Abstract
Mitogen-activated protein kinase (MAPK) pathway signaling plays an important role in the majority of non-small-cell lung cancers (NSCLCs). In a prior microarray analysis of epidermal growth factor receptor (EGFR) inhibition in NSCLC cell lines, we noted that several dual specificity phosphatases (DUSPs) were among the most highly and immediately regulated genes. DUSPs act as natural terminators of MAPK signal transduction and therefore, we hypothesized a tumor suppressive role via feedback mechanisms. In the current study, we focus on the assessment of DUSP6, a cytoplasmic DUSP with high specificity for extracellular signal-regulated kinase (ERK). We demonstrate that DUSP6 expression tracks in tandem with ERK inhibition and that regulation of DUSP6 is mediated at the promoter level by ETS1, a well-known nuclear target of activated ERK. Small interfering RNA knockdown in DUSP6-high H441 lung cancer cells significantly increased ERK activation and cellular proliferation, whereas plasmid-driven overexpression in DUSP6-low H1975 lung cancer cells significantly reduced ERK activation and cellular proliferation and promoted apoptosis. Also, DUSP6 overexpression synergized with EGFR inhibitor treatment in EGFR-mutant HCC827 cells. Our results indicate that DUSP6 expression is regulated by ERK signaling and that DUSP6 exerts antitumor effects via negative feedback regulation, pointing to an important feedback loop in NSCLC. Further studies assessing the tumor suppressive role of DUSP6 and strategies aimed at modulation of its activity are warranted.
Introduction
Lung cancer, in particular non-small-cell lung cancer (NSCLC) remains the leading cause of cancer deaths in both men and women in the USA (1). Despite recent progress in the diagnosis and treatment of NSCLC, survival remains poor (2). Improved outcomes are expected from better understanding of the molecular mechanisms underlying tumorigenesis. The extracellular signal-regulated kinase (ERK) pathway plays an important role in oncogenesis and its overactivation is present in the majority of NSCLC, particularly those with epidermal growth factor receptor (EGFR) and K-RAS mutations (3). EGFR belongs to the HER (or ErbB) family of growth factor receptor tyrosine kinases. Upon ligand binding, these receptors homodimerize or heterodimerize, resulting in autophosphorylation, activation and subsequent activation of intracellular signaling cascades, most notably the RAS-RAF-MEK-ERK pathway. Small-molecule EGFR tyrosine kinase inhibitors (TKIs), such as gefitinib and erlotinib, have shown benefit in patients with advanced NSCLC (4,5). The majority of patients with EGFR TKI-responsive tumors carry activating mutations of EGFR, such as L858R or exon 19 deletions (6–8). Depending on the population studied, EGFR mutations occur on average in 10–20% of patients and identify a specific subset of patients highly dependent on oncogenic EGFR signaling (9). K-RAS mutations, which appear to be mutually exclusive of EGFR mutations occur in ∼20 to 30% of adenocarcinomas and their oncogenic potential is principally mediated via overactivation of ERK (10). Therefore, ERK signaling appears important or critical in at least 30–50% of NSCLC. However, little is known regarding regulation of ERK signaling. ERK1/2 is activated by dual threonine and tyrosine phosphorylation of a TEY motif by the mitogen-activated protein kinases (MAPKs), mitogen-activated protein kinase kinase 1 (MEK1) and mitogen-activated protein kinase kinase 2 (MEK2). Inactivation of ERK1/2 is achieved by dephosphorylation of this TEY motif by distinct members of the dual specificity phosphatase (DUSP) family, including both cytoplasmic (DUSP6, 7 and 9) and nuclear DUSPs (DUSP5) (11). The specific feedback regulatory mechanisms of ERK signaling in lung cancer cells have not been defined. Defects of feedback regulation are posited to contribute to oncogenesis, and an understanding of these mechanisms could provide novel strategies for biomarker and treatment development.
Clinical experience has shown that the majority of patients who initially respond to EGFR TKI treatment eventually develop resistance, most commonly via secondary mutations in EGFR such as T790M (12,13). Irreversible EGFR inhibitors, such as CL-387,785 or HKI-272 can overcome the resistance conferred by this secondary mutation (14–16). H1975, an NSCLC cell line harboring the EGFR-T790M mutation, is highly resistant to gefitinib/erlotinib but sensitive to CL-387,785. Using microarray transcriptional profiling of H1975 cells exposed to CL-387,785 or gefitinib, we identified candidate downstream effectors of oncogenic EGFR signaling, specifically demonstrating that the transcription of several DUSPs is highly suppressed at 6 h by CL-387,785 but not by gefitinib (17). There are ∼65 genes encoding a heterogeneous group of phosphatases broadly described as DUSPs (18). The structure of DUSP proteins confers activity for both phosphoserine/threonine and phosphotyrosine residues. DUSPs are characterized by a common structure, comprising a C-terminal catalytic domain and an N-terminal non-catalytic domain. These enzymes are defined by the active-site signature motif HCX5R, in which the cysteine residue functions as a nucleophile essential to catalysis. A subgroup of DUSPs, mitogen-activated protein kinase-specific phosphatases (MKPs) display distinct patterns of induction, subcellular localization and specificity for individual MAPKs and constitute a response network of phosphatases which attenuate MAPK-dependent signaling (11).
DUSP6 (previously called MKP-3) is a prototypical member of a subfamily of cytoplasmic MKPs, which includes DUSP7 and DUSP9 as well. These enzymes all display a high degree of substrate selectivity for ERK1/2 (19). DUSP6 has been shown to act as a central feedback regulator attenuating ERK levels in developmental programs (20,21). The cytoplasmic localization of DUSP6 is mediated by a chromosome region maintenance-1-dependent nuclear export pathway. DUSP6 appears to play a role in determining the subcellular localization of ERK by serving as a bona fide cytoplasmic anchor for ERK, thereby mediating a spatio-temporal mechanism of ERK signaling regulation. Cytoplasmic retention of ERK requires both a functional kinase interaction motif and nuclear export site. DUSP6 null mice demonstrate enhanced ERK1/2 phosphorylation leading to increased myocyte proliferation and cardiac hypercellularity (22). A recent in vivo study identified DUSP6 as a negative feedback regulator of fibroblast growth factor-stimulated ERK signaling during murine development (21). Several in vitro studies have demonstrated that DUSP6 acts as a negative regulator of fibroblast growth factor receptor signaling and endothelial cell platelet-derived growth factor receptor signaling via termination of ERK activation (23,24). The DUSP6 gene is localized to 12q21–q22, a chromosomal region showing frequent loss of heterozygosity in pancreatic cancer (25). Immunohistochemical staining demonstrated reduced DUSP6 expression in about half of all invasive pancreatic carcinomas, whereas expression was preserved in precursor lesions suggesting that loss of DUSP6 plays a role in tumor progression (26,27). Similarly, loss of DUSP6 expression mediated by oxidative stress-mediated degradation was also noted in ovarian cancer and correlated with high ERK1/2 activity (28). The functional and clinical significance of DUSP6-mediated regulation of ERK signaling in lung cancer has not been carefully investigated. In the current study, we examine effects on DUSP6 expression by EGFR/ERK inhibition and study its negative feedback regulation of ERK activation in NSCLC cell lines.
Materials and methods
Cell lines
The following NSCLC cell lines were obtained from American Type Tissue Collection (Manassas, VA): HCC827, PC9, H1975, A549, H441, H358, Calu-3, H1838, H1650, H125, H1703, H23, H2228, Calu-1, Calu-6, SW900, SK-LU-1, H1993, H1734, H520, SK-MES-1, H157, H460 and H3255. Normal human airway epithelial cell line NuLi-1 cells were provided as a gift from Dr Jeffrey Kern and cultured in F-12/Dulbecco’s modified Eagle’s medium (1:1 ratio) media with 10% fetal bovine serum (FBS). Lung cancer cells were grown in RPMI 1640 supplemented with 10% FBS and 1× Antibiotic/Antimycotic (Invitrogen, Carlsbad, CA) and were in the logarithmic growth phase at initiation of all experiments. EGFR inhibitor erlotinib was obtained from Selleck Chemicals (Houston, TX); irreversible EGFR inhibitor CL-387,785 and MEK1/2 inhibitor U0126 were purchased from Calbiochem (San Diego, CA). Drugs were dissolved in dimethyl sulfoxide (DMSO) at 10 mM and stored at −20°C. The final DMSO concentration in all experiments was <0.5% in medium.
Immunoblotting
Cells were serum-starved overnight and whole cell lysates were analyzed by western blotting as described previously (14). Antibodies to DUSP6, ETS1, ETS2 and hemagglutinin (HA) tag were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against phosphor-EGFR at different tyrosine sites, total-EGFR, phosphorylated-ERK1/2, total-ERK1/2, poly ADP ribose polymerase (PARP) and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were purchased from Cell Signaling Technology (Boston, MA).
Immunohistochemistry
Formalin-fixed primary lung tumor tissue sections were deparaffinized and rehydrated and incubated with 0.6% hydrogen peroxide in methanol, followed by staining using the R.T.U Vectastain Universal Quick Kit (Vector Laboratories, Burlingame, CA). Antigen retrieval treatment with sodium citrate (10 mM, pH 6.0) was used for the detection of DUSP6, whereas a specific retrieving reagent (Dako, target retrieval solution, pH 9.0) was used for the detection of P-ERK1/2. Rabbit polyclonal antibody specific against DUSP6 (Santa Cruz Biotechnology) was used at a dilution of 1:100 (optimal dilution determined in serial dilution studies) and a rabbit antibody against P-ERK1/2 was used (Cell Signaling Technology) at a dilution of 1:1000 for overnight incubation at 4°C followed by hematoxylin nuclear counterstaining. The intensity of the staining of DUSP6/P-ERK was scored by a pulmonary pathologist (A.C.B.) as 0/1 (non-detectable/weak) versus 2 (strong staining).
Quantitative reverse transcription–polymerase chain reaction assay
Total RNA was collected from cultured cells using PureLink Micro-to-Midi Total RNA Purification kit (Invitrogen). Complementary DNA was synthesized and reverse transcription–polymerase chain reaction (PCR) was performed as described previously (29). The primers used for DUSP6 quantitative reverse transcription–PCR were sense 5′-GAGTCTGACCTTGACCGAGACCCCAA-3′ and antisense 5′-TTCCTCCAACACGTCCAAGTTGGTGGAGTC-3′.
Plasmid constructs and cellular transfection
Original vector containing the complementary DNA sequence of human DUSP6 was purchased from OPEN Biosystems (Huntsville, AL) and modified with an HA-tag at the C-terminus by overlapping PCR to distinguish plasmid derived from native protein. An enzyme-dead DUSP6 expression construct was generated via 293 Cysteine to Glycine (C293G) point mutation (30,31) using the QuikChange Site-Directed Mutagenesis XL II kit (Stratagene, La Jolla, CA). The accuracy of all constructs was confirmed by direct DNA sequencing. The C293G mutation was constructed using the following oligonucleotides: sense 5′-TGGTGTCTTGGTACATGGCTTGGCTGGCATTAGCC-3′ and antisense 5′-GGCTAATGCCAGCCAAGCCATGTACCAAGACACCA-3′. Both variants were subcloned into the pcDNA3.1 backbone vector. COS7 cells were transiently transfected with one of three expression vectors as follows: wild-type DUSP6 (pcDNA3.1-DUSP6), enzyme-dead DUSP6 (pcDNA3.1-DUSP6-CG) with C293G mutation or empty pcDNA3.1 vector (pcDNA3.1-EV), using Fugene 6 according to the manufacturer's protocol (Roche, Indianapolis, IN). Whole cell lysates for immunoblotting were collected at indicated time points after transfection to confirm appropriate plasmid DUSP6 expression. H1975 and HCC827 cells were transfected by identical means. Stably transfected subclones were selected with G418 at a concentration of 500 μg/ml starting 48 h posttransfection.
MTS cell growth assay
H1975 stable transfectants were seeded at a density of 6000 cells per well in 96-well plates in RPMI 1640 containing 10% FBS overnight and then maintained in 0.5% FBS media for 3 days. Viable cell numbers were determined using MTS assay kit according to the manufacturer's protocol (Promega, Madison, WI). Each assay consisted of five replicate wells.
BrdU and annexin/propidium iodide assays
For both assays, samples were analyzed on a fluorescence-activated cell scan cytometer EPICS XL MCL (Beckman Coulter, Miami, FL). Bromo-deoxyuridine (BrdU) cell proliferation assay was performed according to manufacturer's instructions (FITC BrdU Flow Kit, BD Pharmingen, San Diego CA). Briefly, H1975 stably transfected subclones were cultured in the 0.5% serum media for 3 days, then pulse labeled for 60 min with 10 μM BrdU, collected by trypsinization and washed with phosphate-buffered saline, stained with fluorescent anti-BrdU antibody, counterstained with 7-amino-actinomycin D for total DNA content and analyzed by flow cytometry. Annexin/propidium iodide (PI) apoptosis assay was performed according to the manufacturer's instructions (Annexin V-FLUOS staining kit, Roche). Briefly, H1975 stably transfected subclones were cultured in 0.5% serum media for 3 days, collected by trypsinization and washed with phosphate-buffered saline, stained with annexin/PI and analyzed by flow cytometry. In synergism studies, HCC827 cells were transiently transfected with plasmid constructs 24 h prior to 48 h treatment with erlotinib, followed by annexin/PI staining as above.
Small interfering RNA knockdown
Knockdown of DUSP6 or ETS1 was performed using specific small interfering RNA (siRNA) pools targeting DUSP6 or ETS1 (SMARTpool) purchased from Dharmacon RNAi Technologies (Thermo, Rockford, IL). SiGENOME Non-targeting siRNA Pools and siGLO Lamin A/C Control siRNA served as negative and positive control, respectively. Introduction of siRNA was performed with DharmaFect1 according to the manufacturer's instructions (Thermo). Levels of DUSP6 or ETS1 knockdown at different time points were assessed by immunoblot analysis in pools of transfected cells.
Luciferase reporter assay
HCC827 cells were grown to 40–50% confluence in triplicates on six-well plates and then transfected using Fugene HD (Roche) with 50 ng of pGL4.74-Renilla luciferase and one of the following plasmids: 0.5 μg of pGL3Basic-DUSP6-Firefly luciferase construct containing 508 bp promoter sequence (−359 to −866) upstream of the DUSP6 gene start codon, generously provided by Dr Stephen M.Keyse, Ninewells Hospital and Medical School, Dundee, UK (23), or 0.5 μg of pGL3Basic empty vector. After 24 h of transfection, cells were treated with 1 μM erlotinib or DMSO as a vehicle control for 6 h. Cell extracts were prepared and luciferase assays were run as described previously (29).
Chromatin immunoprecipitation assay
HCC827 cells were starved with serum-free media overnight, then treated with 1 μM erlotinib or 0.01% DMSO control for 6 h, followed by chromatin immunoprecipitation assay according to manufacturer's instructions (Upstate Biotechnology, Lake Placid, NY). Briefly, cells were cross-linked with 1% formaldehyde, then chromatin was extracted, sonicated and immunoprecipitated with 5 μg of ETS1 or ETS2 antibody (Santa Cruz Biotechnology) at 4°C overnight with rotation. Prior to immunoprecipitation, 10% of each nuclear extract was set aside as input chromatin DNA for use in assay controls. Cross-linking of immunoprecipitated and input samples was reversed by heating at 65°C in the presence of 5 M NaCl for 4 h followed by DNA isolation using QIAquick PCR Purification Kit (QIAGEN, Valencia, CA). Immune complexes were collected by incubation with supplied protein A agarose/Salmon Sperm DNA beads for 1 h at 4°C with rotation. Binding of ETS1/2 to the DUSP6 promoter was assessed by nested PCR and with primer sets amplifying ETS1/2-binding site regions spanning −844 to −404 bp (441 bp in size) and −743 to −487 bp (257 bp in size) of the DUSP6 promoter. An unrelated anti-HA antibody and an unrelated primer pair was used to amplify the sequence downstream of the ETS-binding site, +995 to +1311 bp (317 bp) to serve as control. PCR products were analyzed using a 1% agarose gel run in 1× TAE and stained with ethidium bromide.
Electrophoretic mobility shift assay
All oligonucleotides probes were obtained from Invitrogen. 5′-Biotin-labeled and identical unlabeled oligonucleotide probes, corresponding to the ETS1/2-binding sequences in the DUSP6 promoter region, were used as follows: (sense) 5′-GGCTTATCCGGAGCGGAAATTCCTTTC and (antisense) 5′-GAAAGGAATTTCCGCTCCGGATAAGCC. Two mutant oligonucleotide probes were generated by introduction of mutations within the 508 bp DUSP6 promoter sequence as follows (23): core ETS-binding site GGA (underlined) mutated to TGA; palindromic ETS-binding site TCC (underlined) mutated to GAA. Nuclear extracts were collected from HCC827 cells in presence of erlotinib (1 μM) or 0.01% DMSO control for 6 h. Double-stranded DNA annealing was achieved by incubating complementary pairs of oligonucleotide probes at 95°C for 5 min followed by slow cooling to room temperature. Binding reactions were performed using Pierce LightShift Chemiluminescent EMSA Kit as described previously (29).
Statistical analysis
Fisher's exact test was used to estimate association between categorical measurements. Differences in a continuous measurement between two or more groups were examined by χ2 test. All tests were two sided and P-value < 0.05 was considered statistically significant.
Results
DUSP6 protein expression correlates with ERK signaling activation in lung cancer cell lines and primary NSCLCs
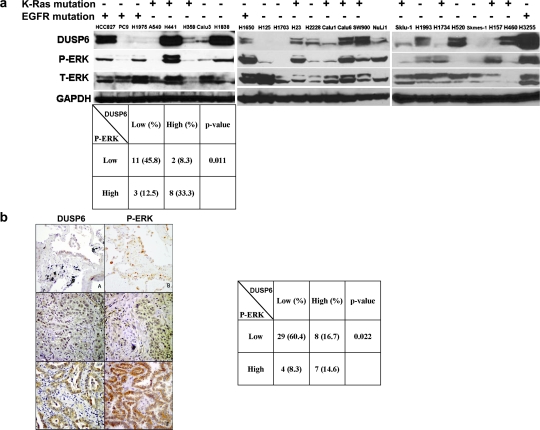
We assayed cellular expression of DUSP6 by immunoblot using an anti-DUSP6 antibody in 24 NSCLC cell lines and a normal human airway epithelial cell line (NuLi-1), correlated to total and P-ERK expression to assess ERK-activation (Figure 1a). We observed DUSP6 expression in the majority of lung cancer cell lines. Although DUSP6 levels varied widely among different cell lines, a positive association between DUSP6 expression and ERK activity was observed (P = 0.011). Only 15.4% (2/13) of cell lines with low P-ERK expression had high expression of DUSP6, whereas 72.7% (8/11) of cell lines with high P-ERK expression had high expression of DUSP6. Next, we performed an immunohistochemical study of DUSP6 and P-ERK expression on 48 primary, human non-small cell lung tumors. As expected, no nuclear expression of DUSP6 was detected consistent with exclusive cytoplasmic localization of this protein. Altogether, 15/48 (31%) of tumors showed strong expression of DUSP6, whereas 11/48 (23%) of the tumors demonstrated strong cytoplasmic and 19/48 (40%) strong nuclear P-ERK staining (Figure 1b). Analogous to our cell line data, a statistically significant correlation was found between cytoplasmic P-ERK and DUSP6 expression (P = 0.022), whereas a trend was also observed between DUSP6 expression and nuclear P-ERK (P = 0.051). Most strikingly, 13/15 (87%) specimens with strong DUSP6 expression showed strong P-ERK staining in either cytoplasmic or nuclear localization as compared with only 11/34 (32%) samples with weak DUSP6 had strong cytoplasmic or nuclear P-ERK (P < 0.001). These results altogether show a close correlation between ERK pathway activation and DUSP6 expression in NSCLC.
Fig. 1.
DUSP6 expression in NSCLC cell lines and primary patient samples. (a) Cells were plated and starved with serum-free media overnight, then lysed with 10% trichloroacetic acid lysis buffer, followed by immunoblotting for DUSP6, phosphor-ERK, total-ERK and GAPDH. Mutation status of EGFR and K-Ras was annotated for all the NSCLC cell lines. (b) Expression of DUSP6 and phospho-ERK in 48 primary NSCLC tumors. The expression of DUSP6 and P-ERK in normal lung epithelia (panels A and B) and primary lung cancers was examined by immunohistochemistry. Cytoplasmic immunoreactivity of DUSP6 and P-ERK expression were subdivided into two categories: low (panels C and D) and high (panels E and F, A–F, diaminobenzidene immunohistochemistry, original magnification ×100). Fisher's exact test was used for the statistical analysis of the correlation between DUSP6 and P-ERK levels for both the cell lines and patient samples.
DUSP6 expression is downregulated by EGFR inhibition in EGFR-dependent lung cancer cell lines
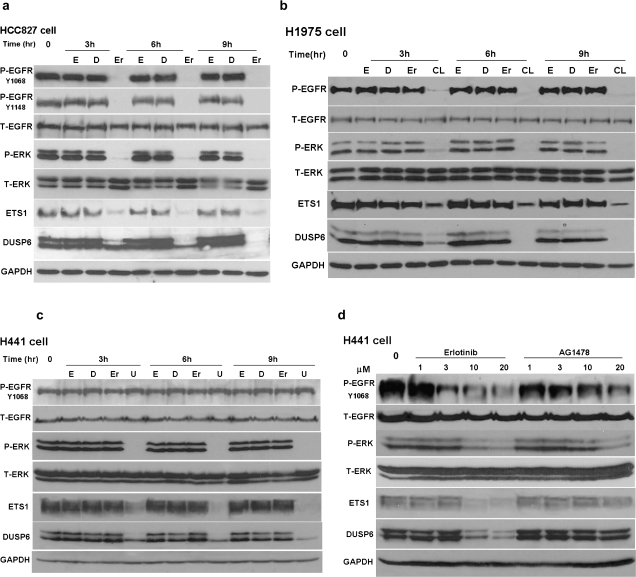
EGFR L858R-mutant H1975 cells also and PC9 are NSCLC cell lines that carry an EGFR exon 19 deletion and thereby are highly sensitive to gefitinib or erlotinib. EGFR L858R-mutant H1975 cells also harbor the T790M resistance mutation but are sensitive to the irreversible EGFR inhibitor CL-387,785. Downregulation of DUSP6 strongly correlated with effective inhibition of EGFR as well as ERK activation demonstrated by diminished P-EGFR and P-ERK expression in EGFR-dependent cell lines (Figure 2a and b and supplementary Figure 1 is available at Carcinogenesis Online). Of note is that the anti-DUSP6 antibody detects two distinct protein bands corresponding in size to translation products initiating at the first ATG and the second ATG (Met14) as previously reported (32). Our studies reveal that the larger of these two proteins is more rapidly suppressed by TKI treatment; the mechanism is currently unclear. Quantitative reverse transcription–PCR demonstrated downregulation of DUSP6 transcription following erlotinib treatment in both HCC827 and PC9 cells (supplementary Figure 2 is available at Carcinogenesis Online), in line with our prior transcriptional profiling findings in CL-387,785 treated H1975 cells (17). DUSP6 downregulation was also observed upon treatment of HCC827 cells with the MEK1/2 inhibitor U0126 accompanied by effective inhibition of P-ERK but no changes in EGFR activation status suggesting that the regulation of DUSP6 expression occurs downstream of EGFR (supplementary Figure 1 is available at Carcinogenesis Online).
Fig. 2.
DUSP6 is regulated by EGFR/ERK inhibition in NSCLC cell lines. Protein expression levels were assayed by immunoblot for phosphor-EGFR at indicated tyrosine sites, total-EGFR, phosphor-ERK, total-ERK, ETS1, DUSP6 and GAPDH demonstrating suppression of DUSP6 following inhibition of activated ERK (P-ERK) and ETS1 levels in the presence of appropriate drug for each of the following cell lines: (a) HCC827 cells treated with erlotinib; (b) H1975 cells treated with erlotinib or CL-387,785 (an irreversible EGFR inhibitor); H441 cells treated with (c) erlotinib or U0126 (an MEK1/2 inhibitor) and (d) erlotinib or AG1478 (specific EGFR inhibitor) (d). Cells were starved overnight with serum-free media and then treated with drug as indicated by the following abbreviations: (E) 100 ng/ml epidermal growth factor; (D) 0.01% DMSO control; (Er) 1 μM erlotinib, (CL) 1 μM CL-387,785, (U) 20 μM U0126 or AG1478. Whole cell lysates were obtained using 10% trichloroacetic acid lysis buffer and immunoblotting was performed at indicated time points.
DUSP6 expression is downregulated by MEK inhibition but not EGFR TKI treatment in EGFR-independent lung cancer cell lines
As demonstrated above, DUSP6 expression is strongly downregulated by EGFR TKI treatment in cell lines harboring activating EGFR mutations. Next, we assessed DUSP regulation in two TKI-resistant NSCLC cell lines with wild-type EGFR, H441 and A549. Erlotinib treatment had no effects on DUSP6 expression or ERK-activation in these cell lines, whereas the MEK1/2 inhibitor U0126 reduced both ERK activation and DUSP6 expression (Figure 2c and d and A549 data: supplementary Figure 1 is available at Carcinogenesis Online). Inhibition of DUSP6 expression paralleled ERK inactivation, either through TKI treatment in EGFR-dependent NSCLC cell lines or through MEK1/2 inhibition in EGFR-independent cell lines, indicating that DUSP6 is an immediately regulated target of ERK signaling. Epidermal growth factor treatment alone, which would drive ERK activation, did not affect DUSP6 protein expression in any of the cell lines studied (Figure 2 and supplementary Figure 1 is available at Carcinogenesis Online), suggesting that DUSP6 expression in these lung cancer cell lines may already be maximally saturated at baseline.
DUSP6 expression is regulated by the ERK-responsive transcription factor, ETS1
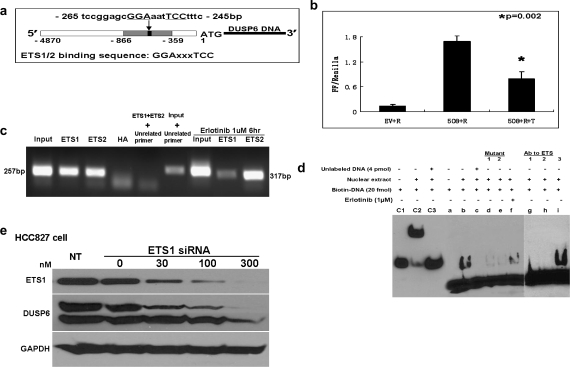
Several in vitro studies have demonstrated that DUSP6 is a negative regulator of fibroblast growth factor receptor signaling and endothelial platelet-derived growth factor receptor signaling via ERK inactivation (21,24). In a recent study, DUSP6 expression in response to fibroblast growth factor receptor signaling in fibroblasts was shown to be mediated by ETS1/2 transcription factor binding to the DUSP6 gene promoter at a consensus binding sequence (Figure 3a) within a 508 bp promoter region upstream of the DUSP6 gene start codon (−359 to −866), which is highly conserved in Xenopus, Fugu, zebrafish, mouse, rat and human (23). We hypothesized that in analogous fashion, ERK signaling may regulate DUSP6 expression in lung cancer cells through ETS1/2 factor binding of the same promoter sequence. In order to test this hypothesis, we compared promoter activity of the DUSP6 gene in the presence or absence of the EGFR inhibitor erlotinib using EGFR-dependent HCC827 cells transfected with a luciferase reporter construct (pGL3Basic-508-Firefly) containing this highly conserved sequence. Luciferase reporter assays demonstrated significantly suppressed DUSP6 promoter activity in the presence of erlotinib, not seen in the presence of DMSO vehicle control (Figure 3b). We then sought to confirm physical binding of ETS1/2 and the DUSP6 promoter sequence in HCC827 cells using chromatin immunoprecipitation assay. A 259 bp band was amplified from either ETS1 or ETS2 immunoprecipitant from HCC827 cells by nested PCR using primers spanning the specific ETS-binding site, indicating direct binding of ETS1/2 to the DUSP6 promoter sequence (Figure 3c). Erlotinib treatment reduced binding of ETS1 but not ETS2 in these assays suggestive of the direct involvement of ETS1 in erlotinib-mediated regulation of promoter activity. In order to further demonstrate erlotinib-induced attenuation of ETS1/2 promoter binding, we performed electrophoretic mobility shift assays using a labeled double-stranded oligonucleotide probe spanning the DUSP6 promoter sequence (Figure 3d). Strong detection of a specific DNA–protein complex was demonstrated in the absence of erlotinib using nuclear extracts from HCC287 cells and the biotin-labeled oligonucleotide probe, whereas diminished binding was seen in the presence of erlotinib. Oligonucleotide probes with targeted mutations led to dramatically reduced complex formation (Mutant 1 and 2), and no complex was seen in the presence of excess unlabeled probe confirming binding specificity of the assay. Antibodies targeting ETS1 and ETS2 led to disappearance of the binding complex, whereas complex formation was unimpeded in the presence of anti-Foxa2 antibody, a control transcription factor again suggestive of physical binding of ETS1/2 to the binding site oligonucleotide. Supershifts in the presence of ETS1/2 antibodies were not observable in the electrophoretic mobility shift assay, potentially due to steric interference between antibody and oligonucleotide binding preventing formation of the antibody–DNA–protein super-complex, as has been previously reported for ETS1/2 by other groups (33). Western blotting studies demonstrated expression of ETS1 in HCC827 cells, whereas ETS2 expression was not detectable and ETS1 expression was also found to closely track the activation status of ERK in HCC827, H1975 and H441 cells (Figure 2). To further corroborate that indeed ETS1 is the critical mediator of DUSP6 regulation in these cells, we pursued siRNA knockdown studies of ETS1 and indeed ETS1 knockdown is accompanied by a marked reduction in DUSP6 expression confirming DUSP6 regulation by ETS1 (Figure 3e)
Fig. 3.
Downregulation of DUSP6 by erlotinib is mediated by the ERK-responsive ETS family transcription factor, ETS1 through direct binding to the DUSP6 gene promoter. (a) A simplified structural map of the DUSP6 gene indicating the ETS1/2-binding site. (b) HCC827 cells were cotransfected with pGL4.74-renilla luciferase reporter (R) plus DUSP6 promoter-conjugated luciferase reporter, pGL3Basic-508-luciferase (508) or empty pGL3Basic reporter (EV) as a control. Twenty-four hours following transfection, cells were treated with 1 μM erlotinib or DMSO for an additional 6 h. Dual luciferase reporter assays were conducted and Firefly luciferase was normalized to Renilla luciferase activity. (c) Chromatin immunoprecipitation assay performed by nested PCR amplified a 257 bp fragment in ETS1 or ETS2 immunoprecipitant. Anti-HA antibody was used as a negative control and pre-immunoprecipitant input as a positive control. An unrelated pair of primers amplified a 317 bp band in the positive control input but not in the presence of ETS1 and ETS2 immunoprecipitant. (d) Electrophoretic mobility shift assay using nuclear extracts from HCC827 cells and biotin-labeled oligonucleotide probes spanning the ETS-binding site domain of the DUSP6 gene promoter: lane b demonstrates detection of a specific DNA–protein complex in the absence of erlotinib, whereas lane f demonstrates diminished detection of complex in the presence of erlotinib; lanes d and e show highly diminished DNA–protein complex detection when targeted mutations of the oligonucleotide probe are introduced (Mutant 1 and 2) confirming binding specificity; lane c demonstrates lack of detectable complex in the presence of competing unlabeled probe and lanes g and h demonstrate lack of detectable complex in the presence of competing antibodies targeting ETS1 and ETS2; lane i demonstrates unimpeded complex formation in the presence of anti-Foxa2 antibody, a control transcription factor; lanes c1, c2 and c3 show the Epstein-Barr virus nuclear antigen control system containing (c1) biotin-labeled EBNA-binding sequence alone, or (c2) with EBNA-1 protein extract or (c3) with protein extract as well as 200-fold excess unlabeled EBNA-binding sequence. (e) Knockdown of ETS1 by siRNA inhibits DUSP6 expression in HCC827 cells in a dose-dependent manner.
DUSP6 protein localizes to the cytoplasm and functions as a negative regulator for ERK activity
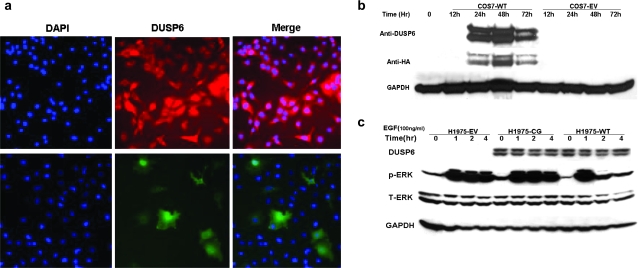
DUSP activity is in part regulated by intracellular localization (34). For example, DUSP6 is primarily cytoplasmic and DUSP1 primarily nuclear, whereas DUSP16 shuttles between compartments (26,35,36). Overexpression of DUSP6 has been reported to result in aberrant accumulation within the nucleus (37). In order to assess endogenous DUSP6 localization in lung cancer cells and confirm appropriate localization of HA-tagged exogenous DUSP6, we performed immunocytochemistry by fluorescent staining using DUSP6-high HCC827 and DUSP6-low H1975 cells. Anti-DUSP6 antibody was used to detect endogenous DUSP6 in naive HCC827 cells and anti-HA antibody was used to detect exogenous DUSP6 in H1975 transfectants, demonstrating appropriate cytoplasmic localization of HA-tagged exogenous DUSP6 (Figure 4a).
Fig. 4.
DUSP6 protein localizes to the cytoplasm and functions as a negative regulator for ERK activity. (a) DUSP6 protein is present in the cytoplasm. Upper row: endogenous DUSP6 in HCC827 cells stained with fluorescent-conjugated secondary antibody after anti-DUSP6 binding, middle panel Red Alexa 560, 4′,6-diamidino-2-phenylindole blue nuclear stain left, merged image right. Lower row: exogenous DUSP6 in H1975 cells expressing DUSP6 stained with anti-HA antibody followed by fluorescent-conjugated secondary antibody, middle panel Green Alexa 488, 4′,6-diamidino-2-phenylindole left, merge right. Amplification ×200. (b) Appropriate expression time course of the pcDNA3.1-DUSP6-HA construct was confirmed in COS7 cells by transient transfection, followed by immunoblot with anti-DUSP6 or anti-HA tag antibody, anti-GAPDH as a control. (c) Stably transfected H1975 subclones (WT) wild-type DUSP6 plasmid, (CG) enzyme-dead C293G mutant DUSP6 plasmid and (EV) empty vector pcDNA3.1 tested by immunoblot for exogenous DUSP6 (anti-HA), phosphor-ERK, total-ERK and GAPDH control at the indicated time points following epidermal growth factor (EGF) stimulation.
H1975 cells are optimal for DUSP6 functional studies in an overexpression system, given that these cells have very low baseline levels of DUSP6 protein (Figure 1b), and were also used in our prior transcriptional profiling study, which identified DUSP6 regulation by EGFR signaling (17). Stable pcDNA3.1-DUSP6-HA subclonal transfectants of H1975 cells with wild-type (WT), C293G-mutated enzyme-dead (CG) and empty vector pcDNA3.1 controls (EV) were generated. Appropriate plasmid expression was confirmed in transient transfection experiments of COS7 cells, which lack endogenous DUSP6 (Figure 4b). To test whether HA-tagged exogenous DUSP6 preserves the functional properties expected of a DUSP, we measured mitogen-stimulated ERK1/2 phosphorylation levels and observed significant reduction in P-ERK levels 2 h post-epidermal growth factor stimulation (Figure 4c), confirming functional activity of plasmid-driven DUSP6 expression, whereas similar changes were not observed for enzyme dead (CG) and empty vector (EV) transfectants. A recent paper also corroborates our findings by reporting analogous results by retroviral transfection of DUSP6 into immortalized normal human airway epithelial cell line NHBE-T and three NSCLC cell lines including A549, H1299 and TKB1 (38).
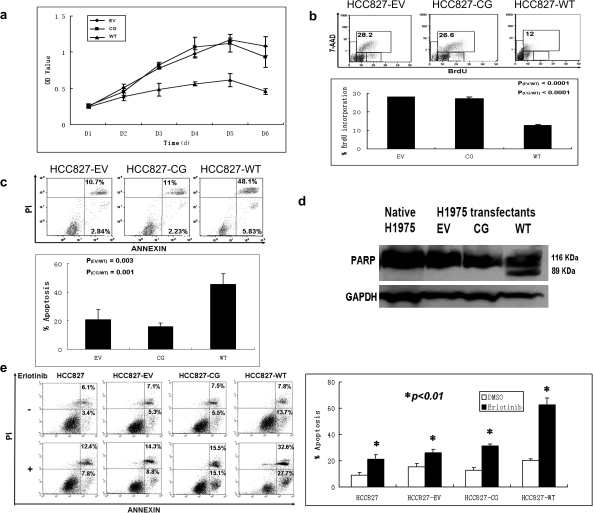
Plasmid-driven DUSP6 overexpression decreases viability via increased apoptosis and growth arrest and synergizes with EGFR inhibitor treatment
We next examined the functional effect of overexpressed DUSP6 on cellular growth of stably transfected DUSP6-low H1975 cells. Of note is that in these studies, two independent clones of WT-DUSP6 transfectants were used and demonstrated identical findings. As determined by MTS assay under low serum culture conditions (0.5% FBS), the growth of wild-type H1975 transfectants (WT) was significantly inhibited as compared with C293G-mutated enzyme-dead (CG) and empty vector (EV) transfected cells (Figure 5a). Interestingly, under higher serum culture conditions (2–10% FBS) differences in growth were not observed suggestive of less critical dependence on ERK signaling at such overstimulated conditions. In order to determine whether reduced viability was a result of decreased proliferation, increased apoptosis or a combination of both, cellular proliferation and apoptosis assays were performed under low serum conditions. We found that wild-type DUSP6 H1975 transfectants (WT) displayed a marked reduction of cells in S-phase by BrdU proliferation assay (Figure 5b and supplementary Table 2 is available at Carcinogenesis Online) and increased apoptosis by annexin/PI assay (Figure 5c). Immunoblot studies also revealed the presence of cleaved PARP product, a marker of caspase-mediated apoptosis in wild-type transfectants (Figure 5d), in marked contrast to the C293G-mutated enzyme-dead (CG) and empty vector (EV) transfected H1975 cells further confirming the induction of apoptosis by DUSP6 expression in these cells. Next, we performed synergism studies by treating stably transfected H1975 cells with CL-387,785. These studies did not demonstrate any difference in cell proliferation rates between the different clonal variants (data not shown). To rule out the possibility that this could be related to compensatory changes in other feedback mechanisms in these long-term cultured cells, we pursued analogous studies using transient transfection of HCC827 cells with EV, CG-mutant and WT-DUSP6 plasmid constructs followed by erlotinib treatment. These studies showed marked synergism with erlotinib treatment in WT-transfected but not in EV or CG-transfected cells (Figure 5e).
Fig. 5.
DUSP6 overexpression inhibited H1975 cell growth through a combination of increased apoptosis and decreased cell proliferation. Stably transfected H1975 subclones: (EV) empty vector pcDNA3.1; (CG) enzyme-dead C293G mut+ DUSP6; (WT) wild-type DUSP6. (a) Cellular growth curve of H1975 subclones by MTS assay. (b) BrdU assay: H1975 subclones were cultured in 0.5% serum media for 3 days and then pulse labeled for 60 min with 10 μM BrdU, followed by staining with fluorescent anti-BrdU antibody and counterstaining with 7-amino-actinomycin D (7-AAD) for total DNA, then analyzed by flow cytometry; (top) representative flow histograms, (bottom) chart showing percentage of cells in S-phases of the cell cycle by subclone. (c) Annexin/PI assay: H1975 subclones were grown in 0.5% serum media for 3 days, stained with Annexin V and PI, then analyzed by flow cytometry, results with percentage of cells listed for each quadrant; left lower—viable cells; right lower—early apoptosis; right upper—late apoptosis; (top) representative flow histograms, (bottom) chart showing percentage of cells in apoptosis by subclone. (d) PARP assay: apoptosis in wild type DUSP6 overexpressing H1975 subclone confirmed by immunoblot detection of the 89 kD PARP cleavage product. (e) Overexpression of DUSP6 synergized with erlotinib treatment to induce apoptosis in HCC827 cells by flow cytometry analysis. HCC827 cells were transiently transfected with the indicated expression plasmids as used for above H1975 cells for 48 h before addition of 1 μM erlotinib for 24 h, followed by detection of cellular apoptosis by Annexin/PI staining by flow cytometry. Annexin/PI assay (left): representative flow histograms, (right) chart showing calculated percentage of cells in apoptosis (early and late) as derived from the flow cytometry analysis.
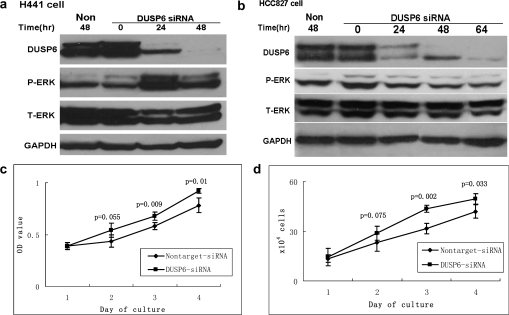
siRNA knockdown in DUSP6-high H441 cells enhances viability through prolonged ERK activation
In order to complement the overexpression experiments in cells with low expression levels of DUSP6, we pursued siRNA-mediated knockdown experiments in H441 and HCC827 cell lines. Knockdown efficiency was demonstrated by reduced DUSP6 levels in both cell lines but only H441 cells demonstrated a substantial increase in ERK activation 24 h following siRNA introduction (Figure 6a and b). In H441 cells, MTS assay demonstrated increased proliferation as compared with non-targeted siRNA control, indicating that DUSP6 knockdown led to enhanced cell growth by sustained ERK activation (Figure 6c and d). HCC827 cells did not demonstrate increased proliferation on MTS assay, consistent with the unchanged level of ERK activation seen on immunoblot (Figure 6b). This suggests comparative variability in ERK-signaling feedback regulation in different NSCLC cell lines.
Fig. 6.
Introduction of DUSP6 siRNA reduced DUSP6 protein levels resulting in increased ERK activity and cellular proliferation. (a and b) DUSP6 siRNA was transfected into HCC827 and H441 cells; immunoblotting of cell lysates was performed at the indicated time points to assess levels of DUSP6, P-ERK, total-ERK and GAPDH as control. (c) MTS assay measuring H441 cell growth at indicated time points following siRNA knockdown. (d) H441 cells were plated in 6 cm dishes at an initial concentration of 6 × 104/ml and counted at indicated time points following introduction of DUSP6 siRNA.
Discussion
DUSP6 is a cytoplasmic DUSP with high specificity for ERK which functions as a negative feedback regulator of ERK activation in normal developmental programs (21) and was also identified in previous studies as highly and immediately regulated by EGFR inhibitor treatment in EGFR-mutant NSCLC cells (17). In the current study, we characterize a feedback regulation loop involving DUSP6 expression and ERK signaling in NSCLC. First, we screened multiple NSCLC cell lines and primary human tumor specimens demonstrated that DUSP6 expression tracks in tandem to ERK activation. Next, we demonstrated that pharmacologic inhibition of ERK activity leads to dramatic downregulation of DUSP6 expression, both in an EGFR-dependent and EGFR-independent manner. We then conducted functional studies with plasmid-driven DUSP6 overexpression in stably transfected DUSP6-low lung cancer cells, demonstrating attenuation of ERK activation, which resulted in growth arrest and apoptosis as well as synergy with EGFR inhibitor treatment. Conversely, we found that siRNA knockdown in DUSP6-high lung cancer cells resulted in enhanced ERK signaling and cellular proliferation. Finally, utilizing luciferase reporter, chromatin immunoprecipitation and electrophoretic mobility shift assays, we demonstrated that regulation of DUSP6 is mediated at the promoter level by ETS family transcription factors, well-known nuclear targets of activated ERK, more specifically ETS1 induction through activation of the ERK pathway. Taken together, these findings indicate that DUSP6 expression is tightly regulated by ERK signaling in NSCLC and exerts antitumor effects via negative feedback mechanisms, pointing to an important feedback loop in NSCLC, which may be prone to dysregulation in tumorigenesis.
K-RAS mutations are common oncogenic events detectable in the majority of pancreatic cancers and 20–30% of NSCLCs. K-RAS mutation results in increased ERK-signaling output, which in turn leads to increased expression of inducible DUSPs, including DUSP6, resulting in negative feedback inhibition of ERK signaling. Similarly, NSCLCs with constitutive activation of cell surface growth factor receptor pathways, such as EGFR or mesenchymal-epithelial transition factor are also at least in part dependent on overactivation of the ERK pathway. Somatic genetic changes leading to inactivation of DUSP6 or other DUSPs and loss of negative feedback regulation of the ERK pathway could certainly represent steps in the progression of such cancers. Previous reports of DUSP6 inactivation via promoter methylation in pancreatic cancers suggest DUSP6 as a candidate tumor suppressor in that disease (39). A recent manuscript reported dowregulation of DUSP6 expression in lung cancer cell lines as well as primary lung cancer specimens correlating with proliferative index. No mutations of DUSP6 and a fairly low loss of heterozygosity rate (17.2%) of the DUSP6 locus were found (38). Our cell line studies demonstrate that in many NSCLC cells, DUSP6 regulation appears intact and responsive to increased ERK activation, arguing against DUSP6 loss as a frequent oncogenic event in NSCLC. More comprehensive studies will be needed to assess whether DUSP6 abrogation is indeed present in specific subsets of patients. The recent generation of knockout mice lacking the murine DUSP6/MKP3 gene will allow systematic evaluation of its potential role in oncogenesis, particularly for model systems of K-RAS and EGFR-driven lung cancers.
Multiple studies demonstrate that among ERK-regulated genes, DUSP6 is one of the most rapidly and significantly regulated. DUSP6 was identified as one of only three genes significantly overexpressed in myeloma cells harboring a constitutively active mutant N-RAS gene and is also overexpressed in H-RAS driven human breast epithelial cells and in human melanoma cell lines harboring potent activating mutations in B-RAF (40). Interestingly, in a study of platelet-derived growth factor receptor signaling, DUSP6 was found to be rapidly phosphorylated on Ser174 and Ser300 leading to platelet-derived growth factor-induced degradation, whereas ERK activation led to DUSP6 induction resulting in restoration of DUSP6 levels within 1–2 h, and DUSP6 knockdown led to increased ERK activation and mitogenic response. These results suggest that DUSP6 is an important regulator of platelet-derived growth factor-induced ERK phosphorylation acting in both a rapid positive feed-forward and a more delayed negative feedback loop (24). Other studies suggested that DUSP6 protein expression is also affected by posttranslational regulation, principally mammalian target of rapamycin-mediated phosphorylation of Ser159 and degradation of DUSP6 (41). Our studies as well as others point to a critical role for ETS-mediated regulation of the DUSP6 promoter in the control of DUSP6 expression (23,42). DUSP6 was reported to be one of the most highly regulated genes in chronic myeloid leukemia cells upon imatinib treatment (43) and similarly DUSP6 is overexpressed upon inducible expression of the EGFRvIII oncogene in glioblastoma cells (44). These data suggest that DUSP6 overexpression may be a fairly uniform phenomenon in oncogenic pathways relying on ERK activation. A systems biological approach identified DUSP6 as a critical regulator shaping the activity of the MAPK pathway during cellular transformation by oncogenic RAS (45). DUSP6 expression was rapidly increased by inducible expression of oncogenic RAS in immortalized rat fibroblasts dampening the initial hyperactivation of ERK (46). Very interestingly, elevated DUSP6 RNA expression was reported to be a major negative predictor of survival in patients with resected NSCLC as part of a five gene signature model (47). The authors did not report K-RAS or EGFR mutational data in these patients. As our data suggests, DUSP6 expression correlates with ERK activation, and we hypothesize that tumors with DUSP6 overexpression represent a group of tumors with excessive activation of the ERK pathway explaining their poorer prognosis. This observation indeed suggests that DUSP6 expression may potentially serve as a biomarker for tumors sensitive to inhibition of the RAS-RAF-MAPK pathway.
A large number of small molecule inhibitors of the MAPK pathway are currently in development. For cancer therapy, the most relevant are inhibitors of RAF kinase and MEK1/2. Initial studies with such agents demonstrated only limited success despite the apparent importance of oncogenic ERK signaling in cancer. The unpredictable cellular and clinical responses seen with such drugs belie a greater complexity surrounding signal regulation than previously thought. Given the critical physiological role of ERK signaling in normal tissue maintenance and proliferation, it is not yet clear whether direct inhibition will indeed turn out to be a successful strategy. Therefore, MAPK drug discovery will probably need to expand to include targets that are not classically druggable, though important modulators of MAPK function nonetheless. Our data suggest that modulation of DUSP-mediated feedback mechanisms in ERK signaling may provide one such avenue for future drug development.
Supplementary material
Supplementary Tables 1 and 2 and Figures 1–6 can be found at http://carcin.oxfordjournals.org/
Funding
Flight Attendant Medical Research Institute; American Cancer Society (RSG-08-303-01-TBE) to B.H; National Institutes of Health/National Cancer Institute R00CA126026 to S.K.
Supplementary Material
Acknowledgments
We are grateful to Dr Jeff Kern for providing an aliquot of Nuli-1 cells and Dr Stephen M.Keyse for providing the luciferase reporter construct.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BrdU
bromo-deoxyuridine
- DMSO
dimethyl sulfoxide
- DUSP
dual specificity phosphatase
- EGFR
epidermal growth factor receptor
- ERK
extracellular signal-regulated kinase
- FBS
fetal bovine serum
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- HA
hemagglutinin
- MAPK
mitogen-activated protein kinase
- MKP
mitogen-activated protein kinase-specific phosphatase
- NSCLC
non-small-cell lung cancer
- PARP
poly ADP ribose polymerase
- PCR
polymerase chain reaction
- PI
propidium iodide
- siRNA
small interfering RNA
- TKI
tyrosine kinase inhibitor
References
- 1.Jemal A, et al. Cancer statistics, 2008. CA Cancer J. Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, et al. Annual report to the nation on the status of cancer, 1975–2005, featuring trends in lung cancer, tobacco use, and tobacco control. J. Natl Cancer Inst. 2008;100:1672–1694. doi: 10.1093/jnci/djn389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhillon AS, et al. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 4.Shepherd FA, et al. Erlotinib in previously treated non-small-cell lung cancer. N. Engl. J. Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 5.Ciardiello F, et al. EGFR antagonists in cancer treatment. N. Engl. J. Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 6.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 7.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 8.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl Acad. Sci. USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar A, et al. Structure and clinical relevance of the epidermal growth factor receptor in human cancer. J. Clin. Oncol. 2008;26:1742–1751. doi: 10.1200/JCO.2007.12.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pao W, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeffrey KL, et al. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat. Rev. Drug Discov. 2007;6:391–403. doi: 10.1038/nrd2289. [DOI] [PubMed] [Google Scholar]
- 12.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 14.Yu Z, et al. Resistance to an irreversible epidermal growth factor receptor (EGFR) inhibitor in EGFR-mutant lung cancer reveals novel treatment strategies. Cancer Res. 2007;67:10417–10427. doi: 10.1158/0008-5472.CAN-07-1248. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi S, et al. An alternative inhibitor overcomes resistance caused by a mutation of the epidermal growth factor receptor. Cancer Res. 2005;65:7096–7101. doi: 10.1158/0008-5472.CAN-05-1346. [DOI] [PubMed] [Google Scholar]
- 16.Kwak EL, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc. Natl Acad. Sci. USA. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi S, et al. Transcriptional profiling identifies cyclin D1 as a critical downstream effector of mutant epidermal growth factor receptor signaling. Cancer Res. 2006;66:11389–11398. doi: 10.1158/0008-5472.CAN-06-2318. [DOI] [PubMed] [Google Scholar]
- 18.Patterson KI, et al. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem. J. 2009;418:475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 19.Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 2008;27:253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- 20.Echevarria D, et al. Mkp3 is a negative feedback modulator of Fgf8 signaling in the mammalian isthmic organizer. Dev. Biol. 2005;277:114–128. doi: 10.1016/j.ydbio.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Li C, et al. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development. 2007;134:167–176. doi: 10.1242/dev.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maillet M, et al. DUSP6 (MKP3) null mice show enhanced ERK1/2 phosphorylation at baseline and increased myocyte proliferation in the heart affecting disease susceptibility. J. Biol. Chem. 2008;283:31246–31255. doi: 10.1074/jbc.M806085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ekerot M, et al. Negative-feedback regulation of FGF signalling by DUSP6/MKP-3 is driven by ERK1/2 and mediated by Ets factor binding to a conserved site within the DUSP6/MKP-3 gene promoter. Biochem. J. 2008;412:287–298. doi: 10.1042/BJ20071512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurek A, et al. Negative and positive regulation of MAP kinase phosphatase 3 controls PDGF-induced Erk activation. J. Biol. Chem. 2009;284:4626–4634. doi: 10.1074/jbc.M808490200. [DOI] [PubMed] [Google Scholar]
- 25.Furukawa T, et al. Genomic analysis of DUSP6, a dual specificity MAP kinase phosphatase, in pancreatic cancer. Cytogenet. Cell Genet. 1998;82:156–159. doi: 10.1159/000015091. [DOI] [PubMed] [Google Scholar]
- 26.Muda M, et al. MKP-3, a novel cytosolic protein-tyrosine phosphatase that exemplifies a new class of mitogen-activated protein kinase phosphatase. J. Biol. Chem. 1996;271:4319–4326. doi: 10.1074/jbc.271.8.4319. [DOI] [PubMed] [Google Scholar]
- 27.Muda M, et al. The mitogen-activated protein kinase phosphatase-3 N-terminal noncatalytic region is responsible for tight substrate binding and enzymatic specificity. J. Biol. Chem. 1998;273:9323–9329. doi: 10.1074/jbc.273.15.9323. [DOI] [PubMed] [Google Scholar]
- 28.Chan DW, et al. Loss of MKP3 mediated by oxidative stress enhances tumorigenicity and chemoresistance of ovarian cancer cells. Carcinogenesis. 2008;29:1742–1750. doi: 10.1093/carcin/bgn167. [DOI] [PubMed] [Google Scholar]
- 29.Huang G, et al. 15-Hydroxyprostaglandin dehydrogenase is a target of hepatocyte nuclear factor 3beta and a tumor suppressor in lung cancer. Cancer Res. 2008;68:5040–5048. doi: 10.1158/0008-5472.CAN-07-6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou B, et al. Mapping ERK2-MKP3 binding interfaces by hydrogen/deuterium exchange mass spectrometry. J. Biol. Chem. 2006;281:38834–38844. doi: 10.1074/jbc.M608916200. [DOI] [PubMed] [Google Scholar]
- 31.Wishart MJ, et al. A single mutation converts a novel phosphotyrosine binding domain into a dual-specificity phosphatase. J. Biol. Chem. 1995;270:26782–26785. doi: 10.1074/jbc.270.45.26782. [DOI] [PubMed] [Google Scholar]
- 32.Dowd S, et al. Isolation of the human genes encoding the pyst1 and Pyst2 phosphatases: characterisation of Pyst2 as a cytosolic dual-specificity MAP kinase phosphatase and its catalytic activation by both MAP and SAP kinases. J. Cell Sci. 1998;111(Pt 22):3389–3399. doi: 10.1242/jcs.111.22.3389. [DOI] [PubMed] [Google Scholar]
- 33.Myers E, et al. Associations and interactions between Ets-1 and Ets-2 and coregulatory proteins, SRC-1, AIB1, and NCoR in breast cancer. Clin. Cancer Res. 2005;11:2111–2122. doi: 10.1158/1078-0432.CCR-04-1192. [DOI] [PubMed] [Google Scholar]
- 34.Farooq A, et al. Structure and regulation of MAPK phosphatases. Cell. Signal. 2004;16:769–779. doi: 10.1016/j.cellsig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Wu JJ, et al. The noncatalytic amino terminus of mitogen-activated protein kinase phosphatase 1 directs nuclear targeting and serum response element transcriptional regulation. Mol. Cell. Biol. 2005;25:4792–4803. doi: 10.1128/MCB.25.11.4792-4803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda K, et al. MKP-7, a novel mitogen-activated protein kinase phosphatase, functions as a shuttle protein. J. Biol. Chem. 2001;276:39002–39011. doi: 10.1074/jbc.M104600200. [DOI] [PubMed] [Google Scholar]
- 37.Bettini ML, et al. MAP kinase phosphatase activity sets the threshold for thymocyte positive selection. Proc. Natl Acad. Sci. USA. 2007;104:16257–16262. doi: 10.1073/pnas.0705321104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okudela K, et al. Down-regulation of DUSP6 expression in lung cancer: its mechanism and potential role in carcinogenesis. Am. J. Pathol. 2009;175:867–881. doi: 10.2353/ajpath.2009.080489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu S, et al. Abrogation of DUSP6 by hypermethylation in human pancreatic cancer. J. Hum. Genet. 2005;50:159–167. doi: 10.1007/s10038-005-0235-y. [DOI] [PubMed] [Google Scholar]
- 40.Bloethner S, et al. Effect of common BRAF and N-RAS mutations on global gene expression in melanoma cell lines. Carcinogenesis. 2005;26:1224–1232. doi: 10.1093/carcin/bgi066. [DOI] [PubMed] [Google Scholar]
- 41.Bermudez O, et al. Post-translational regulation of the ERK phosphatase DUSP6/MKP3 by the mTOR pathway. Oncogene. 2008;27:3685–3691. doi: 10.1038/sj.onc.1211040. [DOI] [PubMed] [Google Scholar]
- 42.Furukawa T, et al. Feedback regulation of DUSP6 transcription responding to MAPK1 via ETS2 in human cells. Biochem. Biophys. Res. Commun. 2008;377:317–320. doi: 10.1016/j.bbrc.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Hakansson P, et al. Gene expression analysis of BCR/ABL1-dependent transcriptional response reveals enrichment for genes involved in negative feedback regulation. Genes Chromosomes Cancer. 2008;47:267–275. doi: 10.1002/gcc.20528. [DOI] [PubMed] [Google Scholar]
- 44.Ramnarain DB, et al. Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res. 2006;66:867–874. doi: 10.1158/0008-5472.CAN-05-2753. [DOI] [PubMed] [Google Scholar]
- 45.Warmka JK, et al. Mitogen-activated protein kinase phosphatase-3 is a tumor promoter target in initiated cells that express oncogenic Ras. J. Biol. Chem. 2004;279:33085–33092. doi: 10.1074/jbc.M403120200. [DOI] [PubMed] [Google Scholar]
- 46.Marchetti S, et al. Inducible expression of a MAP kinase phosphatase-3-GFP chimera specifically blunts fibroblast growth and ras-dependent tumor formation in nude mice. J. Cell. Physiol. 2004;199:441–450. doi: 10.1002/jcp.10465. [DOI] [PubMed] [Google Scholar]
- 47.Chen HY, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N. Engl. J. Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.