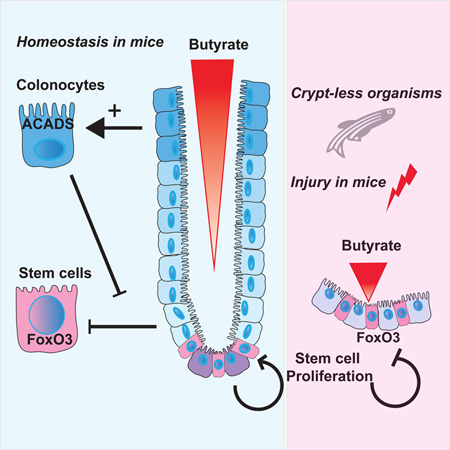
Summary
In the mammalian intestine, crypts of Leiberkühn house intestinal epithelial stem/progenitor cells at their base. The mammalian intestine also harbors a diverse array of microbial metabolite compounds that potentially modulate stem/progenitor cell activity. Unbiased screening identified butyrate, a prominent bacterial metabolite, as a potent inhibitor of intestinal stem/progenitor proliferation at physiologic concentrations. During homeostasis, differentiated colonocytes metabolized butyrate likely preventing it from reaching proliferating epithelial stem/progenitor cells within the crypt. Exposure of stem/progenitor cells in vivo to butyrate through either mucosal injury or application to a naturally crypt-less host organism led to inhibition of proliferation and delayed wound repair. The mechanism of butyrate action depended on the transcription factor Foxo3. Our findings indicate that mammalian crypt architecture protects stem/progenitor cell proliferation in part through a metabolic barrier formed by differentiated colonocytes that consume butyrate, and stimulate future studies on the interplay of host anatomy and microbiome metabolism.
Graphical Abstract
Introduction
The mammalian intestinal epithelium undergoes rapid and perpetual renewal throughout the life of the organism (Stappenbeck et al., 1998). Stem and progenitor cells that drive this process give rise to all the differentiated cell types and are housed near the base of invaginations into the intestinal wall called crypts of Lieberkühn (discovered in 1745) (van der Flier and Clevers, 2009). Host genetic programs involving Wnt, Hedgehog and Noggin signals influence the development and turnover of these stem cells (Haramis et al., 2004; Lickert et al., 2000; Wang et al., 2002). Despite knowledge of their existence for nearly three centuries, the function of the crypt structure remains unclear. It has been broadly inferred that crypts may protect rapidly dividing stem and progenitor cells from potentially damaging luminal factors, including pathogenic invasive microbes and genotoxic agents (Cheng and Leblond, 1974). However, evidence to support this idea is lacking.
The host factors regulating intestinal stem cells and their differentiated progeny include molecules commonly involved in the development of many tissues. For active, Lgr5-positive intestinal epithelial stem cells, canonical Wnts and R-spondins are critical host factors for their maintenance. (Barker et al., 2007; de Lau et al., 2011; Sato et al., 2009; van der Flier et al., 2009). BMP signaling limits the number of crypts (Haramis et al., 2004). The Notch pathway affects cell fate decisions (VanDussen and Samuelson, 2010; Yang et al., 2001). In sum, these classical host pathways interact to drive stem cell turnover and dictate cell differentiation of the intestinal epithelium. An open question is how the neighboring microbiota modulates stem cell function.
A variety of host functions including metabolism, immunity, as well as neuronal and vascular development are regulated by the intestinal microbiota (Erny et al., 2015; Kabat et al., 2014; Kaiko and Stappenbeck, 2014; Ridaura et al., 2013; Stappenbeck et al., 2002). Important mediators of these interactions can be microbial metabolites. These are small, diffusible factors capable of engaging host cells, which could facilitate their ability to modulate basic physiologic processes (Donia and Fischbach, 2015). Specific molecules influence important aspects of host metabolism (Tolhurst et al., 2012), pathogenesis of atherosclerosis (Koeth et al., 2013) and the development of immune cell subsets (Arpaia et al., 2013; Furusawa et al., 2013; Mazmanian et al., 2005; Smith et al., 2013).
Broadly, the microbiota affects the intestinal epithelium during damage. Several studies have proposed a role for the microbiota through immune cell-epithelial cross-talk in promoting intestinal epithelial repair. These pathways include important contributions from Toll-like and formyl peptide receptors in detecting broad bacterial ligands (Leoni et al., 2013; Pull et al., 2005; Rakoff-Nahoum et al., 2004). Yet, how specific microbiota-derived signals directly influence the stem/progenitor cells of the intestinal crypt remains unknown. We hypothesized that the crypt structure may act to protect stem/progenitor cells from soluble microbiota-derived signals present in the intestinal lumen. To test this idea, we took a reductionist approach to understand interactions between microbes and stem cells. Over the past decade, various approaches to study intestinal stem cells have been developed, including derivation of these cells from induced pluripotent stem cells (Spence et al., 2011) and isolating crypts for perpetual culture by adding recombinant stem cell factors including Wnt3a and R-spondin-3 (Sato et al., 2009). These approaches have led to critical breakthroughs in advancing our understanding of stem cell maintenance. However, these approaches tend to utilize heterogeneous populations of cells (both stem and differentiated) and have a low rate of turnover. Recently, we developed a system to culture large numbers of primary intestinal stem and progenitor cells (Miyoshi et al., 2012; Miyoshi and Stappenbeck, 2013), which has now enabled us to conduct high throughput functional screens.
To determine how intestinal epithelial progenitors are influenced by surrounding microbiota and their soluble metabolites, we utilized a set of metabolites that were identified as induced or produced by the microbiota in wild-type mice (Matsumoto et al., 2012). We screened these metabolites and known pathogen-associated molecular patterns (PAMPs) for their impact on stem and progenitor cell activity and discovered that the fiber fermentation product, butyrate, potently inhibited proliferation of these cells through a Foxo3-dependent mechanism. This effect was dependent on the access of butyrate to the intestinal crypt base. Butyrate-mediated suppression was only observed in mouse models of crypt perturbation or mucosal injury (involving removal of the overlying epithelium from the crypt) and in the naturally crypt-less zebrafish. We show that the overlying layer of differentiated colonocytes breaks down butyrate and shuttles it for oxidative phosphorylation. Taken together, our results lead us to propose that the mammalian crypt may have evolved, in part due to the presence of microbial metabolites, to afford protection to stem/progenitor cells via its action as a metabolic barrier.
Results
Microbial metabolite screen reveals potent suppressors of colonic epithelial stem/progenitor proliferation
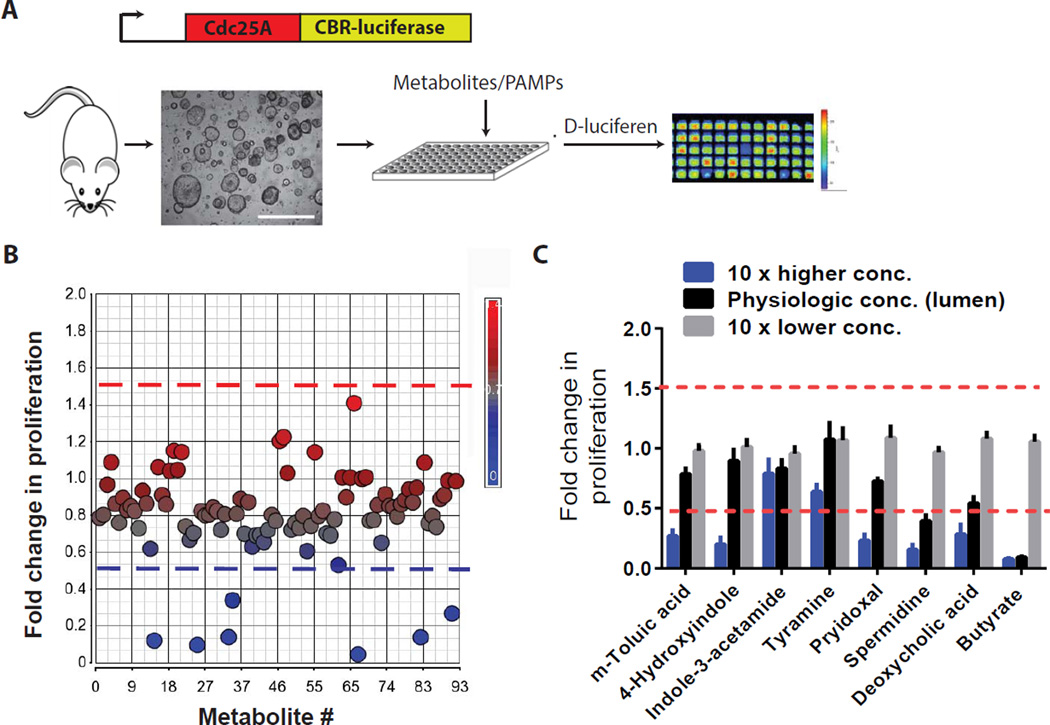
We screened a catalogue of intestinal microbial metabolites and PAMPs (92 molecules) for their effects on intestinal stem and progenitor cell activity. These metabolites are either induced or produced by the intestinal microbiota of wild-type mice (Matsumoto et al., 2012) (Table S1). They were screened on primary colonic epithelial cells isolated from Cdc25A-click beetle red luciferase reporter mice (Sun et al., 2015) grown under conditions enriched for rapidly dividing Lgr5+ stem/progenitor cells (Miyoshi et al., 2012) (Figure 1A) Cdc25A is a cell cycle phosphatase with peak protein levels during mitosis (Boutros et al., 2006); luminescence intensity correlates with cell proliferation and this can be modulated by host genetic factors (Sun et al., 2015). The initial screen revealed eight candidates that suppressed epithelial proliferation (Figure 1B). All eight metabolites suppressed proliferation under conditions enriched for stem (high Wnt signaling) and progenitor cells (diminished Wnt signaling) (Figure S1A). A secondary screen used a dose response analysis for each of the eight metabolites, based on the luminal concentration of each individual metabolite (Figure S1B). Of the eight metabolites, butyrate most potently suppressed epithelial proliferation at its physiologic concentration using three distinct measurements of proliferation (Figure 1C, 2A, 2B, S1C, S1D).
Figure 1. Large-scale screen identifies a microbial-derived metabolite as a potent suppressor of colonic epithelial stem/progenitor cells.
(A) Assay schematic for screen. Colonic stem/progenitor cells from Cdc25a-luciferase mice were cultured with individual microbial-associated metabolites and PAMPs and luminescence assayed. (B) Scatter plot displaying fold-change luminescence (24 hrs) for each individual metabolite/PAMP versus vehicle. Dashed lines indicate significance thresholds (>1.5-fold and <0.5-fold; N=4/metabolite) (C) Dose plots for each candidate metabolite identified in B. (N=4 experiments). All values, mean±SEM. See Figure S1 and Supplementary Table 1.
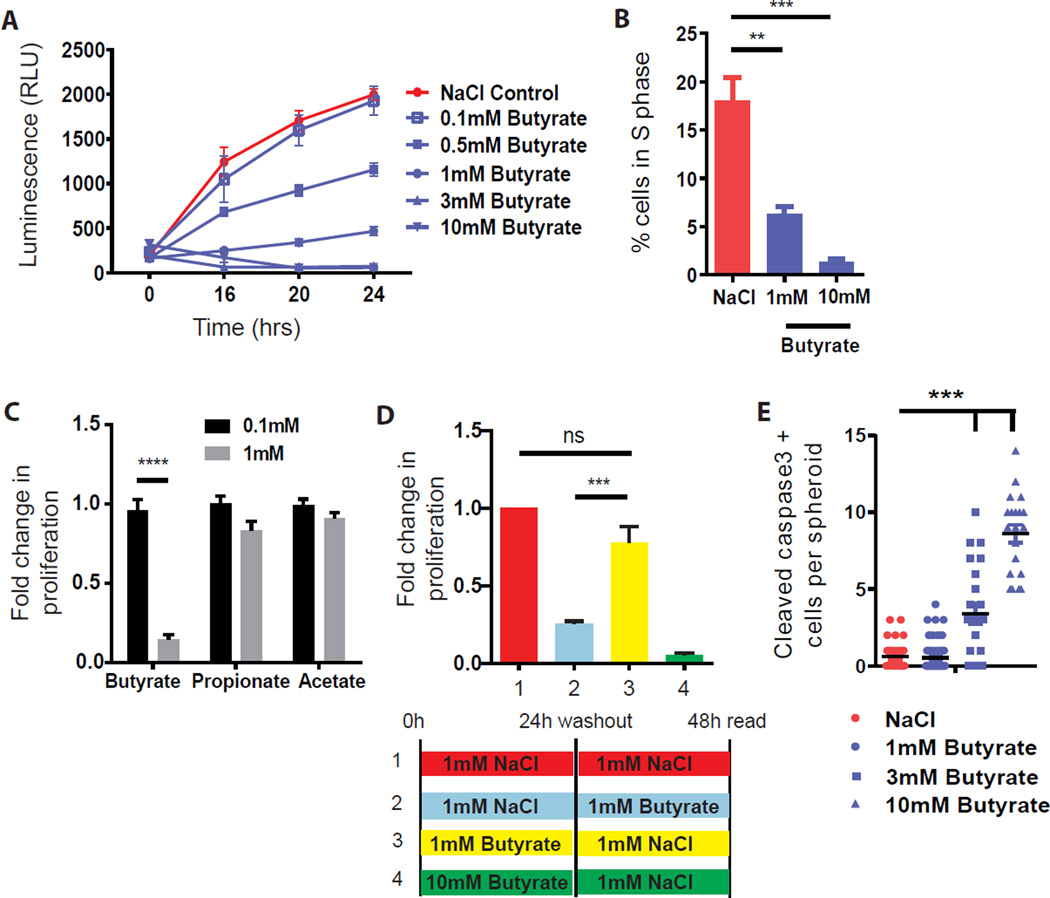
Figure 2. Butyrate but no other SCFAs suppress colonic epithelial stem/progenitor cell proliferation.
(A) Dose curve of butyrate on proliferation (N=8–20 experiments). (B) Percentage of S-phase cells 24 hrs post-treatment with NaCl or butyrate (N=3), one-way ANOVA ***p<0.001, **p<0.01. (C) Fold-change in proliferation (24 hrs) post-treatment with short chain fatty acids (butyrate, propionate, and acetate) (N=4 experiments), ANOVA ****p<0.0001 (D) Stem/progenitor cells were pre-treated for 24h with either 1mM NaCl, 1mM butyrate, or 10mM butyrate followed by a wash out, then re-incubated in NaCl or butyrate, proliferation was measured 24h later. Fold change in proliferation compared to the NaCl control (N=10 experiments), one-way ANOVA ***p<0.001, ns; not significant. (E) Number of cleaved caspase 3 positive cells/spheroid after 24 hr treatment with NaCl or butyrate (N=4–6), one-way ANOVA ***p<0.001. All values, mean±SEM. See Figure S1.
Butyrate is a product of bacterial fermentation of dietary fiber and is one of the most abundant metabolites found in the mammalian colonic lumen (~5 mM in mice and ~70 mM in humans) (Louis and Flint, 2007). The effect of butyrate on epithelial proliferation was specific amongst short chain fatty acids (SCFAs) as propionate and acetate had no effect (Figure 2C). The effect of butyrate was reversible at a dose of 1mM but became permanent at higher concentrations (3–10mM) through induction of apoptosis (Figure 2D, 2E, S1C, S1D). These findings may oppose the beneficial roles of butyrate attributed to its anti-inflammatory effects on immune cells (Arpaia et al., 2013; Chang et al., 2014; Furusawa et al., 2013; Smith et al., 2013) and suggest that it has a suppressive effect on epithelial stem/progenitor cells of the colon.
The crypt structure protects colonic stem/progenitor cells from butyrate-mediated suppression
We hypothesized that the crypt structure protects stem/progenitor cells by sequestering them from the high concentration of luminal butyrate. Alteration of luminal butyrate in mice by direct enema (increase) or antibiotics (decrease by eliminating butyrate-producing microbes) showed no effect on intestinal proliferation (Figure 3A, S2A). The latter experiment confirmed previous studies using germ-free mice (Pull et al., 2005).
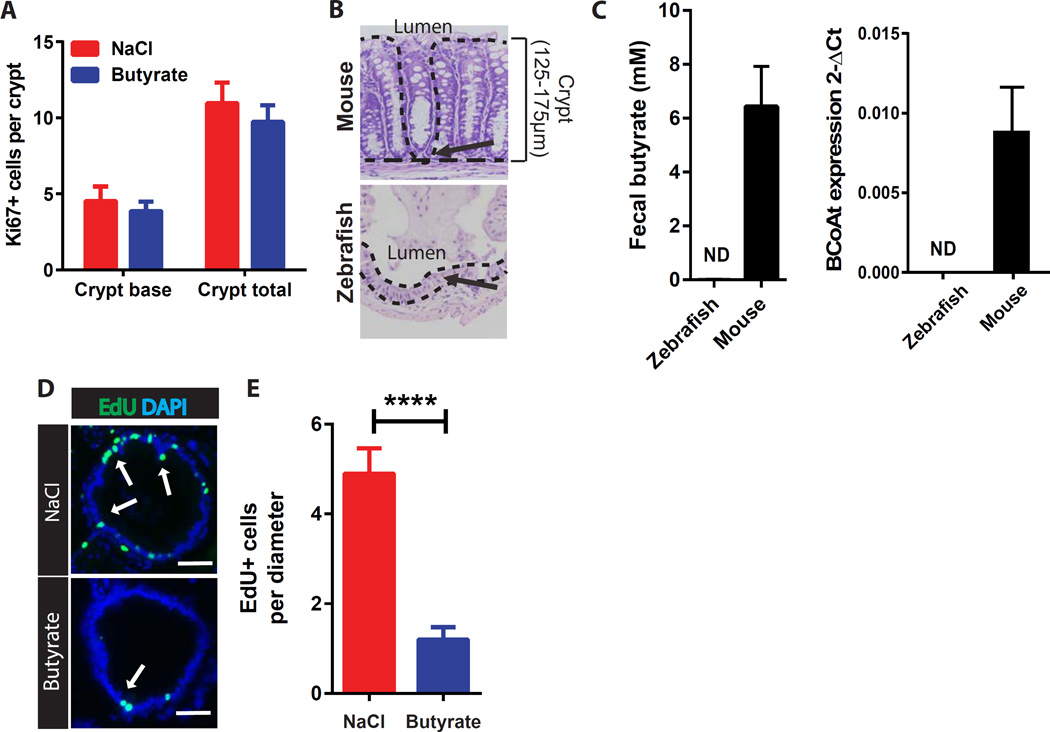
Figure 3. Colonic crypt structure protects epithelial stem/progenitor cells from butyrate-suppression.
(A) Number of Ki67+ cells per crypt at the colonic crypt base or total crypt in mice treated with enemas of NaCl or butyrate (N=5). (B) Representative images of mouse crypts and zebrafish inter-villus regions. Dashed lines indicate the epithelial architecture/crypt structure in mouse, which is absent in the zebrafish. Arrows depict the localization of stem/progenitor cells. Crypt height/distance from the lumen is also indicated. (C) Fecal butyrate concentration and level of butyryl-Coenzyme A (CoA) CoA transferase (BCoAt) in fecal samples of each organism (N=9 mice, and 5 samples pooled from 15 zebrafish). ND; not detected. (D) Representative images of EdU staining in intestinal bulge of zebrafish treated with NaCl or butyrate. White arrows indicate EdU+ epithelial cells (green). (E) Average number of EdU+ epithelial cells per intestinal diameter in zebrafish treated with 50mM NaCl or butyrate (N=8–12 zebrafish), unpaired t test ***p<0.001. All values, mean±SEM. Bars=50µm.
We next examined an experimental model system that lacks crypts but still has high epithelial turnover. Unlike mice and other mammals, zebrafish do not have crypts (Ng et al., 2005) and their intestinal stem/progenitor cells are exposed to the lumen (Figure 3B). Importantly, zebrafish also lack the bacterial organisms and enzymes that produce butyrate (Figure 3C). We observed a potent suppressive effect on epithelial proliferation within the intestinal bulge region of zebrafish exposed to butyrate (Figure 3D, 3E, S2B). This supports a role for crypts in preventing the effects of butyrate on stem/progenitor cell proliferation.
Based on the dichotomous effects of butyrate in mice and zebrafish, we hypothesized that crypts limit access of butyrate to stem/progenitor cells. To test this hypothesis in mice, we focally removed the epithelium by two methods to expose stem/progenitor cells to luminal butyrate. In mice treated with dextran sodium sulfate (DSS) to induce colonic ulcers (Pull et al., 2005), exogenous butyrate diminished epithelial proliferation in crypts adjacent to ulcers (Figure 4A 4B). This effect was also associated with an increased ulcer size, number of atrophic crypts (Figure S2C–F) and the number of CD44-positive cells in crypts surrounding ulcers (Figure S2G). This treatment did not affect epithelial apoptosis or secretory differentiation status (Figure S2H, S2I). In separate experiments, mice pretreated with metronidazole to eliminate butyrate-producing organisms (Louis and Flint, 2007) showed a decreased ulcer size with DSS treatment (Figure S2J, S2K). The addition of exogenous butyrate or fecal transplant that included butyrate-producing bacteria reversed this effect (Figure S2J, S2K). We next injured the intestinal lining with biopsy forceps to remove small, ~1mm2 fragments of the colonic mucosa (Seno et al., 2009). This model revealed similar suppressive effects of exogenous butyrate on epithelial stem/progenitor cell proliferation in crypts adjacent to the wound area (Figure 4C–E). Thus, butyrate suppresses stem/progenitor cell proliferation upon exposure.
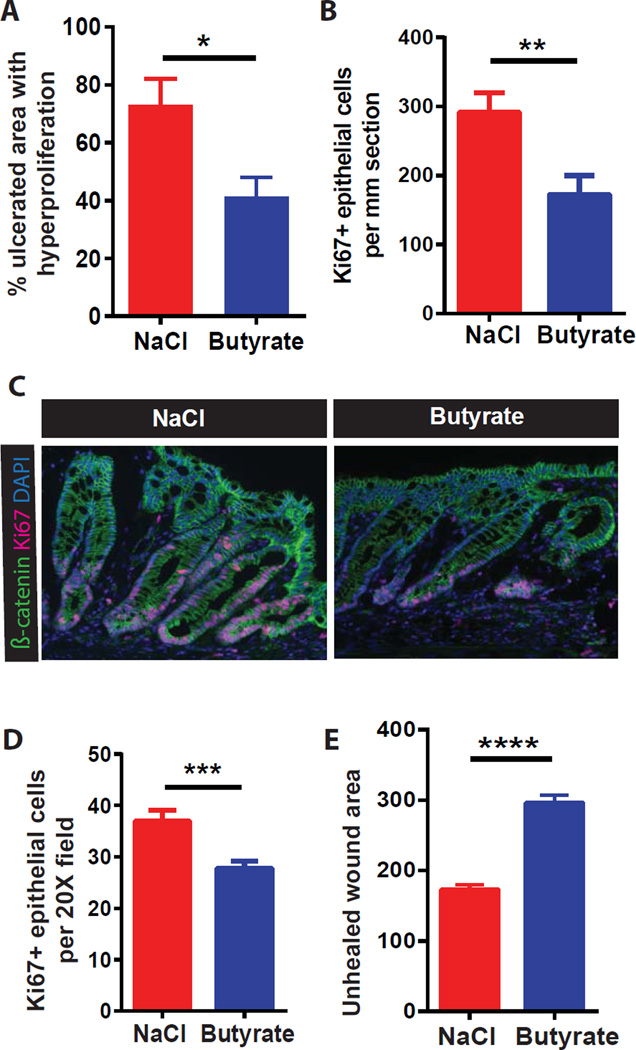
Figure 4. Mucosal injury exposes stem/progenitor cells to butyrate leading to suppression of proliferation.
(A–B) Percentage area of epithelial hyperproliferation surrounding ulcers (A) and number of Ki67+ cells (B) in DSS-treated mice +/− butyrate enema (N=8 mice/group in 2 experiments). Unpaired t-test *p<0.05, **p<0.01. (C–E) Representative images (C), number of wound-adjacent Ki67+ cells (D), and unhealed wound area (E) in mice injured by colonic biopsy +/− butyrate enema. (N=12 mice/group in 3 experiments). Unpaired t-test ***p<0.001, ****p<0.0001. All values, mean±SEM. See Figure S2.
Development of in vitro differentiated colonocytes
Our data suggest a crypt-shielding model of stem/progenitor cells that limits butyrate access to the crypt base. Colonocytes residing at the surface of the crypts are the most abundant differentiated epithelial cell type in this organ and potentially metabolize butyrate (Ruppin et al., 1980). To test if colonocytes could drive crypt protection in vitro, we developed a method to differentiate stem cells into enriched cultures of colonocytes. These in vitro differentiated cells were polarized, post-mitotic (Figure S3A) and expressed an array of differentiation markers including Aquaporin8, Carbonic anhydrase 4 (Car4), F-actin and Alkaline phosphatase (Figure S3B, S4A–D; Table S2). In addition, the cell shape and organelle composition were morphologically similar to in vivo colonocytes by TEM analysis (Figure S4E).
Colonocytes metabolize butyrate through oxidation and shield stem/progenitor cells from butyrate
In order to test whether colonocytes protect stem/progenitor cells from butyrate in vitro, we performed a supernatant transfer experiment. Cell culture media supplemented with butyrate was pre-incubated with cells grown as colonocytes or stem/progenitor. Supernatant transferred to Cdc25A-luciferase expressing stem cells showed that pre-incubation with colonocytes, but not with stem/progenitor cells, significantly decreased the amount of butyrate in the media and reversed the suppressive effect on proliferation (Figure 5A, S5A). A ~30% reduction in the high concentration of butyrate incubated with the colonocytes was sufficient to induce a greater than 2-fold reversal in suppression of proliferation. As a control, addition of the amount of butyrate consumed by the colonocytes reversed this effect (Figure 5A, S5A). Furthermore, butyrate had no effect on colonocyte apoptosis (Figure S5B). These data suggest that colonocytes protect stem/progenitor cells from butyrate.
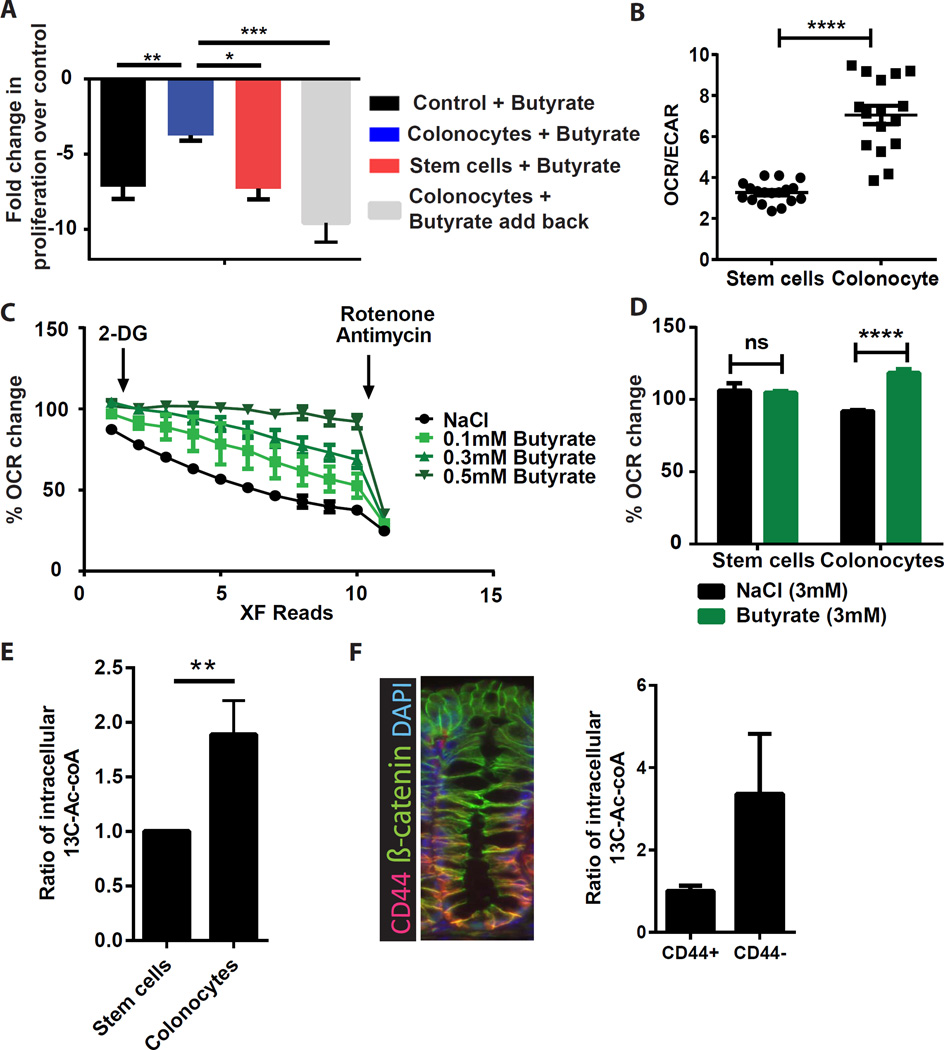
Figure 5. Colonocytes protect stem/progenitor cells by metabolic breakdown of butyrate.
(A) Fold change in proliferation at 24 hrs in Cdc25A-luciferase colonic stem/progenitor cells upon treatment with butyrate-containing supernatant pre-incubated with either: no cells, colonocytes, stem cells, or colonocytes with add back of metabolized butyrate. Fold change for each group is expressed relative to NaCl-containing supernatant pre-incubated with the relevant cell type (N=4–12), one-way ANOVA *p<0.05, **p<0.01 ***p<0.001. (B) Oxygen consumption rate (OCR) to extra cellular acidification rate (ECAR) ratio in stem/progenitor cells and colonocytes (N=18 replicates/group), unpaired t test ****p<0.0001. Data are representative of 5 experiments. (C) OCR measured in colonocytes with NaCl or butyrate. Percentage OCR changes upon 2-deoxyglucose (2-DG) and Rotenone/Antimycin injections (representative of 4 experiments). (D) Percentage change in OCR in stem/progenitor cells and colonocytes upon injection of NaCl or butyrate (N=6 replicates/group; representative of 4 experiments), 2-way ANOVA Sidak multiple comparisons ****p<0.0001. (E) Ratio of intracellular 13C2-acetyl-CoA in stem/progenitor cells versus colonocytes after incubation with 13C4-butyrate (N=12), Unpaired t-test **P<0.01. (F) Immunofluorescence image of colonic crypt stained with CD44 (red), β-catenin (green, epithelium) and DAPI (blue, nuclei). Mice received enema of 13C4-butyrate and intracellular 13C2-acetyl-CoA was detected in CD44+ and CD44− colonic epithelial fractions (N=8–10). See Figure S3, S4, and S5.
We tested the hypothesis that colonocytes mediated their protective effect by metabolizing butyrate. First, analysis of microarray data (Figure S5C, S5D) showed colonocytes were highly enriched in mRNAs encoding enzymes in the TCA cycle and lipid metabolism, suggesting that they were equipped to oxidize fatty acids, such as butyrate. Second, metabolic profiles (Figure 5B) showed that colonocytes had a significantly higher ratio of oxygen consumption rate (OCR, an indicator of oxidative phosphorylation or OXPHOS) to extracellular acidification rate (ECAR, an indicator of glycolysis) compared to stem cells. Higher OCR/ECAR ratios suggest that colonocytes have a greater reliance on OXPHOS rather than on glycolysis for ATP generation compared to stem/progenitor cells. Third, colonocytes but not stem/progenitor cells utilized butyrate as a substrate for OXPHOS. In vehicle-treated control cells, the OCR steadily dropped upon inhibition of glycolysis by 2-DG treatment. Addition of butyrate rescued and maintained OCR/OXPHOS in a dose-dependent manner. The inhibitory effect of Rotenone and Antimycin on oxygen consumption in this assay confirmed that it was due to mitochondrial OXPHOS (Figure 5C, D). This effect was specific for butyrate, as acetate and propionate had minimal effect (Figure S5E). Fourth, isotope-tracing using in vitro13C-labelled butyrate showed that colonocytes contained higher levels of 13C-labelled acetyl-CoA, an end product of fatty acid oxidation, which is subsequently utilized in the TCA cycle (Figure 5E). In vivo tracing analysis of colonic injection of 13C-butyrate showed 3-fold higher level of 13C-labeled acetyl-CoA present in the cells at the top of the crypt compared to stem/progenitor cells at the base (Figure 5F, S5F). Taken together, these data suggest that differentiated colonocytes located at the top of crypts can metabolize butyrate as an energy source, thus potentially preventing exposure of the stem cell niche to high levels of luminal butyrate.
Colonocytes shield stem/progenitor cells from butyrate through Acads-dependent oxidation
Laws of diffusion would naturally setup a butyrate gradient in the crypt, resulting in lower levels of exposure at the base as compared to the surface. However, we hypothesized that this was not the only mechanism involved in protecting stem/progenitor cells at the crypt base; rather the differentiated colonocytes could also further reduce the level of luminal butyrate reaching the crypt base by actively metabolizing butyrate. To test whether butyrate metabolism is required to protect stem/progenitor cells in vivo, we utilized mice deficient in acyl-CoA dehydrogenase (Acads), the key enzyme that converts butyrate to acetyl-CoA (Kelly and Wood, 1996). Acads is expressed in colonocytes in vivo and in vitro (Figure 6A, 6B). Metabolic profiling demonstrated a significant reduction in butyrate oxidation in Acads−/− colonocytes compared to WT cells in vitro (Figure 6C). Epithelial proliferation in Acads−/− mice showed a significant decrease in the proliferative zone in the crypt compared to WT mice. Exogenous butyrate administration further diminished the proliferative zone in Acads−/− mice in contrast to WT mice even in the absence of injury (Figure 6D–F). During DSS injury, Acads−/− mice demonstrated exaggerated suppression of stem/progenitor cell proliferation in crypts surrounding ulcers. This occurred both with and without exogenous butyrate treatment (Figure 6G, 6H, S5G). This was independent of Acads expression in stem cells as WT and Acads−/− stem cells did not metabolize butyrate and exhibited a similar level of suppression in proliferation in response to butyrate treatment (Figure S5H–I). These data suggest that the butyrate oxidation pathway in colonocytes was required to limit exposure of stem/progenitor cells to luminal butyrate.
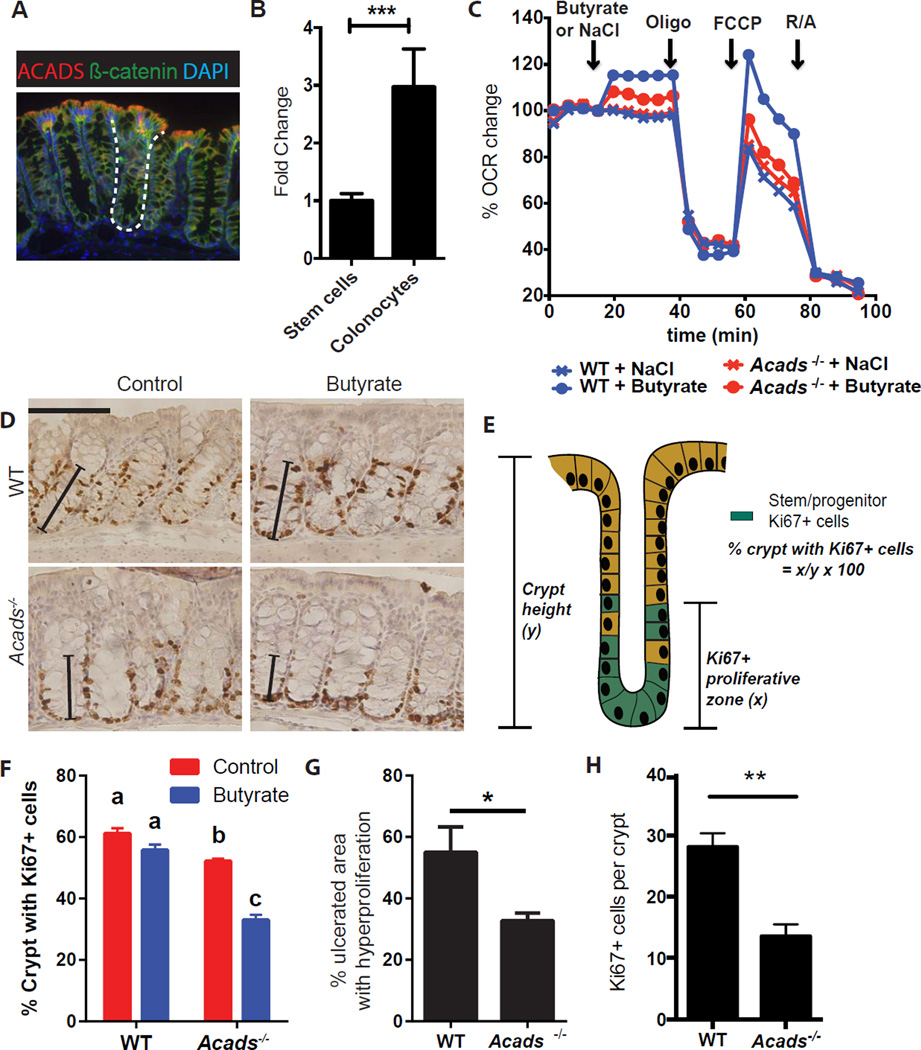
Figure 6. Colonocytes metabolize butyrate to protect underlying stem/progenitor cells through an Acads-dependent mechanism.
(A) Image of acyl CoA-dehydrogenase (Acads) (red) localization in the colon (β-catenin, green, epithelial cells; DAPI, nuclei). Dashed line marks the crypt. (B) Relative expression of Acads mRNA in stem/progenitor cells versus colonocytes generated in vitro (N=3), unpaired t test ***p<0.001. All values are mean±SEM. (C) OCR percentage in WT and Acads−/− colonocytes at basal conditions and after NaCl or butyrate, Oligomycin (Oligo), FCCP, and rotenone/antimycin (R/A) injections. Data are representative of 2 experiments. (D–F) Crypt proliferation in WT and Acads−/− mice. Images (D) and percentage of crypts with Ki67+ epithelial cells (F) in WT and Acads−/− mice at baseline and after butyrate enema. Solid line indicates proliferative zone with Ki67+ cells. Bar=100µm. (Two way ANOVA, Means with different letters are significantly different by Tukey's multiple comparison test). Percentage of crypt with Ki67+ cells calculated as the distance between crypt base and Ki67+ cells at the highest position (E). (G–H) Percentage area of hyperproliferation around the ulcers (G) and number of Ki67+ cells (H) in DSS-treated WT and Acads−/− mice with butyrate enemas on days 5–7 (N=8 mice/group, 2 experiments). Unpaired t test *p<0.05. All values, mean±SEM. See Figure S5.
Butyrate suppresses stem/progenitor cells through a Foxo3-dependent mechanism
To investigate the mechanism of proliferation suppression by butyrate, we examined two candidate pathways: i) stimulation of G-protein coupled receptors (GPCRs) or ii) inhibition of histone deacetylase (HDAC) enzymes (Arpaia et al., 2013; Furusawa et al., 2013; Mohammad, 2015). First, GPCR signaling was blocked with Gi (Pertussis toxin) and Gq signaling (U73122) inhibitors (Figure S6A). Secondly, colonic stem/progenitor cells were generated from mice that genetically over-express an endogenous GPCR inhibitor (Figure S6B) (Regard et al., 2007). In both cases, butyrate suppression of proliferation was not altered (Figure S6A, S6B). These results suggest that the effect of butyrate was GPCR-independent.
In contrast, we found that butyrate directly inhibited nucleic HDAC activity in a cell free system in a dose-dependent manner (Figure 7A). Supporting this finding, butyrate increased acetylation in stem/progenitor cells at both the histone H3K27 and H3K9 sites (Figure 7B). In addition, an expanded panel of HDAC inhibitors phenocopied the suppressive effect of butyrate on stem/progenitor cells (Figure S6C). The prototypical pan-HDAC inhibitor, Trichostatin A also suppressed epithelial proliferation in zebrafish, suggesting that the mechanism of butyrate-mediated suppression through HDAC inhibition was conserved (Figure S6D). To explore this mechanism further, we sought to identify a master regulator or transcription factor driving the effect of butyrate on stem cells. We performed genome wide ChIP-seq and FAIRE-seq of butyrate-treated colonic stem/progenitor cells to identify sites with increased H3K27 acetylation suggestive of more accessible chromatin. We found ~900 such regions within 2kb of transcriptional start sites (Figure 7C). Parallel analysis of gene expression showed that butyrate treatment was associated with alterations in transcript abundance for >2400 genes (Figure 7D). We then used predictive algorithms to identify the common DNA binding motifs enriched in both the upstream promoter regions identified in the ChIP-seq, and the promoter regions of genes with altered expression after butyrate treatment (Figure S6E).
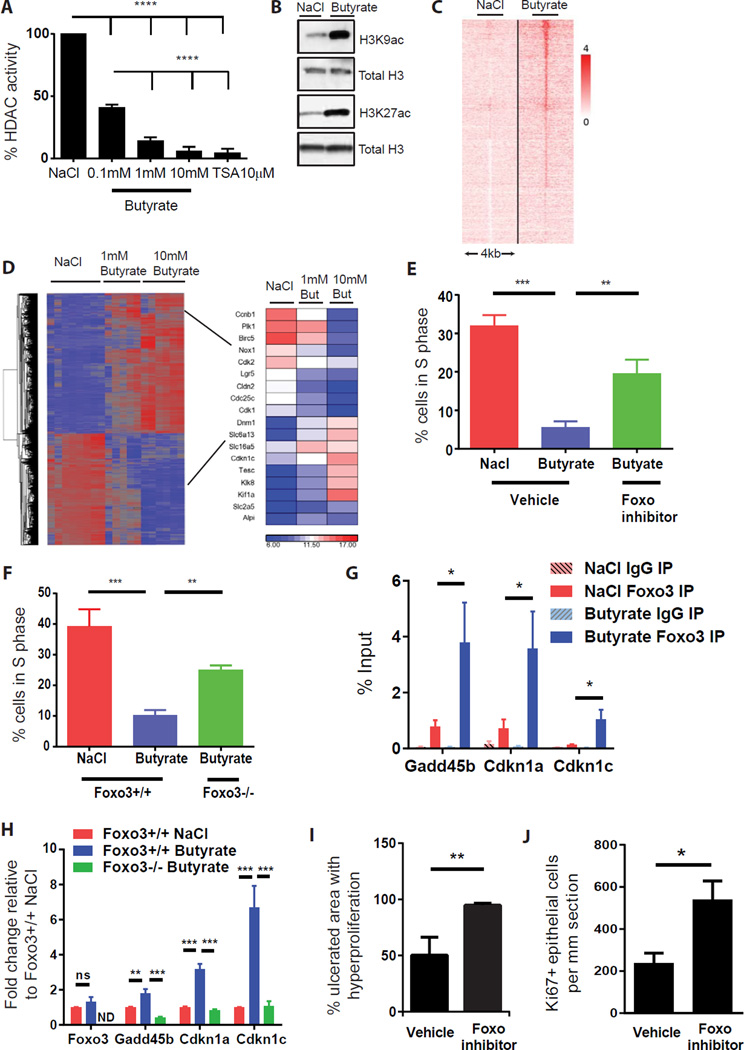
Figure 7. Butyrate suppresses stem and progenitor cell proliferation through a Foxo3-dependent mechanism.
(A) HDAC activity in colonic stem/progenitor cell nuclei after treatment with butyrate or HDAC inhibitor Trichostatin A (TSA; 10µM) (N=5 experiments) one-way ANOVA ****p<0.0001. (B) Immunoblot of acetylated (ac) Histone H3K27 and H3K9 residues in stem/progenitor cells post-treatment with 1mM butyrate or NaCl. Total H3 is loading control (representative of 3 experiments). (C) H3K27acetylation peaks at 938 sites (2 kb upstream or 2 kb downstream of any TSS) were significantly altered by butyrate (1mM) treatment of stem/progenitor cells. (D) Hierarchical clustering and heatmap analysis (most up- or down-regulated gene changes) from gene expression arrays for stem/progenitor cells treated with NaCl (1mM) or butyrate (1mM or 10mM). (E) Percentage of stem/progenitor cells in S-phase after treatment with butyrate or NaCl (1mM) +/− Foxo inhibitor (1µM) (N=6 experiments), one-way ANOVA **p<0.01. (F) Percentage of cells in S phase in Foxo3-deficient cells upon butyrate treatment (1mM) (N=5 experiments), one-way ANOVA **p<0.01. (G) ChIP pull down using anti-Foxo3 or isotype control IgG post-treatment with NaCl or butyrate (1mM) followed by qPCR of indicated gene promoter regions. (H) mRNA expression of the indicated genes in Foxo3flox/flox and Foxo3-deficient cells post-NaCl or butyrate treatment. ns: not significant, ND: not detected. (I–J) Percentage area of hyperproliferation (I) and number of Ki67+ cells (J) around ulcers in DSS-treated WT mice receiving enemas of butyrate +/− Foxo inhibitor (N=5–9 mice/group in 2 experiments). Unpaired t test *p<0.05,**p<0.01. All values mean±SEM. See Figure S6 and S7.
We focused on transcription factors that regulate cell cycle genes. This analysis identified Foxo1 and Foxo3 transcription factors as highly predicted candidates. We functionally validated these factors by pharmacologic and genetic inhibition. A compound that inhibits both Foxo1 and Foxo3 activity (Nagashima et al., 2010) significantly reversed the effects of butyrate (Figures 7E, S7A). In addition, colonic stem/progenitor cells with genetic ablation of Foxo3, but not Foxo1, showed strong resistance to the effects of butyrate (Figures 7F, S7B). This effect appeared to be specific to the Foxo3 transcription factor as inhibition of other known negative cell cycle regulators such as p53, TGF-β, p300/CBP, retinoic acid receptor-β, and microrna-34 were all unable to reverse the suppressive effects of butyrate (Figures S7C, S7D). Furthermore, inhibition of PI3kinase (the upstream negative regulator of Foxo3) also led to a reduction in proliferation in these stem cells (Figure S7E). Butyrate did not affect the levels of nuclear Foxo3 (Figure S7F). This suggested that butyrate instead increased the activity and potentially the promoter binding of this transcription factor. We confirmed the role of Foxo3 by ChIP assays identifying increased binding of Foxo3 to the promoter regions of the negative cell cycle regulators Cdkn1a, Cdkn1c, and Gadd45b following butyrate treatment (Figure 7G). This increased binding led to a corresponding increase in mRNA expression for all 3 genes following butyrate treatment of colonic stem/progenitor cell, which was reversed in Foxo3-deficient cells. (Figure 7H). Finally, to test the role of this transcription factor in vivo we treated mice undergoing mucosal DSS injury with enemas of the Foxo inhibitor and showed that this reversed the effect of butyrate on epithelial proliferation (Figure 7I, 7J). These data show that butyrate acts on stem/progenitor cells to acetylate histones and induces a Foxo3-dependent suppression of proliferation.
Discussion
By screening metabolites using a primary intestinal stem cell system, this study has assessed the function of intestinal microbial metabolites in stem cell biology. We discovered that butyrate potently suppresses proliferation by acting as a HDAC inhibitor and enabling increased promoter activity for the negative cell cycle regulator Foxo3. The anti-proliferative effect was dependent on exposure of the stem cell niche (i.e. disruption of crypts by injury or in organisms that lack crypts) to luminal butyrate as the presence of the intact crypt structure prevented this effect. Metabolic analyses revealed that colonocytes metabolized butyrate through Acads-dependent oxidative phosphorylation to protect the underlying stem/progenitor cells from butyrate exposure. This study suggests that certain organisms have an architectural mechanism to prevent detrimental effects of microbial cues on stem/progenitor cells, while utilizing them as an energy source through metabolic synergy. Given the correlation between the presence of the butyrate-producing bacteria and the presence of the crypt structure, it is intriguing to speculate that this prominent microbial metabolite may have exerted a selective pressure on the host developmental programs to generate crypts during co-evolution of the microbiota with its host.
Over the past decade, many advances have been made in understanding the composition and structure of the microbiome (Integrative, 2014). Only recently however, has attention switched to understanding the function of the individual components of the microbiota, that is, the microorganisms themselves and the molecules they produce. Several studies have shown important effects of individual members of the commensal community, as well as families of bacteria (Atarashi et al., 2013; Ivanov et al., 2009). A handful of studies have also examined the function of a small number of highly abundant metabolites produced by the microbiota (Trompette et al., 2014). Here, we used an unbiased screen to examine the function of individual microbial metabolites that are detectable in the intestinal lumen. We anticipate the discovery of additional gut microbial metabolites that will be of utility in future studies. The catalog of metabolites utilized here consisted of those that were either induced in the host by the presence of the microbiota, or produced and secreted directly by the microbiota itself (Matsumoto et al., 2012). To screen these molecules, we developed a high throughput assay using the primary epithelial culture system developed in our lab (Miyoshi et al., 2012). The ability to generate large numbers of enriched stem/progenitor cells as well as differentiated colonocytes was essential to understand the epithelial response to specific metabolites. We believe that this system serves as a platform that may help guide future efforts to investigate the function of individual components of the microbiota.
This study identified butyrate as the most potent microbial metabolite affecting stem/progenitor proliferation. This metabolite is a single component of a complex network of host-microbial interactions. Yet, the effect of this abundant metabolite occurs despite the pro-proliferative effects of global microbiota recognition by pattern recognition receptors (Pull et al., 2005; Rakoff-Nahoum et al., 2004). We found that butyrate delayed the stem cell expansion in the crypt base during mucosal injury, where the overlying colonocytes are damaged. Although this negatively affects wound repair in the short term, it may actually benefit the host in the long-term. By suppressing the rapid expansion of stem cells following mucosal damage of the epithelium, butyrate may prevent stem cells from dividing while in direct contact with genotoxic luminal contents. Therefore, this would reduce the risk of cancerous transformation of colonic stem cells (Barker et al., 2009). The initial delay in stem cell expansion would allow an epithelial layer to cover the wound/ulcer (Miyoshi et al., 2012) while blocking exposure to the luminal contents thus creating a safe environment for the expansion of stem cells.
We show that colonocytes break down butyrate using the oxidative phosphorylation machinery to provide a major source of their energy requirement. During homeostasis, this process helps to protect stem cell turnover from butyrate-mediated suppression, thus maintaining the epithelial barrier. However, this process is not binary. Rather, it is a process of titration whereby the surface colonocytes significantly reduce the level of butyrate (but not eliminate it) before it reaches the stem cell base and immune cells of the lamina propria. We found that even a 3-fold reduction in butyrate completely protected stem cells. In this manner, the surface layer of differentiated colonocytes appears to act as gatekeepers absorbing the majority of the abundant butyrate present at the surface. Our data suggests that active metabolism of butyrate by colonocytes, together with a natural limiting gradient by diffusion along the crypt axis, protects the stem/progenitor cell base. Only relatively small amounts of butyrate pass through these cells to exert beneficial anti-inflammatory and immune tolerogenic effects on such cells as Tregs and macrophages (Arpaia et al., 2013; Chang et al., 2014; Furusawa et al., 2013; Smith et al., 2013).
As colonocytes occupy an enormous surface area with fast turnover (3–4 days in mice) (Stappenbeck et al., 1998), this represents a large energy demand on the host that would act as a large sinkhole for glucose, sequestering it away from other vital systemic organs. To resolve this issue, it is likely that the mammalian host developed colonocytes and crypts with the ability to source their tremendous energy demands from butyrate produced by the neighboring microbiota. Studies in germ-free mice suggest that the colonic epithelial layer is under metabolic stress and upregulates the autophagy pathway for survival (Donohoe et al., 2011). Administration of butyrate reverses autophagy in colonocytes. Our study may provide insights as to why colonocytes show high specificity for preferential breakdown of butyrate rather than the other SCFAs propionate and acetate, which are also highly abundant in the colon. We speculate that although all three of these SCFAs can be used as substrates for the TCA cycle, colonocytes mainly utilize butyrate as its levels need to be titrated down to prevent their effect on stem cells. Whereas butyrate potently suppressed stem cell proliferation, propionate and acetate were not able to do so. Therefore, it appears likely that colonocytes upregulate the enzymes specifically involved in the metabolism of butyrate as opposed to those involved in the metabolism of propionate and acetate. From our data, this appears to be the case. For example, enzymes specifically involved in the breakdown of propionate to produce acetyl-CoA, such as aldehyde dehydrogenase family 6, subfamily A1 (Aldh6a1), are expressed at relatively low levels in the colonocyte cultures compared to butyrate-metabolizing enzymes. Furthermore, there is also a need to allow larger concentrations of propionate and acetate to pass through the epithelial barrier as these molecules contribute heavily to metabolic homeostasis in the liver and other systemic organs (Weidemann et al., 1970).
Using global ChIP-seq and transcriptional analysis, we identified that the Foxo3 transcription factor, in part, mediated the suppressive effect of butyrate on stem cells. Butyrate treatment led to increased binding of Foxo3 to several key cell cycle genes. Furthermore, as butyrate acted to inhibit HDAC activity, it is plausible that this increased acetylation of histones facilitated the increased access of Foxo3 to alter cell cycle gene expression. Foxo3 is known to play a key role in halting cell cycle progression (Paik et al., 2007; Ramaswamy et al., 2002). For example, the Foxo family of transcription factors mediates quiescence and enhanced survival in hematopoietic stem cells (Tothova et al., 2007). This suppressive function of Foxo3 is important to maintain the long-term regenerative potential of stem cells in the hematopoietic compartment. Finally, SNPs in Foxo3 have recently been implicated as a genetic correlate in the progression of inflammatory bowel disease, suggesting that this is a key transcription factor in the maintenance of intestinal homeostasis (Lee et al., 2013). These known activities of Foxo3 support our overall model. Given the suppressive effects of butyrate on stem cells during injury, Foxo3 would be capable of slowing cell cycling in order to reduce potentially genotoxic damage from oxidative stress.
Based on extensive microbial profiling of a variety of eukaryotic organisms, a major theory was developed that microbes and their products played an important role in the evolution of their hosts (McFall-Ngai et al., 2013). In particular, intestinal development, with its regional specification, was predicted to be a result of the host organ’s interactions with microbes that cohabit the space. Our study with a microbial metabolite, butyrate, which perturbs crypt cell proliferation, may support this larger biological question. Based on the results of our experiments in mice and zebrafish, we speculate a model whereby butyrate may have influenced crypt development during evolution. Future applications of such experimental systems will permit us to probe more deeply into the theory of host-microbial co-evolution.
Experimental Procedures
Mice
All experimental procedures were performed under approval by Washington University’s Animal Studies Committee. C57BL/6J, BALB/cJ, BALB/cBYJ (Acads−/−), Foxo1-floxed, and Foxo3-floxed mice were purchased from Jackson Laboratories and bred in house. All experiments used littermate controls.
Mouse treatments
Mice received ad libitum antibiotics cocktail of vancomycin, neomycin, ampicillin and metronidazole (VNAM), or metronidazole alone. For DSS experiments, mice received 2.5% DSS in drinking water for 7 days. Mucosal colorectal injuries were created using an endoscopic-guided biopsy system as previously described (Seno et al., 2009). Mice received intra-rectal administration of sodium chloride or sodium butyrate (150 µmoles; twice a day) from day 5–7 (DSS) or day 1–4 (biopsy). Detailed methods are described in Supplemental Experimental Procedures.
Primary intestinal epithelial cell culture
Mouse and human colonic and small intestinal crypts were isolated and cultured in 3D Matrigel as previously described (Miyoshi et al., 2012; Miyoshi and Stappenbeck, 2013). For colonocyte differentiation, culture media was switched to differentiation media 24 hours after passage. Detailed methods are described in Supplemental Experimental Procedures.
Metabolic assays
Metabolic profiling of primary epithelial cells was performed using XF96e analyzer (Seahorse Bioscience). Detailed methods are described in Supplemental Experimental Procedures.
Zebrafish experiments
Day 5 post-fertilization AB* WT larvae were treated with sodium chloride or sodium butyrate as indicated. Epithelial proliferation was quantified by EdU staining as described in Supplemental Experimental Procedures.
ChIP-seq
ChIP-seq was performed to identify promoter sites with increased H3K27 acetylation in epithelial stem cells using anti-acetylated H3K27 antibody. ChIP-seq data available at GEO accession number GSE74601. Microarray data available at ArrayExpress accession number EMTAB-4005. Detailed methods are described in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
CCFA, NIH (DK071619) and WashU DDRCC (P30DK052574) supported this work. We thank Wandy Beatty (EM), Maxene Ilagan (screening, P30CA91842), Brad Evans (Danforth, mass spectrometry, Dallas Donohoe (U. Tenn. scientific discussion), William Sly (SLU, Car4 antibody) and Stephen Canter (zebrafish).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors with no conflict of interest.
Author Contributions
GEK, SHR, LSK, EJP, ELP, EMO and TSS designed experiments, analyzed results, and prepared the manuscript. OIK and PLC designed experiments and analyzed the results.
References
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Boutros R, Dozier C, Ducommun B. The when and wheres of CDC25 phosphatases. Current opinion in cell biology. 2006;18:185–191. doi: 10.1016/j.ceb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. The American journal of anatomy. 1974;141:537–561. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- Donia MS, Fischbach MA. HUMAN MICROBIOTA Small molecules from the human microbiota. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, Bultman SJ. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell metabolism. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015;18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Haramis AP, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJ, Clevers H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004;303:1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- Integrative HMPRNC. The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell host & microbe. 2014;16:276–289. doi: 10.1016/j.chom.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat AM, Srinivasan N, Maloy KJ. Modulation of immune development and function by intestinal microbiota. Trends Immunol. 2014;35:507–517. doi: 10.1016/j.it.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiko GE, Stappenbeck TS. Host-microbe interactions shaping the gastrointestinal environment. Trends Immunol. 2014;35:538–548. doi: 10.1016/j.it.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly CL, Wood PA. Cloning and characterization of the mouse short-chain acyl-CoA dehydrogenase gene. Mammalian genome : official journal of the International Mammalian Genome Society. 1996;7:262–264. doi: 10.1007/s003359900078. [DOI] [PubMed] [Google Scholar]
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature medicine. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Espeli M, Anderson CA, Linterman MA, Pocock JM, Williams NJ, Roberts R, Viatte S, Fu B, Peshu N, et al. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell. 2013;155:57–69. doi: 10.1016/j.cell.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni G, Alam A, Neumann PA, Lambeth JD, Cheng G, McCoy J, Hilgarth RS, Kundu K, Murthy N, Kusters D, et al. Annexin A1, formyl peptide receptor, and NOX1 orchestrate epithelial repair. The Journal of clinical investigation. 2013;123:443–454. doi: 10.1172/JCI65831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Domon C, Huls G, Wehrle C, Duluc I, Clevers H, Meyer BI, Freund JN, Kemler R. Wnt/(beta)-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development. 2000;127:3805–3813. doi: 10.1242/dev.127.17.3805. [DOI] [PubMed] [Google Scholar]
- Louis P, Flint HJ. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol. 2007;73:2009–2012. doi: 10.1128/AEM.02561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Kibe R, Ooga T, Aiba Y, Kurihara S, Sawaki E, Koga Y, Benno Y. Impact of intestinal microbiota on intestinal luminal metabolome. Sci Rep. 2012;2:233. doi: 10.1038/srep00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, et al. Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Ajima R, Luo CT, Yamaguchi TP, Stappenbeck TS. Wnt5a potentiates TGF-beta signaling to promote colonic crypt regeneration after tissue injury. Science. 2012;338:108–113. doi: 10.1126/science.1223821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad S. Role of Free Fatty Acid Receptor 2 (FFAR2) in the Regulation of Metabolic Homeostasis. Curr Drug Targets. 2015;16:771–775. doi: 10.2174/1389450116666150408103557. [DOI] [PubMed] [Google Scholar]
- Nagashima T, Shigematsu N, Maruki R, Urano Y, Tanaka H, Shimaya A, Shimokawa T, Shibasaki M. Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Molecular pharmacology. 2010;78:961–970. doi: 10.1124/mol.110.065714. [DOI] [PubMed] [Google Scholar]
- Ng AN, de Jong-Curtain TA, Mawdsley DJ, White SJ, Shin J, Appel B, Dong PD, Stainier DY, Heath JK. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Developmental biology. 2005;286:114–135. doi: 10.1016/j.ydbio.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer cell. 2002;2:81–91. doi: 10.1016/s1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- Regard JB, Kataoka H, Cano DA, Camerer E, Yin L, Zheng YW, Scanlan TS, Hebrok M, Coughlin SR. Probing cell type-specific functions of Gi in vivo identifies GPCR regulators of insulin secretion. The Journal of clinical investigation. 2007;117:4034–4043. doi: 10.1172/JCI32994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppin H, Bar-Meir S, Soergel KH, Wood CM, Schmitt MG., Jr Absorption of short-chain fatty acids by the colon. Gastroenterology. 1980;78:1500–1507. [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Seno H, Miyoshi H, Brown SL, Geske MJ, Colonna M, Stappenbeck TS. Efficient colonic mucosal wound repair requires Trem2 signaling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:256–261. doi: 10.1073/pnas.0803343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science (New York, NY) 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck TS, Wong MH, Saam JR, Mysorekar IU, Gordon JI. Notes from some crypt watchers: regulation of renewal in the mouse intestinal epithelium. Current opinion in cell biology. 1998;10:702–709. doi: 10.1016/s0955-0674(98)80110-5. [DOI] [PubMed] [Google Scholar]
- Sun L, Miyoshi H, Origanti S, Nice TJ, Barger AC, Manieri NA, Fogel LA, French AR, Piwnica-Worms D, Piwnica-Worms H, et al. Type I interferons link viral infection to enhanced epithelial turnover and repair. Cell host & microbe. 2015;17:85–97. doi: 10.1016/j.chom.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61:364–371. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nature medicine. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- VanDussen KL, Samuelson LC. Mouse atonal homolog 1 directs intestinal progenitors to secretory cell rather than absorptive cell fate. Developmental biology. 2010;346:215–223. doi: 10.1016/j.ydbio.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LC, Nassir F, Liu ZY, Ling L, Kuo F, Crowell T, Olson D, Davidson NO, Burkly LC. Disruption of hedgehog signaling reveals a novel role in intestinal morphogenesis and intestinal-specific lipid metabolism in mice. Gastroenterology. 2002;122:469–482. doi: 10.1053/gast.2002.31102. [DOI] [PubMed] [Google Scholar]
- Weidemann MJ, Hems R, Williams DL, Spray GH, Krebs HA. Gluconeogenesis from propionate in kidney and liver of the vitamin B12-deficient rat. The Biochemical journal. 1970;117:177–181. doi: 10.1042/bj1170177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.