Abstract
The etiology of acute respiratory tract illnesses is sometimes unclear due to limitations of diagnostic tests or the existence of as-yet-unidentified pathogens. Here we describe the identification and characterization of a not previously recognized coronavirus obtained from an 8-mo-old boy suffering from pneumonia. This coronavirus replicated efficiently in tertiary monkey kidney cells and Vero cells, in contrast to human coronaviruses (HCoV) 229E and OC43. The entire cDNA genome sequence of the previously undescribed coronavirus was determined, revealing that it is most closely related to porcine epidemic diarrhea virus and HCoV 229E. The maximum amino acid sequence identity between ORFs of the newly discovered coronavirus and related group 1 coronaviruses ranged from 43% to 67%. Real-time RT-PCR assays were designed to test for the prevalence of the previously undescribed coronavirus in humans. Using these tests, the virus was detected in four of 139 individuals (3%) who were suffering from respiratory illness with unknown etiology. All four patients suffered from fever, runny nose, and dry cough, and all four had underlying or additional morbidity. Our data will enable the development of diagnostic tests to study the prevalence and clinical impact of this virus in humans in more detail. Moreover, it will be important to discriminate this previously undescribed coronavirus from HCoV 229E and OC43 and the severe acute respiratory syndrome coronavirus.
Acute respiratory tract infections are responsible for considerable morbidity and mortality in humans and animals, and the costs attributable to acute respiratory tract illnesses (RTI) in humans are an important burden on national health care budgets (1). A variety of viruses, bacteria, and fungi are associated with RTI (2), with most of the viruses belonging to the families of Paramyxoviridae, Orthomyxoviridae, Picornaviridae, Adenoviridae, and Coronaviridae. In a relatively large proportion of samples obtained from persons suffering from RTI, no pathogens can be detected despite the use of a wide range of sensitive diagnostic assays; even in the most comprehensive studies, a causative agent (either viral or nonviral) can be identified in only 85% of the patients (3). In part, this may be due to the limitations of diagnostic assays, but a proportion of RTI may be caused by still unknown pathogens. The recent discovery of the human metapneumovirus, a significant human pathogen, is an example of the long-undetected circulation of an unidentified RTI-associated viral pathogen in the human population (4).
Over the past decades, our virus diagnostic laboratory has tested clinical specimens collected from individuals suffering from RTI for the presence of viruses. Virus isolation was performed initially on tertiary monkey kidney (tMK) and subsequently in a variety of cell lines such as Vero, HeLa, and HEp-G2 cells. Although for many cases the etiological agent could be identified, some specimens yielded cytopathic effects (CPE) in cell cultures but were negative for known viruses (4). Here we describe the isolation and characterization of a recently discovered coronavirus, obtained from an 8-mo-old boy suffering from pneumonia in The Netherlands in 1988. The complete genomic cDNA sequence revealed that this virus is related to human and animal group-1 coronaviruses. Using a real-time RT-PCR assay, we detected the virus in four additional children who were suffering from RTI.
Methods
Virus Isolation, Propagation, and Characterization. Virus isolation was performed in tMK cells in Eagle's MEM with Hanks' salt (BioWhittaker), supplemented with 0.0001% trypsin (BioWhittaker) and without serum. After inoculation, the plates were incubated at 37°C for a maximum of 14 days, during which time the medium was changed twice a week and cultures were checked daily for CPE. Vero cells were grown in Iscove's modified Dulbecco's medium (BioWhittaker) supplemented with 10% FBS/100 units/ml penicillin/100 mg/ml streptomycin/2 mM glutamine. For virus replication, Vero cells were grown in DMEM (BioWhittaker) supplemented with 0.3% BSA fraction V/100 units/ml penicillin/100 mg/ml streptomycin/2 mM glutamine/0.0004% trypsin. Of note, the addition of trypsin to the cultures is a standard procedure, and the trypsin dependence of the virus isolate was not investigated. Infected cell culture supernatants were inspected by negative contrast electron microscopy after potassium tungstat acid staining (5).
Arbitrarily Primed PCR and RT-PCR. RNA was isolated from infected Vero cell supernatants by using a High Pure RNA Isolation kit according to instructions from the manufacturer (Roche Diagnostics, Almere, The Netherlands). RT-PCR assays for HCoV-OC43, HCoV-229E were performed as described (6). Broad reacting primer sets specific for the Coronaviridae family were also used (7, 8). RNA arbitrarily primed PCR was performed essentially as described (4, 9, 10), and primer sequences are available on request. PCR fragments were purified from agarose gels with a QIAquick Gel Extraction kit (Qiagen, Leusden, The Netherlands), cloned in vector pCR2.1 (Invitrogen) according to instructions from the manufacturer and sequenced with M13-specific oligonucleotides. Sequencing was performed by using the Big Dye terminator sequencing kit version 3.1 (Amersham Pharmacia Biotech) and an ABI Prism 3100 genetic analyzer (Applied BioSystems) according to the instructions of the manufacturer.
Genome Sequence Analysis. The complete cDNA sequence of the viral genome was determined by a combination of shotgun sequencing, RT-PCR, and RACE. Virus was concentrated from infected Vero cell supernatants by using sucrose gradients and purified viral RNA was converted to cDNA by using the SuperScript Choice system (Invitrogen) by random priming according to the manufacturer's instructions. Blunt-ended double-stranded cDNA fragments were size-selected on agarose gel to include fragments ranging from 750 bp to 4 kb. After purification with spin columns (Zymo Research, Orange, CA), cDNA fragments were ligated into pSMART-HCAmp (Lucigen, Middleton, WI). The resulting library was electroporated into DH10B ElectroMAX cells (Invitrogen). Inserts from the shotgun library were amplified from individual colonies by using pSMART AmpL1 and AmpR1 primers. In total, inserts were amplified from 175 different colonies. Gaps that remained after initial assembly of shotgun clones were closed by amplification and sequencing of RT-PCR products that were obtained from viral RNA by using specific forward and reverse primers and either the RNA LA PCR or BcaBEST RNA PCR kit (Takara Bio, Shiga, Japan). The 3′ end of the viral genome was determined by using RACE, with the First Choice RLM-RACE kit (Ambion, Austin TX). To determine the 5′ terminal sequence, viral RNA was reverse-transcribed by using specific primers proximal to the putative 5′ end with the RNA LA PCR kit (Takara Bio), after which single-stranded cDNA fragments were elongated with terminal transferase and dCTP and amplified by using a polydG primer and specific reverse primers. All PCR fragments were sequenced by using BigDye Ver. 3.1 chemistry (Amersham Pharmacia Biotech). Primer sequences are available on request. Sequence reactions were cleaned by using Sephadex plates and run on an ABI 3730 machine (Applied Biosystems). A total of 290 sequence runs was assembled into a single contig sequence of 27,555 bp, including the polyA tail.
Phylogenetic Trees. Nucleotide sequences were aligned by using the clustalw package running under bioedit Ver. 5.0.9 (11) (available from www.mbio.ncsu.edu/BioEdit). Phylogenetic trees were made with the dnaml software of the phylip Ver. 3.6 software package (12) (available from http://evolution.genetics.washington.edu/phylip.html) by using 100 bootstraps and three jumbles. The consensus tree was used as usertree in dnaml to recalculate branch lengths. Accession numbers for the sequences were as follows: AY518894 (HCoV-NL, this work); AF304460 (HCoV-229E); AF353511 (porcine epidemic diarrhea virus); AJ271965 (transmissible gastroenteritis virus); AF220295 (bovine coronavirus); NC_005147 (HCoV-OC43), AF201929 (murine hepatitis virus); M95169 and M27569 (avian infectious bronchitis virus); AY278741 [severe acute respiratory syndrome (SARS) coronavirus]; Z24675 (porcine respiratory coronavirus); AY078417 (porcine hemagglutinating encephalomyelitis virus); AF207551 (rat coronavirus); AY204704 (feline coronavirus); and D13096 (canine coronavirus).
Real-Time RT-PCR. Three sets of TaqMan primers and probes were designed based on the N gene of HCoV-NL: (i) set 1 forward (5′-TGT-CAA-CGA-GGT-TTT-GCA-TTA-AAT-3′), set 1 reverse (5′-ACT-GGC-CTA-CCA-TTG-TGT-GTA-AGA-3′), and set 1 probe (5′-fluorescein-AAC-CCC-ATG-CGT-TTA-GCG-CAT-GA-tetramethylrhodamine-3′); (ii) set 2 forward (5′-AAG-GGT-TTT-CCA-CAG-CTT-GCT-3′), set 2 reverse (5′-ATC-ACC-CAC-TTC-ATC-AGT-GCT-AAC-3′), and set 2 probe (5′-f luorescein-TCA-CTA-TCA-A AG-A AT-A AC-GCA-GCC-TGA-TTA-GGA-A-tetramethylrhodamine-3′); and (iii) set 3 forward (5′-AGG-ACC-TTA-AAT-TCA-GAC-AAC-GTT-CT-3′), set 3 reverse (5′-GAT-TAC-GTT-TGC-GAT-TAC-CAA-GAC-T-3′), and set 3 probe (5′-fluorescein-TAA-CAG-TTT-TAG-CAC-CTT-CCT-TAG-CAA-CCC-AAA-CA-tetramethylrhodamine-3′).
The real-time RT-PCR were performed as one-step reactions in a final volume of 50 μl by using 5 μl of RNA with the EZ recombinant thermus thermophilus kit and were run on an ABI 7000 machine (Applied Biosystems). The assays were optimized by using serial 10-fold dilutions of a virus stock of HCoV-NL, resulting in the following optimal amounts of oligonucleotides per reaction: set 1 forward, set 1 reverse, set 3 forward, and set 3 reverse, 0.4 pmol; set 1 probe, set 2 forward, and set 2 reverse, 0.2 pmol; and set 2 and set 3 probes, 0.15 pmol. Cycling conditions were 2 min, 50°C; 30 min, 60°C; 7 min, 95°C once; 20 sec, 94°C; 1 min, 60°C repeated 40 times. None of the three assays reacted with HCoV-229E, HCoV-OC43, and SARS coronavirus under these conditions. Set 3 displayed the best performance based on the threshold cycle of detection and linearity of the assay.
Clinical Specimens. Respiratory specimens were collected between November 2000 and January 2002 at different wards of the Erasmus Medical Center in Rotterdam, as described (13). We tested 139 samples collected from individuals suffering from RTI but negative for influenza A and B viruses, respiratory syncytial virus, human metapneumovirus, human parainfluenza virus types 1–4, adenovirus, rhinovirus, HCoV-229E, and HCoV-OC43. Of the patients tested, 48% were between 0 and 1 yr of age, 16% between 1 and 2 yr, 9% between 2 and 5 yr, 14% between 5 and 16 yr, 8% between 16 and 40 yr, and 5% between 40 and 66 yr of age (average 6.4 yr, range 4 days to 66 yr). Routine virological testing for respiratory pathogens was performed by using a combination of direct immunofluorescence on cells present in the respiratory specimen, virus isolation in cell cultures, and immunofluorescence. Cell lines used for virus isolation included human embryonal kidney cells, tMK cells, Madin–Darby canine kidney cells, Vero cells, and HEp-G2 cells. All samples were kept at +4°C during processing and were subsequently stored for prolonged periods at -70°C. For HCoV-NL testing, RNA was isolated from a 40- to 200-μl sample by using a high pure RNA isolation kit (Roche Diagnostics) according to instructions from the manufacturer.
Results
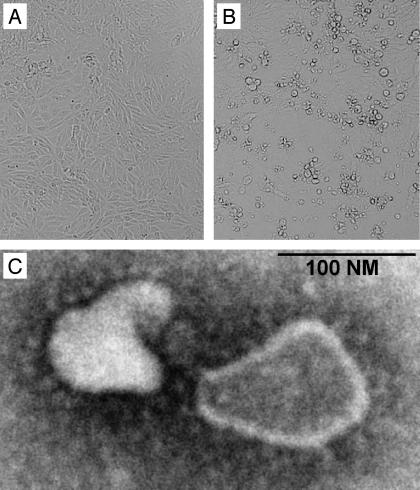
Virus Isolation and Characterization. Within our diagnostic virology setting, we isolated an unidentifiable virus from a nose swab sample collected from an 8-mo-old boy suffering from pneumonia in The Netherlands in April 1988. The unidentified virus was isolated in tMK cells, in which it caused sporadic foci of CPE after 7 days in culture and affected ≈50% of cells after 13 days. The virus could subsequently be passaged in tMK and Vero cells but not in HeLa or HEp-G2 cells, as judged by the presence of CPE in these cultures. After initial isolation in tMK cells, the virus proved to replicate in Vero cells, with CPE appearance from day 4 postinoculation onward, and virus titers of ≈105 50% tissue culture infectious doses per milliliter. The CPE induced by this virus isolate in Vero cells is shown in Fig. 1 and is characterized by rounding of the cells and subsequent detachment from the culture plate. The virus did not react with chicken and guinea pig erythrocytes in hemadsorption or hemagglutination tests. Supernatants of infected tMK and Vero cells were used for negative contrast electron microscopic analysis, revealing the presence of coronavirus-like particles with an average size of 140 nm (range, 95–275 nm) and average envelope projections of 20 nm (range, 12–24 nm) (Fig. 1C). We isolated RNA from the supernatant of Vero cells infected with the unidentified virus isolate for RT-PCR analyses using primer sets specific for the known human coronaviruses OC43 and 229E (6). Whereas the control viruses reacted positively with the respective virus-specific primers, the previously undescribed virus isolate failed to do so (data not shown).
Fig. 1.
Characteristics of the previously undescribed Dutch HCoV. CPE caused by HCoV-NL at 7 days after inoculation of Vero cells are shown (B) with mock-infected cells as controls (A). The cell-free supernatant from these cells revealed the presence of coronavirus-like particles by negative contrast electron microscopy at ×140,000 (C).
The recent discovery of a coronavirus causing SARS in humans (7, 8, 14–19) prompted us to analyze the as-yet-undefined HCoV from The Netherlands (HCoV-NL) further. Using arbitrarily primed RT-PCR (4, 9, 10) and RT-PCR with primers specific for the coronavirus family (7, 8), cDNA fragments were generated and sequenced. Although the nucleotide blast searches did not reveal matches with sequences available from public databases, translated blast searches revealed low homology with ORFs 1a and 1b of HCoV-229E and, to a lesser extent, other coronaviruses.
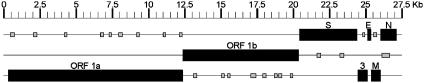
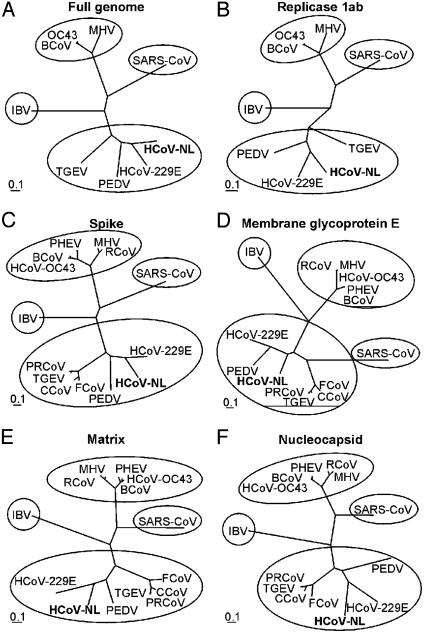
Sequence Analysis of Viral cDNA. By a combination of shotgun sequencing, RT-PCR, and rapid amplification of cDNA ends, the complete cDNA sequence of the viral genome was determined. The data from a total of 290 sequence runs were assembled into a single contig sequence of 27,555 bp, including the polyA tail (GenBank accession no. AY518894). The overall content of G and C residues in the HCoV-NL was 34%, which is lower than the 37–42% reported for other coronaviruses (17). The genome organization of HCoV-NL was found to resemble that of porcine epidemic diarrhea virus (Fig. 2). The order of the genes shared by all coronaviruses (5′-replicase, spike, envelope, membrane, and nucleocapsid-3′) was also found for HCoV-NL; the gene encoding hemagglutinin-esterase, present between the replicase and spike genes in group 2 and some group 3 coronaviruses, was absent in the genome of HCoV-NL. The coronavirus replicase polyprotein is synthesized by a -1 frameshift at a conserved “slippery” site (UUUAAAC) in the overlap of ORF1a and -1b (20). The putative ribosomal frameshift site in the HCoV-NL is located at nucleotide positions 12419–12425. The putative replicase, spike, envelope, and nucleocapsid ORFs revealed the highest-percentage amino acid sequence identity with HCoV-229E (66.6%, 54.7%, 46.7%, and 43.2%, respectively), whereas the matrix ORF revealed the highest identity with porcine epidemic diarrhea virus (65.0%) and somewhat lower identity with HCoV-229E (61.5%) (Table 1). In phylogenetic trees, HCoV-NL is found at the same branch as porcine epidemic diarrhea virus and HCoV-229E, being genetically more closely related to HCoV-229E (Fig. 3). These combined results indicate that HCoV-NL is a previously undescribed group 1 coronavirus. In addition to the relatively low sequence identity between HCoV-229E and HCoV-NL, there are two interesting differences. First, the spike ORF of HCoV-NL is 182 amino acid residues longer than the spike ORF of HCoV-229E, due to an extension at the N terminus. Other group 1 coronaviruses, such as porcine epidemic diarrhea virus, transmissible gastroenteritis virus, canine coronavirus, and feline coronavirus, also have relatively long N termini, whereas porcine respiratory coronavirus does not. Second, whereas HCoV-229E has two ORFs between the spike and envelope genes, ORFs 4a and 4b, HCoV-NL has only one 226-amino acid ORF (Fig. 2), which was tentatively named ORF3 because of its position in the genome. Other group 1 coronaviruses such as porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine respiratory coronavirus contain similarly large ORFs between spike and envelope, named ORF3, -3–1, or -3a/b/c, but the percentage amino acid sequence identity to HCoV-NL ORF3 (33.9%, 27.2%, and 26.4%, respectively) is lower than the identity between the combined 4a and 4b ORFs of HCoV-229E and HCoV-NL ORF3 (42.4%). We observed sequence variation in ORF3 of HCoV-NL; in addition to RT-PCR fragments having a stretch of nine T residues from nucleotide position 24820 to 24828, we obtained RT-PCR fragments with deletions of one or two T residues. Considerable sequence variation was also observed in ORF3 of porcine epidemic diarrhea virus, potentially because this ORF is dispensable for, or incompatible with, virus replication in vitro (21). Analysis of nucleotide sequences of ORF3 homologues derived from other group 1 coronaviruses isolated in cell cultures revealed significant sequence variation as well.
Fig. 2.
Genome organization of HCoV-NL. All putative ORFs of 50 or more amino acid residues in the HCoV-NL genome are shown, with known homologues in HCoV-229E shown in black and the additional 24 small ORFs shown in gray.
Table 1. Percentage amino acid sequence identity between ORFs of HCoV-NL and other known coronaviruses after pairwise alignment of sequences.
| Group virus* | Rep 1ab | S | E | M | N |
|---|---|---|---|---|---|
| 1 HCoV-229E | 66.6 | 54.7 | 46.7 | 61.5 | 43.2 |
| PEDV | 60.7 | 44.4 | 41.5 | 65 | 38 |
| TGEV | 50.7 | 40.6 | 30.4 | 44.4 | 34.8 |
| 2 HCoV-OC43 | 34.7 | 22.4 | 17.6 | 33 | 20.6 |
| BCoV | 33.2 | 22.3 | 17.6 | 33.9 | 21.1 |
| MHV | 34.7 | 21.6 | 13.6 | 32.4 | 21.1 |
| 3 IBV | 32.8 | 23.1 | 10.1 | 24.3 | 20.3 |
| 4 SARS-CoV | 35.4 | 20.2 | 17.9 | 29 | 24.3 |
Rep 1ab, replicase 1ab; S, spike; E, envelope; M, matrix; N, nucleocapsid.
See Fig. 3 legend for abbreviations of virus names
Fig. 3.
Phylogenetic analysis of HCoV-NL and other coronaviruses. The full-length genome (A), replicase 1ab (B), spike (C), membrane glycoprotein E (D), matrix protein (E), and nucleocapsid protein (F) nucleotide sequences were aligned with those of known coronaviruses and maximum likelihood trees were constructed by using 100 bootstraps and three jumbles. Bars roughly represent 10% nucleotide sequence differences between closely related strains. PEDV, porcine epidemic diarrhea virus; TGEV, transmissible gastroenteritis virus; BCoV, bovine coronavirus; MHV, murine hepatitis virus; IBV, infectious bronchitis virus; SARS-CoV, SARS coronavirus; PRCoV, porcine respiratory coronavirus; PHEV, porcine hemagglutinating encephalomyelitis virus; RCoV, rat coronavirus; FCoV, feline coronavirus; CCoV, canine coronavirus.
In addition to the replicase, spike, ORF3, envelope, matrix, and nucleocapsid genes, there were 24 putative ORFs of 50–97 amino acids scattered throughout the HCoV-NL genome and overlapping with the major genes (Fig. 2). The 5′ untranslated region of HCoV-NL was 70% identical to the 5′ untranslated region of HCoV-229E and 58.3% identical to that of porcine epidemic diarrhea virus (22). Based on the similarity of these 5′ untranslated regions, we predict that the core sequence of the transcription-regulating sequence is CUAAAC. Homologues of the core sequence were found upstream of each of the major ORFs of HCoV-NL (Table 2) but not the additional 24 small ORFs. The exact core sequence of the transcription-regulating sequence and the expression of the different mRNA species and proteins must be identified experimentally in the future.
Table 2. Alignment of putative transcriptional regulatory sequences in the HCoV-NL genome.
| Nucleotide* | ORF† | Sequence‡ |
|---|---|---|
| 69 | 5′UTR | ACTCCTCTCAACTAAACGAA |
| 20454 | S | TT-AG----------ATG |
| 24482 | 3 | TTGTGGT----------TTG (N34) ATG |
| 25176 | E | --------------T-----GATG |
| 25421 | M | -AATG---A---------ATG |
| 26110 | N | TT-AA---A--------A--ATG |
Nucleotide position of the first C residue of the CUAAAC sequence
S, spike; 3, ORF3; E, envelope; M, matrix; N, nucleocapsid
Sequences are aligned with the leader sequence, and identical positions are indicated with a dash. The putative initiation codons are underlined, with the conserved putative core sequence shown in bold
Detection of HCoV-NL in Clinical Specimens. We next tested whether the virus that was isolated from a child in 1988 was still circulating in humans. To this end, we designed three real-time RT-PCR assays based on different parts of the nucleocapsid gene of HCoV-NL, thereby reducing the chance of false-negative results based on genetic variation. With these assays, we screened respiratory tract specimens collected from individuals suffering from RTI within the wards of Erasmus Medical Center in Rotterdam between November 2000 and January 2002, which were negative for known viral pathogens (13). We detected the previously undescribed coronavirus in 4 of 139 (2.9%) samples tested with all three RT-PCR assays. The positive samples were collected from girls aged 3 mo, 4 mo, 4 mo, and 10 yr from November 2000 to January 2001. All patients suffered from fever, runny nose, and dry cough, and nasal aspirate samples were collected for diagnostic virology because infection with respiratory syncytial virus was suspected. Three children had severe underlying disease, respectively trisomy 21 with cardiac malformations, giant cell hepatitis, or Rett syndrome. One 4-mo-old child had concomitant pertussis (see Table 3 for further details on these patients).
Table 3. Patients suffering from RTI associated with HCoV-NL infection.
| Age | Gender | Sample date | Symptoms | Underlying disease |
|---|---|---|---|---|
| 5 mo | Male | January 12, 1988 | Pneumonia | Unknown |
| 3 mo | Female | November 1, 2000 | Fever (39.4°C) | Giant cell hepatitis |
| Runny nose | ||||
| 4 mo | Female | December 19, 2000 | Subfebrile (37.6°C) | Trisomy-21 |
| Runny nose | AVSD | |||
| 4 mo | Female | January 18, 2001 | Subfebrile (37.8°C) | Pertussis |
| Sever cough | ||||
| 10 yr | Female | January 18, 2001 | Fever (38.6°C) | Rett syndrome |
| Runny nose | Epilepsia | |||
| Dry cough |
AVSD, atrioventricular septum defect.
From two PCR-positive samples, a 926-nucleotide nucleocapsid gene fragment, was generated with the outer real-time PCR primers (set 3 forward primer and set 1 reverse primer) and subsequently sequenced. The nucleocapsid gene was found to be highly conserved, with up to eight nucleotide substitutions, all of which were silent.
Discussion
Here we identified a previously unrecognized group 1 coronavirus isolated from an 8-mo-old child suffering from pneumonia, which was subsequently detected in four additional children suffering from RTI. The coronaviruses (order Nidovirales, family Coronaviridae, genus coronavirus) are large-enveloped positive-stranded RNA viruses causing RTI and enteric diseases in humans and animals (23–25). Until recently, the coronaviruses were distinguished in three groups or serotypes. With the discovery of SARS coronavirus, a tentative fourth group was identified, based on the distinct genome organization and phylogenetic analyses of this virus (17, 18). Whereas group 3 contains only avian infectious bronchitis virus and group 4 only the SARS coronavirus, groups 1 and 2 contain a variety of human and animal pathogens. Group 1 contains the HCoV-229E and animal coronaviruses porcine epidemic diarrhea virus, transmissible gastroenteritis virus, porcine respiratory coronavirus, feline coronavirus, and canine coronavirus, whereas group 2 contains the HCoV-OC43 and animal coronaviruses porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, murine hepatitis virus, and rat sialodacryoadenitis coronavirus.
HCoV-229E and -OC43 can infect people of all age groups and cause severe lower RTI primarily in frail patients, such as young children and elderly individuals. HCoV infections may account for ≈35% of common colds in adults and ≈2% of influenza-like illnesses in patients of all age groups consulting their general physician (23–27). The patients in whom HCoV-NL was detected suffered from relatively severe RTI, requiring hospitalization. Because the majority of the HCoV-NL-infected children had underlying disease, the clinical symptoms associated with HCoV infections need to be established further. The availability of the HCoV-NL genome sequence will facilitate the development of a variety of diagnostic assays to enable analyses of the prevalence and clinical impact of HCoV-NL infections in humans. In studies on coronavirus infections in humans and animals, including infections with SARS coronavirus, the circulation of HCoV-NL or closely related viruses should be taken into account. With the threat of the reemergence of SARS coronavirus, it is important to note that both HCoV-NL and SARS coronavirus replicate efficiently in tMK and Vero cells, and that HCoV-NL may be associated with severe disease. Thus, laboratory tests based on virus isolation in cell cultures, observation of CPE, and electron microscopy may not be sufficient to discriminate between infection with SARS coronavirus and HCoV-NL. Therefore, the development and evaluation of more specific diagnostic tests and reagents are urgently needed.
Note Added in Proof. L. van der Hoek et al. recently described essentially the same virus in an independent investigation (28).
Acknowledgments
We thank Leo Sprong for technical assistance. R.A.M.F. is a fellow of the Royal Dutch Academy of Arts and Sciences. This work was supported by CoroNovative B.V. Sequence analysis was performed in part by BaseClear B.V. (Leiden, The Netherlands).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RTI, respiratory tract illnesses; tMK, tertiary monkey kidney; CPE, cytopathic effects; HCoV, human coronavirus; HCoV-NL, HCoV from The Netherlands; SARS, severe acute respiratory syndrome.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY518894, AY563107, and AY563108).
References
- 1.Bertino, J. S. (2002) Am. J. Med. 112, 42S-49S. [DOI] [PubMed] [Google Scholar]
- 2.Monto, A. S. & Sullivan, K. M. (1993) Epidemiol. Infect. 110, 145-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juven, T., Mertsola, J., Waris, M., Leinonen, M., Meurman, O., Roivainen, M., Eskola, J., Saikku, P. & Ruuskanen, O. (2000) Pediatr. Infect. Dis. J. 19, 293-298. [DOI] [PubMed] [Google Scholar]
- 4.van den Hoogen, B. G., de Jong, J. C., Groen, J., Kuiken, T., de Groot, R., Fouchier, R. A. & Osterhaus, A. D. (2001) Nat. Med. 7, 719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madeley, C. R. & Field, A. M. (1988) in Virus Morphology, eds. Madeley, C. R. & Field, A. M. (Chruchill Livingstone, New York).
- 6.Myint, S., Johnston, S., Sanderson, G. & Simpson, H. (1994) Mol. Cell Probes 8, 357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ksiazek, T. G., Erdman, D., Goldsmith, C., Zaki, S. R., Peret, T., Emery, S., Tong, S., Urbani, C., Comer, J. A., Lim, W., et al. (2003) N. Engl. J. Med. 348, 1953-1966. [DOI] [PubMed] [Google Scholar]
- 8.Drosten, C., Gunther, S., Preiser, W., Van Der Werf, S., Brodt, H. R., Becker, S., Rabenau, H., Panning, M., Kolesnikova, L., Fouchier, R. A., et al. (2003) N. Engl. J. Med. 348, 1967-1976. [DOI] [PubMed] [Google Scholar]
- 9.Ralph, D., McClelland, M. & Welsh, J. (1993) Proc. Natl. Acad. Sci. USA 90, 10710-10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welsh, J. & McClelland, M. (1990) Nucleic Acids Res. 18, 7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall, A. (1999) Nucleic Acids. Symp. Ser. 41, 95-98. [Google Scholar]
- 12.Felsenstein, J. (1989) Cladistics 5, 164-166. [Google Scholar]
- 13.Van Den Hoogen, B. G., Van Doornum, G. J., Fockens, J. C., Cornelissen, J. J., Beyer, W. E., Groot Rd, R., Osterhaus, A. D. & Fouchier, R. A. (2003) J. Infect. Dis. 188, 1571-1577. [DOI] [PubMed] [Google Scholar]
- 14.Peiris, J. S. M., Lai, S. T., Poon, L. L. M., Guan, Y., Yam, L. Y. C., Lim, W., Nicholls, J., Yee, W. K. S., Yan, W. W., Cheung, M. T., et al.. (2003) Lancet 361, 1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouchier, R. A., Kuiken, T., Schutten, M., Van Amerongen, G., Van Doornum, G. J., Van Den Hoogen, B. G., Peiris, M., Lim, W., Stohr, K. & Osterhaus, A. D. (2003) Nature 423, 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poutanen, S. M., Low, D. E., Henry, B., Finkelstein, S., Rose, D., Green, K., Tellier, R., Draker, R., Adachi, D., Ayers, M., et al. (2003) N. Engl. J. Med. 348, 1995-2005.. [DOI] [PubMed] [Google Scholar]
- 17.Rota, P. A., Oberste, M. S., Monroe, S. S., Nix, W. A., Campagnoli, R., Icenogle, J. P., Penaranda, S., Bankamp, B., Maher, K., Chen, M. H., et al. (2003) Science 300, 1394-1399. [DOI] [PubMed] [Google Scholar]
- 18.Marra, M. A., Jones, S. J., Astell, C. R., Holt, R. A., Brooks-Wilson, A., Butterfield, Y. S., Khattra, J., Asano, J. K., Barber, S. A., Chan, S. Y., et al. (2003) Science 300, 1399-1404. [DOI] [PubMed] [Google Scholar]
- 19.Kuiken, T., Fouchier, R. A., Schutten, M., Rimmelzwaan, G. F., van Amerongen, G., van Riel, D., Laman, J. D., de Jong, T., van Doornum, G., Lim, W., et al.(2003) Lancet 362, 263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brierley, I., Boursnell, M. E., Binns, M. M., Bilimoria, B., Blok, V. C., Brown, T. D. & Inglis, S. C. (1987) EMBO J. 6, 3779-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duarte, M., Tobler, K., Bridgen, A., Rasschaert, D., Ackermann, M. & Laude, H. (1994) Virology 198, 466-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tobler, K. & Ackermann, M. (1995) Adv. Exp. Med. Biol. 380, 541-542. [DOI] [PubMed] [Google Scholar]
- 23.Holmes, K. V. & Lai, M. C. (1996) in Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott–Raven, Philadelphia), Vol. 1, pp. 1075-1093. [Google Scholar]
- 24.McIntosh, K. (1996) in Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott–Raven, Philadelphia), Vol. 1, pp. 1095-1103. [Google Scholar]
- 25.Enjuanes, L., Brian, D., Cavanagh, D., Holmes, K., Lai, M. M. C., Laude, H., Masters, P., Rottier, P. J. M., Siddell, S. G., Spaan, W. J. M., et al. (2000) in Virus Taxonomy, eds. van Regenmortel, M. H. V., Fauquet, C. M., Bishop, D. H. L., Carstens, E. B., Estes, M. K., Lemon, S. M., Maniloff, J., Mayo, M. A., McGeoch, D. J., Pringle, C. R., et al. (Academic, San Diego), pp. 835-849.
- 26.El-Sahly, H. M., Atmar, R. L., Glezen, W. P. & Greenberg, S. B. (2000) Clin. Infect. Dis. 31, 96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glezen, W. P., Greenberg, S. B., Atmar, R. L., Piedra, P. A. & Couch, R. B. (2000) J. Am. Med. Assoc. 283, 499-505. [DOI] [PubMed] [Google Scholar]
- 28.van der Hoek, L., Pyrc, K., Jebbink, M. F., Vermeulen-Oost, W., Berkhout, R. J. M., Wolthers, K. C., Wertheim-van Dillen, P. M. E., Kaandorp, J., Spaargaren, J. & Berkhout, B. (2004) Nat. Med. 10, 368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]