.T. COVID-19 and postinfection immunity: limited evidence, many remaining questions. JAMA. 2020 doi: 10.1001/jama.2020.7869. Published online May 11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch T., Dahlke C., Fathi A., Kupke A., Krähling V., Okba N.M.A., Halwe S., Rohde C., Eickmann M., Volz A., et al. Safety and immunogenicity of a modified vaccinia virus Ankara vector vaccine candidate for Middle East respiratory syndrome: an open-label, phase 1 trial. Lancet Infect. Dis. 2020;20:827–838. doi: 10.1016/S1473-3099(20)30248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law H.K.W., Cheung C.Y., Ng H.Y., Sia S.F., Chan Y.O., Luk W., Nicholls J.M., Peiris J.S.M., Lau Y.L. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood. 2005;106:2366–2374. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F., et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- Lee N., Chan P.K.S., Ip M., Wong E., Ho J., Ho C., Cockram C.S., Hui D.S. Anti-SARS-CoV IgG response in relation to disease severity of severe acute respiratory syndrome. J. Clin. Virol. 2006;35:179–184. doi: 10.1016/j.jcv.2005.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew T.W.K., Kwek T.-K., Tai D., Earnest A., Loo S., Singh K., Kwan K.M., Chan Y., Yim C.F., Bek S.L., et al. Acute respiratory distress syndrome in critically ill patients with severe acute respiratory syndrome. JAMA. 2003;290:374–380. doi: 10.1001/jama.290.3.374. [DOI] [PubMed] [Google Scholar]
- Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M., Tang X., Temperton N.J., Weiss R.A., Brenchley J.M., et al. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Wohlford-Lenane C.L., Channappanavar R., Park J.-E., Earnest J.T., Bair T.B., Bates A.M., Brogden K.A., Flaherty H.A., Gallagher T., et al. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc. Natl. Acad. Sci. USA. 2017;114:E3119–E3128. doi: 10.1073/pnas.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-T., Zhang J.-S., Su N., Xu J.-G., Wang N., Chen J.-T., Chen X., Liu Y.-X., Gao H., Jia Y.-P., et al. Safety and immunogenicity from a phase I trial of inactivated severe acute respiratory syndrome coronavirus vaccine. Antivir. Ther. (Lond.) 2007;12:1107–1113. [PubMed] [Google Scholar]
- Lin M.T., Hinton D.R., Marten N.W., Bergmann C.C., Stohlman S.A. Antibody prevents virus reactivation within the central nervous system. J. Immunol. 1999;162:7358–7368. [PubMed] [Google Scholar]
- Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z., et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.J., Zhao M., Liu K., Xu K., Wong G., Tan W., Gao G.F. T-cell immunity of SARS-CoV: Implications for vaccine development against MERS-CoV. Antiviral Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Hage A., Schindewolf C., Rajsbaum R., Menachery V.D. SARS-CoV-2 is sensitive to type I interferon pretreatment. bioRxiv. 2020 doi: 10.1101/2020.03.07.982264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Mair-Jenkins J., Saavedra-Campos M., Baillie J.K., Cleary P., Khaw F.-M., Lim W.S., Makki S., Rooney K.D., Nguyen-Van-Tam J.S., Beck C.R., Convalescent Plasma Study Group The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.E., Louder M.K., Holman L.A., Gordon I.J., Enama M.E., Larkin B.D., Andrews C.A., Vogel L., Koup R.A., Roederer M., et al. VRC 301 Study Team A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine. 2008;26:6338–6343. doi: 10.1016/j.vaccine.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews A.E., Weiss S.R., Shlomchik M.J., Hannum L.G., Gombold J.L., Paterson Y. Antibody is required for clearance of infectious murine hepatitis virus A59 from the central nervous system, but not the liver. J. Immunol. 2001;167:5254–5263. doi: 10.4049/jimmunol.167.9.5254. [DOI] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K., Dees J.H., Becker W.B., Kapikian A.Z., Chanock R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. USA. 1967;57:933–940. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Perlman S., Van Kerkhove M.D., Zumla A. Middle East respiratory syndrome. Lancet. 2020;395:1063–1077. doi: 10.1016/S0140-6736(19)33221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Josset L., Gralinski L.E., Scobey T., Agnihothram S., Katze M.G., Baric R.S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-o-methyltransferase activity. J. Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Schäfer A., Burnum-Johnson K.E., Mitchell H.D., Eisfeld A.J., Walters K.B., Nicora C.D., Purvine S.O., Casey C.P., Monroe M.E., et al. MERS-CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proc. Natl. Acad. Sci. USA. 2018;115:E1012–E1021. doi: 10.1073/pnas.1706928115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min C.-K., Cheon S., Ha N.-Y., Sohn K.M., Kim Y., Aigerim A., Shin H.M., Choi J.-Y., Inn K.-S., Kim J.-H., et al. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci. Rep. 2016;6:25359. doi: 10.1038/srep25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modjarrad K., Roberts C.C., Mills K.T., Castellano A.R., Paolino K., Muthumani K., Reuschel E.L., Robb M.L., Racine T., Oh M.D., et al. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect. Dis. 2019;19:1013–1022. doi: 10.1016/S1473-3099(19)30266-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monto A.S., Lim S.K. The Tecumseh study of respiratory illness. VI. Frequency of and relationship between outbreaks of coronavirus infection. J. Infect. Dis. 1974;129:271–276. doi: 10.1093/infdis/129.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020:1–7. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D.L., Al Hosani F., Keating M.K., Gerber S.I., Jones T.L., Metcalfe M.G., Tong S., Tao Y., Alami N.N., Haynes L.M., et al. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am. J. Pathol. 2016;186:652–658. doi: 10.1016/j.ajpath.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng O.-W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., Tan Y.-J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., Chu C.M., Hui P.K., Mak K.L., Lim W., et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H.-L.J., Chia A., Chang C.X.L., Leong H.N., Ling K.L., Grotenbreg G.M., Gehring A.J., Tan Y.J., Bertoletti A. Engineering T cells specific for a dominant severe acute respiratory syndrome coronavirus CD8 T cell epitope. J. Virol. 2011;85:10464–10471. doi: 10.1128/JVI.05039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omrani A.S., Saad M.M., Baig K., Bahloul A., Abdul-Matin M., Alaidaroos A.Y., Almakhlafi G.A., Albarrak M.M., Memish Z.A., Albarrak A.M. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect. Dis. 2014;14:1090–1095. doi: 10.1016/S1473-3099(14)70920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page C., Goicochea L., Matthews K., Zhang Y., Klover P., Holtzman M.J., Hennighausen L., Frieman M. Induction of alternatively activated macrophages enhances pathogenesis during severe acute respiratory syndrome coronavirus infection. J. Virol. 2012;86:13334–13349. doi: 10.1128/JVI.01689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Sestak K., Hodgins D.C., Shoup D.I., Ward L.A., Jackwood D.J., Saif L.J. Immune response of sows vaccinated with attenuated transmissible gastroenteritis virus (TGEV) and recombinant TGEV spike protein vaccines and protection of their suckling pigs against virulent TGEV challenge exposure. Am. J. Vet. Res. 1998;59:1002–1008. [PubMed] [Google Scholar]
- Park W.B., Perera R.A.P.M., Choe P.G., Lau E.H.Y., Choi S.J., Chun J.Y., Oh H.S., Song K.-H., Bang J.H., Kim E.S., et al. Kinetics of serologic responses to MERS coronavirus infection in humans, South Korea. Emerg. Infect. Dis. 2015;21:2186–2189. doi: 10.3201/eid2112.151421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Yuen K.Y., Osterhaus A.D.M.E., Stöhr K. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- Peiris J.S.M., Chu C.M., Cheng V.C.C., Chan K.S., Hung I.F.N., Poon L.L.M., Law K.I., Tang B.S.F., Hon T.Y.W., Chan C.S., et al. HKU/UCH SARS Study Group Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10(12, Suppl):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., McIntosh K. In: Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. Ninth Edition. Bennett J.E., Dolin R., Blaser M.J., editors. Elsevier; 2019. Coronaviruses, including severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) pp. 2072–2080.e2. [Google Scholar]
- Pewe L., Perlman S. Cutting edge: CD8 T cell-mediated demyelination is IFN-γ dependent in mice infected with a neurotropic coronavirus. J. Immunol. 2002;168:1547–1551. doi: 10.4049/jimmunol.168.4.1547. [DOI] [PubMed] [Google Scholar]
- Pewe L., Haring J., Perlman S. CD4 T-cell-mediated demyelination is increased in the absence of gamma interferon in mice infected with mouse hepatitis virus. J. Virol. 2002;76:7329–7333. doi: 10.1128/JVI.76.14.7329-7333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., et al. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2011400. Published online May 13, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M., Shi Y., Guo Z., Chen Z., He R., Chen R., Zhou D., Dai E., Wang X., Si B., et al. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect. 2005;7:882–889. doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan B.D., Mou H., Zhang L., Guo Y., He W., Ojha A., Parcells M.S., Luo G., Li W., Zhong G., et al. The SARS-CoV-2 receptor-binding domain elicits a potent neutralizing response without antibody-dependent enhancement. bioRxiv. 2020 doi: 10.1101/2020.04.10.036418. [DOI] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna C., Bergmann C.C., Atkinson R., Stohlman S.A. Control of central nervous system viral persistence by neutralizing antibody. J. Virol. 2003;77:4670–4678. doi: 10.1128/JVI.77.8.4670-4678.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reghunathan R., Jayapal M., Hsu L.-Y., Chng H.-H., Tai D., Leung B.P., Melendez A.J. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6:2. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020 doi: 10.1038/s41586-020-2456-9. Published online June 18, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Paddock C., Vogel L., Butler E., Zaki S., Subbarao K. Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J. Virol. 2005;79:5833–5838. doi: 10.1128/JVI.79.9.5833-5838.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L., Herman B.D., Sheahan T., Heise M., Genrich G.L., et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A., et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Cross J.K., Bender S.J., Weiss S.R. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 2008;82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottier P.J.M., Nakamura K., Schellen P., Volders H., Haijema B.J. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J. Virol. 2005;79:14122–14130. doi: 10.1128/JVI.79.22.14122-14130.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif L.J. Bovine respiratory coronavirus. Vet. Clin. North Am. Food Anim. Pract. 2010;26:349–364. doi: 10.1016/j.cvfa.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif L.J., Bohl E.H., Gupta R.K. Isolation of porcine immunoglobulins and determination of the immunoglobulin classes of transmissible gastroenteritis viral antibodies. Infect. Immun. 1972;6:600–609. doi: 10.1128/iai.6.4.600-609.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif L.J., Wang Q., Vlasova A.N., Jung K., Xiao S. Diseases of Swine. John Wiley & Sons; 2019. Coronaviruses; pp. 488–523. [Google Scholar]
- Sánchez C.M., Gebauer F., Suñé C., Mendez A., Dopazo J., Enjuanes L. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology. 1992;190:92–105. doi: 10.1016/0042-6822(92)91195-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarin C., Bergmann C.C., Hinton D.R., Ransohoff R.M., Stohlman S.A. Memory CD4+ T-cell-mediated protection from lethal coronavirus encephalomyelitis. J. Virol. 2008;82:12432–12440. doi: 10.1128/JVI.01267-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuplein V.A., Seifried J., Malczyk A.H., Miller L., Höcker L., Vergara-Alert J., Dolnik O., Zielecki F., Becker B., Spreitzer I., et al. High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus. J. Virol. 2015;89:3859–3869. doi: 10.1128/JVI.03607-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O.W., Allan I.D., Cooney M.K., Foy H.M., Fox J.P. Rises in titers of antibody to human coronaviruses OC43 and 229E in Seattle families during 1975-1979. Am. J. Epidemiol. 1986;123:862–868. doi: 10.1093/oxfordjournals.aje.a114315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Wessels C., Herrler G. Transmissible gastroenteritis virus infection: a vanishing specter. Dtsch. Tierarztl. Wochenschr. 2006;113:157–159. [PubMed] [Google Scholar]
- Shaman J., Galanti M. Direct measurement of rates of asymptomatic infection and clinical care-seeking for seasonal coronavirus. medRxiv. 2020 doi: 10.1101/2020.01.30.20019612. [DOI] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.-S., Kim Y., Kim G., Lee J.Y., Jeong I., Joh J.-S., Kim H., Chang E., Sim S.Y., Park J.-S., Lim D.G. Immune responses to Middle East respiratory syndrome coronavirus during the acute and convalescent phases of human infection. Clin. Infect. Dis. 2019;68:984–992. doi: 10.1093/cid/ciy595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.-M., Chin A.W.H., Fung K., Choy K.-T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020 doi: 10.1038/s41586-020-2342-5. Published online May 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton R., Etchart N., Hou S., Hyland L. Inability to evoke a long-lasting protective immune response to respiratory syncytial virus infection in mice correlates with ineffective nasal antibody responses. J. Virol. 2003;77:11303–11311. doi: 10.1128/JVI.77.21.11303-11311.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.C., Sexton N.R., Denison M.R. Thinking outside the triangle: replication fidelity of the largest RNA viruses. Annu. Rev. Virol. 2014;1:111–132. doi: 10.1146/annurev-virology-031413-085507. [DOI] [PubMed] [Google Scholar]
- Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.-J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhuang Z., Zheng J., Li K., Wong R.L.-Y., Liu D., Huang J., He J., Zhu A., Zhao J., et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020 doi: 10.1016/j.cell.2020.06.010. Published online June 10, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.-H., Chen Q., Gu H.-J., Yang G., Wang Y.-X., Huang X.-Y., Liu S.-S., Zhang N.-N., Li X.-F., Xiong R., et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28:124–133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., et al. HCA Lung Biological Network SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., Wang Y., Fett C.A., Zhao G., Li F., Perlman S., Jiang S., Zhou Y., Du L. Recombinant receptor-binding domains of multiple Middle East respiratory syndrome coronaviruses (MERS-CoVs) induce cross-neutralizing antibodies against divergent human and camel MERS-CoVs and antibody escape mutants. J. Virol. 2016;91 doi: 10.1128/JVI.01651-16. e01651-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.-Q., Wang Q., Miao H. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct. Target. Ther. 2020;5:1–3. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Quan Y., Xin Z.-T., Wrammert J., Ma M.-J., Lv H., Wang T.-B., Yang H., Richardus J.H., Liu W., Cao W.C. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- Tang X.-C., Agnihothram S.S., Jiao Y., Stanhope J., Graham R.L., Peterson E.C., Avnir Y., Tallarico A.S.C., Sheehan J., Zhu Q., et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl. Acad. Sci. USA. 2014;111:E2018–E2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton S.P., Kim T.S., O’Malley K., Perlman S. Maturation and localization of macrophages and microglia during infection with a neurotropic murine coronavirus. Brain Pathol. 2008;18:40–51. doi: 10.1111/j.1750-3639.2007.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trandem K., Zhao J., Fleming E., Perlman S. Highly activated cytotoxic CD8 T cells express protective IL-10 at the peak of coronavirus-induced encephalitis. J. Immunol. 2011;186:3642–3652. doi: 10.4049/jimmunol.1003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.-T.K., Huang C., Newman P., Wang N., Narayanan K., Watts D.M., Makino S., Packard M.M., Zaki S.R., Chan T.S., Peters C.J. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J. Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynell J., Westenius V., Rönkkö E., Munster V.J., Melén K., Österlund P., Julkunen I. Middle East respiratory syndrome coronavirus shows poor replication but significant induction of antiviral responses in human monocyte-derived macrophages and dendritic cells. J. Gen. Virol. 2016;97:344–355. doi: 10.1099/jgv.0.000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell D.A.J., Bynoe M.L. Cultivation of a novel type of common-cold virus in organ cultures. BMJ. 1965;1:1467–1470. doi: 10.1136/bmj.1.5448.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., de Groot R.J., Harbour D.A., Dalderup M., Gruffydd-Jones T., Horzinek M.C., Spaan W.J. Early death after feline infectious peritonitis virus challenge due to recombinant vaccinia virus immunization. J. Virol. 1990;64:1407–1409. doi: 10.1128/jvi.64.3.1407-1409.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay R., Hua X., Meyerholz D.K., Miki Y., Yamamoto K., Gelb M., Murakami M., Perlman S. Critical role of phospholipase A2 group IID in age-related susceptibility to severe acute respiratory syndrome-CoV infection. J. Exp. Med. 2015;212:1851–1868. doi: 10.1084/jem.20150632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.I., Stohlman S.A., Fleming J.O. Demyelination induced by murine hepatitis virus JHM strain (MHV-4) is immunologically mediated. J. Neuroimmunol. 1990;30:31–41. doi: 10.1016/0165-5728(90)90050-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shi W., Chappell J.D., Joyce M.G., Zhang Y., Kanekiyo M., Becker M.M., van Doremalen N., Fischer R., Wang N., et al. Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on the Middle East respiratory syndrome coronavirus spike glycoprotein to avoid neutralization escape. J. Virol. 2018;92 doi: 10.1128/JVI.02002-17. e02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R.C., Scott F.W. Antibody-mediated enhancement of disease in feline infectious peritonitis: comparisons with dengue hemorrhagic fever. Comp. Immunol. Microbiol. Infect. Dis. 1981;4:175–189. doi: 10.1016/0147-9571(81)90003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley R. Neutralizing antibody decay and lack of contact transmission after inoculation of 3- and 4-day-old piglets with porcine respiratory coronavirus. J. Vet. Diagn. Invest. 2002;14:525–527. doi: 10.1177/104063870201400617. [DOI] [PubMed] [Google Scholar]
- Wesley R.D., Woods R.D. Induction of protective immunity against transmissible gastroenteritis virus after exposure of neonatal pigs to porcine respiratory coronavirus. Am. J. Vet. Res. 1996;57:157–162. [PubMed] [Google Scholar]
- Williamson J.S., Stohlman S.A. Effective clearance of mouse hepatitis virus from the central nervous system requires both CD4+ and CD8+ T cells. J. Virol. 1990;64:4589–4592. doi: 10.1128/jvi.64.9.4589-4592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson B.N., Feldmann F., Schwarz B., Meade-White K., Porter D.P., Schulz J., van Doremalen N., Leighton I., Yinda C.K., Pérez-Pérez L., et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020:1–7. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Rasmussen A.L., Falzarano D., Bushmaker T., Feldmann F., Brining D.L., Fischer E.R., Martellaro C., Okumura A., Chang J., et al. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc. Natl. Acad. Sci. USA. 2013;110:16598–16603. doi: 10.1073/pnas.1310744110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W.K., et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Coronavirus disease (COVID-19) dashboard.https://covid19.who.int/ [Google Scholar]
- World Health Organization . 2020. MERS situation update, January 2020.http://www.emro.who.int/pandemic-epidemic-diseases/mers-cov/mers-situation-update-january-2020.html [Google Scholar]
- World Health Organization . 2020. Draft landscape of COVID-19 candidate vaccines.https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines [Google Scholar]
- Wu G.F., Dandekar A.A., Pewe L., Perlman S. CD4 and CD8 T cells have redundant but not identical roles in virus-induced demyelination. J. Immunol. 2000;165:2278–2286. doi: 10.4049/jimmunol.165.4.2278. [DOI] [PubMed] [Google Scholar]
- Wu L.-P., Wang N.-C., Chang Y.-H., Tian X.-Y., Na D.-Y., Zhang L.-Y., Zheng L., Lan T., Wang L.-F., Liang G.-D. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 2007;13:1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Gu H., Zhao Z., Wang W., Cao B., Lai C., Yang X., Zhang L., Duan Y., Zhang S., et al. Angiotensin-converting enzyme 2 (ACE2) mediates influenza H7N9 virus-induced acute lung injury. Sci. Rep. 2014;4:7027. doi: 10.1038/srep07027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Shen C., Li J., Yuan J., Wei J., Huang F., Wang F., Li G., Li Y., Xing L., et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 2020;146:119–127.e4. doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen Y.-T., Liao F., Hsiao C.-H., Kao C.-L., Chen Y.-C., Wu-Hsieh B.A. Modeling the early events of severe acute respiratory syndrome coronavirus infection in vitro. J. Virol. 2006;80:2684–2693. doi: 10.1128/JVI.80.6.2684-2693.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilla M., Harcourt B.H., Hickman C.J., McGrew M., Tamin A., Goldsmith C.S., Bellini W.J., Anderson L.J. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res. 2005;107:93–101. doi: 10.1016/j.virusres.2004.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T., Hill T., Li K., Peters C.J., Tseng C.-T.K. Severe acute respiratory syndrome (SARS) coronavirus-induced lung epithelial cytokines exacerbate SARS pathogenesis by modulating intrinsic functions of monocyte-derived macrophages and dendritic cells. J. Virol. 2009;83:3039–3048. doi: 10.1128/JVI.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.Y., Zhang Y.C., Han C.W., Wang P., Xue X.J., Cong Y.L. [Change of T lymphocyte and its activated subsets in SARS patients] Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2003;25:542–546. [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhang F., Yu W., He T., Yu J., Yi C.E., Ba L., Li W., Farzan M., Chen Z., et al. Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J. Med. Virol. 2006;78:1–8. doi: 10.1002/jmv.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li J., Zhan Y., Wu L., Yu X., Zhang W., Ye L., Xu S., Sun R., Wang Y., Lou J. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect. Immun. 2004;72:4410–4415. doi: 10.1128/IAI.72.8.4410-4415.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang H.-N., Wang T., Fan W.-Q., Zhang A.-Y., Wei K., Tian G.-B., Yang X. Complete genome sequence and recombination analysis of infectious bronchitis virus attenuated vaccine strain H120. Virus Genes. 2010;41:377–388. doi: 10.1007/s11262-010-0517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Van Rooijen N., Perlman S. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 2009;5:e1000636. doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 2010;84:9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Fett C., Trandem K., Fleming E., Perlman S. IFN-γ- and IL-10-expressing virus epitope-specific Foxp3(+) T reg cells in the central nervous system during encephalomyelitis. J. Exp. Med. 2011;208:1571–1577. doi: 10.1084/jem.20110236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Legge K., Perlman S. Age-related increases in PGD(2) expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J. Clin. Invest. 2011;121:4921–4930. doi: 10.1172/JCI59777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Wohlford-Lenane C., Zhao J., Fleming E., Lane T.E., McCray P.B., Jr., Perlman S. Intranasal treatment with poly(I•C) protects aged mice from lethal respiratory virus infections. J. Virol. 2012;86:11416–11424. doi: 10.1128/JVI.01410-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Perlman S. Virus-specific regulatory T cells ameliorate encephalitis by repressing effector T cell functions from priming to effector stages. PLoS Pathog. 2014;10:e1004279. doi: 10.1371/journal.ppat.1004279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J., Gale M.J., Jr., Baric R.S., Enjuanes L., Gallagher T., et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. USA. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway memory CD4(+) T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Alshukairi A.N., Baharoon S.A., Ahmed W.A., Bokhari A.A., Nehdi A.M., Layqah L.A., Alghamdi M.G., Al Gethamy M.M., Dada A.M., et al. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci. Immunol. 2017;2:2. doi: 10.1126/sciimmunol.aan5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Jha B.K., Wu A., Elliott R., Ziebuhr J., Gorbalenya A.E., Silverman R.H., Weiss S.R. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Hassan S., Alagaili A.N., Alshukairi A.N., Amor N.M.S., Mukhtar N., Nazeer I.M., Tahir Z., Akhter N., Perlman S., Yaqub T. Middle East respiratory syndrome coronavirus seropositivity in camel handlers and their families, Pakistan. Emerg. Infect. Dis. 2019;25:25. doi: 10.3201/eid2512.191169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Yang H., Guo Z.-F., Sin W.-Y.F., Chen W., Xu J., Fu L., Wu J., Mak C.-K.G., Cheng C.-S.S., et al. B-cell responses in patients who have recovered from severe acute respiratory syndrome target a dominant site in the S2 domain of the surface spike glycoprotein. J. Virol. 2005;79:3401–3408. doi: 10.1128/JVI.79.6.3401-3408.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Chu H., Li C., Wong B.H.-Y., Cheng Z.-S., Poon V.K.-M., Sun T., Lau C.C.-Y., Wong K.K.-Y., Chan J.Y.-W., et al. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J. Infect. Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Yan Y., Shu Y., Gao R., Sun Y., Li X., Ju X., Liang Z., Liu Q., Zhao Y., et al. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat. Commun. 2014;5:3594. doi: 10.1038/ncomms4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S., et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]