Significance
Leishmania parasites can be infected with Leishmaniavirus (LRV1), a double-stranded RNA virus whose presence in Leishmania guyanensis parasites exacerbates disease severity in both mouse models and humans. Studies of the role of the virus on parasite biology and virulence are hampered by the dearth of isogenic lines bearing and lacking LRV, particularly in the clinically important species Leishmania braziliensis. Here, we describe a method to systematically generate LRV1-free Leishmania parasites using the parasite RNA interference (RNAi) pathway. The ability of transgene-driven RNAi to overcome the ability of LRV1 to withstand the endogenous RNAi attack suggests a third paradigm of virus–RNAi interaction where RNAi and virus replication exist in balance to maintain persistent infection.
Keywords: dsRNA virus, protozoan parasite, endosymbiont, trypanosomatid protozoa, microbial pathogenesis
Abstract
Many Leishmania (Viannia) parasites harbor the double-stranded RNA virus Leishmania RNA virus 1 (LRV1), which has been associated with increased disease severity in animal models and humans and with drug treatment failures in humans. Remarkably, LRV1 survives in the presence of an active RNAi pathway, which in many organisms controls RNA viruses. We found significant levels (0.4 to 2.5%) of small RNAs derived from LRV1 in both Leishmania braziliensis and Leishmania guyanensis, mapping across both strands and with properties consistent with Dicer-mediated cleavage of the dsRNA genome. LRV1 lacks cis- or trans-acting RNAi inhibitory activities, suggesting that virus retention must be maintained by a balance between RNAi activity and LRV1 replication. To tilt this balance toward elimination, we targeted LRV1 using long-hairpin/stem-loop constructs similar to those effective against chromosomal genes. LRV1 was completely eliminated, at high efficiency, accompanied by a massive overproduction of LRV1-specific siRNAs, representing as much as 87% of the total. For both L. braziliensis and L. guyanensis, RNAi-derived LRV1-negative lines were no longer able to induce a Toll-like receptor 3–dependent hyperinflammatory cytokine response in infected macrophages. We demonstrate in vitro a role for LRV1 in virulence of L. braziliensis, the Leishmania species responsible for the vast majority of mucocutaneous leishmaniasis cases. These findings establish a targeted method for elimination of LRV1, and potentially of other Leishmania viruses, which will facilitate mechanistic dissection of the role of LRV1-mediated virulence. Moreover, our data establish a third paradigm for RNAi–viral relationships in evolution: one of balance rather than elimination.
The early-diverging protozoan parasite genus Leishmania causes the disease leishmaniasis in many regions of the world, with an estimated 12 million symptomatic cases, at least 120 million asymptomatic cases, and nearly 1.7 billion at risk (1–5). The disease has three predominant clinical manifestations, ranging from the relatively mild cutaneous form to mucocutaneous disease (where parasites metastasize to, and cause destruction of, mucous membranes of the nose, mouth, and throat) and fatal visceral disease. Disease phenotypes segregate primarily with the infecting species; however, it is not fully understood which parasite factors affect severity and disease manifestations.
One recently identified parasite factor contributing to disease severity in Leishmania guyanensis is the RNA virus Leishmaniavirus (6, 7). This virus is a member of the Totiviridae family and consists of a single-segmented dsRNA genome that encodes only a capsid protein and an RNA-dependent RNA polymerase (RDRP) (8, 9). It is most frequently found (as LRV1) in New World parasite species in the subgenus Viannia, such as Leishmania braziliensis (Lbr) and L. guyanensis (Lgy), which cause both cutaneous and mucocutaneous disease (6), but it has also been found sporadically in Old World subgenus Leishmania species (as LRV2) (10, 11). Like most totiviruses, LRV1 is neither shed nor infectious, and it thus can be viewed as a long-term evolutionary endosymbiont whose activities on the mammalian host arise indirectly through the parasite, rather than by direct infection of the mammalian host by the virus (6). Previous work has shown that mice infected with LRV1-bearing strains of Lgy exhibit greater footpad swelling and higher parasitemia than mice infected with LRV1-negative Lgy (7). Similarly, macrophages infected in vitro with LRV1+ Lgy or LRV2+ Leishmania aethiopica release higher levels of cytokines, phenotypes that were dependent on Toll-like receptor 3 (7, 10). The assignment of the LRV1 specificity of these phenotypes benefited greatly from the availability of a single isogenic LRV1-free line of Lgy (12). Importantly, recent studies have shown that disease severity is increased in patients infected with LRV1+ Lgy, relative to LRV1-negative parasites (13).
In humans, Lbr is associated with cutaneous leishmaniasis, as well as the larger share of the more debilitating mucocutaneous leishmaniasis (MCL) (14, 15). Although in some studies LRV1 was not correlated with MCL (16, 17), in others there was a strong association (6, 18, 19). Recent studies show that LRV1 in Lbr and Lgy clinical isolates correlates with drug treatment failure (16, 20). Thus, although other parasite or host factors may play a significant role in the development of MCL (21, 22), current data support a role for LRV1 in exacerbating the pathogenesis of human leishmaniasis caused by Lbr and Lgy. A similar role in pathogenicity has been proposed for the Trichomonas vaginalis totiviruses (23). In contrast, endobiont viruses in other systems more often impair the host or have no known effect on disease. Hypoviruses of Cryphonectria parasitica are associated with decreased virulence of their fungal host whereas the L-A totivirus of Saccharomyces cerevisiae is not thought to affect pathogenicity, instead contributing to intermicrobial competition (24–27).
Research into the role of LRV1 in Lbr disease is hampered by the fact that animal models are less well-developed than for other Leishmania (28) and by the absence of isogenic lines bearing or lacking LRV1. Because reverse genetic systems for Totiviridae do not exist and attempts to stably transfer LRV1 have proven unsuccessful (29), we asked whether RNA-interference (RNAi) could be used to generate LRV1-free isogenic isolates. Unlike Old World Leishmania, species of the Viannia subgenus, including Lbr and Lgy, retain an active endogenous RNAi pathway (30). The RNAi pathway converts double-stranded RNA into siRNAs, which trigger the degradation of an mRNA with complementary sequence (31). Importantly, the RNAi pathway acts as a defense against RNA viruses in plants and some animals, leading to great reductions or complete elimination (32, 33). Further, introduction of RNAi pathway proteins from Saccharomyces castellii into the naturally RNAi-null S. cerevisiae resulted in greatly decreased levels of persistently infecting L-A totivirus (26). In mammals, siRNA-mediated RNAi activity seems to play a smaller direct role in antiviral responses in adult mice (34, 35) although evidence of a direct response has been found in embryonic stem cells and young animals (36, 37).
Here, we explored further the interactions of the RNAi pathway with LRV1 in both Lbr and Lgy and show first that LRV1 is indeed seen by the endogenous RNAi pathway, as judged by the presence of significant levels of antiviral small RNAs (sRNAs). Thus, and different from other systems, RNAi and viral replication seem to be balanced. However, by increased siRNA expression, RNAi could be used to efficiently eliminate the virus. Importantly, these LRV1-negative transfectants recapitulate the in vitro macrophage cytokine release defect seen in naturally occurring LRV1-negative lines, suggesting that the engineered LRV1-negative isogenic lines will be valuable in studying the role of LRV1-mediated biology and virulence.
Results
Naturally Abundant siRNAs Directed Against LRV1 of L. braziliensis and L. guyanensis.
Previous siRNA studies in Leishmania analyzed RNAs using a tagged Argonaute inserted into an δago1− knockout of Lbr M2903, which lacks LRV1 (9, 29, 38, 39). Because the lines bearing LRV1 studied here had not been similarly modified, we sequenced total small RNAs (sRNAs) as an alternative. Lbr siRNAs bear a 5′-P and 3′-OH, reflecting their origin through the action of cellular Dicer nucleases (39), and we used these properties to make siRNA-focused sRNA (<42 nt) libraries for next-generation sequencing (Table S1). For Lgy, we chose the established LRV1+ Lgy M4147 strain (7), and three different Lbr shown to bear LRV1 by PCR and/or anti-dsRNA antibody tests (40).
Table S1.
Summary of sRNA reads and mapping
| Sample | Total reads (raw) | Total trimmed reads aligned to the Leish. genomes + viruses (% total) | Aligned 33-nt peak reads to Leishmania (% alignable reads) | Aligned 23-nt peak reads to Leishmania (% alignable reads) | Aligned 23-nt peak reads to LRV1 (% alignable reads) | % with 3′ extension (Leishmania) | % with 3′ extension (LRV1) | Genome-mapping reads 3′ extension base A-T-C-G, % |
| Total reads | ||||||||
| Lbr M2903* | 29,391,347 | 19,447,509 (66.2%) | 1,827,623 (9.40%) | 15,147,603 (77.9%) | n/a | 21 | n/a | 43-41-8-8 |
| Lbr LEM2700 | 40,384,483 | 29,473,443 (73.0%) | 8,055,506 (27.3%) | 15,540,670 (52.7%) | 59,287 (0.20%) | 19 | 20 | 43-41-7.9-8.1 |
| Lbr LEM2780† | 48,615,815 | 34,959,518 (71.9%) | 5,121,894 (14.7%) | 25,361,713 (72.5%) | 326,021 (0.93%) | 20 | 20 | 43-40-7.8-8.2 |
| Lbr LEM3874 | 36,543,649 | 25,591,362 (70.0%) | 4,052,707 (15.8%) | 16,756,188 (65.5%) | 347,022 (1.36%) | 19 | 19 | 44-41-6.6-7.4 |
| Lgy M4147 | 55,220,664 | 37,159,548 (67.3%) | 1,261,927 (3.40%) | 25,489,186 (68.6%) | 660,143 (1.78%) | 15 | 13 | 32-33-19-15 |
| Collapsed reads | ||||||||
| Lbr M2903* | 2,327,188 | 1,038,131 (44.6%) | 68,895 (6.64%) | 776,204 (74.8%) | n/a | 37 | n/a | 35-35-15-15 |
| Lbr LEM2700 | 2,948,017 | 1,206,587 (40.9%) | 127,998 (10.6%) | 758,664 (62.9%) | 18,642 (1.54%) | 35 | 36 | 34-34-16-16 |
| Lbr LEM2780† | 3,439,169 | 1,777,142 (51.7%) | 150,229 (8.45%) | 1,050,094 (59.1%) | 61,753 (3.47%) | 33 | 34 | 34-34-16-16 |
| Lbr LEM3874 | 2,379,761 | 1,224,741 (51.5%) | 105,696 (8.63%) | 632,961 (51.7%) | 48,287 (3.94%) | 31 | 32 | 35-35-15-15 |
| Lgy M4147 | 1,437,673 | 752,464 (52.3%) | 152,059 (20.2%) | 358,039 (47.6%) | 43,099 (5.73%) | 22 | 22 | 30-28-22-20 |
The 23-nt siRNA analysis to Leishmania genome and LRV1. For Lbr, Lbr M2904 reference genome was used, and, for Lgy, an M4147 draft genome (BioProject PRJEB168, accession nos. HG800646–HG802771) was used. References for viral genomes are sequences reported in this work. n/a, Not applicable.
Lbr M2903 SSU:IR2-LUCSR.
The sum of reads mapping to LRV1-LbrLEM2780(a) and -(b) are shown, which map quantitatively to similar levels.
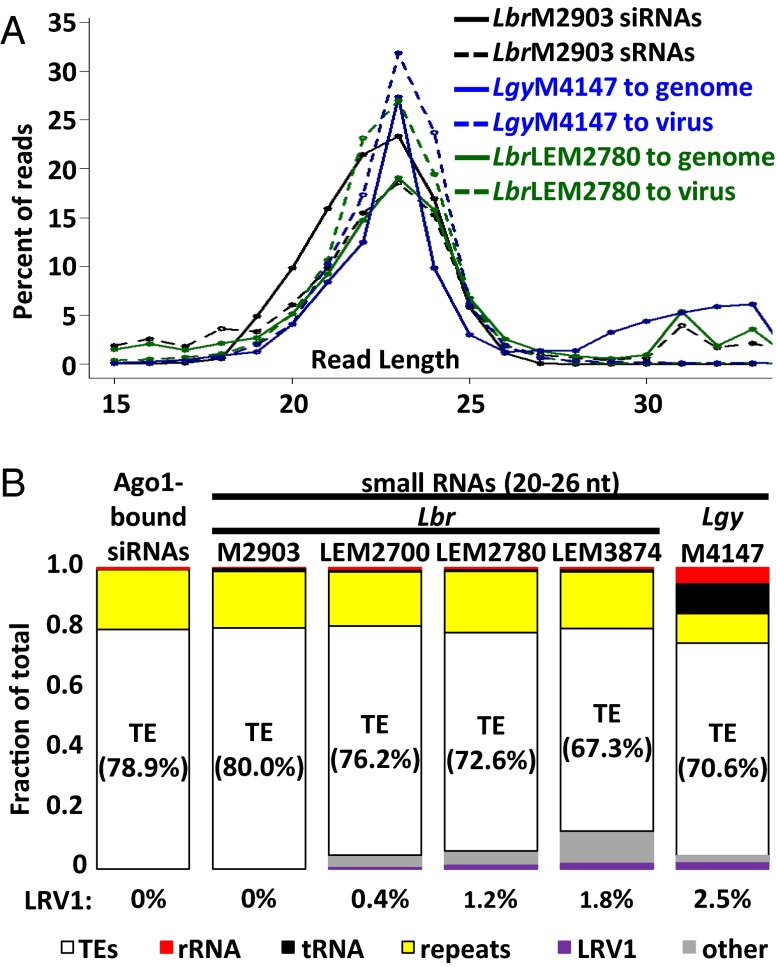
For sRNAs from Lbr M2903 mapping to the Lbr reference genome, read length displayed a biphasic distribution, with a major peak centered around 23 nt (20 to 26 nt, 77.9% of total mapped reads) and a minor one around 33 nt (30 to 36 nt, 9.4% of total mapped reads) (Fig. 1A and Tables S1 and S2). The 33-nt peak reads mapped primarily to structural RNA loci (62% of mapped reads) (Table S2), similar to an sRNA class described in many eukaryotes, including trypanosomes and Leishmania lacking the RNAi pathway (41–44). In contrast, reads from the 23-nt peak showed properties similar to AGO1-bound siRNAs (39), including their size and the presence of one to two untemplated nucleotides at the 3′ end in about 21% of the reads (Fig. 1A and Table S1). The 3′ untemplated bases likely arise from the action of cellular terminal transferases because Leishmania sp. lack the HEN1 methyltransferase that normally blocks their action (39). When both AGO1-bound siRNAs and the 23-nt sRNA peak reads were mapped to the Lbr genome, their distributions were very similar, with the vast majority mapping to transposable elements (TEs) (Fig. 1B and Table S2; and see Fig. S1A) (39). We concluded that the 23-nt peak sRNAs (23-nt sRNAs) provide a reasonable proxy for siRNAs.
Fig. 1.
Properties of Lbr siRNAs and sRNAs from Lbr and Lgy. (A) Distributions of read lengths of siRNAs or sRNAs mapping to Leishmania genomes or LRV1s. Shown are (i) AGO1-bound siRNAs (black, solid) or sRNAs (black, dashed) from WT Lbr M2903 mapping to the Lbr genome, (ii) Lgy M4147 sRNAs mapped to the Lgy genome (blue, solid) or LRV1-LgyM4147 (blue, dashed), and (iii) Lbr LEM2780 sRNAs mapped to the Lbr genome (green, solid) or LRV1-LbrLEM2780 (green, dashed). (B) Percentage of 23-nt sRNA reads (20 to 26 nt) mapping to transposable elements (TEs) (white), rRNA (red), tRNAs (black), genomic repeat regions (yellow), LRV1 (purple), and other Leishmania genomic regions (other, gray).
Table S2.
Genomic mapping of 23- and 33-nt “peak” sRNA fractions from Lbr and Lgy
| Feature | Lbr M2903 | Lgy M4147 | |||
| AGO1-bound siRNAs | 23-nt (20 to 26 nt) sRNAs | 33-nt (30 to 36 nt) sRNAs | 23-nt (20 to 26 nt) sRNAs | 33-nt (30 to 36 nt) sRNAs | |
| Alignable reads | 20,029,304 | 19,447,509 | 1,827,623 | 37,159,548 | 1,261,927 |
| Percent mapping to transposable elements | |||||
| SLACS | 33.9 | 26.9 | 1.2 | 59.8 | 0.25 |
| TATE | 45.1 | 53.1 | 5.2 | 10.7 | 0.32 |
| Percent mapping to repeats | |||||
| Misc. | 5.8 | 5.0 | 7.7 | 1.1 | 2.4 |
| TAR | 4.7 | 4.2 | 0.08 | 4.0 | 0.38 |
| TAS | 4.2 | 5.1 | 20.3 | 4.8 | 6.5 |
| CIR | 5.1 | 4.6 | 0.09 | 0.0 | 0.0 |
| Percent mapping to structural RNAs | |||||
| tRNAs | 0.12 | 0.75 | 32.1 | 9.9 | 75.4 |
| rRNAs | 0.42 | 0.47 | 29.4 | 5.5 | 14.6 |
Fig. S1.
Properties of Lbr siRNAs and 23-nt sRNAs from Lbr and Lgy. This figure shows mapping of the indicated small RNAs after collapsing the data to remove duplicate reads. Shown are the percentages of 23-nt sRNA reads (20 to 26 nt) mapping to transposable elements (TEs) (white), rRNA (red), tRNAs (black), genomic repeat regions (yellow), LRV1 (purple), and other Leishmania genomic regions (other, gray). (A) As in Fig. 1B, mappings in WT parent lines. (B) As in Fig. 4A, comparing parental lines with capsid StL Lgy M4147 (Left) or capsid-RDRP StL Lbr LEM3874 (Right).
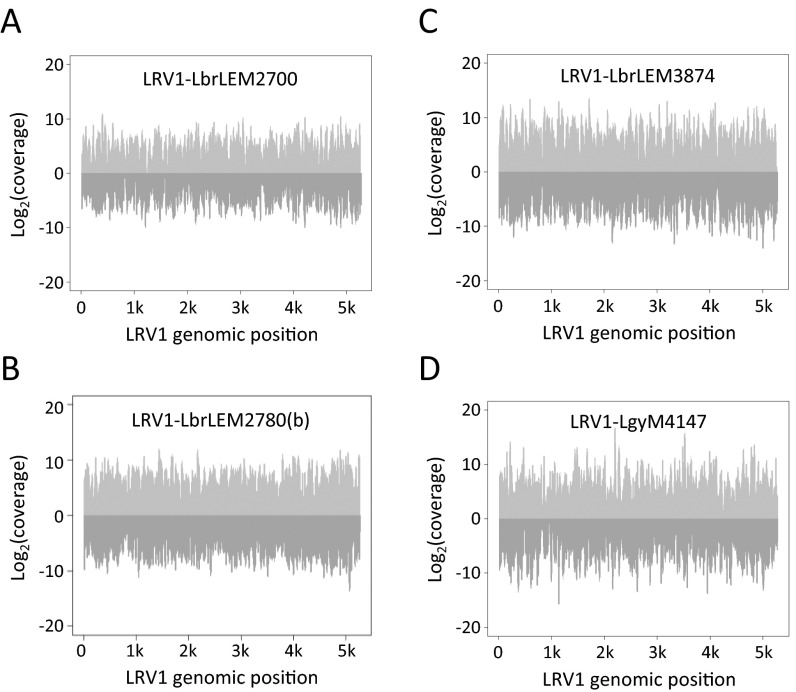
The properties of sRNAs from the LRV1-bearing Lgy M4147 and Lbr LEM2700, LEM2780, and LEM3874 mapping to the Lgy or Lbr reference genomes were similar to those of Lbr M2903, including the 23- and 33-nt sRNA peaks, genomic mappings, and the presence and level of 3′-nt extensions in the 23-nt sRNAs (Fig. 1, Fig. S1, and Tables S1 and S2). Importantly, a substantial fraction of sRNA reads obtained from the LRV1+ Lgy and Lbr lines mapped to the LRV1 genomes, ranging from 0.4 to 2.5% of the 23-nt mapped reads (Fig. 1B). Unlike those aligned to the nuclear genome, LRV1-mapped reads showed a single size distribution centered around 23 nt (Fig. 1A), with about 20% again showing short 3′ extensions (Table S1), typical of Lbr siRNAs and 23-nt sRNAs (39). LRV1-mapping 23-nt sRNAs showed no consistent strand- or region-specific biases in all four strains (Fig. S2), suggesting that they likely originated from the action of DICERs on the viral dsRNA genome.
Fig. S2.
Mapping of 23-nt sRNA reads (20 to 26 nt) from the respective parasite lines to LRV1-LbrLEM2700 (A), LRV1-LbrLEM2780 (B) (B), LRV1-LbrLEM3874 (C), and LRV1-LgyM4147 (D). Reads mapping to the positive strand (light gray) and negative strand (dark gray).
We previously showed that LRV1 does not encode a trans-acting inhibitor of RNAi activity (30), and the presence of high levels of LRV1-directed sRNAs similarly suggests that it does not encode a strong cis-acting inhibitor. Importantly, the levels of 23-nt sRNAs mapping to LRV1s were in the same range as siRNAs mapping to an efficiently silenced Luciferase reporter (0.4 to 2.5% vs. 0.8% targeted by the long-hairpin/stem-loop transgene) (30, 39). Thus, LRV1 is able to persist in the face of a significant RNAi response, as judged by 23-nt sRNA levels.
LRV1 Can Be Efficiently Targeted by Transgenic RNAi.
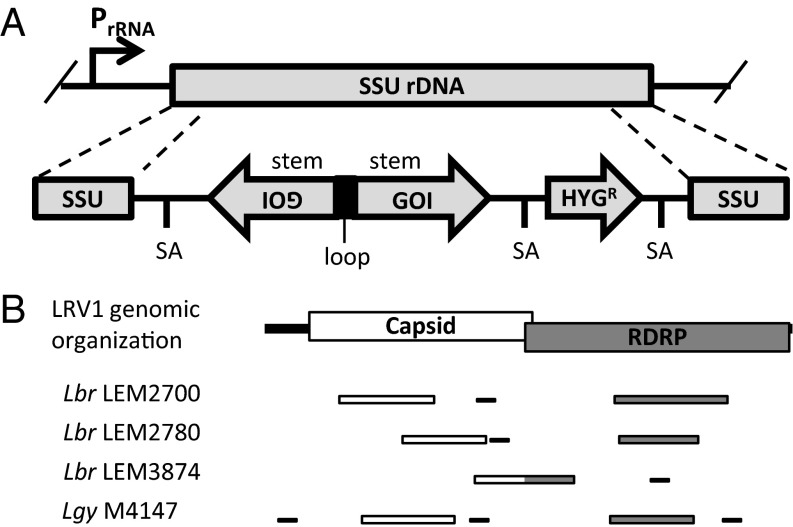
These data are consistent with a model where RNAi activity and LRV1 replication have achieved a “balance” between viral synthesis and degradation, which might be shifted by increasing or decreasing RNAi activity. With an eye toward virus elimination, we focused on increasing LRV1-targeting siRNA levels through the use of transgenic RNAi methods developed previously (30), in which long-hairpin RNA is expressed at high levels from a stem-loop (StL) construct containing LRV1 sequences integrated into the ribosomal RNA locus (Fig. 2A). We targeted regions of LRV1 from the capsid or RDRP ORFs (Lgy M4147, Lbr LEM2700, and LEM2780), or a region that spanned them (Lbr LEM3874), ranging in length from 794 to 1,143 bp (Fig. 2B and Table S3); because the two viral genes reside within the same RNA segment, targeting either one should lead to degradation of the entire LRV1 RNA. Because LRV1 sequences diverge substantially between parasite strains (69 to 90% nt identity), “stems” specific for each species/strain were used. To assess nonspecific effects, we integrated an StL construct for an AT-rich GFP (GFP65 StL), which efficiently silences expression of GFP65 (30). The untransfected parental lines served as LRV1+ controls, and Lbr M2903 or Lgy M4147/HYG (12) served as LRV1-negative controls.
Fig. 2.
RNAi constructs for LRV1 elimination. (A) Schematic of an RNAi “stem-loop” (StL) construct. Each construct includes an inverted repeated sequence containing 800 to 1,200 bp of the target gene (gene of interest, GOI) and a hygromycin drug resistance marker (HYGR). The construct is flanked with the sequence of the small subunit ribosomal RNA gene, which allows it to integrate into this locus, where it is transcribed at high levels. Splice acceptor (SA) signals within the construct allow for polyadenylylation and processing. (B) Schematic showing LRV1 genome organization and regions targeted for RNAi StL constructions (thick bars) from Lbr LEM2700, LEM2780, and LEM3874, and Lgy M4147 targeted by RNAi (white, capsid; gray, RDRP). The locations of qPCR amplicons for quantification of LRV1 levels are shown (thin black bars).
Table S3.
Primer sequences used to amplify regions of LRV1 for cloning into stem-loop (StL) constructs
| Parasite strain | Target | Construct ID | Name | Stem length, bp | Sequence* |
| Lbr LEM2700 | Capsid | B6910 | pIR2HYG-LRV1_LbrLEM2700_CapsidStL(A) | 943 | 5′-CGCTAGTCTAGAATACTACAGCAAACATGTTTCG |
| 5′-CGCTAGTCTAGACAAGGTGTCTGTTGGGTTCGAT | |||||
| RDRP | B6908 | pIR2HYG-LRV1_LbrLEM2700_RDRPStL(A) | 1,143 | 5′-CGCTAGTCTAGAATGTGCTTCAAACTTGAAGATG | |
| 5′-CGCTAGTCTAGATAGCAGCAATCTAACGACCTGC | |||||
| Lbr LEM2780 | Capsid | B7061 | pIR2HYG-LRV1_LbrLEM2780_CapsidStL(A) | 835 | 5′-CCAGCTTGGGATCAATTTGCGG |
| 5′-GGACATCTCCATCAGCCGATGA | |||||
| RDRP | B7062 | pIR2HYG-LRV1_LbrLEM2780_RDRPStL(A) | 794 | 5′-GTGAGGATGAGTTGCGCGCTGC | |
| 5′-ATTGCTAAGTAGACTGTTTGCG | |||||
| Lbr LEM3874 | Capsid/RDRP | B7268 | pIR2HYG-LRV1_LbrLEM3874_StL(A) | 1,000 | 5′-GGCTAGTCTAGA GTCGTGCGATCTATTCCATCCT |
| 5′-GGCTAGTCTAGATTAGTGCTTATGTTAGGATCAG | |||||
| Lgy M4147 | Capsid | B7066 | pIR2HYG-LRV1_LgyM4147_CapsidStL(A) | 926 | 5′-CTTCTCCTTTACGTGCCAGC |
| 5′-GCGCATTGTTGTCCACTCAA | |||||
| RDRP | B7063 | pIR2HYG-LRV1_LgyM4147_RDRPStL(A) | 829 | 5′-CTTGCTAGGTCGTGGGGTGA | |
| 5′-ACCAACATGCATAGACGTGG |
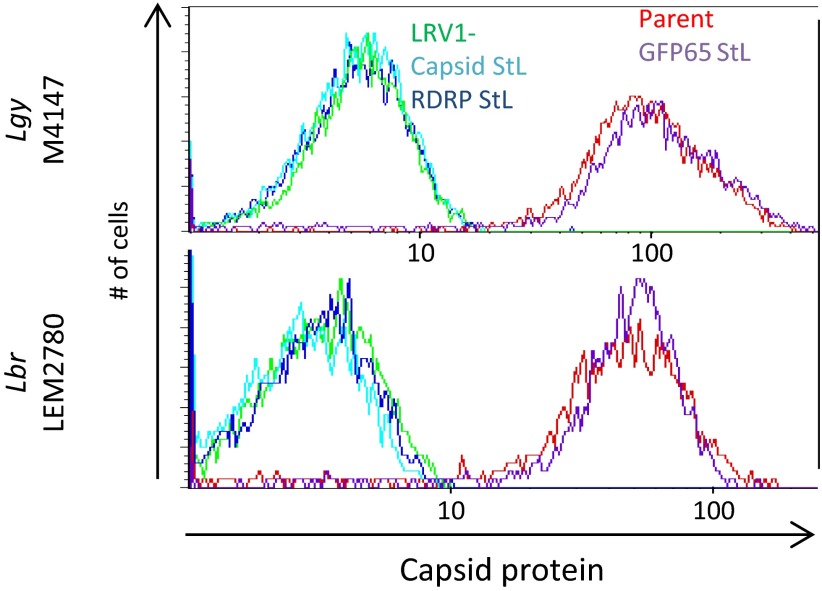
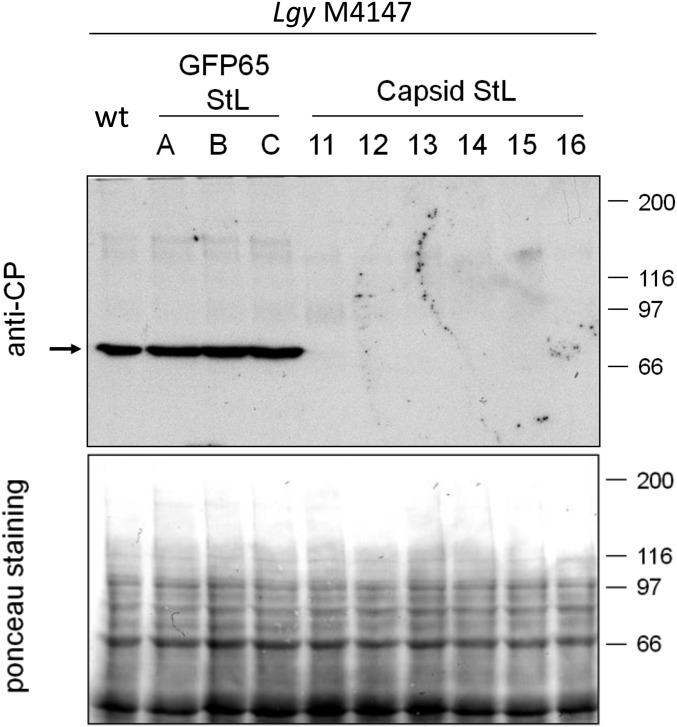
To screen for loss of LRV1, StL transfectants were analyzed by flow cytometry of fixed, permeabilized cells using an antibody raised against the Lgy M4147 LRV1 capsid (45), which cross-reacts with Lbr LRV1. For both Lgy M4147 (Fig. 3, Top) and Lbr LEM2780 (Fig. 3, Bottom), there was a clear separation in capsid staining between the LRV1-positive (Fig. 3, red) and LRV1-negative controls (Fig. 3, green). Although control GFP65 StL lines (Fig. 3, purple) had capsid protein levels similar to WT, capsid protein was undetectable in LRV1-targeted StL lines (Fig. 3, light and dark blue), indistinguishable from the LRV1-negative control. This result was observed whether the capsid or RDRP was targeted (Fig. 3). Similar results were obtained with LRV1 StL transfectants from Lbr LEM2700 and Lbr LEM3874. In support of the flow cytometry data, Western blot analysis with an anti-capsid antibody showed high LRV1 levels in the Lgy parental line and GFP65 StL transfectants whereas capsid protein was undetectable in the capsid StL transfectants (Fig. S3).
Fig. 3.
Loss of LRV1 induced by RNAi anti-capsid flow cytometry analysis of LRV1-knockdown lines in Lgy M4147 and Lbr LEM2780 (Top and Bottom, respectively). LRV1 capsid protein levels are unchanged in GFP65 StL lines whereas LRV1 StL lines have undetectable capsid protein. Red, parent lines; purple, GFP65 StLs (off target control); green, LRV1-negative controls; light blue, capsid StL; dark blue, RDRP StL.
Fig. S3.
Capsid protein is lost in Lgy M4147 capsid StL transfectants. Three GFP65 StL control clones and six capsid StL clones were evaluated. (Top) Western blot analysis was performed using g018d53 anti-capsid polyclonal antibody (35). The arrow marks the location of the capsid protein band. (Bottom) Ponceau S stain of protein gel.
StL Constructs Result in High Levels of siRNAs Mapping to the LRV1 Stem.
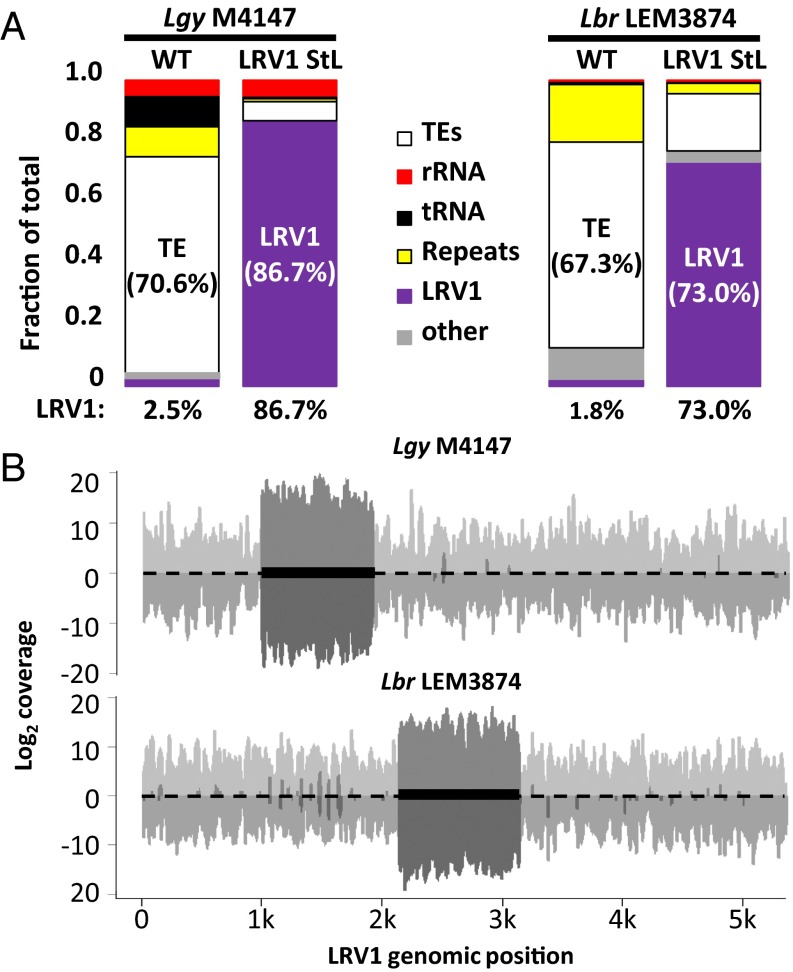
Despite the insensitivity of LRV1 to “natural” levels of RNAi, as judged by the abundance of 23-nt sRNAs, introduction of StL constructs targeting LRV1 resulted in great reduction in LRV1 levels. To understand the basis for this reduction, we analyzed 23-nt sRNA peak reads mapping to the nuclear and LRV1 genomes, for one LRV1 StL transfectant of each species (Fig. 4). Remarkably, the percentage of total 23-nt sRNAs mapping to LRV1 had increased greatly from that seen in the WT parent, from 2.5 to 86.7% for Lgy and from 1.8 to 73.0% for Lbr LEM3874 (Fig. 4A and Fig. S1B). Concomitantly, the percentages of 23-nt sRNAs mapping to the nuclear genome was proportionately reduced, with some variability among loci and/or lines (for example, rRNA reads were unchanged in both species whereas tRNA reads decreased in Lgy) (Fig. 4A and Fig. S1B). Although we did not measure the absolute levels of sRNAs, previous studies have shown that these levels are tightly controlled by the level of Argonaute 1 and thus are unlikely to differ significantly (39). Essentially, all LRV1-mapping sRNAs in LRV1 StL lines now mapped only to the RNAi-targeted stem region (Fig. 4B, dark gray), as expected, because LRV1 had been eliminated (as discussed in Complete Virus Elimination Following RNAi of LRV1), which also argues against the occurrence of “transitive” siRNA formation (46, 47).
Fig. 4.
Overexpression of LRV1-mapping 23-nt sRNAs in LRV1 StL transfectants. (A) Genomic mapping of 23-nt sRNA reads from sRNA sequencing of parental or capsid StL Lgy M4147 (Left) or capsid-RDRP StL Lbr LEM3874 (Right) mapping to transposable elements (TEs) (white), rRNA (red), tRNAs (black), genomic repeat regions (yellow), LRV1 (purple), and other Leishmania genomic regions (other, gray). (B) LRV1 mapping of 23-nt sRNA reads from LRV1StL lines described in A (Lgy M4147, Top; Lbr LEM3874, Bottom). Light gray trace indicates parental read distributions; dark gray trace indicates LRV1 StL read distributions. The dark box indicates the region targeted by the StL stems.
The levels of LRV1 23-nt sRNAs (76 to 87%) in LRV1 StL transfectants were much greater than those seen with siRNAs mapping to the LUC ORF/stem targeted using the same StL transfection construct (0.8%) (39). To rule out the possibility that this result arose from reliance on 23-nt sRNAs, we analyzed these 23-nt sRNAs from a line bearing the LUC StL RNAi reporter used in the siRNA studies [IR2-LUCStL(b)-LUC(a)]. From this analysis, 1.14% of the 23-nt sRNA peak reads mapped to the LUC ORF/stem, suggesting that use of 23-nt sRNAs vs. siRNAs did not significantly impact quantitation. To assess the target-specific effects, we compared these results with those quantitating 23-nt peak sRNAs after RNAi StL targeting of a panel of 10 chromosomal genes. For this group, 1.5 to 34% of 23-nt sRNAs mapped to the RNAi-targeted gene, compared with less than 0.02% basally. Thus, the StL-bearing vectors generate a high but variable level of sRNAs for all genes tested, with the LUC reporter being at the low end and LRV1 at the high end, which may reflect the fact that, although the LRV1 target is typically eliminated by RNAi (Fig. 3 and Complete Virus Elimination Following RNAi of LRV1), chromosomal RNAi targets continuously transcribe mRNAs. In other organisms, studies have shown that the presence of a cognate target facilitates the turnover of sRNAs; thus, the absence of an LRV1 target may lead to higher levels of siRNAs (48, 49). Future studies should address the factors contributing to the differences in sRNA levels among genes and to the very high steady-state levels of LRV1-directed 23-nt sRNAs seen here.
Complete Virus Elimination Following RNAi of LRV1.
RNAi-mediated LRV1 knockdown would be most useful as a tool if it resulted in a complete elimination of LRV1. To achieve a sensitivity beyond that of flow cytometry (∼20-fold) or Western blotting (∼100-fold), we validated a sensitive quantitative RT-PCR assay (qRT-PCR) for LRV1, using strain- and LRV1-specific primers to amplify a region located outside the stem regions (Fig. 2B and Table S4). Because the melting temperatures of PCR amplicons are sequence- and length-dependent, comparison of dissociation (melt) curves facilitated discrimination between specific and nonspecific amplification.
Table S4.
Primer sequences used in measurement of LRV1 RNA levels by qRT-PCR
| Name | Primer orientation | Sequence |
| KMP-11 | F | 5′-GCCTGGATGAGGAGTTCAACA |
| R | 5′-GTGCTCCTTCATCTCGGG | |
| L. braziliensis LEM2700 | F | 5′-CATCCTGCTGAGTTGACTTCATAC |
| R | 5′-GTCACACCTTGTGATGACATTGC | |
| L. braziliensis LEM2780 | F | 5′-GTCATTACGAGGTGTGATGGAAT |
| R | 5′-GGTAACGCGCCATCACACAGT | |
| L. braziliensis LEM3874 | F | 5′-GAATATGCTCTCCGACCGGTTG |
| R | 5′-AATTCTCGCAGCCACCCCACAG | |
| L. guyanensis M4147 | Set 1 F | 5′-CTGACTGGACGGGGGGTAAT |
| Set 1 R | 5′-CAAAACACTCCCTTACGC | |
| Set 2 F | 5′-CACGCTAGATGAGTACATCTGG | |
| Set 2 R | 5′-GTAGTTGCGGAATCTGACG | |
| Set 3 F | 5′-GGTAATATCACGCAGTGTAAGC | |
| Set 3 R | 5′-GACACCACCTCTAAGACACG |
F, forward; R, reverse.
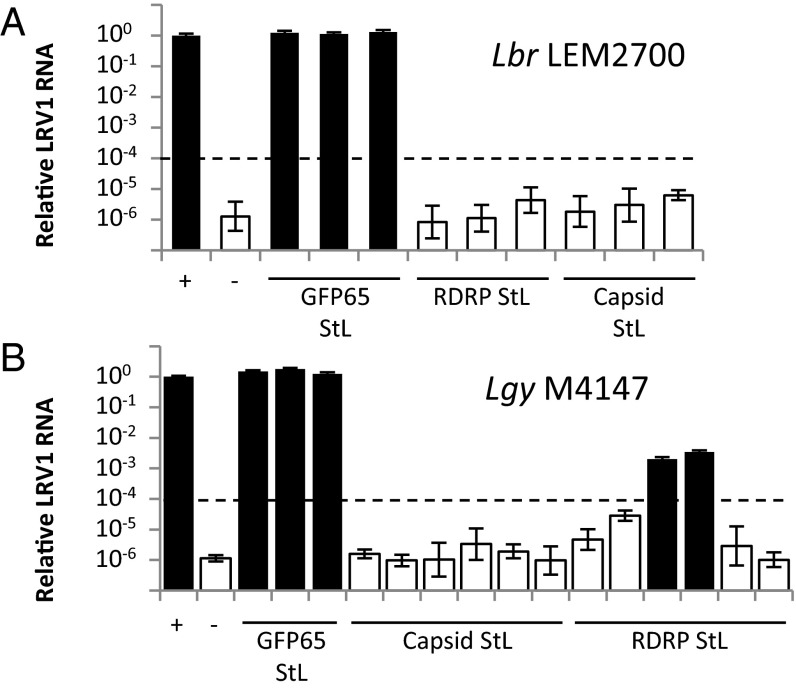
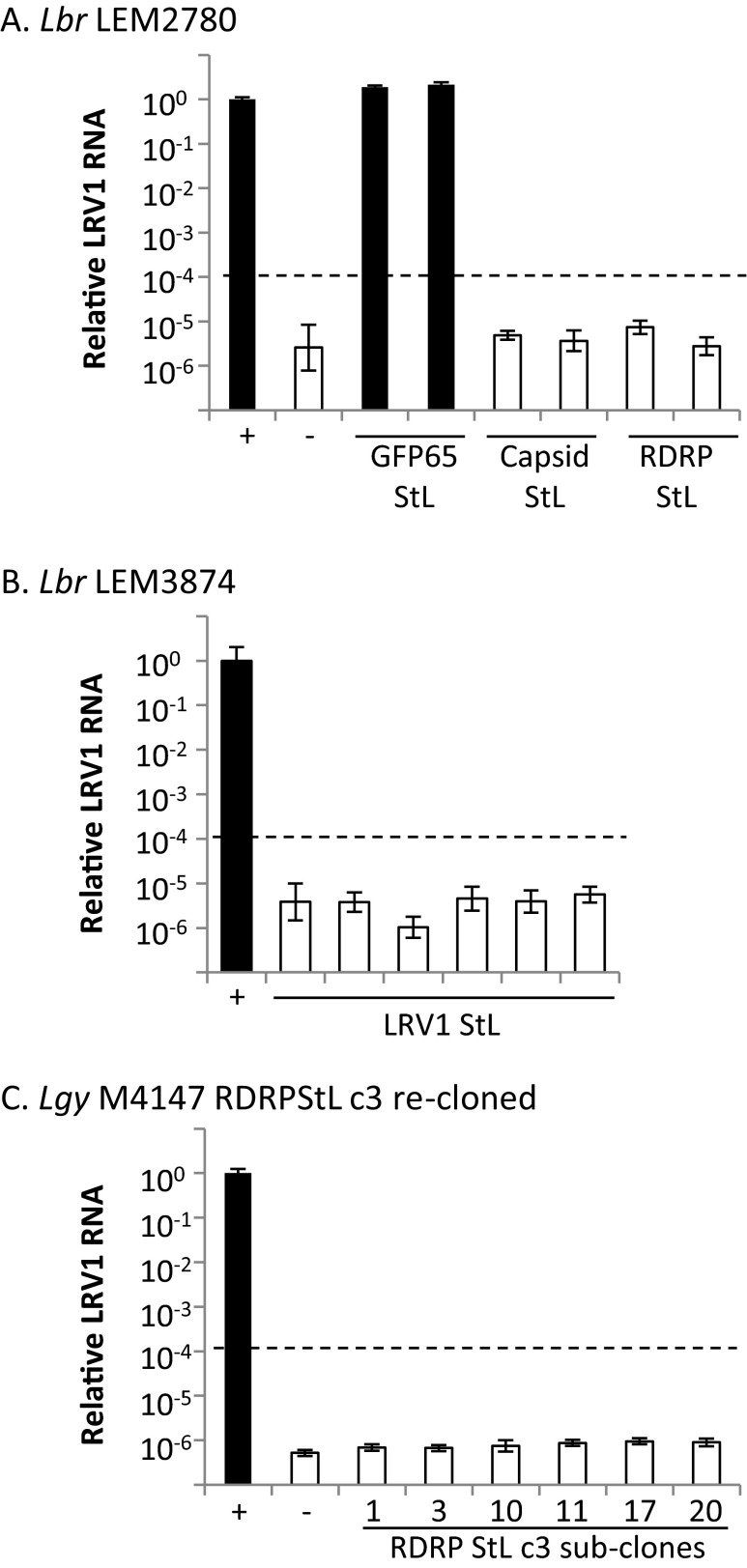
Because LRV1 copy number was estimated to be ∼100 per cell (50), a cutoff for classification as LRV1-negative was set at 104-fold below WT. Analysis of Lbr qPCR data by the ΔΔCt method (51) showed that most LRV1 StL transfectants had LRV1 RNA levels more than 105-fold lower than WT (Fig. 5A and Fig. S4 A and B). Raw Ct values for LRV1 StL lines with LRV1-specific primers were indistinguishable from mock cDNA preparations, and ΔCt values were indistinguishable from those of negative controls. Melt curves showed that products seen at Ct arose from nonspecific amplification (Fig. 5A and Fig. S4; white bars). As expected for control GFP65 StL lines, LRV1 RNA levels were similar to those in WT (Fig. 5A and Fig. S4; black bars).
Fig. 5.
The LRV1 genome is completely lost in most LRV1-StL transfectants. qPCR analysis of LRV1 RNA levels in LRV1 StL transfectant clones of Lbr LEM2700 (A) and Lgy M4147 (B), along with positive and negative controls (+ and –, respectively) and control GFP65 StL transfectants. White bars denote a nonspecific qPCR product whereas black bars denote an LRV1-specific amplicons (melt curve analysis). Dashed line indicates the cutoff for designating a clone as LRV1-negative. Error bars are the SD of three technical replicates for each line.
Fig. S4.
qPCR analysis of LRV1 RNA levels in LRV1 StL clones of L. braziliensis strain LEM2780 (A), L. braziliensis strain LEM3874 (B), and recloned L. guyanensis M4147 RDRP StL c3 (C). White bars denote a nonspecific product; black bars denote an LRV1-specific product (melt curve analysis). Dashed line indicates cutoff for designating a clone as LRV1-negative. Error bars are the SD of three technical replicates for each line.
Similar results were obtained with RNAi of LRV1 in Lgy M4147, with most transfectants showing reductions below the 104-fold cutoff (Fig. 5B). However, low levels of LRV1 remained in two lines where the RDRP was targeted, ∼300- to 500-fold less than the parent line (Fig. 5B, black bars); here, melt curve analysis suggested that these products were LRV1-specific. Alternate primers targeting other regions across the virus gave similar results, suggesting the presence of intact LRV1. We hypothesized that this result was due to heterogeneity in viral load, with most but not all cells lacking LRV1. In support of this hypothesis, we generated and showed that all clonal lines arising from one of the “weakly positive” lines were negative for LRV1 by flow cytometry and satisfied the 104-fold cutoff by qPCR (Fig. S4C). The occasionally incomplete LRV1 elimination is consistent with our prior observation that RNAi was somewhat less efficient in Lgy than in Lbr (30). Nonetheless, even for weakly positive Lgy transfectants, RNAi was sufficiently efficient for the ready isolation of LRV1-negative lines (Fig. 3, top; Fig. 5B; and Fig. S3).
LRV1 Knockdowns Induce Less Cytokine Production in in Vitro Macrophage Infection Assays.
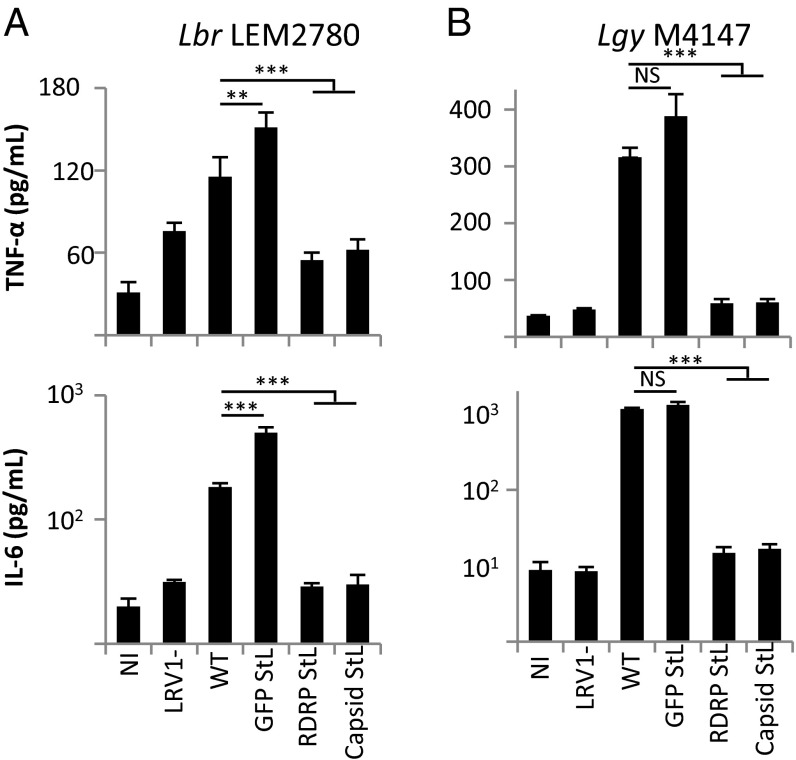
Previous reports showed that LRV1+ Lgy stimulated the TLR3-dependent release of higher levels of cytokines from bone marrow-derived macrophages (BMDMs) than LRV1-negative strains (7). The availability of defined RNAi-derived LRV1-negative lines now allowed tests of this finding in Lbr, as well as confirmation of prior results obtained with a single isogenic LRV1- Lgy. Briefly, BMDMs were infected in vitro with LRV1 StL and GFP65 StL Lbr and Lgy transfectants, as well as positive and negative control lines, and the levels of two cytokines known to be induced by LRV1 (TNF-α and IL-6) (7, 10) were measured.
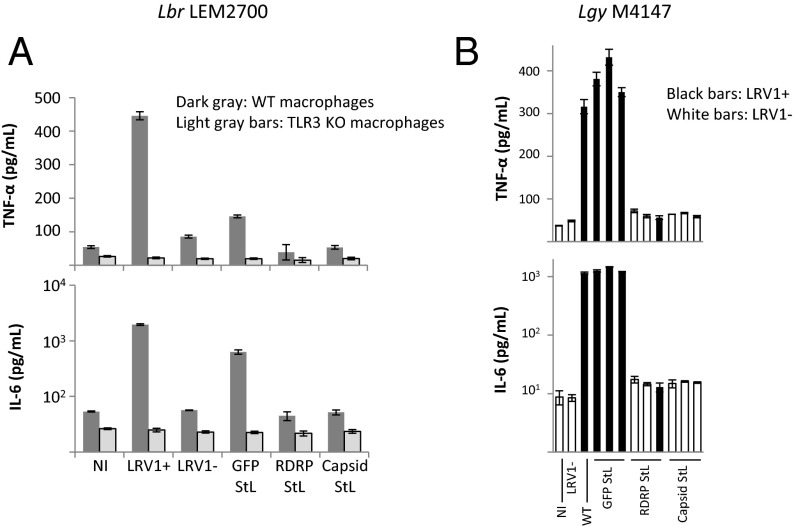
Capsid StL and RDRP StL LRV1-negative lines of both Lbr and Lgy induced significantly lower levels of cytokine production than did the LRV1-positive lines (both parental and GFP65 STL) (Fig. 6 and Fig. S5). Additionally, when macrophages from TLR3-deficient mice were infected with Lbr LEM2700, the LRV1-positive parasites no longer elicited higher levels of cytokine release (Fig. S5). Of note, all Lgy LRV1 StL lines induced background levels of cytokine release, including the two lines that retained low levels of LRV1 (Figs. 5B and 6B and Fig. S5), consistent with the observation that high levels of LRV1 were necessary for cytokine stimulation (7, 10).
Fig. 6.
LRV1 elimination results in decreased release of cytokines from infected macrophages. TNF-α or IL-6 levels were quantified 24 h after infection of macrophages with Lbr LEM2780 (A) or Lg M4147 (B) parent, GFP65 knockdown control, or LRV1-StL transfectants. In both studies, the LRV1-control was Lgy M4147. For A, results are averages of two to three technical replicates for two clones of each line. For B, results are the averages of two technical replicates for three to six clones of each line. NS, not significant; **P < 0.01; ***P < 0.0001 by t test.
Fig. S5.
Infection of macrophages by Lbr LEM2700 (A) and Lgy M4147 (B). TNF-α or IL-6 levels were quantified 24 h after infection of macrophages with Lbr (A) or Lgy (B) parasites. GFP65 StL transfectants; LRV1+ or LRV1−, infected with Lgy M4147 LRV1+ or LRV1-negative cells; NI, not infected; and RDRP StL or capsid StL transfectants. (A) Results are the averages of two technical replicates of two clones per line. Dark gray bars, experiment performed using WT macrophages; light gray bars, experiment performed using TLR3 knockout macrophages. (B) Results are the averages of two technical replicates for each representative clone indicated. Lines found to be LRV1+ by qPCR are denoted by black bars; white bars are lines found to be LRV1-negative by qPCR.
Discussion
In this study, we characterized the endogenous RNAi response in Leishmania bearing the dsRNA virus LRV1 and used these insights to generate virus-negative lines that facilitate the study of the role of LRV1 in parasite biology and host–parasite interactions.
Leishmania LRV1 and the Endogenous RNAi Pathway.
We identified two populations of sRNA in Lbr and Lgy. The less abundant 33-nt sRNAs mapped primarily to genes encoding structural RNAs (Table S2), as seen in other organisms, including trypanosomatids (41–44). In contrast, the more abundant 23-nt sRNA fraction exhibited properties similar to authentic AGO1-bound Lbr siRNAs (39), including size, the presence of 3′ untemplated bases at the same frequency (∼20%), and mapping primarily to transposable elements and repetitive sequences (Fig. 1 and Tables S1 and S2). Only 23-nt sRNA reads mapped to the LRV1 dsRNA genome (Fig. 1A), and these reads also bore 3′ nucleotide extensions at the same frequency, again consistent with an origin via the RNAi pathway (Table S1). Importantly, the levels of 23-nt sRNAs mapping to LRV1 constituted a substantial fraction of total aligned 23-nt sRNAs (Fig. 1B and Table S1), comparable with those targeting an efficiently silenced LUC reporter gene (30, 39). Thus, LRV1 can persist in the face of RNAi pressure that gives rise to sRNA levels comparable with that which efficiently silences a chromosomal target gene.
In other organisms, sRNA/siRNA levels provide a gauge of RNAi pathway recognition and targeting of viruses: When RNAi controls virus replication, as in plants, fungi, and insects (26, 32, 33), high levels of siRNAs accompany viral infections, leading to eradication of the virus. In mammals, quantitatively fewer siRNAs are present, which do not effectively control virus levels, at least in adult somatic tissues (34, 36, 37). In contrast, high levels of siRNA-like 23-nt sRNAs in Leishmania suggest an attack on LRV1 by the RNAi pathway, but the virus persists. Although many viruses encode trans-acting RNAi suppressors, mediating their survival (52), this seems unlikely for LRV1. There is no obvious coding potential for an RNAi suppressor in the compact LRV1 genome. Our studies here suggest that there is no strong cis-acting inhibitory activity, and we showed previously that a luciferase reporter was equally silenced in the LRV1+ and LRV1-negative Lgy studied here (30), which suggests a third model where LRV1 is targeted strongly by the RNAi pathway, but the RNAi-mediated degradation is “balanced” by virus replication or other factors. This balance is perhaps most likely mediated by the slicer activity of Argonaute; however, previous studies examining the role of RNAi in control of viruses frequently raise the possibility of Dicer-mediated control as well (53–55). It is likely that the sequestration of the LRV1 dsRNA genome within the capsid may also contribute by limiting the exposure of the LRV1 dsRNA to the RNAi machinery and other degradative pathways. In yeast, SKI genes act to prevent deleterious effects of L-A viruses toward its fungal host through alterations in mRNA degradation and/or surveillance (27), and homologs for several mRNA degradation and surveillance genes are evident in the Leishmania genome.
In other organisms, persistent viruses can also be maintained in the face of an active RNAi pathway, but at considerably reduced levels (26, 56). Over evolutionary time, this strong pressure likely accounts for the inverse relationship in fungi between virus levels and the activity and/or presence of the RNAi pathway, especially when associated with a selective advantage for viral retention, as seen with the yeast killer factors that are dependent on the L-A virus (26, 57). Similarly, in Leishmania, we had originally proposed that RNAi pressure would be sufficiently strong as to in some cases provide a driving force for loss of RNAi, to maintain LRV1-dependent increases in pathogenicity (30). Given the greater ability of LRV1 to survive in the presence of an active RNAi pathway, our data suggest that the magnitude of this effect may be considerably less than envisioned. However, even small pressure could prove a significant force toward down-regulating pathways impacting on LRV1 levels during evolution.
RNAi as a Tool for Generating LRV1-Negative Lines for Biology.
Following the predictions of the balance hypothesis, we aimed to increase activity against LRV1 through the increased synthesis of siRNAs targeting LRV1, which proved quite successful: The fraction of 23-nt sRNAs targeting LRV1 rose dramatically in lines expressing StL constructs targeting LRV1 (Figs. 1B and 4A). Correspondingly, the fraction of 23-nt sRNAs mapping to the Leishmania genome dropped proportionately, most of which again mapped to TEs and repeats (Fig. 4A). Importantly, LRV1 levels were dramatically reduced for all LRV1 StL transfectants, and, in most cases, the virus was eliminated, as judged by protein and RNA methods (Figs. 3 and 5 and Figs. S3 and S4). Targeting of either the capsid or RDRP gene eliminated LRV1, as was expected given that both are encoded by the same RNA (Fig. 2B). Only in Lgy were some transfectants found that retained low levels of LRV1, which could reflect less RNAi activity in this species, as was seen with reporter genes (30). However, most transfectants had completely lost LRV1.
Viral infection has been reported for Giardiavirus (58), and stable viral transfer for several fungal Totiviruses (59). However, de novo infection and stable viral transfer have been unsuccessful with Lgy (29), and reverse genetic systems have yet to be reported for any Totivirus. Therefore, the ability to reproducibly mediate viral cure by RNAi is of great value for biological studies of LRV1. Previous work used an LRV1-negative Lgy, which was obtained after transfection with an episomal Leishmania vector expressing resistance to hygromycin B, followed by a long period of growth under selection (12); however, this method seems to have been successful only once. Neither have we succeeded with several “stress-related” treatments that have proven effective in curing mycoviruses, such as yeast L-A (60). Our studies establish RNAi as a viable strategy for cure of LRV1 and perhaps other viruses in RNAi-competent Leishmania species.
LRV1+, but not LRV1-negative, Lgy, induces a “hyperinflammatory” cytokine response in infections of BMDMs in vitro, which is TLR3-dependent (6, 7). Infectivity tests of mouse BMDMs in vitro showed that RNAi-generated LRV1-negative Lgy lines likewise failed to induce a substantial cytokine response, as shown for two cytokines (TNF-α and IL-6) known to be diagnostic for an LRV1-driven innate immune response. Interestingly, this result occurred with RNAi-derived lines where LRV1 loss was substantial but incomplete (RDRP StL c3 and 4; 500- and 300-fold below parental levels, respectively) (Figs. 5B and 6B and Fig. S4C), consistent with data from natural Lgy showing low LRV1 levels (7). Thus, a partial reduction in LRV1 levels, similar to that seen in some natural isolates, is sufficient to ameliorate LRV1-dependent virulence, which may facilitate future efforts targeting LRV1 in human disease. Importantly, the continued presence of the integrated StL constructs seemed to have no “off target” effect in the BMDM infections, despite the high levels of transgene-derived 23-nt sRNAs present in these lines (Fig. 4); the LRV1 StL “cured” lines induced the release of cytokines at a level similar to that of StL-negative, LRV1-negative controls, and control GFP65 StL lines that maintained LRV1 induced the release of cytokines at a level similar to the StL-negative, LRV1+ parent (Figs. 3, 5, and 6). Additional studies will be necessary to determine whether this also pertains to other cell types or host infections.
LRV1-Dependent Virulence in L. braziliensis.
Previous studies of LRV1-dependent virulence focused primarily on Lgy; however, in humans, Lbr is associated with the larger share of MCL (14, 15). Our studies extend the generality of LRV1-dependent virulence to Lbr because LRV1+ Lbr likewise induces strong TLR3-dependent cytokine responses. These findings are especially important in light of published work on the association of LRV1 with MCL, with mixed results depending on the geographic region and methods used (6, 16–19). Our data show that, in in vitro infections, LRV1 contributes strongly to the proinflammatory phenotype associated with elevated pathogenicity, as seen in Lgy, which suggests that, in human infections, it may be informative to seek for correlations between LRV1 and the severity of CL in Lbr infections. Indeed, recent studies show that LRV1 in Lbr clinical isolates correlates with drug treatment failure (16), as was also seen in Lgy (20). Thus, although other parasite or host factors may play a significant role in the development of MCL (21, 22), current data now bolstered by our studies of isogenic LRV1+/negative lines support a role for LRV1 in the severity of human leishmaniasis caused by Lbr.
Methods
Parasites and in Vitro Culture.
Lbr LEM2700 (MHOM/BO/90/AN), LEM2780 (MHOM/BO/90/CS), and LEM3874 (MHOM/BO/99/IMT252 n°3) were from Patrick Bastien, Université de Montpellier, Montpellier, France; Lbr M2903 (MHOM/BR/75/M2903) was from Diane McMahon Pratt, Yale School of Public Health, New Haven, CT; and Lgy M4147 (MHOM/BR/78/M4147) and its derivative Lgy M4147/HYG were from Jean Patterson, Southwest Foundation for Biomedical Research, San Antonio, TX. Before introduction of StL constructs, parasites were transfected with the linear SSU-targeting SwaI fragment from B6367 pIR2SAT-LUC(B) (30), and clonal lines were derived, validated, and used. The luciferase-expressing clone of Lbr LEM2780 contained only LRV1-LbrLEM2780(b). Parasites were grown in fresh Schneider’s Insect Medium supplemented with 10% (vol/vol) heat-inactivated FBS, 100 μM adenine, 10 μg/mL hemin, 2 μg/mL biopterin, 2 mM l-glutamine, 500 units/mL penicillin, and 50 μg/mL streptomycin, and selective drugs as indicated below.
RNAi Stem-Loop Constructs.
Regions of interest from LRV1 were screened using the RNAit target selection tool to ensure that there was no homologous sequence in the parasite genome (61), amplified from cDNA by PCR using KlenTaq-LA polymerase, and cloned into the pCR8/GW/TOPO cloning vector (Thermo Fisher Scientific) using the protocol recommended by the manufacturer and a 20-min ligation. The stem segments and PCR primer sequences can be found in Table S3. The stems were transferred from the pCR8/GW/TOPO donor vector to the pIR2HYG-GW(A) (B6365) destination vector (which contains sequences from the parasite rRNA locus to enable integration into the genome and inverted LR recombinase sites for the generation of inverted repeat through Gateway technology) using LR Clonase II (Thermo Fisher) in an overnight reaction at room temperature. Reactions were terminated by incubating with proteinase K for 1 h at 37 °C. Constructs were verified by restriction digest.
Transfections.
Stable transfections were performed as previously described (30, 62). Clonal lines were obtained by plating on semisolid media with 50 µg/mL hygromycin B. After colonies formed, cells were grown to stationary phase in 1 mL of media and passaged thereafter in 10 mL of media with 30 µg/mL hygromycin B.
RNA Preparation and Quantitative Real-Time PCR.
Total RNA was prepared from log-phase cells dissolved in TRIzol reagent (Thermo Fisher) at 3 × 108 cells per milliliter using the Direct-zol kit (Zymo Research) and eluted in 50 µL of nuclease-free water. The RNA was DNaseI-treated (Thermo Fisher) in a 200-µL reaction using the provided buffer and 20 units of enzyme for 1 h at 37 °C, purified using the RNA Clean & Concentrator-25 kit (Zymo Research), and eluted in 50 µL of nuclease-free water. Reverse transcription was performed using the SuperScript III first-strand synthesis kit (Thermo Fisher) according to the manufacturer’s instructions in a 20-µL reaction containing 0.25 μg of purified RNA. Control reactions contained the same amount of RNA but lacked the reverse transcriptase enzyme. For qRT-PCR, primers were designed to amplify ∼100-bp regions of the LRV1 genome that lie outside the stem regions (Table S4). qPCR reactions were performed with cDNA templates in 20 µL of total reaction volume using the Power SYBR Green Master Mix (Thermo Fisher), 5 µL of 10-fold diluted cDNA, and final primer concentrations of 0.2 µM. Reactions were run on the ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Thermo Fisher). PCR amplification conditions were as follows: 50 °C for 2 min and 95 °C for 10 s followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. PCR products were confirmed to be specific by melt curve analysis. All experiments were performed in triplicate. Amplification of KMP-11 was used as an internal control to normalize parallel reactions.
Small RNA Sequencing.
sRNA libraries were generated from total RNA as described (39); briefly, a primer (5′-rApppATCTCGTATGCCGTCTTCTGCTTG/ddC for all samples except Lgy M4147, which used primer rApppTGGAATTCTCGGGTGCCAAGG/ddC) was ligated first to the 3′ end using truncated mutant T4 RNA Ligase (New England Biolabs), and then a second riboprimer (5′-GUUCAGAGUUCUACAGUCCGACGAUC) to the 5′ end with T4 RNA Ligase. cDNA was generated using reverse transcriptase and primer 5′-CAAGCAGAAGACGGCATACGA, and then PCR was performed with this primer in conjunction with primer 5′-AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA. Products corresponding to inserts of 10 to 50 nt were purified and taken for sequencing with Illumina HiSeq2500 technology. Sequences have been deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive (accession SRP082553).
Bioinformatic Analysis of sRNAs.
The 5′ and 3′ adapter sequences were removed from the sRNA reads, those fewer than 15 nt were removed, and the trimmed reads were mapped to homologous LRV1 or Leishmania genomes [Lbr M2904 (63) or a draft Lgy M4147 genome (Bioproject PRJEB82; accession CALQ01000001–CALQ01004013)] using Novoalign software (www.novocraft.com); parameters were set as -F ILMFQ; -H; -g 40; -x 6; -R 5; -r; and -e 1000). A random strategy was used to align reads mapping to multiple regions, and hard clipping of low coverage bases at the 3′ end was performed. sRNA abundance was assessed directly, or after “collapsing” to remove duplicate reads using algorithms within the FASTX toolkit (hannonlab.cshl.edu/fastx_toolkit/index.html). To annotate transposable or repeated elements, we used RepeatMasker (www.repeatmasker.org) to identify known elements and/or BLAST to identify regions corresponding to Leishmania specific elements (SLACS, TAS, and TATE) (63). The annotations were collected in .bed file format for further use. Coverage was calculated by counting the number of reads that align to each strand of the LRV1 genome.
LRV1 Sequences.
From the sRNA sequences we assembled whole or partial LRV1 contigs, which were confirmed and completed by PCR amplification and sequencing. The sequences for LRV1-LbrLEM2700, LRV1-LbrLEM2780(a) and -(b), LRV1-LbrLEM3874, and a revision of the LRV1-LgyM4147 (formerly LRV1-4) (64) genome sequences were deposited in GenBank (accession numbers KX808483–KX808487).
LRV1 Capsid Flow Cytometry.
The development and optimization of the LRV1 capsid flow cytometry protocol will be described elsewhere. Briefly, 1 × 107 cells were fixed at room temperature using 2% (vol/vol) paraformaldehyde (Thermo Fisher) in PBS for 2 min and then incubated in blocking/permeabilization buffer (BPB) [10% (vol/vol) normal goat serum (Vector Laboratories) and 0.2% Triton X-100 in PBS] for 30 min at room temperature. Anti-Lgy LRV1 capsid antibody (45) was added (1:20,000 dilution) and incubated at room temperature for 1 h. After two washes with PBS, cells were resuspended in 200 μL of BPB with Alexa 488-labeled goat anti-rabbit antibody (Thermo Fisher) (1:2,000 dilution) and incubated 1 h at room temperature. After two additional washes with PBS, cells were subjected to flow cytometry, and the data were analyzed using CellQuest software (BD Bioscience).
Western Blot, Macrophage Infections, and Cytokine Assays.
After an initial wash with PBS, 5 × 107 parasites were resuspended in 100 µL of 1× PBS. Then, 1 × 107 cells (20 µL) were lysed with 7 µL of 4× Laemmli’s gel sample buffer. After heating for 5 min at 95 °C, cell lysates were loaded and separated on a 10% polyacrylamide denaturing gel, transferred to a nitrocellulose membrane, and visualized by Ponceau Red staining. The membrane was blocked for 1h in 5% powdered milk diluted in TBS plus 0.05% Tween 20, incubated overnight at 4 °C with the g018d53 anti-capsid polyclonal antibody (1:5,000 in 1% milk TBS-Tween 20), washed 4 × 15 min at room temperature, incubated for 1 h with an anti-rabbit IgG antibody coupled to peroxidase (Promega) (1:2,500 in 1% milk TBS-Tween 20), washed again 4×, and finally revealed by ECL chemiluminescence (Amersham). Infections of BL6 mouse BMDM and cytokine assays were performed as previously described (7, 10).
Statement Identifying Institutional and/or Licensing Committee Approving Animal Experiments.
Animal handling and experimental procedures were undertaken with strict adherence to ethical guidelines relevant in both host countries. These guidelines are set out by the Swiss Federal Veterinary Office (SFVO) and are under inspection by the Department of Security and Environment of the State of Vaud, Switzerland. Experiments were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (65) of the US National Institutes of Health. Animal studies were approved by the Animal Studies Committee at Washington University (protocol 20090086) in accordance with the Office of Laboratory Animal Welfare's guidelines and the Association for Assessment and Accreditation of Laboratory Animal Care International.
Acknowledgments
We thank D. E. Dobson for comments on this manuscript, Florence Prevel for excellent technical support, Jean Patterson (Southwest Foundation for Biomedical Research) for providing Lgy M4147 and anti-capsid antisera, P. Bastien (University of Montpellier) for Lbr strains, and S. P. Calderon-Copete for use of a draft Lgy genome. This work was supported in part by NIGMS Cell and Molecular Biology Training Grant GM:007067 and the Monsanto Excellence Fund for Graduate Fellowships (to E.A.B.), NIH Grants RO1AI029646 and R56AI099364 (to S.M.B.), Grants FNRS 3100A0-116665/1 and IZ70Z0-131421 (to N.F.), and the Division of Infectious Diseases (F.M.K.). Next-generation sequencing was performed at the Washington University School of Medicine, Department of Genetics Genome Technology Access Center (partially supported by NCI Cancer Center Support Grant P30 CA91842 and NCRR ICTS/CTSA Grant UL1 TR000448). S.M.B. would like to dedicate this paper to the late Elisabetta Ullu, a pioneer of trypanosomatid molecular biology, and a very special colleague and friend.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive (accession no. SRP082553) and in the GenBank database (accession nos. KX808483–KX808487).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615085113/-/DCSupplemental.
References
- 1.Alvar J, et al. WHO Leishmaniasis Control Team Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7(5):e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bañuls AL, et al. Clinical pleiomorphism in human leishmaniases, with special mention of asymptomatic infection. Clin Microbiol Infect. 2011;17(10):1451–1461. doi: 10.1111/j.1469-0691.2011.03640.x. [DOI] [PubMed] [Google Scholar]
- 3.Singh OP, Hasker E, Sacks D, Boelaert M, Sundar S. Asymptomatic Leishmania infection: A new challenge for Leishmania control. Clin Infect Dis. 2014;58(10):1424–1429. doi: 10.1093/cid/ciu102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pigott DM, et al. Global distribution maps of the leishmaniases. eLife. 2014;3:3. doi: 10.7554/eLife.02851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO 2010. Control of the Leishmaniases: Report of a Meeting of the WHO Expert Committee on the Control of Leishmaniases, Geneva, 22–26 March 2010 (World Health Organization, Geneva), WHO Technical Report Series No. 949.
- 6.Hartley MA, Drexler S, Ronet C, Beverley SM, Fasel N. The immunological, environmental, and phylogenetic perpetrators of metastatic leishmaniasis. Trends Parasitol. 2014;30(8):412–422. doi: 10.1016/j.pt.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ives A, et al. Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science. 2011;331(6018):775–778. doi: 10.1126/science.1199326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Widmer G, Comeau AM, Furlong DB, Wirth DF, Patterson JL. Characterization of a RNA virus from the parasite Leishmania. Proc Natl Acad Sci USA. 1989;86(15):5979–5982. doi: 10.1073/pnas.86.15.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stuart KD, Weeks R, Guilbride L, Myler PJ. Molecular organization of Leishmania RNA virus 1. Proc Natl Acad Sci USA. 1992;89(18):8596–8600. doi: 10.1073/pnas.89.18.8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zangger H, et al. Leishmania aethiopica field isolates bearing an endosymbiontic dsRNA virus induce pro-inflammatory cytokine response. PLoS Negl Trop Dis. 2014;8(4):e2836. doi: 10.1371/journal.pntd.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheffter SM, Ro YT, Chung IK, Patterson JL. The complete sequence of Leishmania RNA virus LRV2-1, a virus of an Old World parasite strain. Virology. 1995;212(1):84–90. doi: 10.1006/viro.1995.1456. [DOI] [PubMed] [Google Scholar]
- 12.Ro YT, Scheffter SM, Patterson JL. Hygromycin B resistance mediates elimination of Leishmania virus from persistently infected parasites. J Virol. 1997;71(12):8991–8998. doi: 10.1128/jvi.71.12.8991-8998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartley M-A, et al. Leishmaniavirus-Dependent Metastatic Leishmaniasis Is Prevented by Blocking IL-17A. PLoS Pathog. 2016;12(9):e1005852. doi: 10.1371/journal.ppat.1005852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerra JA, et al. Mucosal leishmaniasis caused by Leishmania (Viannia) braziliensis and Leishmania (Viannia) guyanensis in the Brazilian Amazon. PLoS Negl Trop Dis. 2011;5(3):e980. doi: 10.1371/journal.pntd.0000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto H, Lindoso JAL. Current diagnosis and treatment of cutaneous and mucocutaneous leishmaniasis. Expert Rev Anti Infect Ther. 2010;8(4):419–433. doi: 10.1586/eri.10.19. [DOI] [PubMed] [Google Scholar]
- 16.Adaui V, et al. Association of the endobiont double-stranded RNA virus LRV1 with treatment failure for human leishmaniasis caused by Leishmania braziliensis in Peru and Bolivia. J Infect Dis. 2016;213(1):112–121. doi: 10.1093/infdis/jiv354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pereira LdeO, et al. Severity of tegumentary leishmaniasis is not exclusively associated with Leishmania RNA virus 1 infection in Brazil. Mem Inst Oswaldo Cruz. 2013;108(5):665–667. doi: 10.1590/0074-0276108052013021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantanhêde LM, et al. Further evidence of an association between the presence of Leishmania RNA virus 1 and the mucosal manifestations in tegumentary leishmaniasis Patients. PLoS Negl Trop Dis. 2015;9(9):e0004079. doi: 10.1371/journal.pntd.0004079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito MM, et al. Correlation between presence of Leishmania RNA virus 1 and clinical characteristics of nasal mucosal leishmaniosis. Rev Bras Otorrinolaringol (Engl Ed) 2015;81(5):533–540. doi: 10.1016/j.bjorl.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourreau E, et al. Presence of Leishmania RNA virus 1 in Leishmania guyanensis increases the risk of first-line treatment failure and symptomatic relapse. J Infect Dis. 2016;213(1):105–111. doi: 10.1093/infdis/jiv355. [DOI] [PubMed] [Google Scholar]
- 21.Castellucci LC, et al. Host genetic factors in American cutaneous leishmaniasis: A critical appraisal of studies conducted in an endemic area of Brazil. Mem Inst Oswaldo Cruz. 2014;109(3):279–288. doi: 10.1590/0074-0276140028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schriefer A, Wilson ME, Carvalho EM. Recent developments leading toward a paradigm switch in the diagnostic and therapeutic approach to human leishmaniasis. Curr Opin Infect Dis. 2008;21(5):483–488. doi: 10.1097/QCO.0b013e32830d0ee8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fichorova RN, et al. Endobiont viruses sensed by the human host: Beyond conventional antiparasitic therapy. PLoS One. 2012;7(11):e48418. doi: 10.1371/journal.pone.0048418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawe AL, Nuss DL. Hypoviruses and chestnut blight: Exploiting viruses to understand and modulate fungal pathogenesis. Annu Rev Genet. 2001;35:1–29. doi: 10.1146/annurev.genet.35.102401.085929. [DOI] [PubMed] [Google Scholar]
- 25.Dawe AL, Nuss DL. Hypovirus molecular biology: From Koch’s postulates to host self-recognition genes that restrict virus transmission. Adv Virus Res. 2013;86:109–147. doi: 10.1016/B978-0-12-394315-6.00005-2. [DOI] [PubMed] [Google Scholar]
- 26.Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333(6049):1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wickner RB, Fujimura T, Esteban R. Viruses and prions of Saccharomyces cerevisiae. Adv Virus Res. 2013;86:1–36. doi: 10.1016/B978-0-12-394315-6.00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mears ER, Modabber F, Don R, Johnson GE. A review: The current in vivo models for the discovery and utility of new anti-leishmanial drugs targeting cutaneous leishmaniasis. PLoS Negl Trop Dis. 2015;9(9):e0003889. doi: 10.1371/journal.pntd.0003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armstrong TC, Keenan MC, Widmer G, Patterson JL. Successful transient introduction of Leishmania RNA virus into a virally infected and an uninfected strain of Leishmania. Proc Natl Acad Sci USA. 1993;90(5):1736–1740. doi: 10.1073/pnas.90.5.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lye L-F, et al. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010;6(10):e1001161. doi: 10.1371/journal.ppat.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Q, Niu Y, Zhang K, Liu Y, Zhou X. Virus-derived transgenes expressing hairpin RNA give immunity to tobacco mosaic virus and cucumber mosaic virus. Virol J. 2011;8(1):41. doi: 10.1186/1743-422X-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tompkins SM, Lo C-Y, Tumpey TM, Epstein SL. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci USA. 2004;101(23):8682–8686. doi: 10.1073/pnas.0402630101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cullen BR, Cherry S, tenOever BR. Is RNA interference a physiologically relevant innate antiviral immune response in mammals? Cell Host Microbe. 2013;14(4):374–378. doi: 10.1016/j.chom.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 35.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23(10):1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342(6155):231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maillard PV, et al. Antiviral RNA interference in mammalian cells. Science. 2013;342(6155):235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarr PI, et al. LR1: A candidate RNA virus of Leishmania. Proc Natl Acad Sci USA. 1988;85(24):9572–9575. doi: 10.1073/pnas.85.24.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atayde VD, et al. The structure and repertoire of small interfering RNAs in Leishmania (Viannia) braziliensis reveal diversification in the trypanosomatid RNAi pathway. Mol Microbiol. 2013;87(3):580–593. doi: 10.1111/mmi.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zangger H, et al. Detection of Leishmania RNA virus in Leishmania parasites. PLoS Negl Trop Dis. 2013;7(1):e2006. doi: 10.1371/journal.pntd.0002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138(2):215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Silva MR, et al. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol. 2010;171(2):64–73. doi: 10.1016/j.molbiopara.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Franzén O, et al. The short non-coding transcriptome of the protozoan parasite Trypanosoma cruzi. PLoS Negl Trop Dis. 2011;5(8):e1283. doi: 10.1371/journal.pntd.0001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lambertz U, et al. Small RNAs derived from tRNAs and rRNAs are highly enriched in exosomes from both old and new world Leishmania providing evidence for conserved exosomal RNA Packaging. BMC Genomics. 2015;16:151. doi: 10.1186/s12864-015-1260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cadd TL, Keenan MC, Patterson JL. Detection of Leishmania RNA virus 1 proteins. J Virol. 1993;67(9):5647–5650. doi: 10.1128/jvi.67.9.5647-5650.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sijen T, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107(4):465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 47.Vaistij FE, Jones L, Baulcombe DC. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14(4):857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ameres SL, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328(5985):1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baccarini A, et al. Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr Biol. 2011;21(5):369–376. doi: 10.1016/j.cub.2011.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung IK, et al. Generation of the short RNA transcript in Leishmaniavirus correlates with the growth of its parasite host, Leishmania. Mol Cells. 1998;8(1):54–61. [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Wu Q, Wang X, Ding SW. Viral suppressors of RNA-based viral immunity: Host targets. Cell Host Microbe. 2010;8(1):12–15. doi: 10.1016/j.chom.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiba S, Suzuki N. Highly activated RNA silencing via strong induction of dicer by one virus can interfere with the replication of an unrelated virus. Proc Natl Acad Sci USA. 2015;112(35):E4911–E4918. doi: 10.1073/pnas.1509151112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7(6):590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 55.Lambrechts L, et al. Specificity of resistance to dengue virus isolates is associated with genotypes of the mosquito antiviral gene Dicer-2. Proc Biol Sci. 2013;280(1751):20122437. doi: 10.1098/rspb.2012.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodríguez-Cousiño N, Gómez P, Esteban R. L-A-lus, a new variant of the L-A totivirus found in wine yeasts with Klus killer toxin-encoding Mlus double-stranded RNA: Possible role of killer toxin-encoding satellite RNAs in the evolution of their helper viruses. Appl Environ Microbiol. 2013;79(15):4661–4674. doi: 10.1128/AEM.00500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wickner RB, Edskes HK. Yeast killer elements hold their hosts hostage. PLoS Genet. 2015;11(5):e1005139. doi: 10.1371/journal.pgen.1005139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang AL, Wang CC. Viruses of the protozoa. Annu Rev Microbiol. 1991;45:251–263. doi: 10.1146/annurev.mi.45.100191.001343. [DOI] [PubMed] [Google Scholar]
- 59.el-Sherbeini M, Bostian KA. Viruses in fungi: Infection of yeast with the K1 and K2 killer viruses. Proc Natl Acad Sci USA. 1987;84(12):4293–4297. doi: 10.1073/pnas.84.12.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fink GR, Styles CA. Curing of a killer factor in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1972;69(10):2846–2849. doi: 10.1073/pnas.69.10.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Redmond S, Vadivelu J, Field MC. RNAit: An automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol Biochem Parasitol. 2003;128(1):115–118. doi: 10.1016/s0166-6851(03)00045-8. [DOI] [PubMed] [Google Scholar]
- 62.Robinson KA, Beverley SM. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol. 2003;128(2):217–228. doi: 10.1016/s0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- 63.Peacock CS, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39(7):839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams MJ, Lefkowitz EJ, King AM, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2014) Arch Virol. 2014;159(10):2831–2841. doi: 10.1007/s00705-014-2114-3. [DOI] [PubMed] [Google Scholar]
- 65.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]