Significance
Coronaviruses such as Middle East respiratory syndrome coronavirus (MERS-CoV) cause severe respiratory distress with high fatality rates. The spike (S) glycoprotein is a determinant of host range and is the target of neutralizing antibodies and subunit vaccine development. We describe an engineering strategy for stabilization of soluble S proteins in the prefusion conformation, which results in greatly increased expression, conformational homogeneity, and elicitation of potent antibody responses. Cryo-EM structures of the stabilized MERS-CoV S protein in complex with a stem-directed neutralizing antibody provide a molecular basis for host-cell protease requirements and identify a site of immune pressure. We also defined four conformational states of the trimer wherein each receptor-binding domain is either packed together at the membrane-distal apex or rotated into a receptor-accessible conformation.
Keywords: coronavirus, neutralizing antibody, cryo-EM, X-ray crystallography, peplomer
Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) is a lineage C betacoronavirus that since its emergence in 2012 has caused outbreaks in human populations with case-fatality rates of ∼36%. As in other coronaviruses, the spike (S) glycoprotein of MERS-CoV mediates receptor recognition and membrane fusion and is the primary target of the humoral immune response during infection. Here we use structure-based design to develop a generalizable strategy for retaining coronavirus S proteins in the antigenically optimal prefusion conformation and demonstrate that our engineered immunogen is able to elicit high neutralizing antibody titers against MERS-CoV. We also determined high-resolution structures of the trimeric MERS-CoV S ectodomain in complex with G4, a stem-directed neutralizing antibody. The structures reveal that G4 recognizes a glycosylated loop that is variable among coronaviruses and they define four conformational states of the trimer wherein each receptor-binding domain is either tightly packed at the membrane-distal apex or rotated into a receptor-accessible conformation. Our studies suggest a potential mechanism for fusion initiation through sequential receptor-binding events and provide a foundation for the structure-based design of coronavirus vaccines.
Coronaviruses are enveloped, positive-sense, single-stranded RNA viruses. They have the largest genomes (26–32 kb) among known RNA viruses and are phylogenetically divided into four genera (α, β, γ, and δ), with betacoronaviruses further subdivided into four lineages (A, B, C, and D). Coronaviruses infect a wide range of avian and mammalian species, including humans (1). Of the six known human coronaviruses, four of them (HCoV-OC43, HCoV-229E, HCoV-HKU1, and HCoV-NL63) circulate annually in humans and generally cause mild respiratory diseases, although severity can be greater in infants, the elderly, and the immunocompromised. In contrast, the Middle East respiratory syndrome coronavirus (MERS-CoV) and the severe acute respiratory syndrome coronavirus (SARS-CoV), belonging to betacoronavirus lineages C and B, respectively, are highly pathogenic. Both viruses emerged into the human population from animal reservoirs within the last 15 y and caused outbreaks with high case-fatality rates (2, 3).
MERS-CoV was isolated in 2012 from a patient in Saudi Arabia and is still circulating across the Arabian Peninsula (3, 4). Primary transmission, most likely from camels, is now considered to be the most common route of transmission (5–7), and camels are thought to be a secondary or intermediate reservoir for MERS-CoV, with bats serving as the primary reservoir (8). Human-to-human transmission, especially as a result of close contact between patients and hospital workers within health-care settings, is another important route of transmission (9) and was responsible for an outbreak of MERS-CoV in South Korea (10). The high pathogenicity and airborne transmissibility of SARS-CoV and MERS-CoV have raised concern about the potential for another coronavirus pandemic. Unfortunately, vaccines against individual coronaviruses are not available, much less one that broadly protects against multiple coronaviruses.
Like all coronaviruses, MERS-CoV uses a large surface spike (S) glycoprotein for receptor recognition and entry into target cells (11, 12). The MERS-CoV S protein is synthesized as a single-chain inactive precursor that is cleaved by furin-like host proteases in the producing cell into two noncovalently associated subunits, S1 and S2 (13). The S1 subunit contains the receptor-binding domain (RBD), which recognizes the host-cell receptor DPP4 (CD26) (14–16). The S2 subunit contains the fusion peptide, two heptad repeats, and a transmembrane domain, all of which are required to mediate fusion of the viral and host-cell membranes by undergoing a large conformational rearrangement. The S1 and S2 subunits trimerize to form a large prefusion spike (∼600 kDa) with ∼25 N-linked glycans per monomer. Recent cryo-EM structures of trimeric prefusion S protein ectodomains from murine hepatitis virus (MHV), HCoV-HKU1, and HCoV-NL63 have revealed an overall mushroom-like architecture, with three identical S1 subunits forming an interwoven cap that rests atop the spring-loaded S2 stem (17–19). Interestingly, in this conformation the RBDs at the C terminus of S1 are not accessible for receptor binding, suggesting that a conformational change is required to expose the RBDs. Recently, a partial cryo-EM structure of the SARS-CoV S protein ectodomain was obtained that contained one of the three RBDs rotated into a receptor-accessible conformation (20). This configuration of the trimer may represent an initial intermediate state, although many questions remain unanswered, such as why only one of the RBDs is rotated.
As the primary glycoprotein on the surface of the viral envelope, S proteins are the major target of neutralizing antibodies elicited by natural infection and are key antigens in experimental vaccine candidates (11, 21, 22). However, the S protein ectodomain from MERS-CoV is less stable and more difficult to produce than other S proteins, and soluble constructs of the RBD have been the main focus of structural studies (14, 16), antibody isolation efforts (21, 23–25), and subunit vaccine development (26). A drawback of this approach is that coronaviruses can readily generate antibody-escape mutations in the RBD (23, 27, 28). Thus, the use of a mixture of antibodies, including some directed against non-RBD epitopes, is a preferred strategy (29) and has been used successfully for the treatment of Ebola virus disease (30). However, due to the difficulty in producing prefusion-stabilized MERS-CoV S proteins, few non-RBD antibodies have been described, and less is known about their epitopes. Antibodies against the prefusion conformation of the S2 stem are particularly attractive because the stem is more conserved than the S1 cap. Therefore, the ability to produce prefusion-stabilized S protein ectodomains from highly pathogenic coronaviruses, combined with the structural characterization of non-RBD epitopes that are recognized by potent neutralizing antibodies (31), would greatly facilitate the development of broadly protective interventions for current and emerging coronaviruses. Similar approaches are currently being developed for HIV-1 and influenza (32–34). Notably, the identification of stem-directed antibodies against influenza HA (35) represented a paradigm shift in the approach to develop a universal influenza vaccine, with recent studies demonstrating substantial promise (33, 34).
In this study, we rationally designed a general strategy to retain betacoronavirus S proteins in the prefusion conformation. The prefusion-stabilized MERS-CoV S protein (MERS S-2P) retained high-affinity binding to its dimeric receptor DPP4 and a panel of neutralizing antibodies, and elicited high titers of neutralizing antibodies in mice. The MERS S-2P construct also facilitated single-particle cryo-EM studies on a complex with G4, the first identified MERS-CoV S2-directed antibody (31). G4 was isolated from immunized mice and shown to be neutralizing, yet its epitope on S2 was unknown. The structures revealed that G4 recognizes the membrane-proximal surface of a variable, glycosylated loop in the S2 connector domain and avoids the glycosylation via its angle of approach, which is directed up from the viral membrane. The structures also defined four configurations of the trimer apex that represent the receptor-inaccessible ground state and three receptor-accessible intermediates. Collectively, these results advance our understanding of MERS-CoV entry and antibody-mediated neutralization and provide a foundation for the structure-based design of vaccine antigens for highly pathogenic coronaviruses, including those expected to emerge in the future.
Results
Engineering of Coronavirus S Proteins That Retain the Prefusion Conformation.
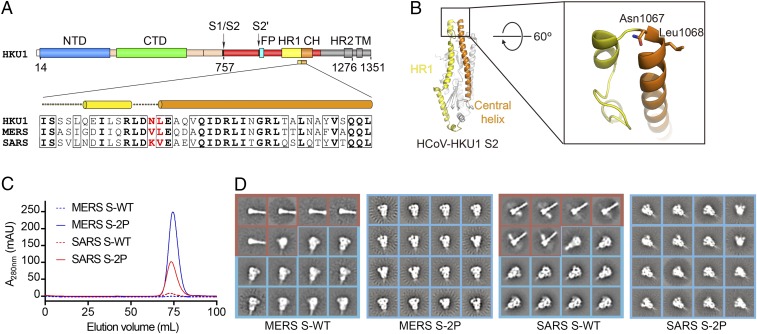
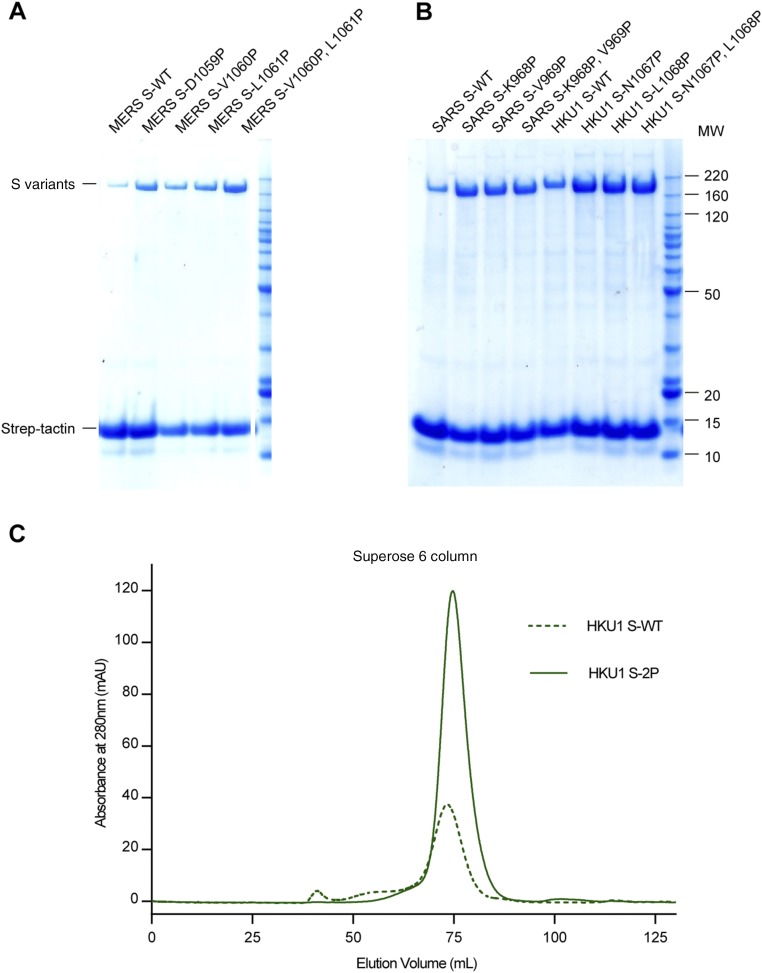
To improve the expression and conformational homogeneity of the prefusion MERS-CoV S protein ectodomain we engineered variants based upon our previously determined structure of the trimeric S protein from the lineage A betacoronavirus HCoV-HKU1 (18) (Fig. 1 A and B). Recent work on the fusion proteins from HIV-1 and respiratory syncytial virus (RSV) (32, 36, 37) has demonstrated that proline substitutions in the loop between the first heptad repeat (HR1) and the central helix restrict premature triggering of the fusion protein and often increase expression yields of prefusion ectodomains. Introduction of single proline substitutions into a similar region in the MERS-CoV S2 subunit dramatically increased expression levels of the ectodomains, and two consecutive proline substitutions at residues V1060 and L1061 (hereafter referred to as “2P”) resulted in a >50-fold improvement in yield (Fig. 1C and Fig. S1A). As evidenced by negative-stain EM, the 2P variant maintained prototypic prefusion morphology (Fig. 1D). Homologous substitutions in the S proteins from SARS-CoV (Fig. 1 C and D and Fig. S1B) and HCoV-HKU1 (Fig. S1 B and C) also increased the expression levels of the ectodomains and improved conformational homogeneity. Thus, the introduction of two consecutive proline residues at the beginning of the central helix seems to be a general strategy for retaining betacoronavirus S proteins in the prototypical prefusion conformation.
Fig. 1.
Structure-based engineering of MERS-CoV and SARS-CoV S proteins. (A) Domain architecture of the HCoV-HKU1 S protein and sequence alignment of the helix-turn-helix between heptad repeat 1 (HR1) and the central helix (CH). The two residues colored red are those mutated to proline to retain S2 in the prefusion conformation. FP, fusion peptide; HR2, heptad repeat 2; TM, transmembrane domain. (B) Structure of HCoV-HKU1 S2. Residues shown in sticks in magnified region are those mutated to proline in the 2P variants. (C) Gel-filtration profiles of WT (dashed lines) and 2P-engineered (solid lines) S protein ectodomains from MERS-CoV (blue) and SARS-CoV (red). Each protein was produced from a 1-L transient transfection. All four proteins were expressed with a C-terminal T4 fibritin trimerization domain. The S1/S2 furin site was mutated in MERS S-WT and MERS S-2P. (D) Two-dimensional class averages of negative stained MERS S-WT, MERS S-2P, SARS S-WT, and SARS S-2P. All particles are included. For WT constructs both the prefusion (blue boxes) and postfusion (red boxes) conformations are visible, whereas for the 2P mutants only the prefusion conformation is observed.
Fig. S1.
Proline substitutions in S2 increase expression levels of coronavirus S proteins. (A and B) Expression levels of WT and proline-substituted S proteins from MERS-CoV (A) and SARS-CoV and HCoV-HKU1 (B) as assessed by SDS/PAGE. (C) Gel-filtration profiles of WT HCoV-HKU1 S (HKU1 S-WT) and stabilized HCoV-HKU1 S (HKU1 S-2P). Each protein was produced from 500 mL FreeStyle 293-F cells.
To investigate the effect of the 2P substitutions on S protein function, MERS-CoV pseudoviruses were generated with WT or 2P-containing S proteins. In contrast to WT pseudoviruses, which were highly infectious in DPP4-expressing Huh7.5 cells, pseudoviruses containing the 2P substitutions in the S protein were essentially noninfectious (Fig. 2A). This lack of infectivity is likely due to the ability of the introduced prolines to prevent conformational rearrangements, which presumably increases the activation energy required for fusion. However, to rule out the possibility of local misfolding as a result of the 2P substitutions, full-length WT and 2P-containing S proteins were expressed on 293T cells, and the reactivity of polyclonal sera and conformation-dependent antibodies was assessed by flow cytometry (Fig. 2B). The polyclonal sera, as well as neutralizing antibodies against the N-terminal domain (NTD) (mAb G2), RBD (mAb D12), and S2 subunit (mAb G4), reacted equally well to cells expressing WT or 2P-containing S proteins. Control cells expressing the RBD fused to a transmembrane domain only reacted with the polyclonal sera and D12 antibody, as expected. All three antibodies also bound to the recombinant MERS-CoV S protein ectodomains containing the 2P substitutions (MERS S-2P) via pull-down (Fig. 2C). We also determined that MERS S-2P bound to a soluble version of its receptor, DPP4, with a Kd of 9.4 nM (Fig. 2D), which is similar to reported values for the binding of DPP4 to the isolated RBD (14). Collectively, these data demonstrate that the 2P substitutions prevent fusion from occurring but do not alter the conformation of the S protein.
Fig. 2.
Characterization of MERS S-2P. (A) MERS-CoV pseudoviruses encoding a luciferase reporter gene were generated with WT (S WT, blue) or 2P (S-2P, red) S proteins. Mock pseudoviruses (gray), expressing no S protein, served as a control. Infectivity in Huh7.5 cells was determined by measuring RLU. The dotted line represents background RLU. (B) Binding of cell-surface expressed MERS-CoV WT and 2P S proteins, as well as membrane-tethered RBD, to polyclonal sera (Poly) and monoclonal antibodies measured by flow cytometry; 101F is an RSV F-specific antibody. (C) SDS/PAGE analysis of copurified complexes of untagged MERS S-2P and monoclonal antibodies. AM14 is an RSV F-specific antibody. (D) Binding of soluble DPP4 to immobilized MERS S-2P measured by surface plasmon resonance. Best fit of the data to a 1:1 binding model is shown in red.
Immunogenicity of MERS S-2P.
To determine the immunogenicity of the MERS S-2P trimer we vaccinated mice and compared the responses to those generated by monomeric S1 protein and WT S trimer (31). Each of the immunogens elicited neutralizing antibodies to the autologous England1 strain of MERS-CoV in a dose-dependent manner. S1 monomers and S WT trimers had a steeper dose effect than the S-2P trimer, which reached a plateau of neutralizing activity at a lower dose (Fig. 3A). At the 0.1-μg dose, MERS S-2P elicited significantly more robust neutralizing activity than S1 against four of the six homologous MERS-CoV pseudoviruses tested. The S-2P vaccination also elicited greater neutralization than S WT against three of the six homologous MERS-CoV pseudoviruses tested (Fig. 3B). These data demonstrate that retaining MERS-CoV S in its prefusion conformation increases the breadth and potency of the neutralizing activity elicited by vaccination.
Fig. 3.
Immunogenicity of MERS S-2P in mice. (A) Reciprocal serum IC90 neutralizing activity against autologous MERS England1 pseudotyped lentivirus reporter plotted against vaccine dose. (B) Reciprocal serum IC90 neutralizing activity against multiple homologous MERS-CoV pseudoviruses of sera from mice immunized with 0.1 μg of protein. For both panels, the geometric mean IC90 titer (GMT) of each group is represented by (A) symbols or (B) bars. Error bars represent geometric SDs. P values denoted as *P < 0.05 and **P < 0.01. The limit of detection for the assay is represented by dotted lines; for sera below the limit of detection a reciprocal IC90 titer of 10 was assigned.
Structure of MERS S-2P Bound to Antibody G4.
We combined MERS S-2P with the S2-directed G4 Fab and conducted single-particle cryo-EM of the resulting complexes to structurally characterize our immunogen and provide atomic-level information needed for future engineering efforts (Fig. 4A, Fig. S2, and Tables S1 and S2). We observed several distinct subpopulations of S proteins that differed in the arrangement of the S1 apex, and these subpopulations were processed separately as described in more detail below. In addition, the cryo-EM density maps were less well-resolved in regions of the S1 NTD, as was the case in the recently determined structure of the SARS-CoV S protein (20). We consequently crystallized this domain and determined its structure to 2.0 Å using a portion of the cryo-EM map as a “search model” for molecular replacement (Fig. S3 and Table S3). We also crystallized and determined the structure of the unbound G4 Fab to 1.57 Å (Table S3), and both of these X-ray structures were used in further refinement of the cryo-EM–derived models.
Fig. 4.
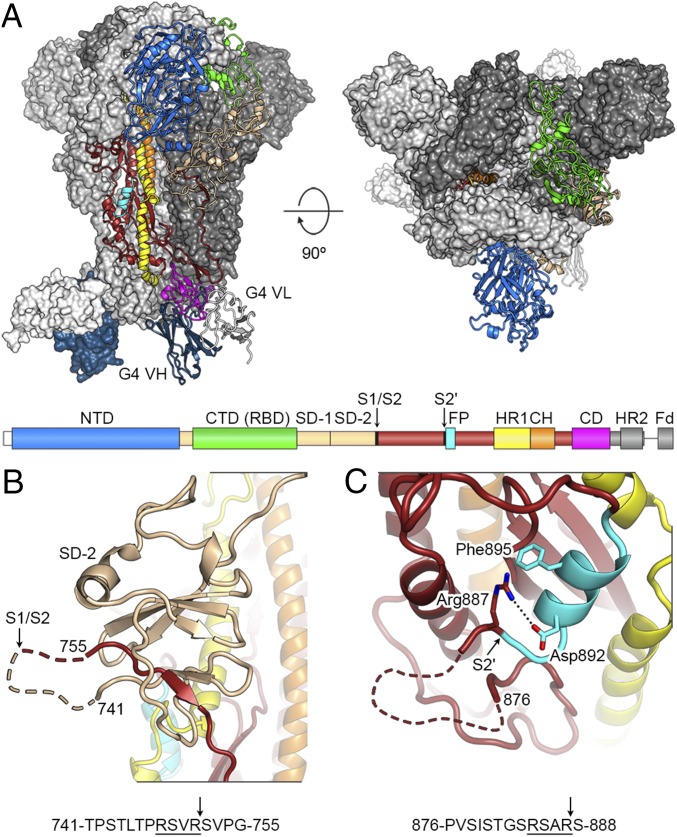
Structure of MERS-CoV S-2P in complex with G4 Fab. (A) Structure of MERS S-2P ectodomain in complex with G4 Fab as viewed along (Left) and above (Right) the viral membrane. A single protomer of the trimeric S protein is shown in ribbon representation and colored as in the primary structure diagram. The two remaining protomers are shown as molecular surfaces and colored white and gray. CD, connector domain; CH, central helix; Fd, Foldon trimerization domain; SD-1, subdomain 1; SD-2, subdomain 2. (B and C) Magnified view of the S1/S2 (B) and S2′ (C) protease sites. Dashed lines represent disordered residues. Arrows indicate position of protease cleavage.
Fig. S2.
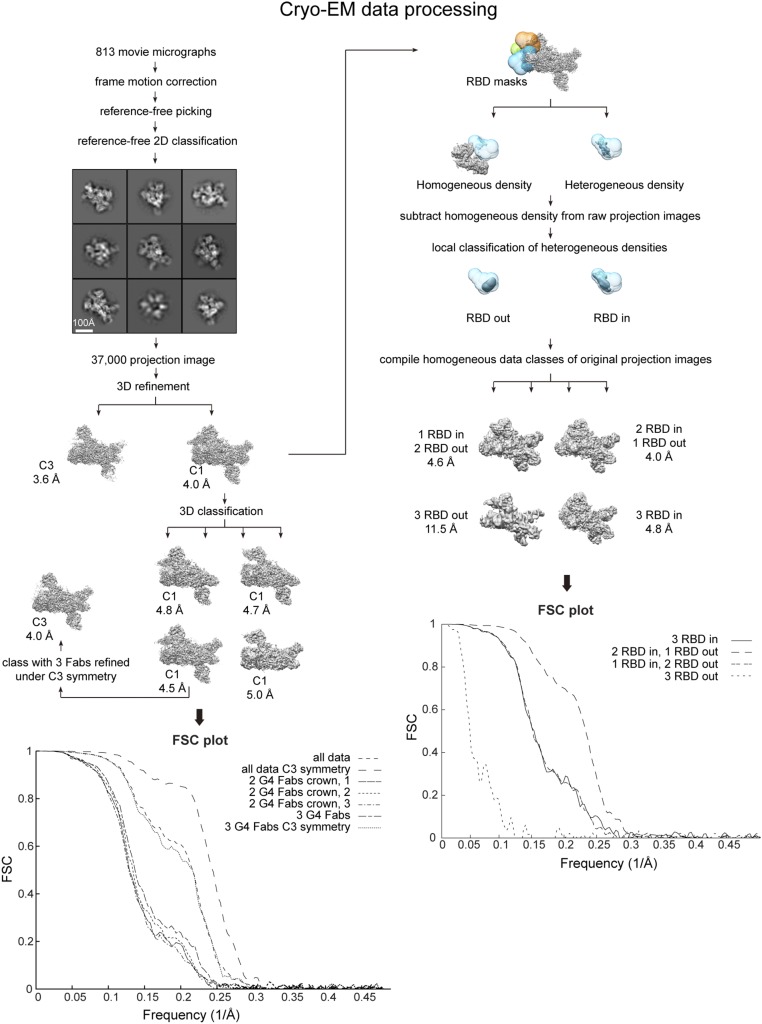
Cryo-EM data processing. Data were sorted and refined according to the flowchart. In particular, to resolve the heterogeneity in RBD configurations in our dataset we used a method of data sorting that involves subtraction of constant density regions from our raw data followed by local classification of the resulting projection images.
Table S1.
Cryo-EM data collection
| Microscope | FEI Titan Krios |
| Voltage, kV | 300 |
| Detector | Gatan K2 Summit |
| Recording mode | Counting |
| Magnification (including postmagnification) | 49,020 |
| Movie micrograph pixel size, Å | 1.02 |
| Dose rate, e−/[(camera pixel)*s] | 10 |
| No. of frames per movie micrograph | 35 |
| Frame exposure time, ms | 200 |
| Movie micrograph exposure time, s | 7 |
| Total dose, e−/Å2 | 66 |
| Defocus range, µm | 0.7–2.5 |
Table S2.
Cryo-EM processing and model validation
| Map | |||||||||||
| C3 | C1 | 2_G4_1 | 2_G4_2 | 2_G4_3 | 3_G4 | 3_G4_C3 | 3in_C3 | 2in_1out | 1in_2out | 3out_C3 | |
| EMDB accession code | EMD-8784 | EMD-8792 | EMD-8787 | EMD-8788 | EMD-8789 | EMD-8790 | EMD-8791 | EMD-8785 | EMD-8783 | EMD-8786 | EMD-8793 |
| PDB ID code | 5W9I | 5W9L | 5W9M | 5W9N | 5W9O | 5W9P | 5W9J | 5W9H | 5W9K | ||
| EM data processing | |||||||||||
| No. of movie micrographs | 813 | 813 | 813 | 813 | 813 | 813 | 813 | 813 | 813 | 813 | 813 |
| No. of molecular projection images in map | 37,180 | 37,180 | 8,496 | 9,333 | 8,133 | 11,218 | 11,218 | 1,988 | 24,250 | 10,544 | 117 |
| Symmetry | C3 | C1 | C1 | C1 | C1 | C1 | C3 | C3 | C1 | C1 | C3 |
| Map resolution (FSC 0.143, Å) | 3.6 | 4.0 | 4.8 | 4.7 | 5.0 | 4.5 | 4.0 | 4.8 | 4.0 | 4.6 | 11.5 |
| Map sharpening B-factor, Å2 | −88 | −96 | −127 | −111 | −43 | −111 | −93 | −75 | −94 | −129 | −81 |
| Structure building and validation | N/A | N/A | |||||||||
| No. of atoms in deposited model | 28,863 | 31,077 | 31,085 | 31,085 | 32,916 | 27,981 | 32,916 | 33,798 | 32,835 | ||
| S1 | 4,010 | 5,655 | 5,655 | 5,655 | 5,655 | 4,010 | 5,655 | 5,655 | 5,642 | ||
| S2 | 3,535 | 3,535 | 3,535 | 3,535 | 3,535 | 3,535 | 3,535 | 3,535 | 3,521 | ||
| G4 VH | 947 | 947 | 947 | 947 | 947 | 947 | 947 | 947 | 947 | ||
| G4 VL | 835 | 835 | 835 | 835 | 835 | 835 | 835 | 835 | 835 | ||
| Glycans | 294 | 294 | |||||||||
| MolProbity score | 1.52 (95%) | 1.51 (95%) | 1.37 (98%) | 1.32 (98%) | 1.65 (91%) | 1.44 (96%) | 1.67 (90%) | 1.39 (97%) | 1.45 (96%) | ||
| Clashscore | 3.81 | 3.50 | 3.12 | 2.57 | 5.95 | 2.75 | 5.70 | 3.10 | 3.68 | ||
| EMRinger score | 2.59 | 0.67 | 0.89 | 0.62 | 1.36 | 2.11 | 0.62 | 1.88 | 1.34 | ||
| Deviations from ideal | |||||||||||
| Bond length | 3 (0.02%) | 2 (0.01%) | 0 (0.00%) | 0 (0%) | 2 (0.01%) | 0 (0%) | 0 (0%) | 0 (0.0%) | 2 (0.01%) | ||
| Bond angles | 93 (0.52%) | 147 (0.73%) | 136 (0.68%) | 126 (0.63%) | 142 (0.67%) | 75 (0.42%) | 108 (0.51%) | 127 (0.60%) | 122 (0.57%) | ||
| Ramachandran plot | |||||||||||
| Favored, % | 95.0 | 94.7 | 96.1 | 95.9 | 95.4 | 94.5 | 94.8 | 95.8 | 95.7 | ||
| Allowed, % | 4.2 | 4.5 | 3.4 | 3.4 | 3.9 | 4.4 | 4.0 | 3.6 | 3.7 | ||
| Outliers, % | 0.9 | 0.9 | 0.6 | 0.6 | 0.7 | 1.2 | 1.2 | 0.6 | 0.6 | ||
N/A, not assessed.
Fig. S3.
Crystal structure of MERS-CoV S1 NTD. (A) The structure of the MERS-CoV S1 NTD can be separated into top, core and bottom regions (86). The two β-sheets of the NTD core are colored green and magenta, and the rest of the NTD is colored cyan. (B) The NTD structure is colored as a rainbow from blue to red, N to C terminus, respectively. (C) Extracted ion chromatogram (extracted theoretical mass 442.15 at 5 ppm), MS, and MS/MS spectra of ligand isolated from purified MERS-CoV S NTD. Concentrated NTD was heated at 98 °C for 10 min, and the denatured protein was pelleted via centrifugation. The supernatant was desalted and analyzed as previously described (87) on a Q Exactive Plus mass spectrometer with the modification that +1 ions were also selected for fragmentation. Data analysis was performed in QualBrowser. (D) Extracted ion chromatogram (extracted theoretical mass 442.15 at 5 ppm), MS, and MS/MS spectra of purified folic acid (Sigma) resuspended in 5% methanol and 1% formic acid and analyzed as described above. (E) The chemical structure of folic acid. (F) Crystal structure of MERS-CoV S NTD shown as a ribbon with the Fo-Fc map before placement and refinement of folic acid shown at 2.0 sigma (gray mesh). Folic acid (shown in sticks) was modeled into this density and used for further structural refinement. Oxygen atoms are red and nitrogen atoms are blue.
Table S3.
Crystallographic data collection and refinement statistics
| MERS-CoV S1 NTD | G4 Fab | |
| PDB ID code | 5VYH | 5VZR |
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell constants | ||
| a, b, c, Å | 49.8 84.4 108.0 | 80.7, 88.8, 128.5 |
| α, β, γ, ° | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength, Å | 1.181 | 0.979 |
| Resolution, Å | 54.0–2.0 (2.05–2.00)* | 88.8–1.57 (1.60–1.57) |
| Rmerge | 0.186 (0.700) | 0.128 (1.641) |
| Rpim | 0.075 (0.325) | 0.056 (0.692) |
| I/σI | 6.1 (2.0) | 7.8 (1.7) |
| CC(1/2) | 0.985 (0.731) | 0.994 (0.601) |
| Completeness (%) | 99.3 (92.5) | 99.8 (99.8) |
| Redundancy | 6.9 (5.4) | 6.1 (6.3) |
| Refinement | ||
| Resolution, Å | 45.5–2.00 (2.06–2.00) | 73.0–1.57 (1.59–1.57) |
| Unique reflections | 31,250 (2,595) | 128,598 (4,250) |
| Rwork/Rfree, % | 19.1/22.2 | 18.2/21.5 |
| No. of atoms | ||
| Protein | 2,703 | 6,573 |
| Ligand/ion | 157 | 6 |
| Water | 411 | 953 |
| B-factors, Å2 | ||
| Protein | 25.7 | 25.8 |
| Ligand/Ion | 44.0 | 35.0 |
| Water | 35.5 | 38.3 |
| Rmsd | ||
| Bond lengths, Å | 0.004 | 0.006 |
| Bond angles, ° | 0.77 | 0.85 |
| Ramachandran | ||
| Favored regions, % | 97.4 | 98.2 |
| Allowed regions, % | 2.6 | 1.8 |
| Disallowed regions, % | 0.0 | 0.0 |
Values in parentheses are for highest-resolution shell.
Overall, the structure of the MERS S-2P protein is similar to the previously determined prefusion structures of alpha- and betacoronavirus S proteins (17–20). Our models consist of residues 18–1223 and like other structures are missing the second heptad repeat in S2, which may be flexible in the absence of a lipid bilayer. The MERS-CoV S2 subunit is arranged similarly to other coronavirus S2 subunits, with rmsd values of ∼1.5 Å for a shared core of ∼260 Cα atoms. For efficient infection of target cells, the MERS-CoV S protein requires a two-step, protease-mediated activation to facilitate membrane fusion. Furin cleavage at the S1/S2 junction occurs in the virus-producing cell, whereas cleavage at the S2ʹ site, upstream of the fusion peptide, occurs during viral entry at the cell surface or in endosomes and can be mediated by several proteases, including furin, TMPRSS2, and cathepsin L (11, 13, 38). However, it has not been understood why furin can access the S1/S2 site but not the S2ʹ site during protein biosynthesis. The S1/S2 furin site (RSVR), which remains uncleaved in our construct due to mutagenesis (ASVG), is located on an accessible solvent-exposed loop that is disordered in our structures (Fig. 4B). In contrast, the S2ʹ site (RSAR) is less exposed, particularly Arg887 at the P1 position, which interacts with Asp892 and Phe895 in the fusion peptide (Fig. 4C). In our structure, the peptide bond between Arg887 and Ser888 remains inaccessible to proteases, suggesting that S2ʹ cannot be efficiently cleaved until a conformational change occurs in S2 during the fusion process. Refolding of HR1 following DPP4 binding and S1 shedding would cause such a change and link the final proteolytic activation step to host-cell attachment, thus ensuring that irreversible refolding of S2 occurs at the proper time and place. Indeed, incubation of MERS-CoV virions with soluble DPP4 receptor increases the efficiency of furin cleavage at the S2ʹ site (13).
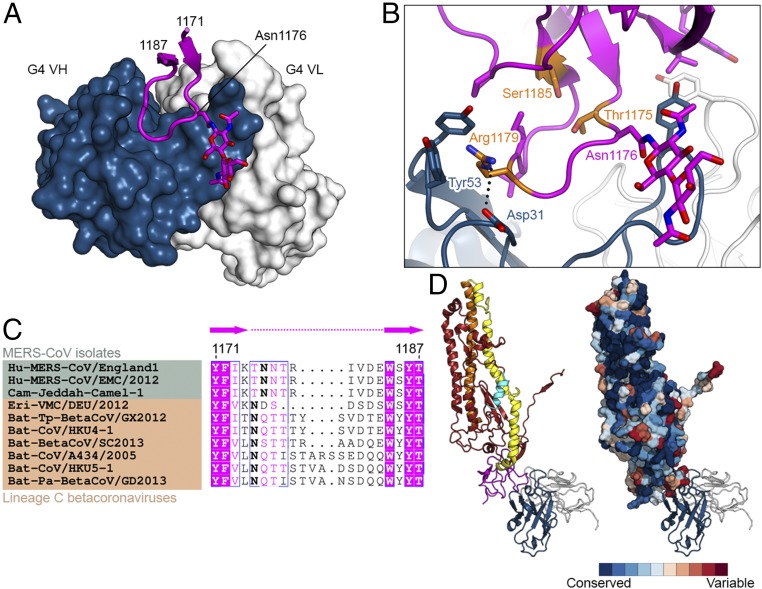
The MERS-CoV S2 structures also contain a well-resolved domain (residues 1152–1223) residing between HR1 and HR2 that was not completely resolved in the related betacoronavirus HCoV-HKU1, MHV, and SARS-CoV S protein structures but was observed in the alphacoronavirus HCoV-NL63 S protein structure (17–20). This connector domain contains the epitope for G4, which is the first reported S2-specific antibody that neutralizes MERS-CoV (31). The local resolution of the maps used for analysis of the antibody interface with S2 exceeds 3.7 Å. The majority of the G4 epitope (710 Å2 of 880 Å2 total buried surface area) consists of a glycosylated, solvent-exposed loop that extends out from two β-strands and is enshrouded by the complementarity-determining regions (CDRs) of G4 (Fig. 5A). The binding of G4 was substoichiometric, allowing comparison of the bound and unbound G4 epitope. In unbound protomers the loop was poorly ordered, suggesting that it is flexible in solution (Fig. S4). Arg1179 is critical for G4 binding, as it forms a cation–π interaction with HCDR2 Tyr53 as well as a salt bridge with HCDR1 Asp31 (Fig. 5B). In addition, G4 binding results in the formation of two hydrophobic cores that include S2 residues outside of the extended loop. These interactions may stabilize the connector domain, which is likely flexible given the poorly resolved density observed in other betacoronavirus S protein structures.
Fig. 5.
G4 recognizes a variable loop in the S2 connector domain. (A) Structure of G4 Fab bound to a variable loop contained within the S2 subunit. Residues 1171–1187 of MERS S-2P are shown as a ribbon, with the side chain of Asn1176 and two attached N-acetylglucosamine moieties shown as sticks. The variable domains of G4 are shown as a molecular surface. (B) G4 binding interface. Side chains of interacting residues are shown as sticks, with residues substituted in G4-escape variants colored orange. Black dotted line indicates a salt bridge. (C) Sequence alignment of MERS-CoV isolates (green) and other lineage C betacoronaviruses (tan). Bold font indicates N-linked glycosylation sites. (D) Side views of one S2 protomer bound to G4 Fab. On the right, S2 is shown as a molecular surface and colored according to sequence conservation as determined by the ConSurf server using 66 diverse coronavirus sequences (85).
Fig. S4.
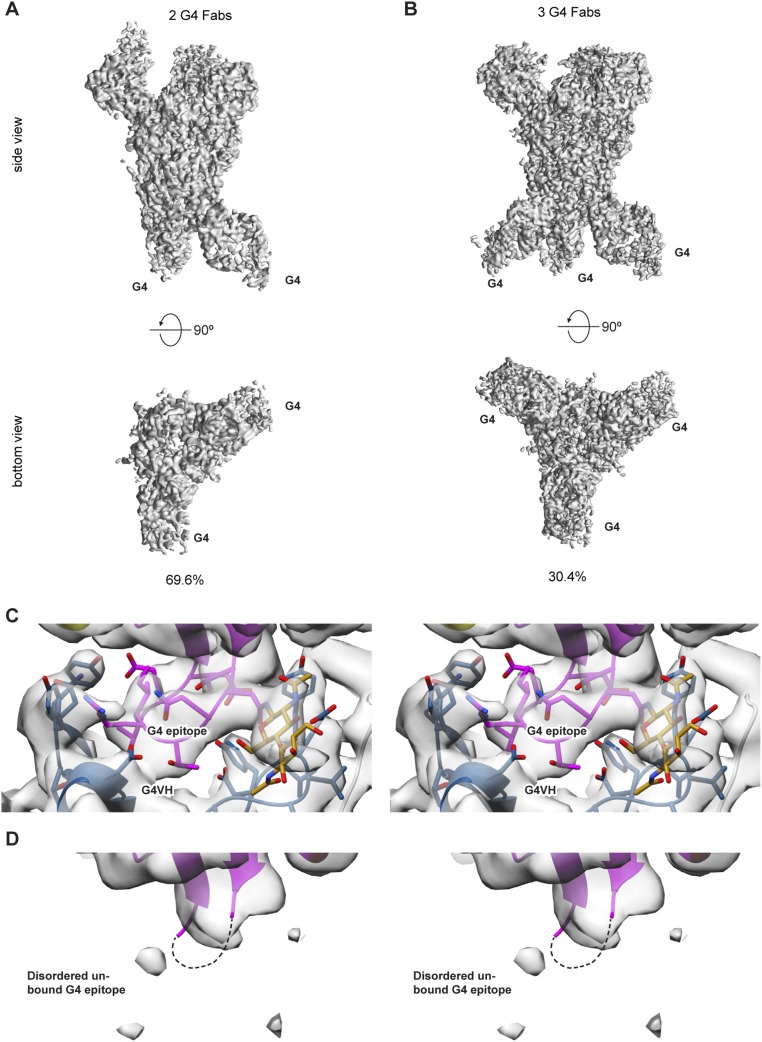
G4 Fab occupancy. (A and B) The MERS S-2P/G4 data contained two stoichiometric G4 classes. A majority of the data (69.6%) contained two G4 Fabs bound to the MERS S-2P spike (A), and the remaining data (30.4%) contained three bound G4 Fabs (B). (C and D) Stereo images of the G4 epitope. Epitopes that have G4 Fabs bound (C) are well-ordered whereas epitopes that remain unbound (D) exhibit a disordered loop.
Although the epitope of G4 is contained within the relatively conserved S2 subunit, the exposed loop to which it primarily binds is variable in both sequence and length, even among lineage C betacoronaviruses (Fig. 5 C and D). The variability in this loop is reminiscent of the variable loops found in HIV-1 gp120, which arise as a result of pressure exerted by the host antibody response. Indeed, we were able to isolate MERS-CoV escape variants by growing the virus in the presence of G4, and the escape mutations (T1175I/P, R1179G/M/T, and S1185L) accumulated within and around this variable loop (Fig. 5B and Table S4). In addition to the sequence and length variability, this loop always contains at least one potential N-linked glycosylation site (Fig. 5C), which, like the HIV-1 gp120 variable loops, helps to shield the exposed loop from antibody recognition. G4 circumvents the glycan mask via its angle of approach, which is directed up from the viral membrane (Fig. 4A). This allows G4 to recognize the membrane-proximal face of the loop and avoid the glycan moieties attached to Asn1176. The requirement for this constrained approach likely comes at a cost to the binding affinity, as G4 has a higher affinity to deglycosylated MERS S-2P (Fig. S5). Because this loop is variable among betacoronaviruses, it should likely be eliminated in vaccine antigens designed to elicit a broadly neutralizing response against S2. As a proof of concept, we expressed a MERS S-2P variant wherein this loop was replaced with a Gly–Ser linker. This variant expressed at comparable levels to the MERS S-2P protein but had greatly reduced binding to G4 (Fig. S5). Addition of one or more N-linked glycosylation sites in the truncated loop would further help to shield this region from antibody recognition. Collectively, these data identify a variable loop in S2 that is likely under immune pressure and can be removed to potentially avoid virus-specific antibody responses in a vaccine antigen designed to elicit protection against multiple coronaviruses.
Table S4.
G4-escape data
| Amino acid change | Domain | Occurrence* | Frequency (% of isolates) |
| R1179T | CD† | 1 | 7.7 |
| T1175P | CD | 1 | 7.7 |
| R1179M | CD | 1 | 7.7 |
| V1150A | CD | 1 | 7.7 |
| A886P | S2′‡ | 1 | 7.7 |
| R1179G | CD | 2 | 15.4 |
| S1185L | CD | 2 | 15.4 |
| T1175I | CD | 5 | 38.5 |
Occurrence is the number of times a given substitution was found in any of the 13 independent G4-resistant isolates. One isolate contained two of the listed substitutions, and thus the total number of occurrences is 14.
Connector domain.
S2′ protease site.
Fig. S5.
G4 Fab affinity as measured by isothermal titration calorimetry. (A–C) ITC data for the binding of G4 Fab to MERS S-2P (A), deglycosylated MERS S-2P (B), and variable-loop-mutated MERS S-2P (C). Red lines represent the best fit of the data to a single-binding-site model. The sequence and secondary structure schematics for the variable loop are shown above the titrations.
Receptor-Accessible S Protein Conformations.
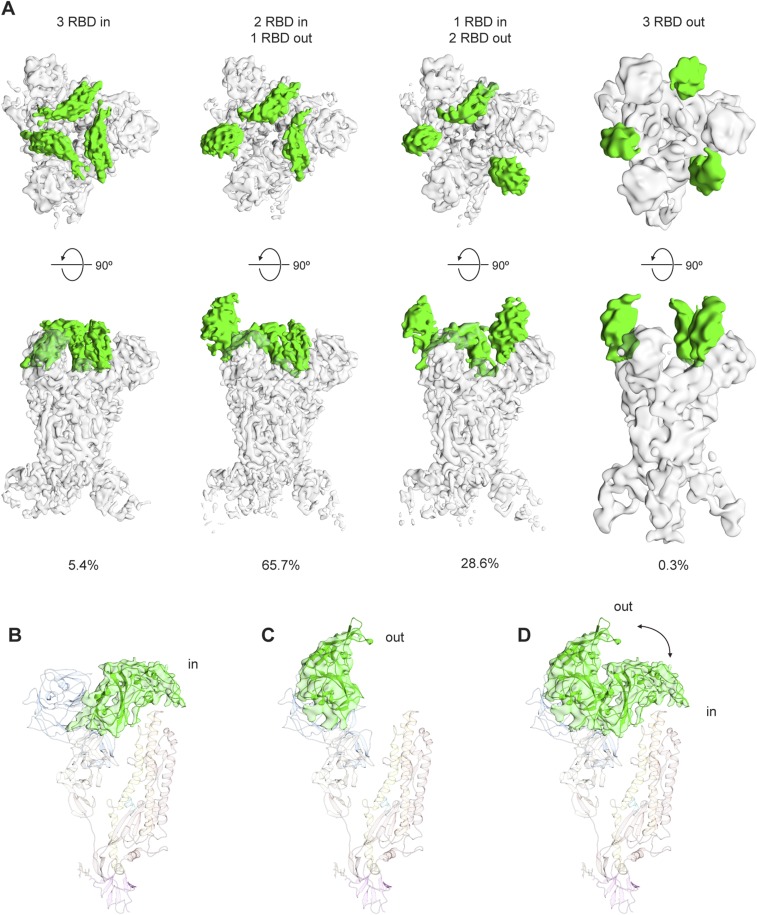
As mentioned above, during our cryo-EM studies we observed distinct subpopulations of S proteins that differed in the arrangement of the S1 apex, and processing these subpopulations separately revealed four distinct S1 crown configurations (Fig. 6A, Fig. S6, and Movies S1 and S2). We identified a small subpopulation of cryo-EM data (5.4%) in the tightly packed “closed” conformation, indicating that neither the 2P substitutions nor G4 binding prevent sampling of this conformation. Indeed, the 2P substitutions do not interact with the RBD and allow Arg1057 in HR1 to maintain its interaction with Tyr577 in the RBD (Fig. 6B). In the remaining three subpopulations of our cryo-EM data (totaling 94.6%), “open” trimers are observed with one, two, or three RBDs in an “out” conformation that extends away from the spike and does not interact with S2 or the glycosylated surface of the neighboring NTD (Fig. 6 A and C). In this extended configuration, the receptor-binding determinants are exposed at the apex of the complex and are fully accessible for interaction with DPP4 (Fig. 6D).
Fig. 6.
RBD conformations observed in the MERS-CoV S protein. (A) Top and side views of the four MERS S-2P structures determined by single particle cryo-EM. Each has a unique arrangement of the three RBDs (green). The percentage of particles in the dataset belonging to each of the four structures is shown below the structures. (B) Interaction between an RBD (green) and the S2 helix–loop–helix spanning the central helix (orange) and HR1 (yellow). The two prolines introduced into S2 are shown as sticks, as are the side chains of interacting residues in the RBD and HR1. Electron density is shown as a transparent surface. (C) Superposition of one protomer with the RBD “in” and another protomer with the RBD “out.” (D) Superposition of the RBD-DPP4 crystal structure (PDB ID code 4KR0) onto trimers with three RBDs in (Left) or two RBDs in and one RBD out (Right). Substantial clashes prevent DPP4 from binding until the RBD rotates outward.
Fig. S6.
MERS-CoV S RBD conformations. (A) MERS-CoV S assumes both open (DPP4-accessible) and closed (DPP4-inaccessible) conformations. A small fraction of the data (5.4%) exhibited the closed conformation characterized by having all three RBDs “in.” The majority of our data (94.6%) exists in an open conformation characterized by having at least one RBD “out.” Only a very small fraction of the data (0.3%) has all three RBDs “out.” (B) Density corresponding to the RBD is colored green and shown “in.” (C) Density corresponding to the RBD is colored green and shown “out.” (D) Superposition of one protomer with the RBD “in” and another protomer with the RBD “out.”
The four trimer configurations observed in our data suggest a potential mechanism for receptor-induced triggering that involves sequential activation of protomers. In this model, we speculate that DPP4 binding to transiently exposed RBDs functions as a molecular ratchet that drives the trimer to the three-RBD-out, open conformation (Fig. 7). This conformation is likely intrinsically unstable because the RBDs no longer help mediate trimerization of S1 or sterically inhibit refolding of S2 by sitting atop the central helix. In support of this model, we note that the three-RBD-out conformation is only observed in 0.3% of our cryo-EM data. Further experiments will be required to test this proposed model of receptor-induced triggering, but sequential activation of protomers in a class-I fusion protein has recently been demonstrated for Moloney murine leukemia virus (39).
Fig. 7.
Simplified model of DPP4 binding leading to MERS-CoV S triggering. The model posits that all three RBDs are in a state of equilibrium between the receptor-accessible “out” conformation and the tightly packed, receptor-inaccessible “in” conformation. DPP4 binding acts as a molecular ratchet that locks the RBD in the “out” conformation until all three RBDs are bound. This open conformation of the trimer is unstable and the S1 subunits ultimately dissociate from S2. Once the S2 subunits are no longer constrained by S1, membrane fusion can proceed by way of a prehairpin intermediate. MERS-CoV S protomers are colored pink, blue, and green and the unresolved HR2 region is depicted as a dashed line. Dimeric DPP4 is colored orange.
Discussion
Approximately one-third of MERS-CoV infections have been fatal, making it the most lethal coronavirus described to date (40). The MERS-CoV S protein, which mediates receptor binding and membrane fusion, is the primary antigenic target for development of coronavirus vaccines (22). The introduction of two consecutive proline substitutions at the beginning of the central helix—our 2P design—presents a general approach to produce soluble prefusion coronavirus S ectodomains and overcomes the first hurdle in subunit vaccine development. Due to restricted backbone torsion angles, proline substitution can disfavor the refolding of the linker between the central helix and HR1, which for class I fusion proteins is a key step in the transition to the postfusion conformation (41). The rigidity of the helix–loop–helix afforded by the prolines impairs or abolishes the membrane fusion activity of the S protein, as evidenced in Fig. 2A. Similar results were obtained when prolines were substituted into influenza HA (42), and because class I fusion proteins have similar membrane fusion machinery, proline substitution has found broad use in subunit vaccine development. For HIV-1 Env, the I559P substitution in gp41 helped to produce a stable ectodomain trimer designated “SOSIP” (32), which facilitated the high-resolution structural analysis of this critical vaccine target (43–45). For RSV, an S215P substitution in the fusion (F) glycoprotein greatly increased the yield and stability of prefusion ectodomain trimers (36). This is consistent with our findings for the MERS-CoV S protein, where the 2P design resulted in a >50-fold increase in the yield of trimeric S protein in its antigenically native prefusion conformation (Fig. 1C). Unlike other structure-based engineering methods, such as the introduction of disulfide bonds or cavity-filling mutations (46), atomic-level information is not as critical for proline-based engineering, and the structure of a homologous protein with low sequence identity can serve as the template (Fig. 1 A and B). Thus, the proline-based strategy should be advantageous for the development of vaccine candidates against emerging coronaviruses for which structures have not yet been determined.
Our studies also characterized the interaction of a first-in-class neutralizing antibody (G4) directed against the S2 subunit, which is more conserved than the S1 cap (31). Prior mapping of neutralizing sites on coronavirus S proteins have focused on the RBD. However, neutralization escape has been described for RBD-specific mAbs, and it has been suggested that having broader epitope coverage outside of the RBD can improve protection from MERS-CoV challenge (31). The G4 epitope is largely conserved between different MERS-CoV isolates and thus G4-like antibodies present an attractive class of MERS-CoV cross-reactive antibodies for therapeutic use. The G4 epitope is, however, variable among the larger family of betacoronaviruses (Fig. 5C), suggesting that it is subject to immune pressure. For other coronaviruses, such as MHV and SARS-CoV, S2-directed antibodies have been isolated that recognize epitopes containing the fusion peptide (47, 48), which is relatively conserved among coronaviruses. An antibody that recognizes this region of the S2 stem, in a way that avoids most of the surrounding sequence variability, may broadly neutralize diverse coronaviruses. Potent antibodies directed against the fusion peptide of HIV-1 Env have recently been described (49, 50), providing hope that similar antibodies for coronaviruses may also be obtained through antigen-specific antibody-isolation efforts. Use of our engineered prefusion S proteins as probes to sort B cells from infected donors should greatly facilitate these efforts and lead to the development of a broadly protective immunotherapeutic.
In our structures of the trimeric prefusion MERS-CoV S protein we observe zero, one, two, or three RBDs rotated to a receptor-accessible “out” position (Fig. 6). We hypothesize that this flexibility in RBD exposure plays a role in the controlled timing of receptor engagement that ultimately leads to triggering of S toward the lower-energy postfusion state (Fig. 7). In addition, transient exposure of the RBDs, which are highly immunogenic, would help hide neutralizing epitopes from the humoral immune system. This “conformational masking” of neutralizing epitopes has also been described for HIV-1 Env (51), which like the coronavirus S proteins exists in several distinct conformations: closed ground state, CD4-accessible, and CCR5-accessible (52). For coronaviruses, this phenomenon was hinted at in the partially resolved structure of the SARS-CoV S protein, wherein a single RBD was observed in the receptor-accessible “out” conformation (20). In the structures of the S proteins from MHV and HCoV-HKU1 all three RBDs were shown to be in a tightly packed, closed configuration (17, 18). The MHV spike may not need to transiently expose the RBDs since it binds to its protein receptor CEACAM1 via the S1 NTD (53–55), and this interaction is thought to be sufficient for entry (56). For HCoV-HKU1, a protein receptor has not yet been identified, but the S1 subunit has been shown to bind O-acetylated sialic acids (57). Additional structural and virological studies are therefore needed to elucidate the molecular mechanisms of S protein-mediated cell entry.
Engineering class-I viral fusion proteins in the prefusion conformation can significantly improve immunogenicity through preservation of neutralization-sensitive conformational and quaternary epitopes. This is exemplified by the failure of postfusion RSV F glycoprotein vaccine antigens and the promise of prefusion-stabilized RSV F glycoproteins (36, 46, 58). Recently, the MERS-CoV S1 monomer has been shown to elicit RBD-specific neutralizing antibodies in mice and protect rhesus macaques from MERS-CoV–induced pneumonia, but protection was improved in animals primed with full-length S antigens that induced neutralizing antibodies directed to non-RBD sites (31). These data, together with the observation that the RBD has positional variability, suggest that the virus has evolved multiple mechanisms to evade neutralization by RBD-specific antibodies. The RBD sequence variability is compounded by the positional variability that allows conformational masking and transient exposure of quaternary surfaces and neutralization-sensitive sites. Our demonstration here that the prefusion-stabilized MERS-CoV S trimer (S-2P) elicits more robust neutralizing antibody responses in mice than S1 monomer or S WT suggests that MERS S-2P is a preferred antigen for vaccine development and is made more attractive due to inclusion of non-RBD epitopes and favorable manufacturing characteristics.
In summary, our 2P design, structures of the MERS-CoV S ectodomain in complex with G4, and demonstration of improved expression and immunogenicity of prefusion S proteins will serve as a basis for further engineering of MERS-CoV vaccine immunogens and provide an important step in the development of broadly protective coronavirus vaccines.
Methods
Production of S Protein Ectodomains.
A mammalian-codon-optimized gene encoding MERS-CoV S (England1 strain) residues 1–1291 with a C-terminal T4 fibritin trimerization domain, an HRV3C cleavage site, an 8xHis-tag and a Twin-Strep-tag was synthesized and subcloned into the eukaryotic-expression vector pαH. The S1/S2 furin-recognition site 748-RSVR-751 was mutated to ASVG to produce a single-chain S0 protein.
A series of proline-substituted variants was generated based on this construct and the resulting plasmids were transfected into 40 mL FreeStyle 293-F cells (Invitrogen). Three hours after transfection, kifunensine was added to a final concentration of 5 μM. Cultures were harvested 6 d later, and secreted protein was purified from the supernatant using 0.5 mL Strep-Tactin resin (IBA). Protein expression levels were then assessed by SDS/PAGE (10 μL of protein-bound resin was boiled and loaded per lane). Similar strategies were used to generate and test proline-substituted variants of SARS-CoV S (Tor2 strain, residues 1–1190) and HCoV-HKU1 S (N5 strain, residues 1–1276).
For large-scale expression, 0.5–1 L FreeStyle 293-F cells were transfected. Three hours after transfection, kifunensine was added to a final concentration of 5 μM. Cultures were harvested after 6 d, and protein was purified from the supernatant using Strep-Tactin resin (IBA). HRV3C protease (1% wt/wt) was added to the protein and the reaction was incubated overnight at 4 °C. The digested protein was further purified using a Superose 6 16/70 column (GE Healthcare Biosciences).
Production of G4 Fab.
The Fab region of the G4 heavy chain was fused with the HRV3C cleavage site and human IgG1 Fc fragment and subcloned into the eukaryotic expression vector pVRC8400. This plasmid was cotransfected with the G4 light chain into Expi293 cells (Invitrogen), and the secreted antibody was purified using Protein A agarose (Fisher). HRV3C protease (1% wt/wt) was added to the protein and the reaction was incubated for 2 h at room temperature. The digested antibody was passed back over Protein A agarose to remove the Fc fragment, and the unbound Fab was additionally purified using a Superdex 75 column (GE Healthcare Biosciences).
Production of MERS S-2P in Complex with G4 Fab.
Purified MERS S-2P was mixed with a 1.5-fold molar excess of G4 Fab. After incubation on ice for 1 h, the complex was separated from excess Fab by size-exclusion chromatography.
Negative-Stain EM.
MERS-CoV and SARS-CoV S-2P proteins were diluted with Tris-buffered saline as necessary and then spotted onto 400-mesh copper grids and stained with 1% uranyl acetate. Grids were imaged in a Tecnai T12 Spirit with a high tension of 120 kV and a Tietz TemCam-F416 complementary metal-oxide semiconductor camera at 52,000× magnification yielding a pixel size of 2.05 Å per pixel at 1.5 μm under focus. Images were collected using Leginon (59) and processed within the Appion workflow (60). Projection images were picked from the raw micrographs using a difference-of-Gaussians approach (61). Images were binned by two and then aligned using reference-free 2D classification with iterative multivariate statistical analysis/multireference alignment (62) to identify and remove amorphous projection images from the image stacks. The image stacks were then realigned into 16 classes representing the entire cleaned stack.
Cryo-EM Data Collection.
Approximately 3 μL of MERS S-2P/G4 Fab complex was mixed with 1 μL of a 0.04% A8-35 amphipol solution (Anatrace) immediately before sample deposition on a CF-2/2-4C C-Flat grid (Protochips; Electron Microscopy Sciences) that had been plasma-cleaned for 5 s using a mixture of Ar/O2 (Gatan Solarus 950 Plasma system). The grid was then blotted and plunged into liquid ethane using a manual freeze plunger. Eight hundred thirteen movie micrographs were collected in one session through the Leginon software solution on an FEI Titan Krios operating at 300 kV and mounted with a Gatan K2 direct electron detector (59). Each micrograph was collected in counting mode at 29,000× nominal magnification resulting in a calibrated pixel size of 1.02 Å at the object level. A dose rate of ∼10 e− per (camera pixel) per s was used and each movie frame was captured from an exposure time of 200 ms. Total dose for each movie micrograph was 66 e−/Å2. The nominal defocus range used was −0.7 to −2.5 μm.
Cryo-EM Data Processing.
Frames in each movie micrograph were aligned and summed using MotionCorr (63). CTF estimation was then performed using CTFFind3 (64), and candidate MERS S-2P/G4 projection images were identified using a difference-of-Gaussians approach (61). Reference-free 2D classification was then performed in RELION version 1.4 (65). After 2D classification, 37,180 good projection images were refined asymmetrically (resolution 4.0 Å) as well as under C3 restraint (resolution 3.6 Å) against a negative-stain reference map rendered at 60-Å resolution. Projection images and the asymmetrically refined map were entered into RELION 3D classification, resulting in three classes each with two copies of G4 Fabs bound but with differing S1 crown configurations (resolutions 4.7 Å, 4.8 Å, and 5.0 Å). A fourth data class was characterized by having three copies of G4 Fabs bound (resolution 4.5 Å). This latter data class was further refined under C3 symmetry constraint (resolution 4.0 Å). As density corresponding to the S1 crown still exhibited more disorder than the otherwise well-ordered data classes, a local classification procedure was pursued (Fig. S2). Here, three masks each encompassing both the “in” and the “out” RBD configuration at respective RBD positions were created. By applying each of these masks in separate parallel classification protocols, separation of a homogeneous density region from a heterogeneous density region was obtained in each case. Subtraction of the homogeneous density from raw projection images was then obtained by projecting the homogeneous density map according to the projection image Euler directions already obtained from refinement. The resulting local RBD density projection images were then subjected to RELION 3D classification using the homogeneous density map (no density in the RBD area that is to be classified) as seed. In each of the three cases this resulted in two data classes corresponding to the RBD “in” or the “out” position. The original raw projection images were then recompiled into four conformationally clean data classes corresponding to three RBD “in”, two RBD “in” and one RBD “out”, one RBD “in” and two RBD “out,” and three RBD “out.” Projection images of the asymmetric classes were rotated into correct alignment (0°, 120°, or 240° Euler rotation) before further refinement. Each of the four classes was individually refined against a reference map at 60 Å simulated from our MERS S-2P coordinate build but without any RBD coordinates included (resulting map resolutions 4.0 Å, 4.6 Å, 4.8 Å, and 11.5 Å). Symmetric classes (three RBD “in” or three RBD “out”) were initially refined asymmetrically to confirm correct classification of “in” and “out” positions before further refinement imposing C3 restraint. All resolutions were estimated by the FSC 0.143 criterion in RELION using a soft-edged mask with a Gaussian fall-off, encompassing entire structures and corrected for mask correlations.
Cryo-EM Model Building and Refinement.
An initial model of MERS S-2P was obtained from the MODELLER homology modeling tool (66) in UCSF Chimera (67) using HCoV-HKU1 S (PDB ID code 5I08) as a template. Significant manual remodeling as well as de novo building of the S2 domain that connects the central helix with HR2 was performed in Coot (68). X-ray structures for MERS S1 NTD and MERS S1 RBD (PDB ID code 4KR0) were used to substitute the respective regions (RBD added only to models where appropriate) and the resulting homology model was combined with X-ray structures of the G4 Fab. The resulting models were then iteratively refined and manually rebuilt against their respective density maps (not remodeling S1 NTD or RBD domains) using Rosetta density-guided iterative local refinement (69) and Coot. Rosetta all-atom refinement was then performed in a modular fashion wherein well-resolved regions were refined under little restraint and gradually higher B-factor regions were refined under gradually stricter restraints. Addition of ligands and further refinements were conducted in Rosetta using the respective density maps as constraints (70) and end-refined in PHENIX (71). Models were evaluated using MolProbity (oneline-analysis command line implementation) (72) and EMRinger (command line implementation) (73) and Privateer (command line implementation) (74) and CARP (web server) (75) where appropriate.
Production of MERS-CoV S1 NTD.
A gene encoding MERS-CoV S1 NTD (residues 1–353) with a C-terminal HRV3C cleavage site and human IgG1 Fc fragment was inserted into the eukaryotic expression vector pαH. Three hours after transient transfection of the plasmid into FreeStyle 293-F cells, kifunensine was added to a final concentration of 5 μM. After 6 d, the supernatant was passed over a Protein A agarose column, and deglycosylation was conducted on-column by adding EndoH (10% wt/wt) at room temperature. After 12 h, the column was washed with PBS and the NTD was eluted by incubating the resin with HRV3C (1% wt/wt). The NTD was further purified using a Superdex 75 column (GE Healthcare Biosciences).
Crystallization and X-Ray Data Collection.
Purified G4 Fab was concentrated to 9.5 mg/mL in TBS (2 mM Tris, pH 8.0, and 50 mM NaCl) for crystallization. Crystals were produced at room temperature using the sitting-drop vapor-diffusion method by mixing 0.1 μL of protein with 0.1 μL of reservoir solution containing 0.1 M MES, pH 6.5, 0.2 M magnesium chloride, and 10% (wt/wt) PEG 4000. Crystals were soaked in reservoir solution supplemented with 20% (vol/vol) glycerol and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at the SBC beamline 19-ID (Advanced Photon Source, Argonne National Laboratory).
Purified MERS-CoV S1 NTD was concentrated to 11.7 mg/mL in TBS for crystallization. Initial hits were obtained in the Wizard Precipitant Synergy screen (76). Crystals were obtained at room temperature using the sitting-drop vapor-diffusion method by mixing 0.1 μL of protein with 0.1 μL of reservoir solution containing 0.1 M imidazole HCl, pH 6.5, 6.6% (wt/wt) PEG 8000, and 1% (vol/vol) 2-methyl-2,4-pentanediol. Crystals were soaked in reservoir solution supplemented with 20% (vol/vol) glycerol and frozen in liquid nitrogen. X-ray diffraction data were collected at the SSRL beamline BL14-1 (Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory).
Crystal Structure Determination and Refinement.
Diffraction data were processed using the CCP4 software suite: data were indexed and integrated in iMOSFLM (77) and scaled and merged with AIMLESS (78). A molecular replacement solution for the G4 Fab data was obtained using PHASER (79) and PDB ID 3QQ9 as the search model. The structure was built manually in Coot (68) and refined using PHENIX (71). Data collection and refinement statistics are presented in Table S3.
For the MERS-CoV S1 NTD dataset, molecular replacement using NTD structures from MHV and bovine coronavirus (BCoV) failed to find a solution. Therefore, a portion of the EM map corresponding to the NTD was extracted and used as the search model for molecular replacement using PHASER-MR in the PHENIX GUI following a recently published protocol (80). The solution contained only one molecule in the asymmetric unit, so noncrystallographic symmetry could not be used to improve the phases. Consequently, the core β-sheet of the BCoV NTD was manually fit into the electron density, and iterative rounds of manual building with Coot and refinement with PHENIX produced a model containing 184 residues. Using this structure as input, MR-ROSETTA (81) was able to produce a model containing 282 residues. This model was then used as input for Buccaneer (82), which produced a model containing all 341 residues. After manually adding N-linked glycans with Coot, additional electron density was observed that did not correspond to any chemicals in the crystallization buffer or cryosolution. We identified the chemical as folic acid using mass spectrometry, and placed the molecule into the density (Fig. S3). The folic acid is thought to have copurified with the NTD from the mammalian expression medium, but whether folic acid, or a closely related chemical analog, has a physiological role in the MERS-CoV infection cycle remains unknown. The final structure was refined using PHENIX, and the data collection and refinement statistics are presented in Table S3. Software used to determine X-ray crystal structures was curated by SBGrid (83).
Protein A Pull-Down Experiments.
MERS S-2P was coexpressed with each antibody by cotransfecting 10 μg of MERS S-2P plasmid with 5 μg of heavy-chain plasmid and 5 μg of light-chain plasmid into 40 mL of FreeStyle 293-F cells (Invitrogen). Three hours after transfection, kifunensine was added to a final concentration of 5 μM. Cultures were harvested after 6 d, and protein was purified from the supernatant using 0.5 mL of Protein A resin (Thermo Fisher). After an extensive wash with PBS, 10 μL of protein-bound resin was boiled and analyzed by SDS/PAGE.
Surface Plasmon Resonance Experiments.
The 8xHis-tagged MERS S-2P protein was captured on an NTA sensor chip to ∼660 response units each cycle using a Biacore X100 (GE Healthcare). The chip was regenerated twice after each cycle using 350 mM EDTA followed by 0.5 mM NiCl2. After three injections of running buffer over both the ligand-bound and reference flow cells, increasing concentrations of soluble DPP4 ectodomain were injected (1.6 nM to 50 nM with a final replicate of 12.5 nM). Data were double-reference subtracted and fit to a 1:1 binding model using Scrubber2 software.
Mouse Immunizations.
Animal experiments were carried out in compliance with all pertinent US National Institutes of Health regulations and policies. The National Institutes of Health, National Institute of Allergy and Infectious Diseases, Vaccine Research Center Animal Care and Use Committee reviewed and approved all animal experiments. Female BALB/cJ mice aged 6–8 wk (Jackson Laboratory) were immunized with MERS S1, MERS S WT, or MERS S-2P protein at 0 and 3 wk. Protein (0.1 μg, 1 μg, or 10 μg) diluted in PBS was mixed 1:1 with 2× Sigma Adjuvant System. Mice were inoculated with 100 μL intramuscularly (50 μL into each hind leg). Two weeks after the final immunization, sera were collected for measurement of antibody responses.
Pseudovirus Production.
Pseudovirus production, infectivity, and neutralization experiments were completed as previously described with minor adaptations (31). We synthesized cDNAs encoding spike protein using the QuikChange XL kit (Stratagene) and introduced divergent amino acids into the parental spike gene (strain England1) predicted from translated sequences of other strains: Bisha1 (GenBank accession no. KF600620), Buraidah1 (GenBank accession no. KF600630), Florida USA2 (GenBank accession no. AIZ48760), Indiana USA1 (GenBank accession no. AHZ58501), JordanN3 (GenBank accession no. KC776174), and Korea002 (GenBank accession no. AKL59401). All constructs were confirmed using sequencing. HEK293T cells were obtained from ATCC and cultured in DMEM supplemented with 10% FBS, 2 mM glutamine, and 1% penicillin/streptomycin at 37 °C and 5% CO2. To produce MERS-CoV pseudoviruses, CMV/R-MERS-CoV spike plasmids were cotransfected into HEK293T cells with packaging plasmid pCMVDR8.2 and transducing plasmid pHR′ CMV-Luc, using Fugene 6 transfection reagent (Promega). Mock pseudoviruses were produced by omitting the MERS-CoV S plasmid. Seventy-two hours posttransfection, supernatants were collected, filtered, and frozen at −80 °C.
Pseudovirus Infectivity and Neutralization Experiments.
Huh7.5 cells were provided by Deborah R. Taylor, US Food and Drug Administration, Silver Spring, MD, and cultured in DMEM supplemented with 10% FBS, 2 mM glutamine, and 1% penicillin/streptomycin at 37 °C and 5% CO2. Pseudovirus infectivity was assessed in Huh7.5 cells plated overnight in 96-well black/white isoplates (PerkinElmer). Twofold serial dilutions of pseudoviruses were added to resting Huh7.5 cells, in triplicate. After a 2-h incubation, fresh medium was added. Cells were lysed at 72 h, and luciferase substrate (Promega) was added. Luciferase activity was measured as relative luciferase units (RLU) at 570 nm on a SpectramaxL (Molecular Devices). For neutralization experiments, serial dilutions of mouse sera (1:40, fourfold, eight dilutions) were mixed with various pseudovirus strains, which were previously titered to target 50,000 RLU. Sigmoidal curves, taking averages of triplicates at each dilution, were generated from RLU readings; 90% neutralization (IC90) titers were calculated considering uninfected cells as 100% neutralization and cells transduced with only virus as 0% neutralization.
Cell-Surface Binding Assay.
Cell-surface binding assays were performed as previously described with minor adaptations (31). HEK 293T cells were plated and maintained overnight to reach 80% confluence. Cells were transfected with plasmids expressing MERS-CoV S-WT, S-2P, or RBD engineered with an influenza HA transmembrane domain, using Fugene 6 transfection reagent (Promega). After 24 h, cells were detached with 4 mM EDTA in PBS, stained with ViViD viability dye (Invitrogen), and then stained with mAbs (10 μg/mL) or polyclonal sera (1:200) obtained from mice vaccinated with MERS-CoV S. Cells were subsequently stained with goat anti-mouse IgG, Alexa-488 (Invitrogen). Cells were sorted with an LSR (BD Biosciences). Data were analyzed with FlowJo software (Tree Star Inc.), using the following gating strategy: size & granularity > single cells > live cells (ViViD negative) > Spike+ (Ab positive). Fluorescence background was calculated using untransfected cells stained with each respective mAb and subtracted from the data.
Isothermal Titration Calorimetry Experiments.
Calorimetric titrations of G4 Fab into MERS S-2P, deglycosylated MERS S-2P, or variable-loop-deleted MERS S-2P were performed using a PEAQ isothermal titration calorimeter (ITC) (Malvern) at 25 °C. All proteins were dialyzed into PBS. Protein concentrations in the sample cell were 3.0–3.7 μM, whereas the concentration of G4 Fab in the injection syringe was 57.5 μM. Titrations consisted of 15 injections. Data were processed with the MicroCal PEAQ-ITC Analysis Software (Malvern) and fit to an independent-binding model.
Viral Resistance to Antibody-Mediated Inhibition of Infectivity.
MERS-CoV variants escaping G4-mediated neutralization of infectivity were selected by serial passage of recombinant MERS-CoV strain EMC/2012 in Vero 81 cell cultures (84) supplemented with progressively escalating concentrations of antibody, eventually reaching 3.6 μg/mL at the terminal passage level, P5. The starting amount of G4, 0.4 μg/mL, corresponded to the antibody concentration required to reduce ∼40 MERS-CoV plaque-forming units by ∼70% in a plaque-reduction neutralization assay using Vero 81 cell monolayers. Culture supernatants were passed onto fresh cells 48 h postinfection and a total of 13 G4-resistant MERS-CoV isolates representing three parallel passage series were plaque-cloned from P5 cultures on Vero 81 cell monolayers in the presence of 1 μg/mL G4. Plaque isolates were expanded in antibody-free cultures of Vero cells in 25-cm2 flasks, followed by total RNA isolation from virally infected cell monolayers using TRIzol reagent (Invitrogen). Two overlapping cDNA amplicons encompassing the entire S gene were generated by RT-PCR using SuperScript III RT (Invitrogen) and Easy-A high-fidelity thermostable DNA polymerase (Agilent Technologies) (40 cycles of RT product amplification). Resultant S-gene PCR products were subjected to dideoxy sequencing using S gene-based primers, and reads were aligned to the native EMC/2012 S-gene sequence (GenBank accession no. JX869059.2) using MacVector to identify mutations associated with neutralization-escape from G4. Cell culture-adaptive mutations in spike identified in antibody-free P10 and P20 cultures of EMC/2012 were excluded from analyses of changes arising under G4 selection.
Supplementary Material
Acknowledgments
We thank members of the J.S.M. and A.B.W. laboratories for critical reading of the manuscript, E. Shipman for assistance with protein expression, Benjamin Cabrera and members of the Vaccine Research Center (VRC) Translational Research Program for technical assistance with immunogenicity experiments, J. Adolf-Bryfogle and S. Rämisch for assistance with Rosetta ligand modeling, and the beamline scientists for X-ray data collection support at SBC 19-ID (Argonne National Laboratory) and BL14-1 SLAC National Accelerator Laboratory. This work was supported by Grants P20GM113132 (to J.S.M.) and R01AI127521 (to J.S.M. and A.B.W.), NIH Contract HHSN261200800001E Agreement 6x142 (to M.R.D. and J.D.C.), and intramural funding from National Institute of Allergy and Infectious Diseases to support work at the VRC (B.S.G.). Argonne is operated by UChicago Argonne, LLC, for the US Department of Energy (DOE), Office of Biological and Environmental Research under Contract DE-AC02-06CH11357. Use of the Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory, is supported by the DOE, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the NIH, National Institute of General Medical Sciences (including P41GM103393).
Footnotes
Conflict of interest statement: J.P., N.W., K.S.C., R.N.K., H.L.T., C.A.C., B.S.G., A.B.W., and J.S.M. are inventors on US patent application no. 62/412,703, entitled “Prefusion Coronavirus Spike Proteins and Their Use.” L.W., W.S., W.-P.K., and B.S.G. are inventors on US patent application no. PCT/US2016/019395, entitled “Middle East Respiratory Syndrome Coronavirus Immunogens, Antibodies and Their Use.”
This article is a PNAS Direct Submission.
Data deposition: Cryo-EM reconstructions have been deposited in the Electron Microscopy Data Bank (EMDB) (accession codes EMD-8783–EMD-8793). Atomic models have been deposited in the Protein Data Bank (PDB ID codes 5W9H–5W9P). Atomic models and structure factors for the crystal structures of MERS-CoV NTD and G4 Fab have also been deposited in the Protein Data Bank (PDB ID codes 5VYH and 5VZR, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707304114/-/DCSupplemental.
References
- 1.Woo PC, Lau SK, Huang Y, Yuen KY. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med (Maywood) 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 2.Peiris JS, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.van Boheemen S, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3:e00473–12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azhar EI, et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 6.Alraddadi BM, et al. Risk factors for primary Middle East respiratory syndrome coronavirus illness in humans, Saudi Arabia, 2014. Emerg Infect Dis. 2016;22:49–55. doi: 10.3201/eid2201.151340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fehr AR, Channappanavar R, Perlman S. Middle East respiratory syndrome: Emergence of a pathogenic human coronavirus. Annu Rev Med. 2017;68:387–399. doi: 10.1146/annurev-med-051215-031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohd HA, Al-Tawfiq JA, Memish ZA. Middle East respiratory syndrome coronavirus (MERS-CoV) origin and animal reservoir. Virol J. 2016;13:87. doi: 10.1186/s12985-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oboho IK, et al. 2014 MERS-CoV outbreak in Jeddah—A link to health care facilities. N Engl J Med. 2015;372:846–854. doi: 10.1056/NEJMoa1408636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ki M. 2015 MERS outbreak in Korea: Hospital-to-hospital transmission. Epidemiol Health. 2015;37:e2015033. doi: 10.4178/epih/e2015033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gierer S, et al. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol. 2013;87:5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci USA. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu G, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raj VS, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang N, et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walls AC, et al. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirchdoerfer RN, et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walls AC, et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol. 2016 doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gui M, et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corti D, et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc Natl Acad Sci USA. 2015;112:10473–10478. doi: 10.1073/pnas.1510199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Modjarrad K, et al. A roadmap for MERS-CoV research and product development: Report from a World Health Organization consultation. Nat Med. 2016;22:701–705. doi: 10.1038/nm.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang XC, et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc Natl Acad Sci USA. 2014;111:E2018–E2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang L, et al. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl Med. 2014;6:234ra259. doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, et al. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res. 2015;25:1237–1249. doi: 10.1038/cr.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang N, Tang J, Lu L, Jiang S, Du L. Receptor-binding domain-based subunit vaccines against MERS-CoV. Virus Res. 2015;202:151–159. doi: 10.1016/j.virusres.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sui J, et al. Broadening of neutralization activity to directly block a dominant antibody-driven SARS-coronavirus evolution pathway. PLoS Pathog. 2008;4:e1000197. doi: 10.1371/journal.ppat.1000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ter Meulen J, et al. Human monoclonal antibody combination against SARS coronavirus: Synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitsuki YY, et al. A single amino acid substitution in the S1 and S2 Spike protein domains determines the neutralization escape phenotype of SARS-CoV. Microbes Infect. 2008;10:908–915. doi: 10.1016/j.micinf.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu X, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat Commun. 2015;6:7712. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders RW, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Impagliazzo A, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 34.Yassine HM, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 35.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krarup A, et al. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun. 2015;6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong L, et al. Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat Commun. 2016;7:12040. doi: 10.1038/ncomms12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirato K, Kawase M, Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol. 2013;87:12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sjoberg M, Loving R, Lindqvist B, Garoff H. Sequential activation of the three protomers in the Moloney murine leukemia virus Env. Proc Natl Acad Sci USA. 2017;114:2723–2728. doi: 10.1073/pnas.1617264114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arabi YM, et al. Middle East respiratory syndrome. N Engl J Med. 2017;376:584–594. doi: 10.1056/NEJMsr1408795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 42.Qiao H, et al. Specific single or double proline substitutions in the “spring-loaded” coiled-coil region of the influenza hemagglutinin impair or abolish membrane fusion activity. J Cell Biol. 1998;141:1335–1347. doi: 10.1083/jcb.141.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pancera M, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyumkis D, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Julien JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLellan JS, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daniel C, et al. Identification of an immunodominant linear neutralization domain on the S2 portion of the murine coronavirus spike glycoprotein and evidence that it forms part of complex tridimensional structure. J Virol. 1993;67:1185–1194. doi: 10.1128/jvi.67.3.1185-1194.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, et al. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J Virol. 2004;78:6938–6945. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kong R, et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science. 2016;352:828–833. doi: 10.1126/science.aae0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 52.Munro JB, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dveksler GS, et al. Cloning of the mouse hepatitis virus (MHV) receptor: Expression in human and hamster cell lines confers susceptibility to MHV. J Virol. 1991;65:6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng G, et al. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc Natl Acad Sci USA. 2011;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kubo H, Yamada YK, Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino-acids of the murine coronavirus spike protein. J Virol. 1994;68:5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsuyama S, Taguchi F. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J Virol. 2009;83:11133–11141. doi: 10.1128/JVI.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang X, et al. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Graham BS. Vaccines against respiratory syncytial virus: The time has finally come. Vaccine. 2016;34:3535–3541. doi: 10.1016/j.vaccine.2016.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 60.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogura T, Iwasaki K, Sato C. Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking. J Struct Biol. 2003;143:185–200. doi: 10.1016/j.jsb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 63.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 65.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2016;86:2 9 1–2 9 37. doi: 10.1002/cpps.20. [DOI] [PubMed] [Google Scholar]
- 67.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 68.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 69.DiMaio F, et al. Atomic-accuracy models from 4.5-A cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using rosetta. J Mol Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 72.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barad BA, et al. EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Agirre J, et al. Privateer: Software for the conformational validation of carbohydrate structures. Nat Struct Mol Biol. 2015;22:833–834. doi: 10.1038/nsmb.3115. [DOI] [PubMed] [Google Scholar]
- 75.Lutteke T, Frank M, von der Lieth CW. Carbohydrate Structure Suite (CSS): Analysis of carbohydrate 3D structures derived from the PDB. Nucleic Acids Res. 2005;33:D242–D246. doi: 10.1093/nar/gki013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Majeed S, et al. Enhancing protein crystallization through precipitant synergy. Structure. 2003;11:1061–1070. doi: 10.1016/s0969-2126(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 77.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jackson RN, McCoy AJ, Terwilliger TC, Read RJ, Wiedenheft B. X-ray structure determination using low-resolution electron microscopy maps for molecular replacement. Nat Protoc. 2015;10:1275–1284. doi: 10.1038/nprot.2015.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.DiMaio F, et al. Improved molecular replacement by density- and energy-guided protein structure optimization. Nature. 2011;473:540–543. doi: 10.1038/nature09964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 83.Morin A, et al. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scobey T, et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ashkenazy H, et al. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peng G, et al. Crystal structure of bovine coronavirus spike protein lectin domain. J Biol Chem. 2012;287:41931–41938. doi: 10.1074/jbc.M112.418210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rusin SF, Schlosser KA, Adamo ME, Kettenbach AN. Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells. Sci Signal. 2015;8:rs12. doi: 10.1126/scisignal.aab3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.