Abstract
In March 2003, a novel coronavirus was isolated from patients exhibiting atypical pneumonia and subsequently proven to be the causative agent of the disease now referred to as severe acute respiratory syndrome (SARS). The complete genome of the SARS coronavirus (SARS-CoV) has since been sequenced. The SARS-CoV nucleocapsid (SARS-CoV N) shares little homology with other members of the coronavirus family. To determine if the N protein is involved in the regulation of cellular signal transduction, an ELISA-based assay on transcription factors was used. We found that the amount of transcription factors binding to promoter sequences of c-Fos, ATF2, CREB-1, and FosB was increased by the expression of SARS-CoV N. Since these factors are related to AP-1 signal transduction pathway, we investigated whether the AP-1 pathway was activated by SARS-CoV N protein using the PathDetect system. The results demonstrated that the expression of N protein, not the membrane protein (M), activated AP-1 pathway. We also found that SARS-CoV N protein does not activate NF-κB pathway, demonstrating that activation of important cellular pathways by SAS-CoV N protein is selective. Thus our data for the first time indicate that SARS-CoV has encoded a strategy to regulate cellular signaling process.
Coronaviruses are enveloped viruses with a single-stranded positive-sense RNA genome approximately 30 kb in length [1]. The two common human coronaviruses, HCoV-229E and HCoV-OC43, are frequently the cause of mild respiratory illnesses. In contrast, the novel coronavirus associated with severe acute respiratory syndrome (SARS-CoV) has been observed to induce fever, edema, and diffuse alveolar damage in severely affected individuals [2]. SARS-CoV is markedly different from other members of the Coronaviridae family in that it is the only coronavirus known to cause severe morbidity and mortality in humans [3], [4].
The gene that encodes the 46 kDa SARS-CoV nucleocapsid protein (N) directly precedes the 3′-UTR of the 29.7 kb viral genome [5]. Several functions including viral packaging, viral core formation, and vRNA synthesis have been attributed to the coronavirus nucleocapsid [6]. However, the SARS-CoV N protein shares little homology with other members of the coronavirus family. It is noted that SARS-CoV N protein contains a short lysine-rich sequence (KTFPPTEPKKDKKKKTDEAQ) near the carboxyl terminus, which has not been found in any other known coronaviruses (aa 362–381) [7]. Yet, it is likely that this region acts as a nuclear localization signal allowing nucleocapsid protein to enter the nucleus by passive diffusion [8]. Indeed, coronavirus nucleocapsid proteins from all three identified Coronaviridae groups have recently been found to localize in the cytoplasm and nucleus in both virus-infected and plasmid-transfected cells [9]. Furthermore, SARS-CoV N protein is also suspected to be extensively phosphorylated after translation, which may allow it to enter the nucleolus at certain stages of the cell cycle [6]. While inside the nucleolus during interphase, the nucleocapsid protein has an opportunity to interact with a wide variety of regulatory complexes and transcription factors.
Many endogenous proteins that localize in the nucleus have been identified as regulators of the cell cycle. Numerous virus species have also evolved strategies in order to induce or inhibit intracellular host cell signaling through this natural protein mechanism. Viruses such as herpes virus and poxvirus encode proteins that can block regulatory enzymes or mimic natural transcription factors [10], [11]. This beneficial control over the host cell cycle can either facilitate the early release of virus progeny or help the emerging virus evade the host immune system. The SARS-CoV nucleocapsid, as discussed above, may also affect host signal transduction pathways resulting in apoptosis, inflammation or other cellular processes. One specific pathway, the activator protein 1 (AP-1) pathway, is a regulator of a wide variety of cellular processes, including cell proliferation, differentiation, and apoptosis [12]. Therefore it is an attractive target for signal transduction modification by viral proteins.
In the current study, the possible regulatory interactions of the SARS-CoV N protein in the AP-1 pathway were investigated through a cis-reporting in vitro microarray and an in vivo inducible-vector assay. The ability of SARS-CoV N protein to affect multiple signal transduction pathways was investigated using the TransFactor assay, an ELISA-based detection with similar sensitivity as gel-mobility shift assays, to map several possible protein–DNA interactions. The PathDetect assay, an alternative method to measure signal transduction changes, was also used to confirm the signal transduction activation found by TransFactor analysis.
Materials and methods
Construction of recombinant vectors. The pcDNA3.1(−) expression vector was obtained from Invitrogen (Carlsbad, CA). The nucleocapsid gene (GenBank Accession #AY274119) was amplified by RT-PCR from the SARS-CoV RNA of patient serum samples. Primers for the amplification are 5′-GTACGAATTCATGTCTGATAATGGACCCCAATC-3′ and 5′-GTACGGATCCGTGGTCATCATGAGTGTTTATG-3′. The amplified product was then purified with MiniElute PCR Purification kit (Qiagen, Valencia, CA). The purified product was subsequently digested with EcoRI and BamHI, and then ligated with pcDNA3.1(−) digested with the same restriction enzymes. The final expression vector for SARS-CoV N is designated as pcDNA-N in this report. The gene for SARS-CoV membrane was amplified with the following primers: 5′-GTACGAATTCATGGCAGACAACGGTA-3′ and GTACGGATCCTTACTGTACTAGCAAAGCA. The PCR product was cloned into pcDNA3.1(−) in EcoRI and BamHI sites as mentioned above. The final expression vector for SARS-CoV M is designated as pcDNA-M in this report.
Cell culture. The African green monkey kidney cell line Vero cells and human hepatocellular carcinoma cell line Huh7 cells were cultured in Dulbecco’s modified Eagle’s medium, supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA). The Huh7 cell line was chosen for the current study due to its high transfection efficiency [13], while Vero cells were selected because they are susceptible to SARS-CoV infection [4]. All cell cultures were maintained in a humidified 5% CO2 incubator at 37 °C.
Cell transfection. As much as 1 × 107 cells were used for transfection using Effectene transfection reagent according to the manufacturer’s protocol (Qiagen, Valencia, CA). Briefly, cells were first washed with serum-free DMEM. Thirty micrograms of plasmid DNA (pcDNA3.1(−), pcDNA3.1(−) vector with nucleocapsid gene (pcDNA3.1(−) + N) or mock transfection) and 240 μl of enhancer were mixed with 4.5 ml of EC buffer and incubated at room temperature for 5 min. For dose–response experiments, the empty vector (pcDNA3−1) was added to hold each individual transfection with the same amount of DNA (30 μg). After addition of 375 μl of the Effectene reagent, the transfection mixture was again incubated for 10 min, followed by dropwise addition to the cell culture. The transfection efficiency is routinely monitored by co-transfecting the cells with pEGFP (Clontech, Palo Alto, CA) In addition, the experiments triplicated for each transfection were repeated at least 3–5 times with SD (Standard Deviation) being less than 10% (see Figure legends for details).
Western blotting. Protein samples were fractionated on 4–12% SDS–PAGE (Invitrogen, Carlsbad, CA) and transferred to PVDF membrane using semi-dry protein transfer apparatus (Bio-Rad, Hercules, CA). The membrane was blocked for 1 h with 5% skimmed milk in TBS buffer (20 mM Tris base, 137 mM NaCl, pH 7.6) containing 0.2% Tween 20. Following the blocking the membrane was probed with chicken IgY antibody against the N protein sequence PKKDKKKKTDEAQPLPQRQK (custom antibody from Sigmagenosystem, The Woodland, TX). Goat anti-chicken HRP-conjugated antibody (Santa Cruz Biotechnologies, Santa Cruz, CA) was added. The results were finally revealed by using Pierce Biotechonlogy SuperSignal West Femto Maximum Sensitivity Substrate (Rockford, IL).
Nuclear extraction. Cells were harvested 48 h after transfection for nuclear extraction using the TransFactor extraction kit (BD Biosciences, Palo Alto, CA) according to the manufacturer’s instruction. Briefly, 150 mm2 flasks of transfected Huh7 cells were washed with PBS and incubated with the provided lysis buffer on ice for approximately 15 min. Lysed cells were then scraped from flasks, transferred to centrifuge tubes and repeatedly disrupted by aspiration through a narrow-gauge needle. The cell sample was then centrifuged and the cytosolic supernatant fraction was removed. Pelleted nuclei fractions were then resuspended in an extraction buffer and again subjected to disruption by needle aspiration. After centrifugation, the nuclear extract supernatant fraction was collected and stored in small aliquots at −20 °C.
TransFactor analysis. To analyze the inflammation-related transcription factors associated with SARS-CoV N, the in vitro TransFactor Inflammation Profiling Kit was used (BD Biosciences, Palo Alto, CA) (Fig. 1A ). The provided positive-control nuclear extracts were used along with the prepared nuclear extracts according to the manufacturer’s standard protocol. Briefly, the provided 96-well TransFactor plate was blocked with a blocking buffer supplied by the manufacturer for 15 min, then removed and replaced with 50 μl of nuclear extract and incubated at room temperature for 1 h. Plates were then washed three times with blocking buffer, incubated with provided primary antibodies, and washed three times. Bound primary antibodies were detected through incubation with HRP-conjugated secondary antibody and the addition of TMB substrate. Absorbance measurements were detected at 655 nm on a standard microplate reader.
Fig. 1.
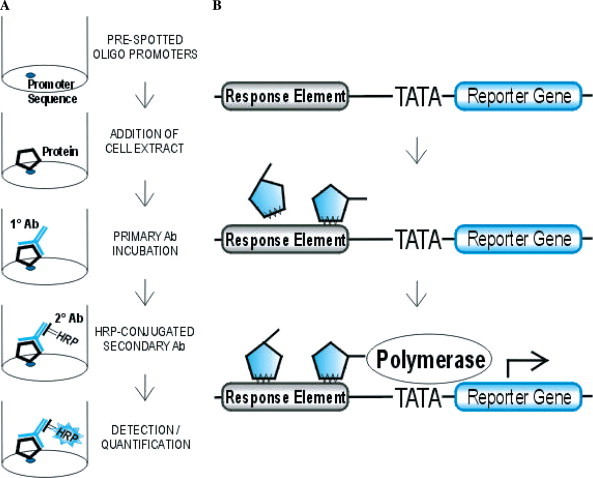
(A) Diagram of TransFactor inflammation profiling kit method. Cell extracts are added to 96-well plates coated with the wildtype DNA elements of individual transcription factors. Bound protein samples are then detected and quantified by primary and secondary antibody incubation. (B) Diagram of the PathDetect method for in vivo signal transduction assessment. The luciferase reporter gene is driven by a basic TATA promoter and an inducible enhancer bound proteins which recognize the AP-1 specific enhancer element are detected by an increase in reporter gene transcription.
PathDetect reporting system. The AP-1 and NF-κB signal transduction pathways were assayed in vivo using the PathDetect AP-1 cis-reporting system (Fig. 1B) according to the manufacturer’s instructions (Stratagene, La Jolla, CA). Briefly, 24-well tissue culture plates were seeded with approximately 1 × 105 Vero and Huh7 cells in 0.5 ml of complete DMEM. After 12 h, cells were cotransfected with 50 ng of the AP-1 reporter vector encoding the luciferase gene, and 500 ng of the constructed vectors pcDNA3.1(−), pcDNA-M, and pcDNA-N. Appropriate controls for transfection efficiency were also transfected as recommended by the manufacturer. After 48 h, the reporter luciferase was extracted by cell lysis and quantitated on a TD-20/20 luminometer (Turner BioSystems, Sunnyvale, CA) using the Luciferase Assay kit (Promega, Madison, WI).
Results and discussion
Identification of SARS-CoV nucleocapsid protein
The expression of NC protein in Huh7 cells was measured by Western blot using chicken IgY antibody against N protein. We transfected cells with pcDNA3.1(−) and pcDNA-N, and harvested the cells 48 h post transfection. The cell lysate samples were subjected to 4–12% SDS–PAGE and transferred to PVDF membrane. Chicken anti-N antibody was used to probe the expression of viral nucleocapsid protein, followed by donkey anti-chicken horseradish peroxidase as secondary antibody. As indicated in Fig. 2 , SARS-CoV N protein expression was only observed in cells transfected by pcDNA-N construct, but not in mock or pcDNA3.1(−) transfected samples. This experiment showed that N was expressed from pcDNA-N construct.
Fig. 2.

Detection of nucleocapsid protein by Western blotting analysis. Expressed proteins were analyzed by SDS–polyacrylamide gel (4–12% Bis–Tris) electrophoresis and visualized by autoradiography after nitrocellulose membrane transfer. Lane 1, mock transfection of Huh7 cells; lane 2, pcDNA3.1(−) transfection vector alone; lane 3, pcDNA3.1(−) transfection vector carrying the SARS-CoV N protein.
TransFactor analysis
The TransFactor profiling kit, which offers a high sensitivity signal, was employed to determine changes of expression levels of several signal transduction factors. Individual plate wells are coated with the wild-type cis-acting DNA elements for several inflammation-related transcription factors. After addition of nuclear extract, cellular transcription factors bind to their consensus sequences and interactions are measured by colorimetric changes. As the standard microplate reader could only register one blank well per TransFactor plate, relative negative controls were used to determine the activity of each experimental sample, with a sample reading of 2-fold higher than the controls being deemed significant. The transcription factors c-Fos, FosB, CREB-1, and ATF2 were all activated by the addition of SARS-CoV N protein to the sample well, with CREB-1 having the most dramatic increase in activity, about five times higher than the vector control (Fig. 3 ). c-Fos, FosB, and ATF-2 all showed increased colorimetric activities as indicated in Fig. 3. These factors are components of AP-1 signal transduction pathway, and their activation suggested that SARS-CoV N protein is involved in AP-1 activation. In contrast, NF-κB sample did not show increased signals, suggesting that the increase of signals of AP-1-related factors is specific.
Fig. 3.
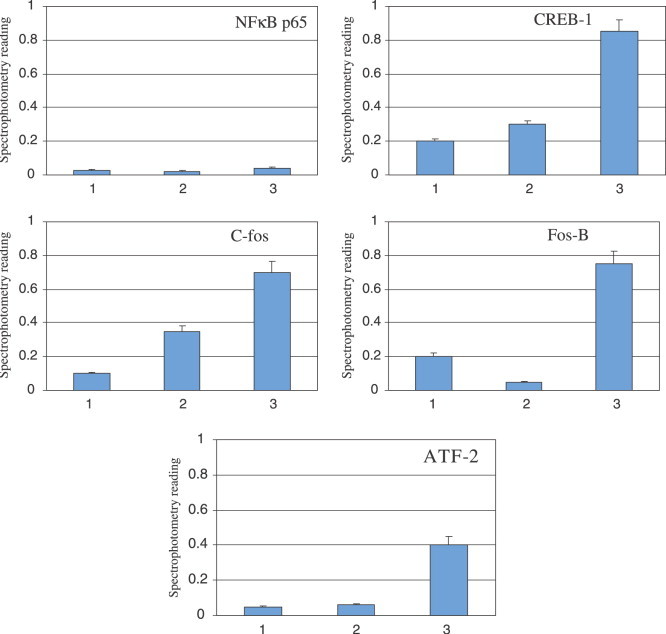
Effect of SARS-CoV N protein on the in vitro activation of pro-inflammatory transcription factors c-Fos, FosB, ATF2, and CREB-1. Absorbance measurements of samples were made at 655 nm after use of the TransFactor Inflammation Profiling Kit (Clontech Laboratories). Lanes 1, mock transfection; lanes 2, pcDNA3.1(−) vector; lanes 3, pcDNA3.1(−) + N.
Determination of AP-1 activation with PathDetect system
To further confirm the activation of AP-1 signal transduction pathway, we employed the PathDetect assay to examine the effect of the nucleocapsid protein on this transcription complex. The PathDetect pAP-1Luc reporter vector uses the inducible cis-enhancer element (TGACTAA)7 to bind potential regulators of AP-1 activity. Two cell lines were used in this assay, Vero cells, and Hu7 cells. We used pcDNA-N as N protein expression vector pcDNA3.1(−), and a construct expressing SARS-CoV membrane protein in pcDNA3.1 (pcDNA-M) to investigate whether NC specifically induces AP-1 signal transduction pathway. We also transfected serial dilutions of the aforementioned constructs to determine if AP-1 activation is dose-dependent. The luminometer reading for the mock transfection was considered blank for this experiment. As shown in Fig. 4A , the results from Vero cells indicated that transfection of pcDNA-N resulted in about four times higher chemiluminescence reading than that in cells transfected with either the pcDNA3.1(−) or pcDNA-M. With a decrease in amount of pcDNA-N, Ap-1 activities decreased accordingly in the pCDNA-N transfected cells, clearly suggesting that induction of AP-1 by SARS-CoV N protein is dose-dependent. Noticeably, transfection of cells with neither the pcDNA-M nor pcDNA3.1 vector was found to affect AP-1 activities. To further determine whether this activation is cell-type specific, the Huh7 cells were used under the same experimental conditions. As shown in Fig. 4B, AP activation by SARS-CoV N protein is four times higher than that observed in cells transfected with either pcDNA3.1(−) or pcDNA-M. Clearly, results obtained from the two cell lines are consistent in terms of AP-1 activation.
Fig. 4.
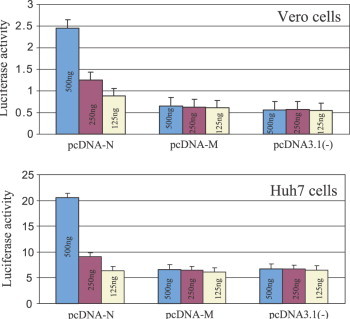
PathDetect assay on AP-1 activation. Vero and Huh7 cells transfected by dilutions of pcDNA3.1(−), pcDNA-3.1, and pcDNA3.1(−) + N. The reporter plasmid pAP-1Luc was co-transfected with the above plasmids. Forty-eight hours after transfection, luciferase activities were measured from the cell lysates of transfected samples. Values shown are means of five experiments subtracted from the blank control.
To further determine that the activation of AP-1 pathway by SARS-CoV N protein is pathway-selective, we performed the PathDetect study using NF-κB luciferase reporter vector. Again the same cell lines were used (Vero and Huh7 cells) for transfection with pcDNA3.1(−), pcDNA-M, and pcDNA-N using conditions described above. The pFC-MEKK vector, supplied by the manufacturer of PathDetect system, was used as the positive control for NF-κB activation. Luciferase assay was conducted to measure the NF-κB activity. As indicated in Fig. 5 , the luciferase activity readings of Vero and Huh7 cells transfected by pcDNA-N vector are at the similar level as cells transfected by pcDNA3.1(−) and pcDNA-M, indicating that SARS-CoV N protein expression does not activate the activation of NF-κB pathway; therefore, the activation of AP-1 by the expression of SARS-CoV N protein is pathway-specific.
Fig. 5.
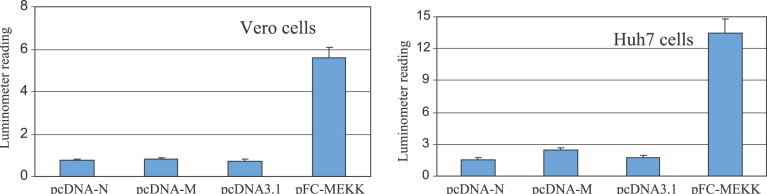
PathDetect assay on AP-1 activation. Vero and Huh7 cells transfected pcDNA3.1(−), pcDNA-3.1, and pcDNA3.1(−) + N. The reporter plasmid pNFκB-Luc was co-transfected with the above plasmids. The plasmid expression MEK kinase (pFC-MEKK) supplied by the manufacture was used as positive control for NFκB activation. Forty-eight hours after transfection, luciferase activities were measured from the cell lysates of transfected samples. Values shown are means of five experiments subtracted from the blank control.
Taken together, we have shown that the coronavirus N protein can activate AP-1 signal transduction pathway. Our data also demonstrated that the AP-1 pathway, not NF-κB signaling, can be activated by SARS-CoV N, suggesting that activation of cellular pathways by SARS-CoV N protein is selective. To our knowledge, this is the first time that a coronavirus N protein has been identified as AP-1 signal transduction activator. In addition, we have also found that the induction of AP-1 by SARS-CoV N protein is not cell type-specific because in both Vero and Huh7 cells AP-1 activities were increased by SARS-CoV N protein expression. Finally, the fact that expression of SARS-CoV M protein did not result in an increase in the level of AP-1 activities not only rules out the possibilities of experimental artifacts but also strengthens the claim of selective activation of AP-1 by SARS-CoV N protein.
The transcription factor AP-1 is composed of homodimers and heterodimers of Fos, Jun, CREB, and ATF subunits which form a specific DNA binding complex for the sequence 5′-TGAGTCA-3′ [14]. Indeed, our data have demonstrated that following SARS-CoV N protein expression, binding of the aforementioned critical components to the AP-1 DNA elements has been substantially enhanced (Fig. 1).
The c-Fos, FosB, and ATF/CREB oncoproteins are members of a family of transcription factors that have been shown to comprise AP-1, a central transcription factor complex that binds to the promoters of numerous genes [15]. C-Fos and FosB are cellular homologues of a viral oncogene carried by murine sarcoma virus, and together with Jun or ATF transcription factors, assemble to form the heterodimer AP-1 complex. The Jun transcription factors can form homodimers with other Jun subunits or heterodimers with any of the AP-1-related factors, and are therefore also central to the AP-1 signal transduction pathway [14].
The mechanism involved in AP-1 signal transduction activation by SARS-CoV N protein currently remains unknown. Interaction of the nucleocapsid proteins from other members of the coronavirus with the AP-1 pathway has not been documented in the literature. Whether SARS-CoV NC directly interacts with components of AP pathway or other intermediate proteins requires vigorous investigation. Furthermore, dissection of the functional domains of SARS-CoV N protein and/or identification of amino acid residue(s) involved in AP-1 activation will provide insightful information regarding pathogenesis of SARS. These studies are currently under way in our laboratories. In conclusion, our study for the first time shows that SARS-CoV can activate important signal transduction pathway with appreciable selectivity since AP-1, not NF-κB, is activated by SARS-CoV N protein, while the SAS-CoV M was found not to activate either AP-1 or NF-κB.
References
- 1.k.V. Holmes, Field’s Virology, vol. I, Lippincott, Williams & Wilkins, Philadelphia, 2001, pp. 1187–1203
- 2.Poutanen S.M., Low D.E., Henry B., Finkelstein S., Rose D., Green K., Tellier R., Draker R., Adachi D., Ayers M., Chan A.K., Skowronski D.M., Salit I., Simor A.E., Slutsky A.S., Doyle P.W., Krajden M., Petric M., Brunham R.C., McGeer A.J. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003;348(20):1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 3.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. SARS working group. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 5.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Hiscox J.A., Wurm T., Wilson L., Britton P., Cavanagh D., Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2001;75(1):506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 8.Rowland R.R., Kervin R., Kuckleburg C., Sperlich A., Benfield D.A. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 1999;64(1):1–12. doi: 10.1016/s0168-1702(99)00048-9. [DOI] [PubMed] [Google Scholar]
- 9.Chen H., Wurm T., Britton P., Brooks G., Hiscox J.A. Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J. Virol. 2002;76(10):5233–5250. doi: 10.1128/JVI.76.10.5233-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith S.A., Kotwal G.J. Immune response to poxvirus infections in various animals. Crit. Rev. Microbiol. 2002;28(3):149–185. doi: 10.1080/1040-840291046722. [DOI] [PubMed] [Google Scholar]
- 11.Kotwal G.J. Poxviral mimicry of complement and chemokine system components: what’s the end game? Immunol. Today. 2000;21(5):242–248. doi: 10.1016/s0167-5699(00)01606-6. [DOI] [PubMed] [Google Scholar]
- 12.Shaulian E., Karin M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 13.Degawa-Yamauchi M., Uotani S., Yamaguchi Y., Takahashi R., Abe T., Kuwahara H., Yamasaki H., Eguchi K. Ethanol inhibits leptin-induced STAT3 activation in Huh7 cells. FEBS Lett. 2002;525(1–3):116–120. doi: 10.1016/s0014-5793(02)03099-5. [DOI] [PubMed] [Google Scholar]
- 14.Wisdom R. AP-1: one switch for many signals. Exp. Cell Res. 1999;253:180–185. doi: 10.1006/excr.1999.4685. [DOI] [PubMed] [Google Scholar]
- 15.Karin M., Liu Z., Zandi E. AP-1 function and regulation. Curr. Opin. Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]


