Abstract
Our immune system has evolved to recognize and eradicate pathogenic microbes. However, we have a symbiotic relationship with multiple species of bacteria that occupy the gut and comprise the natural commensal flora or microbiota. The microbiota is critically important for the breakdown of nutrients, and also assists in preventing colonization by potentially pathogenic bacteria. In addition, the gut commensal bacteria appear to be critical for the development of an optimally functioning immune system. Various studies have shown that individual species of the microbiota can induce very different types of immune cells (e.g., Th17 cells, Foxp3+ regulatory T cells) and responses, suggesting that the composition of the microbiota can have an important influence on the immune response. Although the microbiota resides in the gut, it appears to have a significant impact on the systemic immune response. Indeed, specific gut commensal bacteria have been shown to affect disease development in organs other than the gut, and depending on the species, have been found to have a wide range of effects on diseases from induction and exacerbation to inhibition and protection. In this review, we will focus on the role that the gut microbiota plays in the development and progression of inflammatory/autoimmune disease, and we will also touch upon its role in allergy and cancer.
Keywords: commensal bacteria, microbiota, autoimmunity, allergy, cancer, Treg, Th17, probiotics
Interaction between the Gut Microbiota and the Immune System
The trillions of commensal microorganisms that constitute the intestinal microbiota are primarily composed of five bacterial phyla, Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, and Fusobacteria. Bacteroidetes and Firmicutes predominate and represent ∼90% of the total gut microbiota (i.e., 25 and 65%, respectively; Rajilic-Stojanovic et al., 2007). However, these proportions can vary greatly between individuals and even within single individual over time. Although commensal bacteria inhabit the entire gut, greater numbers reside in the distal part of the small intestine and in the large intestine or colon. Acquisition of the intestinal microbiota occurs in the first year of life and is influenced by the maternal bacteria obtained during vaginal delivery (Bennet and Nord, 1987; Mandar and Mikelsaar, 1996; Penders et al., 2006), versus Cesarean section (Dominguez-Bello et al., 2010), and breastfeeding (Yoshioka et al., 1983; Balmer and Wharton, 1989; Harmsen et al., 2000; Hopkins et al., 2005). Early in life the major shifts in the gut microbiota can be caused by antibiotics, dietary changes, etc. (Koenig et al., 2011) as opposed to a purely stochastic model (Palmer et al., 2007). However, there is a lack of large-scale studies of temporal variation in adults.
The gut microbiota plays a critical role in the breakdown of indigestible complex plant polysaccharides, and provides an important layer of defense against invasion by pathogenic microorganisms. It is also required for proper development of the immune system, as indicated by the fact that germ-free (GF) mice have poorly developed lymphoid tissues. GF mice have spleens with few germinal centers and poorly formed T and B cell zones, hypoplastic Peyer’s patches, lower numbers of lamina propria CD4+ cells and IgA-producing plasma cells (Macpherson and Harris, 2004), and aberrant development and maturation of isolated lymphoid follicles (Bouskra et al., 2008). The lymphoid organs of GF mice exhibit aberrant development of T and B cell subsets, and more specifically, contain CD4+ T cells that are skewed toward an T helper 2 (Th2) phenotype and produce little, if any, IFNγ (Mazmanian et al., 2005), and have fewer and smaller germinal centers (Bauer et al., 1963). There is also evidence that the microbiota plays a key role in the induction of IgA (Klaasen et al., 1993b; Talham et al., 1999) and maintenance of homeostasis of various T cell populations in the gut, including regulatory T cells (Tregs), and T helper 1 (Th1) and 17 (Th17) cells (Gaboriau-Routhiau et al., 2009).
The commensal bacteria that reside in the gut are diverse, and individual species appear, in some cases, to have distinct and opposing roles. Some commensal bacteria appear to drive Treg development preferentially, while others promote Th17 development. In a comparative analysis of terminal ileal tissue from mice maintained in conventional (CV) versus GF housing, tissue from CV mice exhibited 5- to 10-fold higher IL-10 and Foxp3 mRNA expression compared to GF mice, suggesting that the microbiota can drive Foxp3+Treg induction and/or expansion in the intestine (Gaboriau-Routhiau et al., 2009). Furthermore, in the Peyer’s patches and mesenteric lymph nodes (LN) of naive GF mice, Tregs were at a decreased frequency, were less effective at suppressing responder cell proliferation in vitro, and produced lower levels of IL-10 and TGFβ than naive mice housed under specific pathogen-free (SPF) conditions (Ishikawa et al., 2008). The colonization of GF mice with Bacteroides fragilis increases the suppressive capacity of Tregs and induces anti-inflammatory cytokine production by Foxp3+ T cells in the gut (Mazmanian et al., 2008). Similarly, colonization of GF mice with a cocktail of Clostridium strains increased dramatically the frequency of IL-10-producing Helios− Tregs (induced Tregs) in the colonic lamina propria (Atarashi et al., 2011). Numerous studies have also found that Tregs are induced following gavage with gut commensal bacteria of either the Lactobacillus or Bifidobacterium genus (Di Giacinto et al., 2005; O’Mahony et al., 2008; Karimi et al., 2009; Livingston et al., 2010; Zhang et al., 2010). In contrast, Th17 cell responses appear to be induced by a restricted number of microbial species, e.g., segmented filamentous bacteria (SFB). Candidatus arthromitus, commonly called SFB, are Gram-positive bacteria of the Firmicutes phylum and most closely related to the Clostridium genus (Snel et al., 1995). These bacteria colonize the small intestine and have been shown to stimulate the production of secretory IgA (Klaasen et al., 1993b; Talham et al., 1999) and to activate CD8αβ+TCRαβ+ intraepithelial cells in the small intestine (Umesaki et al., 1995). The SFB are thought to play a key role in the maturation of the adaptive mucosal immune response in the gut. Indeed, SFB colonization of GF mice has been shown to induce differentiation of primarily IL-17+ T cells (Ivanov et al., 2008, 2009; Gaboriau-Routhiau et al., 2009) and to a far lesser extent, IFNγ+Th1 and IL-10+Treg cells (Gaboriau-Routhiau et al., 2009). These data indicate that SFB can promote the development of intestinal Th17 cells, a population of T cells that is not only important for fighting bacterial infection, but is also involved in the pathogenesis of a number of inflammatory and autoimmune diseases. A microbiota favoring SFB could, therefore, have an impact on the immune response, and consequently, on the development of Th17-mediated inflammatory/autoimmune diseases in the gut and at distant sites in predisposed individuals. The specific mechanisms that lead to the preferential induction of Tregs versus Th17 cells by various commensal bacteria are currently unknown. However, DNA derived from commensal bacteria has been shown to play a major role in intestinal homeostasis through toll-like receptor 9 (TLR9) engagement, and appears to be involved in controlling the balance between Tregs and effector cells. Indeed, TLR9 deficient (TLR9−/−) mice exhibit increases in intestinal Foxp3+ Tregs, and decreases in IL-17 and IFN-γ production by comparison to wild-type mice (Hall et al., 2008). One could speculate that specific commensal species may differentially modulate the T effector/Treg cell balance via this mechanism, since the ability of gut commensal bacteria to stimulate TLR9 varies and depends on the frequency of CG dinucleotides (Dalpke et al., 2006).
The composition of the microbiota can be influenced by various factors, including diet and exposure to antibiotics. Analysis of the fecal microbiota of humans and 59 other mammalian species has shown that the gut microbiota of humans living a modern lifestyle is typical of omnivorous primates (Ley et al., 2008). Moreover, feeding GF mice colonized with human fecal microbial communities a high-fat, high sugar “Western” diet instead of a low-fat, plant polysaccharide-rich diet significantly altered the microbiota composition, resulting in an increase in Firmicutes and decrease in Bacteroidetes (Turnbaugh et al., 2009). In contrast, a diet very high in fiber has been associated with increases in Bacteroidetes and a much lower abundance of Firmicutes in humans (De Filippo et al., 2010), although other factors may confound these comparisons. Interestingly, fluctuations in the proportions of these types of commensal bacteria have been found in patients and animals with inflammatory/autoimmune diseases (see below). In addition, antibiotic treatment is usually followed by a decrease in the diversity of the microbiota. Although the composition of the microbiota is similar in the days or weeks following the termination of treatment, some bacterial members are lost from the community indefinitely (Jernberg et al., 2007; Jakobsson et al., 2010). Furthermore, different types of antibiotics differentially affect bacteria and, consequently, immune cell development. Antibiotic treatment (ampicillin, gentamicin, metronidazole, neomycin, and vancomycin) that reduced bacteria from the Firmicutes phylum while increasing the proportion of bacteria from the Bacteroidetes, phylum was associated with a reduction in mucosal CD4+T cells expressing IFNγ and IL-17 (Hill et al., 2010). Similarly, vancomycin or ampicillin treatment, both of which inhibit Gram-positive bacteria, dramatically decreased the numbers of Th17 cells in the lamina propria. In contrast, metronidazole or neomycin, which target anaerobes and Gram-negative bacteria, had little effect on the Th17 cell population (Ivanov et al., 2008). Because the microbiota profoundly affects immune system development and maturation, it is not surprising that modulation of the balance between the various commensal bacteria can influence inflammatory/autoimmune disease development and/or progression.
Involvement of Commensal Bacteria in Protection Against Autoimmunity
The less than 100% concordance rate for autoimmune diseases in monozygotic twins (e.g., <40% for type 1 diabetes; Hyttinen et al., 2003), suggests that environmental factors can have a strong influence on inflammatory/autoimmune disease development. The composition of the microbial community that comprises the microbiota varies between individuals including monozygotic twins. Because the gut commensal bacteria influences the development of the immune system, the microbiota could be one of the environmental factors that affects inflammatory/autoimmune disease development in genetically susceptible individuals, i.e., differences in its composition could contribute to the lower than expected concordance rate in monozygotic twins. The composition of the gut microbiota may, therefore, either confer protection or trigger disease in genetically susceptible individuals.
Considerable evidence indicates that the composition of the microbiota can influence the development of intestinal inflammation. Some commensal bacteria may induce intestinal inflammation while others control it. The commensals capable of controlling inflammation in the gut mediate their effect either by balancing the immune response in favor of regulation or by controlling bacteria that may directly mediate intestinal inflammation. With regard to balancing the immune response, commensal bacteria from the Bacteroidetes and Firmicutes phyla appear to induce Tregs that can control Th17 cells that are responsible for intestinal inflammation. In a recent report, colonization of GF mice with the human commensal, B. fragilis, was shown to induce IL-10 production and Foxp3 expression, decrease Th17 cells and prevent colitis development. Furthermore, injection of polysaccharide A (PSA) isolated from B. fragilis was found to recapitulate the effects of colonization with B. fragilis (Mazmanian et al., 2008; Round and Mazmanian, 2009), with PSA-induced signaling through TLR2 expressed by Tregs promoting tolerance (Round et al., 2011). In addition, mice orally inoculated with murine Clostridium species belonging to clusters IV and XIVa (Firmicutes phylum) were resistant to colitis development and exhibited an increase in the frequency of colonic Tregs (Atarashi et al., 2011). Various human Lactobacillus strains (of the Firmicutes phylum) have also been shown to be protective in several animal models of colitis, including the TNBS-induced and IL-10 deficient mouse models of colitis, and were found to mediate their effect via induction of Tregs (Di Giacinto et al., 2005; O’Mahony et al., 2008; Livingston et al., 2010). Commensal bacteria can also affect other potentially pathogenic bacteria; for example, commensal bacteria from the Actinobacteria phylum appear to control the levels of other commensal bacteria that cause intestinal inflammation. Administration of Bifidobacterium animalis subsp. Lactis (Actinobacteria phylum) reduced intestinal inflammation in the T-bet−/−Rag2−/− model of colitis. This was associated with a concomitant reduction in Enterobacteriaceae levels, the commensal bacteria shown to initiate intestinal inflammation in this colitis model. Moreover, the authors reported that the fecal levels of Bifidobacterium in untreated T-bet−/−Rag2−/− mice were lower compared to control mice, and found that the Bifidobacterium fecal levels were inversely related to the colitis score (Veiga et al., 2010). Taken together, these studies demonstrate that several different species of commensal bacteria can control inflammation in the gut via different mechanisms, e.g., by directly regulating the immune response or by affecting the composition of the microbiota and reducing other disease-mediating commensals.
Commensal bacteria can also have a protective effect at sites distant from the gut. The results of one recent study suggest that the microbiota plays an important role in type 1 diabetes in genetically susceptible mice. Type 1 diabetes-susceptible NOD mice that were crossed with MyD88−/− mice (NODMyD88−/−) were found to be protected from type 1 diabetes. This protection appeared to involve changes in the composition of the microbiota, since protection was abrogated when NODMyD88−/− mice were treated with antibiotics or derived and maintained under GF conditions. A comparison of the gut microbiota between NODMyD88 sufficient and NODMyD88−/− mice confirmed that there were differences in the microbiota composition which could account for the differences in disease susceptibility. The NODMyD88−/− mice had a lower Firmicutes/Bacteroidetes ratio than the NODMyD88 sufficient mice, and an enrichment in cecal Lactobacillacae, Rikenellaceae, and Porphyromonadaceae. Finally, wild-type NOD mice that were colonized at birth with microbiota from NODMyD88−/− mice exhibited significantly reduced islet infiltration compared to untreated NOD mice (Wen et al., 2008), i.e., the microbiota from the NODMyD88−/− mice conferred some protection from diabetes to wild-type NOD mice. Moreover, a recent publication has reported that female NOD mice colonized with SFB exhibited a large Th17 population in the lamina propria of the small intestine and were protected from developing type 1 diabetes (Kriegel et al., 2011). These findings corroborate studies showing that Th17 cells do not directly mediate type 1 diabetes in NOD mice (Bending et al., 2009; Martin-Orozco et al., 2009), and in fact, appear to act as regulatory cells that protect against disease development (Han et al., 2010; Nikoopour et al., 2010). Interestingly, a recent study has shown that Th17 cell pathogenicity can be controlled in the small intestine, where pro-inflammatory Th17 cells can acquire a regulatory phenotype (rTh17) that includes in vivo immunosuppressive properties (Esplugues et al., 2011). Taken together, these data suggest that gut microbiota has a significant, if complex, influence on the development of inflammatory/autoimmune diseases. Another study has found that the microbiota plays a protective role in a model of collagen-induced arthritis. The study found that rats maintained in a GF environment developed more severe rheumatoid arthritis (RA) than rats maintained in a conventional environment, suggesting that the microbiota had a suppressive effect on RA development (Breban et al., 1993). Lastly, a series of studies has shown that the microbiota can protect against central nervous system (CNS) disease. Oral administration of antibiotic, leading to a reduction in bacteria from the Firmicutes phylum, but a relative increase in bacteria from the Bacteroidetes phylum, impaired the development of myelin oligodendrocyte glycoprotein (MOG) or proteolipid protein-induced experimental autoimmune encephalitis (EAE) in mice. In this study, protection was associated with diminished pro-inflammatory responses, higher numbers of Tregs in mesenteric LN and potent IL-10-producing Foxp3+ Tregs that were present in the LN (Ochoa-Reparaz et al., 2009, 2010). Moreover, oral administration of PSA isolated from B. fragilis (of the Bacteroidetes phylum) was able to prevent EAE development via enhancement of CD103+ DC that converted naïve T cells into IL-10-producing Foxp3+ Tregs, suggesting that antibiotic-mediated protection is mediated the increase in Bacteroidetes (Ochoa-Reparaz et al., 2009, 2010). Finally, oral administration of probiotics, including the Lactobacillus and Bifidobacterium species, has been shown to protect against development of various autoimmune diseases such as type 1 diabetes (Matsuzaki et al., 1997; Calcinaro et al., 2005), experimental autoimmune encephalomyelitis (Lavasani et al., 2010), experimental RA (Kato et al., 1998; Baharav et al., 2004; So et al., 2008), and systemic lupus erythematosus (personal observation; Alard et al., 2009), via induction of IL-10-producing Tregs and attenuation of Th1 and Th17 cytokines. It is not clear whether the Tregs are induced upon direct interaction with the probiotics and/or via stimulation with tolerogenic antigen presenting cells that have been induced by the probiotics. We have, however, shown recently that dendritic cells cultured with Lactobacillus casei shift their cytokine profile in favor of IL-10, and are able to prevent type 1 diabetes development upon injection into NOD mice. Since a single injection of L. casei-treated DC is sufficient to mediate long-term protection, the mechanism in this case most likely involves induction of regulatory cells (Manirarora et al., 2011). Altogether, these data emphasize the importance of a balanced gut microbiota that can induce a protective immune response capable of suppressing inflammation in organs distant from the gut, via induction of tolerogenic DC and Tregs. How these cells traffic from the gut to the distal lymph nodes and possibly to the peripheral tissue (CNS, joint, pancreas) is currently unknown and remains to be determined.
Studies have indicated that the composition of the microbiota can have a significant effect on the development of inflammatory/autoimmune disease in humans. The composition of the gut microbiota differs greatly in subsets of patients with inflammatory bowel disease (IBD), including Crohn’s disease and ulcerative colitis, compared to non-IBD individuals. The microbiota of IBD patients is characterized by depletion of Bacteroidetes and Firmicutes (Frank et al., 2007). More specifically, recurrence of Crohn’s disease was associated with decreases in the relative proportion of Faecalibacterium prausnitzii, a bacterium belonging to Clostridium cluster IV, in these patients (Sokol et al., 2009). In animal studies, F. prausnitzii has been shown to have anti-inflammatory effects, and oral administration of F. prausnitzii has been found to reduce the severity of TNBS-induced colitis (Sokol et al., 2008). These data suggest that restoration of the microbial balance in IBD patients may be a good strategy to treat this disease, and in fact, administration of VSL#3, a cocktail of several Lactobacillus species (L. Casei, Lactobacillus plantarum, Lactobacillus acidophilus, and Lactobacillus delbrueckii), Bifidobacterium species (Bifidobacterium longum, Bifidobacterium breve, and Bifidobacterium infantis) and a Streptococcus salivarius subsp., Thermophilus, has been shown to decrease ulcerative colitis in patients with mild to moderately active ulcerative colitis (Sood et al., 2009; Tursi et al., 2010). Studies in patients with early RA have also found that Bifidobacteria and B. fragilis in the gut are decreased (Vaahtovuo et al., 2008), suggesting that alteration in the abundance of these two commensal species may influence the pathogenesis of RA. However, in patients with established disease, the possibility that an alteration in the composition of the microbiota may be a consequence rather than a cause of the disease cannot be ruled out.
In conclusion, commensal bacteria belonging to the Bifidobacterium, Bacteroides, Clostridium, and Lactobacillus genera are associated with inflammatory/autoimmune disease protection. Therefore, establishing a balanced microbiota in favor of these protective commensal bacteria may be a good strategy for the prevention and/or treatment of inflammatory/autoimmune diseases in autoimmune-prone individuals.
Involvement of Commensal Bacteria in Triggering Inflammatory/Autoimmune Disease
Microbial pathogens have long been implicated in the etiology of a variety of autoimmune/inflammatory diseases, including IBD (Sanderson and Hermon-Taylor, 1992; Chen et al., 2000), RA (Toivanen, 2003), experimental autoimmune encephalomyelitis (Cermelli and Jacobson, 2000; Buljevac et al., 2005; Gilden, 2005; Farrell et al., 2009), type 1 diabetes (Filippi and von Herrath, 2005), and systemic lupus erythematosus (Cavallo and Granholm, 1990; Zandman-Goddard and Shoenfeld, 2005; Poole et al., 2006). However, a growing body of evidence suggests that specific commensal bacteria may also negatively impact inflammatory and autoimmune diseases in genetically susceptible individuals. Interestingly, rendering mice germ-free (GF) can dramatically alter inflammatory/autoimmune disease incidence and severity, and/or the kinetics of disease onset and progression in animal models of disease. This is not surprising since we know that commensal bacteria play an important role in shaping the host systemic immune response. However, the mechanisms underlying the relationship between commensal microorganisms and the induction/exacerbation of autoimmune/inflammatory diseases are poorly understood. Furthermore, it is unclear how individual microbial species comprising the microbiota contribute to the effects on these diseases. Recent studies focusing on the relationship between the microbiota and induction of autoimmunity have begun to shed some light on these issues.
Most mouse models of colitis are antibiotic-responsive, and patients with IBD often benefit from treatment with antibiotics, suggesting that bacteria play a role in disease pathogenesis. The general consensus is that IBD may be driven by aberrant pro-inflammatory host responses to the commensal microbiota (Packey and Sartor, 2008). A recent study has found that the microbiota can induce inflammation in models of colitis by activating T cells via both innate and adaptive immune mechanisms (Feng et al., 2010). In that study, homeostatic proliferation of transferred T cells was inhibited in GF RAG−/− mice, but restored after reconstitution with Altered Schaedler’s Flora (a standardized cocktail of mouse microbiota), indicating that T cells require the presence of the microbiota to proliferate. Moreover, microbiota-induced production of IL-6 by DC was required to reach the threshold number of IFNγ and IL-17-producing microbiota-specific T cells (i.e., induction of T cell proliferation) that was needed to induce colitis (Feng et al., 2010). In other studies, a number of bacterial species isolated from the intestines of IBD patients have been shown to be capable of inducing intestinal inflammation in rodents, including Helicobacter hepaticus, enterotoxigenic B. fragilis, and Bacteroides vulgatus. H. hepaticus infection is associated with the spontaneous development of colitis in some types of immunodeficient, but not immunocompetent mice (Ward et al., 1996). Furthermore, colonization with enterotoxigenic strains, but not non-toxigenic strains, of B. fragilis have been shown to induce colitis. Enterotoxigenic B. fragilis of human origin mediates colitis in multiple intestinal neoplasia (Min) transgenic mice in a Th17 cell-dependent manner (Wu et al., 2009). Similarly, enterotoxigenic B. fragilis of piglet origin has been shown to enhance DSS-induced colitis in mice (Rabizadeh et al., 2007), and the B. fragilis toxin appears to be essential for disease pathogenesis (Rhee et al., 2009). In an older study, colitis severity was found to be increased in a carrageenan-based model of colitis in guinea pigs after oral administration of B. vulgatus isolated from IBD patients, but not healthy individuals. This disease exacerbation could be transferred to recipients via spleen cells (Onderdonk et al., 1984), suggesting that bacteria isolated from IBD patients were inducing a cell-mediated (most likely T cell) immune response capable of enhancing inflammation. Interestingly, another study has found that a single species of commensal bacteria, filamentous segmented bacteria (SFB), can induce intestinal inflammation in the presence of a limited microbiota (Stepankova et al., 2007). In this study, GF SCID mice were colonized with individual or combinations of strains of anaerobic and aerobic bacteria and SFB, including Enterococcus faecalis, SFB, Fusobacterium mortiferum, Bacteroides distasonis, F. mortiferum + SFB, B. distasonis + SFB, and a SPF bacterial cocktail with limited diversity (SPF cocktail) ±SFB. Using the CD45high transfer model of intestinal inflammation, the study found that only the mice colonized with the SPF cocktail +SFB developed clinical and histological signs of intestinal inflammation following transfer of CD45RBhigh cells (Stepankova et al., 2007). These data indicate that, unlike SFB, most gut bacteria cannot or do not trigger intestinal inflammation in the presence of a limited microbiota, such as the typical SPF bacteria.
The involvement of SFB in inflammatory disease pathogenesis is not limited to the intestine. Two recent studies have shown that SFB may also be involved in development of experimental autoimmune encephalomyelitis (EAE) (Lee et al., 2011) and rheumatoid arthritis (RA) (Wu et al., 2010). In the first study, the authors found that GF mice exhibited decreased severity of MOG-induced EAE compared to mice raised under SPF conditions. The reduced severity was associated with reduced IL-17A and IFNγ production and RORγt transcripts, but elevated levels of CD4+CD25+Foxp3+ cells. Interestingly, whereas transfer of CD4+ T cells from MOG-immunized GF mice into RAG−/− GF mice only induced very mild EAE, transfer of CD4+ T cells from MOG-immunized standard pathogen free (SPF) mice induced severe disease, suggesting that the gut microbiota plays a role in activating T cells capable of inducing EAE. Finally, intestinal colonization of GF mice with SFB alone restored susceptibility to severe EAE induction, and correlated with elevated IL-17 and IFNγ expression in T cells in the spinal cord and small intestine lamina propria as well as a reduction of Foxp3+ T cell levels. This study demonstrates that intestinal colonization with SFB can induce Th17 cells that attack the central nervous system (CNS) (Lee et al., 2011). In the second study, arthritis-prone K/BxN mice raised under GF conditions exhibited attenuated RA that was associated with a reduction in serum autoantibody titers, splenic autoantibody-secreting cells, germinal centers, and IL-17. Interleukin-17 was found to be critical for the pathogenesis of the disease in these mice, as treatment with anti-IL-17 antibodies abrogated RA development in mice raised under SPF conditions. Interestingly, colonization of GF K/BxN mice with a single bacterial species, SFB, resulted in the rapid development of arthritis, which correlated with restoration of IL-17-producing T cells in the lamina propria. These cells could in turn migrate to the spleen where they provided help for germinal center formation and antibody production (Wu et al., 2010). Therefore, SFB, a bacterium known to induce IL-17 production (Ivanov et al., 2009), is capable of triggering RA in arthritis-prone K/BxN mice via activation of Th17 cells (Wu et al., 2010). Colitis, EAE, and RA are all mediated by IL-17-producing cells, and it is perhaps not surprising that a commensal bacteria that induces Th17 cells may be involved in the etiology of these diseases. However, it is surprising that a commensal gut bacteria (such as SFB) can play a role in the development of autoimmune disease at a distant site, i.e., CNS (Lee et al., 2011) or joints (Wu et al., 2010). In fact, evidence of SFB-induced Th17 cells could be found not only in the intestine, but also in the spinal chord and spleen, demonstrating that a single intestinal commensal bacterial species is capable of priming immune responses that are extra-intestinal and extend to peripheral lymphoid organs and tissues. However, the role that the intestinal SFB-induced Th17 cells play at the disease sites, i.e., in the spinal chord or the joints, remains to be determined. Several studies have shown that the presence of IL-17 in the joints enhanced disease (Lubberts et al., 2004; Jacobs et al., 2009), suggesting that Th17 cells could amplify the inflammatory process in the joints. Indeed, IL-17 receptor is expressed by a wide range of synovial cells (Yao et al., 1995; Silva et al., 2003), and IL-17 drives these cells to produce pro-inflammatory mediators such as cytokines (GM-CSF) and chemokines (CXCL2, CXCL8), and induces recruitment of neutrophils (Laan et al., 1999, 2003). Moreover, one could speculate that Th17 cells may also participate in the inflammatory process by producing other cytokines capable of promoting the inflammatory response. For instance, IL-23 and GM-CSF have been shown very recently to play a crucial role in EAE induction. Interleukin-23 appears to induce Th17 or RORγt+ T cells to produce GM-CSF that is required for disease induction; this was demonstrated in experiments showing that GM-CSF-producing IL-17-deficient T cells, but not GM-CSF-deficient Th17 cells, were able to induce EAE (Codarri et al., 2011; El-Behi et al., 2011). Furthermore, GM-CSF seems to target the myeloid cells that are responsible for sustaining inflammation in the CNS (El-Behi et al., 2011). Depending on the type of autoimmune diseases or the site of inflammation, SFB-induced Th17 cells may therefore promote/exacerbate inflammation in tissue through the production of GM-CSF and/or IL-17.
Understanding the role that commensal bacteria play in disease development in humans has been challenging. Studies about the composition of the gut microbiota in patients have identified bacteria with the potential to cause disease. Nonetheless, it is also possible that these microorganisms colonize the gut after disease is established and are not involved in the development of the disease. There is, however, a general consensus that IBD arises from an abnormal immune response to bacterial components of the intestinal microbiota (Packey and Sartor, 2008). Indeed, it has been reported that a subset of people with Crohn’s disease harbor a strain of adherent–invasive Escherichia coli in their small intestine that is potentially pro-inflammatory (Pineton de Chambrun et al., 2008). In addition, RA patients treated with antibiotics such as minocycline exhibit significant disease improvement (Stone et al., 2003), suggesting that the commensal bacteria contributes, at some level, to disease development. Although it has been shown that humans can be colonized with SFB (Klaasen et al., 1993a), there is little evidence yet for involvement of SFB in inflammatory/autoimmune diseases in humans. It would be of great interest to determine whether SFB is more prominent in the microbiota of patients exhibiting certain types of autoimmune diseases, i.e., Th17-mediated. Moreover, multiple issues remain to be addressed concerning the involvement of SFB in autoimmune diseases, including identifying the molecules that mediate Th17 cell induction as well as determining whether other Th17-mediated autoimmune diseases can be triggered by SFB.
Role of the Microbiota in Allergic Diseases
In addition to inflammatory and autoimmune diseases, there are other chronic diseases that may be impacted by the gut microbial community. In particular, allergic disease development has been associated with alterations in the intestinal microbiota. The incidence of allergic diseases has increased over the past 40 years in industrialized countries including the United States, Canada, United Kingdom, Ireland, New Zealand, and Australia, but not in developing countries. These data suggest that environmental changes are a major factor in the development of allergic diseases (Asher, 1998; Mannino et al., 1998; Beasley et al., 2000). The “hygiene hypothesis” suggests that the increase in the prevalence of allergic diseases observed in developed countries could be due to decreased early exposure to infectious agents that may alter the immune response and, in particular, the immunoregulatory compartment (Okada et al., 2010). However, the “microflora hypothesis” has been put forth as an alternative explanation and is based on changes in the intestinal microbiota that are due to antibiotic use and dietary differences (Noverr and Huffnagle, 2005). In either case, the regulatory mechanisms that normally control the Th2 responses generally associated with allergic diseases and asthma would not develop properly. Indeed, treatment of normal 3-week-old mice with the broad-spectrum antibiotic, kanamycin, results in the elimination of all Gram-negative bacteria in the stool, and a subsequent shift to a Th2 response (Oyama et al., 2001), suggesting that intestinal bacteria are involved in controlling the Th2 response. This was further supported in a study showing that treatment of mice with antibiotics combined with oral administration of the commensal fungus, Candida albicans, triggers pulmonary allergic responses (Noverr et al., 2004). Taken together, these studies suggest that the microbiota plays an important role in controlling allergic diseases.
Several studies have reported differences in the composition of the microbiota of infants who develop allergic diseases. Infants with food allergies were found to exhibit an imbalance between “beneficial” and potentially harmful bacteria, i.e., Lactobacilli and Bifidobacteria species were lower while coliforms and Staphylococcus aureus were higher (Bjorksten et al., 1999). Decreased levels of Bifidobacteria were also observed in infants with atopic eczema (Kirjavainen et al., 2001). Moreover, Bifidobacteria and Enterococcus were decreased while Clostridium was increased, resulting in a reduced ratio of Bifidobacteria to Clostridia in infants that developed atopic dermatitis (Bjorksten et al., 2001). These data suggest that specific species of intestinal commensal bacteria can play either a pathogenic or protective role in allergies that occur in the gut or at sites distant from the gut, such as lung and skin.
Prophylactic approaches based on the administration of probiotics to newborns at high risk for allergies have proven successful. The World Health Organization has defined probiotics as “live microorganisms which when administered in adequate amounts confer a health benefit on the host” FAO/WHO (2002). Administration of L. casei GG to the mothers before and after (via breastfeeding) delivery prevents atopic eczema, as well as other atopic diseases that develop later, in children at risk (Kalliomaki et al., 2001, 2003). Protection from allergic diseases provided by oral administration of non-pathogenic E. coli in early life was shown to extend to adult life (Lodinova-Zadnikova et al., 2003). Finally, a number of studies have been performed using probiotics to treat pre-existing atopic disease in infants. These studies found that administration of Lactobacilli was able to decrease the severity of various allergic diseases, including atopic eczema, atopic dermatitis, and food allergy in these children (Majamaa and Isolauri, 1997; Kirjavainen et al., 1999; Rosenfeldt et al., 2003; Viljanen et al., 2005). There is no evidence to date indicating that probiotic therapies have a beneficial effect on the incidence or severity of asthma in children at high risk (Vliagoftis et al., 2008). However, studies using animal models have found that oral administration of certain Lactobacilli and Bifidobacterium species were able to modulate allergic responses in the respiratory tract apparently via induction of regulatory T cells, suggesting that it may be possible to treat asthma using an optimal combination of probiotics (Feleszko et al., 2007; Forsythe et al., 2007; Karimi et al., 2009; Lyons et al., 2010).
As described above for autoimmune diseases, microbiota that favor certain species of commensal bacteria appears to also predispose or protect susceptible individuals to allergy development at various extra-intestinal sites, including lung and skin. The mechanisms by which intestinal immune responses affect inflammatory responses at distant sites are still unknown and remain to be established.
Role of the Microbiota in Cancer
The intestinal microbiota does not only influence autoimmune and allergic diseases, but has also been implicated in the development of cancer, especially colon cancer. Colorectal cancer is highly prevalent in the United States, with an estimated 1 in 20 men and women receiving a diagnosis of colon or rectum cancer sometime during their lifetime (National Cancer Institute Website: http://seer.cancer.gov/statfacts/html/colorect.html). Similarly, colorectal cancer is the second and third most common cancer in women and men, respectively, in European Union countries (Ferlay et al., 2007). This is an important heath problem in developed countries, where the primary risk factor is diet in up to 80% of all colorectal cancers (Bingham, 2000).
Interestingly, accumulating evidence in the last 3 years has underscored the importance of the intestinal microbiota in the development of colorectal cancer (Bingham, 2000; Davis and Milner, 2009; O’Keefe et al., 2009; Uronis et al., 2009). In fact, diet and the composition of the microbiota strongly correlate with the risk of developing colorectal cancer in certain groups of individuals (Bingham, 2000; Davis and Milner, 2009; O’Keefe et al., 2009). One mechanism by which commensal bacteria contribute to the initiation of colorectal cancer involves the induction of inflammation by commensal bacteria. A study using an animal model of colorectal carcinoma, IL-10−/− mice exposed to the carcinogen, azoxymethane (AOM) tested this hypothesis. In this study, AOM-exposed IL-10−/− mice developed colorectal carcinomas when housed conventionally, but not when housed under GF conditions. Furthermore, MyD88 (an adaptor molecule necessary for most TLR signaling) was found to be essential in this process, suggesting that bacterial-induced inflammation in the intestine can play a crucial role in the development of the carcinomas (Uronis et al., 2009). Interestingly, ingestion of lactic acid-producing bacteria has been shown to prevent carcinogen-induced lesions and tumors in animal models (Goldin and Gorbach, 1980; Goldin et al., 1996; Pool-Zobel et al., 1996; Challa et al., 1997; Rowland et al., 1998). However, the epidemiologic studies are controversial in humans; some studies have found no association (Kampman et al., 1994a,b; Kearney et al., 1996), whereas others have found a significant association (Malhotra, 1977; Peters et al., 1992; Boutron et al., 1996) between consumption of fermented milk products and occurrence of colon cancer. Further studies are, therefore, needed to determine if probiotics can be used as protective agents for the prevention of human colon cancer.
Unlike the situation in autoimmune and allergic diseases, there is little evidence that the microbiota directly affects the immune response against tumors, and consequently, somehow impacts tumor progression. One can, however, speculate that the type of immune response generated by the gut commensal bacteria could potentially influence tumor immunity. For example, mice colonized with enterotoxigenic B. fragilis exhibit colonic Th17 inflammatory infiltrates that are involved in induction of colon tumors, as indicated by inhibition of colon tumor formation following treatment with anti-IL-17 antibody (Wu et al., 2009). Moreover, Th17 cells have been found in cellular infiltrations in a variety of tumors, including prostate (Sfanos et al., 2008), pancreatic (Gnerlich et al., 2010), and ovarian cancer (Kryczek et al., 2009a), and in stomach cancer and the lymph nodes draining gastric cancer sites (Zhang et al., 2008). The frequency of Th17 cells correlates with the clinical stage of gastric cancer, i.e., Th17 cells are found at higher levels in late stage versus early stage (Zhang et al., 2008). On the other hand, Th17 cells appear to have a beneficial role in human ovarian cancer (Kryczek et al., 2009a) and murine melanoma, pancreatic, and colon cancer (Muranski et al., 2008; Kryczek et al., 2009b; Gnerlich et al., 2010). It is, therefore, possible that a microbiota favoring commensal bacteria that induces a Th17 response could have differential effects on tumors depending on the type of tumor or the stage of tumor development, and as found for autoimmunity and allergy, could alter the immune response to tumors at extra-intestinal as well as intestinal sites.
Conclusion
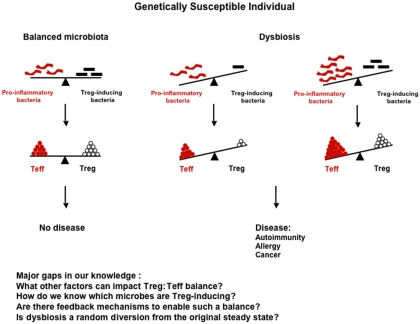
Genetic and environmental factors appear to shape the composition of the gut microbiota, which in turn plays a very important role in shaping the immune response at both intestinal and extra-intestinal sites, and in controlling the development of some types of autoimmune and allergic diseases as well as some forms of cancer. Therefore, any external factor that can alter the gut microbiota balance, such as diet or antibiotic treatment, should be viewed as a potential risk factor for development of these inflammatory diseases. Any alteration of the microbiota that leads to (1) a reduction in commensal bacteria favoring regulatory cells (Foxp3+ or other, e.g., rTh17) or (2) an enrichment in commensal bacteria favoring the induction of potential pathogenic cells may elicit disease in genetically susceptible individuals (Figure 1). However, the relationship between gut microbiota, immunity, and disease is very complex, since the same commensal bacteria can induce a protective response or a pathogenic response depending on the susceptibility of the individual. SFB is the perfect example, since it is protective in type 1 diabetes, but causes disease in mouse models of EAE or arthritis. The specific commensal microorganisms that contribute to either the etiology of or protection from many different types of diseases have not yet been fully identified and described. Future studies aimed at determining the impact of specific commensal bacteria on the immune response and induction of inflammatory diseases are crucial for a better understanding of pathogenesis of these diseases. Furthermore, an understanding of the genetic and environmental factors that shape the composition of the microbiota will provide the basis for strategies that will allow for the manipulation of microbiota in individuals at risk of developing disease. With this information in hand, it should be possible to design novel tailored therapies capable of preventing disease development in high risk individuals and treating established disease in already sick patients. Although not discussed in this review, other microorganisms, such as fungi and viruses, are present in the intestine, and have also been shown to be involved in autoimmune disease or allergic disease pathogenesis in susceptible animals (Noverr et al., 2004; Cadwell et al., 2010). It would, therefore, be very important in the future to extend the studies cited above to these microorganisms rather than limiting them to commensal bacteria only. It may eventually be possible to establish profiles of the microbiota in humans based on the bacterial species composition or enterotypes (Arumugam et al., 2011). This “biological fingerprint” of the future, similar to blood or tissue typing, may be used to predict responses to drugs or diet as well as aid in disease diagnosis/prognosis, and could ultimately lead to the development of personalized therapies.
Figure 1.
Impact of the composition of the flora on the immune response and disease development. Dysbiosis can arise from either (1) a reduction in Treg-inducing bacteria (e.g., Bifidobacteria or Bacteroides) that results in decreased Tregs, but unchanged levels of Teff cells; or (2) an enrichment in pro-inflammatory bacteria (e.g., SFB) that results in increased Teff cells, but unchanged levels of Tregs. In both cases, the Treg:Teff cell balance is biased toward the Teff cells, and the outcome of this imbalance can be development of disease in genetically susceptible individuals.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Alard P., Parnell S. A., Manirarora J. N., Kosiewicz M. M. (2009). Probiotics control lupus progression via induction of regulatory cells and IL-10 production. J. Immunol. 182, 50.30 [Google Scholar]
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D. R., Fernandes G. R., Tap J., Bruls T., Batto J. M., Bertalan M., Borruel N., Casellas F., Fernandez L., Gautier L., Hansen T., Hattori M., Hayashi T., Kleerebezem M., Kurokawa K., Leclerc M., Levenez F., Manichanh C., Nielsen H. B., Nielsen T., Pons N., Poulain J., Qin J., Sicheritz-Ponten T., Tims S., Torrents D., Ugarte E., Zoetendal E. G., Wang J., Guarner F., Pedersen O., De Vos W. M., Brunak S., Dore J., Consortium M., Weissenbach J., Ehrlich S. D., Bork P., Antolin M., Artiguenave F., Blottiere H. M., Almeida M., Brechot C., Cara C., Chervaux C., Cultrone A., Delorme C., Denariaz G., Dervyn R., Foerstner K. U., Friss C., Van De Guchte M., Guedon E., Haimet F., Huber W., Van Hylckama-Vlieg J., Jamet A., Juste C., Kaci G., Knol J., Lakhdari O., Layec S., Le Roux K., Maguin E., Merieux A., Melo Minardi R., M’Rini C., Muller J., Oozeer R., Parkhill J., Renault P., Rescigno M., Sanchez N., Sunagawa S., Torrejon A., Turner K., Vandemeulebrouck G., Varela E., Winogradsky Y., Zeller G. (2011). Enterotypes of the human gut microbiome. Nature 43, 174–180 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher M. I. (1998). Worldwide variations in the prevalence of asthma symptoms: the International Study of Asthma and Allergies in Childhood (ISAAC). Eur. Respir. J. 12, 315–335 [DOI] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., Cheng G., Yamasaki S., Saito T., Ohba Y., Taniguchi T., Takeda K., Hori S., Ivanov Ii., Umesaki Y., Itoh K., Honda K. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharav E., Mor F., Halpern M., Weinberger A. (2004). Lactobacillus GG bacteria ameliorate arthritis in Lewis rats. J. Nutr. 134, 1964–1969 [DOI] [PubMed] [Google Scholar]
- Balmer S. E., Wharton B. A. (1989). Diet and faecal flora in the newborn: breast milk and infant formula. Arch. Dis. Child. 64, 1672–1677 10.1136/adc.64.12.1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H., Horowitz R. E., Levenson S. M., Popper H. (1963). The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am. J. Pathol. 42, 471–483 [PMC free article] [PubMed] [Google Scholar]
- Beasley R., Crane J., Lai C. K., Pearce N. (2000). Prevalence and etiology of asthma. J. Allergy Clin. Immunol. 105, S466–S472 10.1016/S0091-6749(00)90044-7 [DOI] [PubMed] [Google Scholar]
- Bending D., De La Pena H., Veldhoen M., Phillips J. M., Uyttenhove C., Stockinger B., Cooke A. (2009). Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J. Clin. Invest. 119, 565–572 10.1172/JCI37865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennet R., Nord C. E. (1987). Development of the faecal anaerobic microflora after caesarean section and treatment with antibiotics in newborn infants. Infection 15, 332–336 10.1007/BF01647733 [DOI] [PubMed] [Google Scholar]
- Bingham S. A. (2000). Diet and colorectal cancer prevention. Biochem. Soc. Trans. 28, 12–16 [DOI] [PubMed] [Google Scholar]
- Bjorksten B., Naaber P., Sepp E., Mikelsaar M. (1999). The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin. Exp. Allergy 29, 342–346 10.1046/j.1365-2222.1999.00560.x [DOI] [PubMed] [Google Scholar]
- Bjorksten B., Sepp E., Julge K., Voor T., Mikelsaar M. (2001). Allergy development and the intestinal microflora during the first year of life. J. Allergy Clin. Immunol. 108, 516–520 10.1067/mai.2001.118130 [DOI] [PubMed] [Google Scholar]
- Bouskra D., Brezillon C., Berard M., Werts C., Varona R., Boneca I. G., Eberl G. (2008). Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510 10.1038/nature07450 [DOI] [PubMed] [Google Scholar]
- Boutron M. C., Faivre J., Marteau P., Couillault C., Senesse P., Quipourt V. (1996). Calcium, phosphorus, vitamin D, dairy products and colorectal carcinogenesis: a French case – control study. Br. J. Cancer 74, 145–151 10.1038/bjc.1996.330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breban M. A., Moreau M. C., Fournier C., Ducluzeau R., Kahn M. F. (1993). Influence of the bacterial flora on collagen-induced arthritis in susceptible and resistant strains of rats. Clin. Exp. Rheumatol. 11, 61–64 [PubMed] [Google Scholar]
- Buljevac D., Van Doornum G. J., Flach H. Z., Groen J., Osterhaus A. D., Hop W., Van Doorn P. A., Van Der Meche F. G., Hintzen R. Q. (2005). Epstein-Barr virus and disease activity in multiple sclerosis. J. Neurol. Neurosurg. Psychiatr. 76, 1377–1381 10.1136/jnnp.2004.048504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K., Patel K. K., Maloney N. S., Liu T. C., Ng A. C., Storer C. E., Head R. D., Xavier R., Stappenbeck T. S., Virgin H. W. (2010). Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145 10.1016/j.cell.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcinaro F., Dionisi S., Marinaro M., Candeloro P., Bonato V., Marzotti S., Corneli R. B., Ferretti E., Gulino A., Grasso F., De Simone C., Di Mario U., Falorni A., Boirivant M., Dotta F. (2005). Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia 48, 1565–1575 10.1007/s00125-005-1831-2 [DOI] [PubMed] [Google Scholar]
- Cavallo T., Granholm N. A. (1990). Lipopolysaccharide from Gram-negative bacteria enhances polyclonal B cell activation and exacerbates nephritis in MRL/lpr mice. Clin. Exp. Immunol. 82, 515–521 10.1111/j.1365-2249.1990.tb05482.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermelli C., Jacobson S. (2000). Viruses and multiple sclerosis. Viral Immunol. 13, 255–267 10.1089/08828240050144590 [DOI] [PubMed] [Google Scholar]
- Challa A., Rao D. R., Chawan C. B., Shackelford L. (1997). Bifidobacterium longum and lactulose suppress azoxymethane-induced colonic aberrant crypt foci in rats. Carcinogenesis 18, 517–521 [DOI] [PubMed] [Google Scholar]
- Chen W., Li D., Paulus B., Wilson I., Chadwick V. S. (2000). Detection of Listeria monocytogenes by polymerase chain reaction in intestinal mucosal biopsies from patients with inflammatory bowel disease and controls. J. Gastroenterol. Hepatol. 15, 1145–1150 10.1046/j.1440-1746.2000.02095.x [DOI] [PubMed] [Google Scholar]
- Codarri L., Gyulveszi G., Tosevski V., Hesske L., Fontana A., Magnenat L., Suter T., Becher B. (2011). RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat. Immunol. 12, 560–567 10.1038/ni.2027 [DOI] [PubMed] [Google Scholar]
- Dalpke A., Frank J., Peter M., Heeg K. (2006). Activation of toll-like receptor 9 by DNA from different bacterial species. Infect. Immun. 74, 940–946 10.1128/IAI.74.2.940-946.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. D., Milner J. A. (2009). Gastrointestinal microflora, food components and colon cancer prevention. J. Nutr. Biochem. 20, 743–752 10.1016/j.jnutbio.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C., Cavalieri D., Di Paola M., Ramazzotti M., Poullet J. B., Massart S., Collini S., Pieraccini G., Lionetti P. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U.S.A. 107, 14691–14696 10.1073/pnas.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giacinto C., Marinaro M., Sanchez M., Strober W., Boirivant M. (2005). Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-beta-bearing regulatory cells. J. Immunol. 174, 3237–3246 [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello M. G., Costello E. K., Contreras M., Magris M., Hidalgo G., Fierer N., Knight R. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U.S.A. 107, 11971–11975 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Behi M., Ciric B., Dai H., Yan Y., Cullimore M., Safavi F., Zhang G. X., Dittel B. N., Rostami A. (2011). The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat. Immunol. 12, 568–575 10.1038/ni.2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esplugues E., Huber S., Gagliani N., Hauser A. E., Town T., Wan Y. Y., O’Connor W., Rongvaux A., Van Rooijen N., Haberman A. M., Iwakura Y., Kuchroo V. K., Kolls J. K., Bluestone J. A., Herold K. C., Flavell R. A. (2011). Control of T(H)17 cells occurs in the small intestine. Nature 475, 514–518 10.1038/nature10228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO/WHO (2002). Guidelines for the Evaluation of Probiotics in Food. Available at: http://www.who.int/foodsafety/publications/fs_manage ment/probiotics2/en/index.html
- Farrell R. A., Antony D., Wall G. R., Clark D. A., Fisniku L., Swanton J., Khaleeli Z., Schmierer K., Miller D. H., Giovannoni G. (2009). Humoral immune response to EBV in multiple sclerosis is associated with disease activity on MRI. Neurology 73, 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feleszko W., Jaworska J., Rha R. D., Steinhausen S., Avagyan A., Jaudszus A., Ahrens B., Groneberg D. A., Wahn U., Hamelmann E. (2007). Probiotic-induced suppression of allergic sensitization and airway inflammation is associated with an increase of T regulatory-dependent mechanisms in a murine model of asthma. Clin. Exp. Allergy 37, 498–505 10.1111/j.1365-2222.2006.02629.x [DOI] [PubMed] [Google Scholar]
- Feng T., Wang L., Schoeb T. R., Elson C. O., Cong Y. (2010). Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J. Exp. Med. 207, 1321–1332 10.1084/jem.20092253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J., Autier P., Boniol M., Heanue M., Colombet M., Boyle P. (2007). Estimates of the cancer incidence and mortality in Europe in 2006. Ann. Oncol. 18, 581–592 10.1093/annonc/mdl498 [DOI] [PubMed] [Google Scholar]
- Filippi C., von Herrath M. (2005). How viral infections affect the autoimmune process leading to type 1 diabetes. Cell. Immunol. 233, 125–132 10.1016/j.cellimm.2005.04.009 [DOI] [PubMed] [Google Scholar]
- Forsythe P., Inman M. D., Bienenstock J. (2007). Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am. J. Respir. Crit. Care Med. 175, 561–569 10.1164/rccm.200606-821OC [DOI] [PubMed] [Google Scholar]
- Frank D. N., St Amand A. L., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V., Rakotobe S., Lecuyer E., Mulder I., Lan A., Bridonneau C., Rochet V., Pisi A., De Paepe M., Brandi G., Eberl G., Snel J., Kelly D., Cerf-Bensussan N. (2009). The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 10.1016/j.immuni.2009.08.020 [DOI] [PubMed] [Google Scholar]
- Gilden D. H. (2005). Infectious causes of multiple sclerosis. Lancet Neurol. 4, 195–202 10.1016/S1474-4422(05)01017-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerlich J. L., Mitchem J. B., Weir J. S., Sankpal N. V., Kashiwagi H., Belt B. A., Porembka M. R., Herndon J. M., Eberlein T. J., Goedegebuure P., Linehan D. C. (2010). Induction of Th17 cells in the tumor microenvironment improves survival in a murine model of pancreatic cancer. J. Immunol. 185, 4063–4071 10.4049/jimmunol.0902609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin B. R., Gorbach S. L. (1980). Effect of Lactobacillus acidophilus dietary supplements on 1,2-dimethylhydrazine dihydrochloride-induced intestinal cancer in rats. J. Natl. Cancer Inst. 64, 263–265 [DOI] [PubMed] [Google Scholar]
- Goldin B. R., Gualtieri L. J., Moore R. P. (1996). The effect of Lactobacillus GG on the initiation and promotion of DMH-induced intestinal tumors in the rat. Nutr. Cancer 25, 197–204 10.1080/01635589609514442 [DOI] [PubMed] [Google Scholar]
- Hall J. A., Bouladoux N., Sun C. M., Wohlfert E. A., Blank R. B., Zhu Q., Grigg M. E., Berzofsky J. A., Belkaid Y. (2008). Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 29, 637–649 10.1016/j.immuni.2008.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G., Wang R., Chen G., Wang J., Xu R., Wang L., Feng J., Li X., Guo R., Fu L., Shen B., Li Y. (2010). Interleukin-17-producing gammadelta+ T cells protect NOD mice from type 1 diabetes through a mechanism involving transforming growth factor-beta. Immunology 129, 197–206 10.1111/j.1365-2567.2009.03166.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen H. J., Wildeboer-Veloo A. C., Raangs G. C., Wagendorp A. A., Klijn N., Bindels J. G., Welling G. W. (2000). Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30, 61–67 10.1097/00005176-200001000-00019 [DOI] [PubMed] [Google Scholar]
- Hill D. A., Hoffmann C., Abt M. C., Du Y., Kobuley D., Kirn T. J., Bushman F. D., Artis D. (2010). Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 3, 148–158 10.1038/mi.2009.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins M. J., Macfarlane G. T., Furrie E., Fite A., Macfarlane S. (2005). Characterisation of intestinal bacteria in infant stools using real-time PCR and northern hybridisation analyses. FEMS Microbiol. Ecol. 54, 77–85 10.1016/j.femsec.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Hyttinen V., Kaprio J., Kinnunen L., Koskenvuo M., Tuomilehto J. (2003). Genetic liability of type 1 diabetes and the onset age among 22,650 young Finnish twin pairs: a nationwide follow-up study. Diabetes 52, 1052–1055 10.2337/diabetes.52.4.1052 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Tanaka K., Maeda Y., Aiba Y., Hata A., Tsuji N. M., Koga Y., Matsumoto T. (2008). Effect of intestinal microbiota on the induction of regulatory CD25+ CD4+ T cells. Clin. Exp. Immunol. 153, 127–135 10.1111/j.1365-2249.2008.03668.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. I., Atarashi K., Manel N., Brodie E. L., Shima T., Karaoz U., Wei D., Goldfarb K. C., Santee C. A., Lynch S. V., Tanoue T., Imaoka A., Itoh K., Takeda K., Umesaki Y., Honda K., Littman D. R. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. I., Frutos Rde L., Manel N., Yoshinaga K., Rifkin D. B., Sartor R. B., Finlay B. B., Littman D. R. (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 10.1016/j.chom.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J. P., Wu H. J., Benoist C., Mathis D. (2009). IL-17-producing T cells can augment autoantibody-induced arthritis. Proc. Natl. Acad. Sci. U.S.A. 106, 21789–21794 10.1073/pnas.0912152106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson H. E., Jernberg C., Andersson A. F., Sjolund-Karlsson M., Jansson J. K., Engstrand L. (2010). Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE 5, e9836. 10.1371/journal.pone.0009836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernberg C., Lofmark S., Edlund C., Jansson J. K. (2007). Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 1, 56–66 10.1038/ismej.2007.3 [DOI] [PubMed] [Google Scholar]
- Kalliomaki M., Salminen S., Arvilommi H., Kero P., Koskinen P., Isolauri E. (2001). Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357, 1076–1079 10.1016/S0140-6736(00)04259-8 [DOI] [PubMed] [Google Scholar]
- Kalliomaki M., Salminen S., Poussa T., Arvilommi H., Isolauri E. (2003). Probiotics and prevention of atopic disease: 4-year follow-up of a randomised placebo-controlled trial. Lancet 361, 1869–1871 10.1016/S0140-6736(03)13490-3 [DOI] [PubMed] [Google Scholar]
- Kampman E., Goldbohm R. A., Van Den Brandt P. A., Van’ T Veer P. (1994a). Fermented dairy products, calcium, and colorectal cancer in The Netherlands Cohort Study. Cancer Res. 54, 3186–3190 [PubMed] [Google Scholar]
- Kampman E., Van’ T Veer P., Hiddink G. J., Van Aken-Schneijder P., Kok F. J., Hermus R. J. (1994b). Fermented dairy products, dietary calcium and colon cancer: a case-control study in The Netherlands. Int. J. Cancer 59, 170–176 10.1002/ijc.2910590205 [DOI] [PubMed] [Google Scholar]
- Karimi K., Inman M. D., Bienenstock J., Forsythe P. (2009). Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am. J. Respir. Crit. Care Med. 179, 186–193 [DOI] [PubMed] [Google Scholar]
- Kato I., Endo-Tanaka K., Yokokura T. (1998). Suppressive effects of the oral administration of Lactobacillus casei on type II collagen-induced arthritis in DBA/1 mice. Life Sci. 63, 635–644 10.1016/S0024-3205(98)00315-4 [DOI] [PubMed] [Google Scholar]
- Kearney J., Giovannucci E., Rimm E. B., Ascherio A., Stampfer M. J., Colditz G. A., Wing A., Kampman E., Willett W. C. (1996). Calcium, vitamin D, and dairy foods and the occurrence of colon cancer in men. Am. J. Epidemiol. 143, 907–917 [DOI] [PubMed] [Google Scholar]
- Kirjavainen P. V., Apostolou E., Arvola T., Salminen S. J., Gibson G. R., Isolauri E. (2001). Characterizing the composition of intestinal microflora as a prospective treatment target in infant allergic disease. FEMS Immunol. Med. Microbiol. 32, 1–7 10.1111/j.1574-695X.2001.tb00526.x [DOI] [PubMed] [Google Scholar]
- Kirjavainen P. V., Apostolou E., Salminen S. J., Isolauri E. (1999). New aspects of probiotics – a novel approach in the management of food allergy. Allergy 54, 909–915 10.1034/j.1398-9995.1999.00103.x [DOI] [PubMed] [Google Scholar]
- Klaasen H. L., Koopman J. P., Van Den Brink M. E., Bakker M. H., Poelma F. G., Beynen A. C. (1993a). Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab. Anim. 27, 141–150 10.1258/002367793780810441 [DOI] [PubMed] [Google Scholar]
- Klaasen H. L., Van Der Heijden P. J., Stok W., Poelma F. G., Koopman J. P., Van Den Brink M. E., Bakker M. H., Eling W. M., Beynen A. C. (1993b). Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect. Immun. 61, 303–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig J. E., Spor A., Scalfone N., Fricker A. D., Stombaugh J., Knight R., Angenent L. T., Ley R. E. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4578–4585 10.1073/pnas.1000081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel M. A., Sefik E., Hill J. A., Wu H. J., Benoist C., Mathis D. (2011). Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. U.S.A. 108, 11548–11553 10.1073/pnas.1108924108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I., Banerjee M., Cheng P., Vatan L., Szeliga W., Wei S., Huang E., Finlayson E., Simeone D., Welling T. H., Chang A., Coukos G., Liu R., Zou W. (2009a). Phenotype, distribution, generation, and functional and clinical relevance of Th17 cells in the human tumor environments. Blood 114, 1141–1149 10.1182/blood-2009-03-208249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I., Wei S., Szeliga W., Vatan L., Zou W. (2009b). Endogenous IL-17 contributes to reduced tumor growth and metastasis. Blood 114, 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan M., Cui Z. H., Hoshino H., Lotvall J., Sjostrand M., Gruenert D. C., Skoogh B. E., Linden A. (1999). Neutrophil recruitment by human IL-17 via C-X-C chemokine release in the airways. J. Immunol. 162, 2347–2352 [PubMed] [Google Scholar]
- Laan M., Prause O., Miyamoto M., Sjostrand M., Hytonen A. M., Kaneko T., Lotvall J., Linden A. (2003). A role of GM-CSF in the accumulation of neutrophils in the airways caused by IL-17 and TNF-alpha. Eur. Respir. J. 21, 387–393 10.1183/09031936.03.00303503 [DOI] [PubMed] [Google Scholar]
- Lavasani S., Dzhambazov B., Nouri M., Fak F., Buske S., Molin G., Thorlacius H., Alenfall J., Jeppsson B., Westrom B. (2010). A novel probiotic mixture exerts a therapeutic effect on experimental autoimmune encephalomyelitis mediated by IL-10 producing regulatory T cells. PLoS ONE 5, e9009. 10.1371/journal.pone.0009009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. K., Menezes J. S., Umesaki Y., Mazmanian S. K. (2011). Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4615–4622 10.1073/pnas.1100652108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Hamady M., Lozupone C., Turnbaugh P. J., Ramey R. R., Bircher J. S., Schlegel M. L., Tucker T. A., Schrenzel M. D., Knight R., Gordon J. I. (2008). Evolution of mammals and their gut microbes. Science 320, 1647–1651 10.1126/science.1155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston M., Loach D., Wilson M., Tannock G. W., Baird M. (2010). Gut commensal Lactobacillus reuteri 100-23 stimulates an immunoregulatory response. Immunol. Cell Biol. 88, 99–102 10.1038/icb.2009.71 [DOI] [PubMed] [Google Scholar]
- Lodinova-Zadnikova R., Cukrowska B., Tlaskalova-Hogenova H. (2003). Oral administration of probiotic Escherichia coli after birth reduces frequency of allergies and repeated infections later in life (after 10 and 20 years). Int. Arch. Allergy Immunol. 131, 209–211 10.1159/000071488 [DOI] [PubMed] [Google Scholar]
- Lubberts E., Koenders M. I., Oppers-Walgreen B., Van Den Bersselaar L., Coenen-De Roo C. J., Joosten L. A., Van Den Berg W. B. (2004). Treatment with a neutralizing anti-murine interleukin-17 antibody after the onset of collagen-induced arthritis reduces joint inflammation, cartilage destruction, and bone erosion. Arthritis Rheum. 50, 650–659 10.1002/art.20001 [DOI] [PubMed] [Google Scholar]
- Lyons A., O’Mahony D., O’Brien F., Macsharry J., Sheil B., Ceddia M., Russell W. M., Forsythe P., Bienenstock J., Kiely B., Shanahan F., O’Mahony L. (2010). Bacterial strain-specific induction of Foxp3+ T regulatory cells is protective in murine allergy models. Clin. Exp. Allergy 40, 811–819 [DOI] [PubMed] [Google Scholar]
- Macpherson A. J., Harris N. L. (2004). Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4, 478–485 10.1038/nri1373 [DOI] [PubMed] [Google Scholar]
- Majamaa H., Isolauri E. (1997). Probiotics: a novel approach in the management of food allergy. J. Allergy Clin. Immunol. 99, 179–185 10.1016/S0091-6749(97)70093-9 [DOI] [PubMed] [Google Scholar]
- Malhotra S. L. (1977). Dietary factors in a study of cancer colon from cancer registry, with special reference to the role of saliva, milk and fermented milk products and vegetable fibre. Med. Hypotheses 3, 122–126 10.1016/0306-9877(77)90024-X [DOI] [PubMed] [Google Scholar]
- Mandar R., Mikelsaar M. (1996). Transmission of mother’s microflora to the newborn at birth. Biol. Neonate 69, 30–35 10.1159/000244275 [DOI] [PubMed] [Google Scholar]
- Manirarora J. N., Parnell S. A., Hu Y.-H., Kosiewicz M. M., Alard P. (2011). NOD dendritic cells stimulated with lactobacilli preferentially produce IL-10 versus IL-12 and decrease diabetes incidence. Clin. Dev. Immunol. 2011, 630187. 10.1155/2011/630187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannino D. M., Homa D. M., Pertowski C. A., Ashizawa A., Nixon L. L., Johnson C. A., Ball L. B., Jack E., Kang D. S. (1998). Surveillance for asthma – United States, 1960–1995. MMWR CDC Surveill. Summ. 47, 1–27 [PubMed] [Google Scholar]
- Martin-Orozco N., Chung Y., Chang S. H., Wang Y. H., Dong C. (2009). Th17 cells promote pancreatic inflammation but only induce diabetes efficiently in lymphopenic hosts after conversion into Th1 cells. Eur. J. Immunol. 39, 216–224 10.1002/eji.200838475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki T., Nagata Y., Kado S., Uchida K., Kato I., Hashimoto S., Yokokura T. (1997). Prevention of onset in an insulin-dependent diabetes mellitus model, NOD mice, by oral feeding of Lactobacillus casei. APMIS 105, 643–649 10.1111/j.1699-0463.1997.tb05065.x [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K., Liu C. H., Tzianabos A. O., Kasper D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 10.1016/j.cell.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K., Round J. L., Kasper D. L. (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- Muranski P., Boni A., Antony P. A., Cassard L., Irvine K. R., Kaiser A., Paulos C. M., Palmer D. C., Touloukian C. E., Ptak K., Gattinoni L., Wrzesinski C., Hinrichs C. S., Kerstann K. W., Feigenbaum L., Chan C. C., Restifo N. P. (2008). Tumor-specific Th17-polarized cells eradicate large established melanoma. Blood 112, 362–373 10.1182/blood-2007-11-120998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoopour E., Schwartz J. A., Huszarik K., Sandrock C., Krougly O., Lee-Chan E., Singh B. (2010). Th17 polarized cells from nonobese diabetic mice following mycobacterial adjuvant immunotherapy delay type 1 diabetes. J. Immunol. 184, 4779–4788 10.4049/jimmunol.0902822 [DOI] [PubMed] [Google Scholar]
- Noverr M. C., Huffnagle G. B. (2005). The “microflora hypothesis” of allergic diseases. Clin. Exp. Allergy 35, 1511–1520 10.1111/j.1365-2222.2005.02379.x [DOI] [PubMed] [Google Scholar]
- Noverr M. C., Noggle R. M., Toews G. B., Huffnagle G. B. (2004). Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect. Immun. 72, 4996–5003 10.1128/IAI.72.9.4996-5003.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Reparaz J., Mielcarz D. W., Ditrio L. E., Burroughs A. R., Foureau D. M., Haque-Begum S., Kasper L. H. (2009). Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J. Immunol. 183, 6041–6050 10.4049/jimmunol.0900747 [DOI] [PubMed] [Google Scholar]
- Ochoa-Reparaz J., Mielcarz D. W., Wang Y., Begum-Haque S., Dasgupta S., Kasper D. L., Kasper L. H. (2010). A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 3, 487–495 10.1038/mi.2010.29 [DOI] [PubMed] [Google Scholar]
- Okada H., Kuhn C., Feillet H., Bach J. F. (2010). The “hygiene hypothesis” for autoimmune and allergic diseases: an update. Clin. Exp. Immunol. 160, 1–9 10.1111/j.1365-2249.2009.04098.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe S. J., Ou J., Aufreiter S., O’Connor D., Sharma S., Sepulveda J., Fukuwatari T., Shibata K., Mawhinney T. (2009). Products of the colonic microbiota mediate the effects of diet on colon cancer risk. J. Nutr. 139, 2044–2048 10.3945/jn.109.104380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Mahony C., Scully P., O’Mahony D., Murphy S., O’Brien F., Lyons A., Sherlock G., Macsharry J., Kiely B., Shanahan F., O’Mahony L. (2008). Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog. 4, e1000112. 10.1371/journal.ppat.1000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onderdonk A. B., Steeves R. M., Cisneros R. L., Bronson R. T. (1984). Adoptive transfer of immune enhancement of experimental ulcerative colitis. Infect. Immun. 46, 64–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama N., Sudo N., Sogawa H., Kubo C. (2001). Antibiotic use during infancy promotes a shift in the T(H)1/T(H)2 balance toward T(H)2-dominant immunity in mice. J. Allergy Clin. Immunol. 107, 153–159 10.1067/mai.2001.111142 [DOI] [PubMed] [Google Scholar]
- Packey C. D., Sartor R. B. (2008). Interplay of commensal and pathogenic bacteria, genetic mutations, and immunoregulatory defects in the pathogenesis of inflammatory bowel diseases. J. Intern. Med. 263, 597–606 10.1111/j.1365-2796.2008.01962.x [DOI] [PubMed] [Google Scholar]
- Palmer C., Bik E. M., Digiulio D. B., Relman D. A., Brown P. O. (2007). Development of the human infant intestinal microbiota. PLoS Biol. 5, e177. 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J., Thijs C., Vink C., Stelma F. F., Snijders B., Kummeling I., Van Den Brandt P. A., Stobberingh E. E. (2006). Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118, 511–521 10.1542/peds.2005-2824 [DOI] [PubMed] [Google Scholar]
- Peters R. K., Pike M. C., Garabrant D., Mack T. M. (1992). Diet and colon cancer in Los Angeles County, California. Cancer Causes Control 3, 457–473 10.1007/BF00051359 [DOI] [PubMed] [Google Scholar]
- Pineton de Chambrun G., Colombel J. F., Poulain D., Darfeuille-Michaud A. (2008). Pathogenic agents in inflammatory bowel diseases. Curr. Opin. Gastroenterol. 24, 440–447 10.1097/MOG.0b013e3283023be5 [DOI] [PubMed] [Google Scholar]
- Poole B. D., Scofield R. H., Harley J. B., James J. A. (2006). Epstein-Barr virus and molecular mimicry in systemic lupus erythematosus. Autoimmunity 39, 63–70 10.1080/08916930500484849 [DOI] [PubMed] [Google Scholar]
- Pool-Zobel B. L., Neudecker C., Domizlaff I., Ji S., Schillinger U., Rumney C., Moretti M., Vilarini I., Scassellati-Sforzolini R., Rowland I. (1996). Lactobacillus- and Bifidobacterium-mediated antigenotoxicity in the colon of rats. Nutr. Cancer 26, 365–380 10.1080/01635589609514492 [DOI] [PubMed] [Google Scholar]
- Rabizadeh S., Rhee K. J., Wu S., Huso D., Gan C. M., Golub J. E., Wu X., Zhang M., Sears C. L. (2007). Enterotoxigenic Bacteroides fragilis: a potential instigator of colitis. Inflamm. Bowel Dis. 13, 1475–1483 10.1002/ibd.20265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M., Smidt H., De Vos W. M. (2007). Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 9, 2125–2136 10.1111/j.1462-2920.2007.01369.x [DOI] [PubMed] [Google Scholar]
- Rhee K. J., Wu S., Wu X., Huso D. L., Karim B., Franco A. A., Rabizadeh S., Golub J. E., Mathews L. E., Shin J., Sartor R. B., Golenbock D., Hamad A. R., Gan C. M., Housseau F., Sears C. L. (2009). Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect. Immun. 77, 1708–1718 10.1128/IAI.00814-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeldt V., Benfeldt E., Nielsen S. D., Michaelsen K. F., Jeppesen D. L., Valerius N. H., Paerregaard A. (2003). Effect of probiotic Lactobacillus strains in children with atopic dermatitis. J. Allergy Clin. Immunol. 111, 389–395 10.1067/mai.2003.389 [DOI] [PubMed] [Google Scholar]
- Round J. L., Lee S. M., Li J., Tran G., Jabri B., Chatila T. A., Mazmanian S. K. (2011). The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977 10.1126/science.1206095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J. L., Mazmanian S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 10.1038/nri2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland I. R., Rumney C. J., Coutts J. T., Lievense L. C. (1998). Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis 19, 281–285 10.1093/carcin/19.2.281 [DOI] [PubMed] [Google Scholar]
- Sanderson J. D., Hermon-Taylor J. (1992). Mycobacterial diseases of the gut: some impact from molecular biology. Gut 33, 145–147 10.1136/gut.33.7.890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfanos K. S., Bruno T. C., Maris C. H., Xu L., Thoburn C. J., Demarzo A. M., Meeker A. K., Isaacs W. B., Drake C. G. (2008). Phenotypic analysis of prostate-infiltrating lymphocytes reveals TH17 and Treg skewing. Clin. Cancer Res. 14, 3254–3261 10.1158/1078-0432.CCR-07-5164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva W. A., Jr., Covas D. T., Panepucci R. A., Proto-Siqueira R., Siufi J. L., Zanette D. L., Santos A. R., Zago M. A. (2003). The profile of gene expression of human marrow mesenchymal stem cells. Stem Cells 21, 661–669 10.1634/stemcells.21-6-661 [DOI] [PubMed] [Google Scholar]
- Snel J., Heinen P. P., Blok H. J., Carman R. J., Duncan A. J., Allen P. C., Collins M. D. (1995). Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus arthromitus.” Int. J. Syst. Bacteriol. 45, 780–782 10.1099/00207713-45-4-780 [DOI] [PubMed] [Google Scholar]
- So J. S., Kwon H. K., Lee C. G., Yi H. J., Park J. A., Lim S. Y., Hwang K. C., Jeon Y. H., Im S. H. (2008). Lactobacillus casei suppresses experimental arthritis by down-regulating T helper 1 effector functions. Mol. Immunol 45, 2690–2699 10.1016/j.molimm.2007.12.010 [DOI] [PubMed] [Google Scholar]
- Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermudez-Humaran L. G., Gratadoux J. J., Blugeon S., Bridonneau C., Furet J. P., Corthier G., Grangette C., Vasquez N., Pochart P., Trugnan G., Thomas G., Blottiere H. M., Dore J., Marteau P., Seksik P., Langella P. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 105, 16731–16736 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H., Seksik P., Furet J. P., Firmesse O., Nion-Larmurier I., Beaugerie L., Cosnes J., Corthier G., Marteau P., Dore J. (2009). Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 15, 1183–1189 10.1002/ibd.20903 [DOI] [PubMed] [Google Scholar]
- Sood A., Midha V., Makharia G. K., Ahuja V., Singal D., Goswami P., Tandon R. K. (2009). The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin. Gastroenterol. Hepatol. 7, 1202–1209, 1209 e1201. 10.1016/j.cgh.2009.07.016 [DOI] [PubMed] [Google Scholar]
- Stepankova R., Powrie F., Kofronova O., Kozakova H., Hudcovic T., Hrncir T., Uhlig H., Read S., Rehakova Z., Benada O., Heczko P., Strus M., Bland P., Tlaskalova-Hogenova H. (2007). Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm. Bowel Dis. 13, 1202–1211 10.1002/ibd.20221 [DOI] [PubMed] [Google Scholar]
- Stone M., Fortin P. R., Pacheco-Tena C., Inman R. D. (2003). Should tetracycline treatment be used more extensively for rheumatoid arthritis? Metaanalysis demonstrates clinical benefit with reduction in disease activity. J. Rheumatol. 30, 2112–2122 [PubMed] [Google Scholar]
- Talham G. L., Jiang H. Q., Bos N. A., Cebra J. J. (1999). Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect. Immun. 67, 1992–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivanen P. (2003). Normal intestinal microbiota in the aetiopathogenesis of rheumatoid arthritis. Ann. Rheum. Dis. 62, 807–811 10.1136/ard.62.9.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ridaura V. K., Faith J. J., Rey F. E., Knight R., Gordon J. I. (2009). The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14. 10.1126/scitranslmed.3000322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursi A., Brandimarte G., Papa A., Giglio A., Elisei W., Giorgetti G. M., Forti G., Morini S., Hassan C., Pistoia M. A., Modeo M. E., Rodino S., D’Amico T., Sebkova L., Sacca N., Di Giulio E., Luzza F., Imeneo M., Larussa T., Di Rosa S., Annese V., Danese S., Gasbarrini A. (2010). Treatment of relapsing mild-to-moderate ulcerative colitis with the probiotic VSL#3 as adjunctive to a standard pharmaceutical treatment: a double-blind, randomized, placebo-controlled study. Am. J. Gastroenterol. 105, 2218–2227 10.1038/ajg.2010.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesaki Y., Okada Y., Matsumoto S., Imaoka A., Setoyama H. (1995). Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol. Immunol. 39, 555–562 [DOI] [PubMed] [Google Scholar]
- Uronis J. M., Muhlbauer M., Herfarth H. H., Rubinas T. C., Jones G. S., Jobin C. (2009). Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS ONE 4, e6026. 10.1371/journal.pone.0006026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaahtovuo J., Munukka E., Korkeamaki M., Luukkainen R., Toivanen P. (2008). Fecal microbiota in early rheumatoid arthritis. J. Rheumatol. 35, 1500–1505 [PubMed] [Google Scholar]
- Veiga P., Gallini C. A., Beal C., Michaud M., Delaney M. L., Dubois A., Khlebnikov A., Van Hylckama Vlieg J. E., Punit S., Glickman J. N., Onderdonk A., Glimcher L. H., Garrett W. S. (2010). Bifidobacterium animalis subsp. Lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc. Natl. Acad. Sci. U.S.A. 107, 18132–18137 10.1073/pnas.1011737107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viljanen M., Savilahti E., Haahtela T., Juntunen-Backman K., Korpela R., Poussa T., Tuure T., Kuitunen M. (2005). Probiotics in the treatment of atopic eczema/dermatitis syndrome in infants: a double-blind placebo-controlled trial. Allergy 60, 494–500 10.1111/j.1398-9995.2004.00514.x [DOI] [PubMed] [Google Scholar]
- Vliagoftis H., Kouranos V. D., Betsi G. I., Falagas M. E. (2008). Probiotics for the treatment of allergic rhinitis and asthma: systematic review of randomized controlled trials. Ann. Allergy Asthma Immunol. 101, 570–579 10.1016/S1081-1206(10)60219-0 [DOI] [PubMed] [Google Scholar]
- Ward J. M., Anver M. R., Haines D. C., Melhorn J. M., Gorelick P., Yan L., Fox J. G. (1996). Inflammatory large bowel disease in immunodeficient mice naturally infected with Helicobacter hepaticus. Lab. Anim. Sci. 46, 15–20 [PubMed] [Google Scholar]
- Wen L., Ley R. E., Volchkov P. Y., Stranges P. B., Avanesyan L., Stonebraker A. C., Hu C., Wong F. S., Szot G. L., Bluestone J. A., Gordon J. I., Chervonsky A. V. (2008). Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455, 1109–1113 10.1038/nature07336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. J., Ivanov Ii., Darce J., Hattori K., Shima T., Umesaki Y., Littman D. R., Benoist C., Mathis D. (2010). Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 10.1016/j.immuni.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Rhee K. J., Albesiano E., Rabizadeh S., Wu X., Yen H. R., Huso D. L., Brancati F. L., Wick E., Mcallister F., Housseau F., Pardoll D. M., Sears C. L. (2009). A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15, 1016–1022 10.1038/nm.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Fanslow W. C., Seldin M. F., Rousseau A. M., Painter S. L., Comeau M. R., Cohen J. I., Spriggs M. K. (1995). Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity 3, 811–821 10.1016/1074-7613(95)90070-5 [DOI] [PubMed] [Google Scholar]
- Yoshioka H., Iseki K., Fujita K. (1983). Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics 72, 317–321 [PubMed] [Google Scholar]
- Zandman-Goddard G., Shoenfeld Y. (2005). Infections and SLE. Autoimmunity 38, 473–485 10.1080/08916930500285352 [DOI] [PubMed] [Google Scholar]
- Zhang B., Rong G., Wei H., Zhang M., Bi J., Ma L., Xue X., Wei G., Liu X., Fang G. (2008). The prevalence of Th17 cells in patients with gastric cancer. Biochem. Biophys. Res. Commun. 374, 533–537 10.1016/j.bbrc.2008.07.060 [DOI] [PubMed] [Google Scholar]
- Zhang L. L., Chen X., Zheng P. Y., Luo Y., Lu G. F., Liu Z. Q., Huang H., Yang P. C. (2010). Oral Bifidobacterium modulates intestinal immune inflammation in mice with food allergy. J. Gastroenterol. Hepatol. 25, 928–934 10.1111/j.1440-1746.2009.05921.x [DOI] [PubMed] [Google Scholar]