Abstract
A major concern about the ongoing swine-origin H1N1 influenza virus (S-OIV) outbreak is that the virus may be so different from seasonal H1N1 that little immune protection exists in the human population. In this study, we examined the molecular basis for pre-existing immunity against S-OIV, namely the recognition of viral immune epitopes by T cells or B cells/antibodies that have been previously primed by circulating influenza strains. Using data from the Immune Epitope Database, we found that only 31% (8/26) of B-cell epitopes present in recently circulating H1N1 strains are conserved in the S-OIV, with only 17% (1/6) conserved in the hemagglutinin (HA) and neuraminidase (NA) surface proteins. In contrast, 69% (54/78) of the epitopes recognized by CD8+ T cells are completely invariant. We further demonstrate experimentally that some memory T-cell immunity against S-OIV is present in the adult population and that such memory is of similar magnitude as the pre-existing memory against seasonal H1N1 influenza. Because protection from infection is antibody mediated, a new vaccine based on the specific S-OIV HA and NA proteins is likely to be required to prevent infection. However, T cells are known to blunt disease severity. Therefore, the conservation of a large fraction of T-cell epitopes suggests that the severity of an S-OIV infection, as far as it is determined by susceptibility of the virus to immune attack, would not differ much from that of seasonal flu. These results are consistent with reports about disease incidence, severity, and mortality rates associated with human S-OIV.
Keywords: databases, epitopes, meta-analysis, pandemic
The spread of the ongoing human swine-origin H1N1 influenza virus (S-OIV) outbreak meets the World Health Organization (WHO) criteria for a pandemic (1). As this virus contains a unique combination of gene segments from both North American and Eurasian swine lineages, and is antigenically distinct from seasonal human influenza A (2), there is concern that little protective immune memory exists in the general human population. This concern was confirmed by reports that neutralizing antibodies against S-OIV are found nearly exclusively in persons born before 1957, presumably because of their exposure to H1N1 influenza strains that did not circulate after that time (3, 4). Together with reports of differences in pathogenicity of the virus in animal models compared with seasonal human H1N1 strains (3, 5, 6), this raises the specter of a pandemic with major public health consequences. At the same time, the incidence of clinically severe cases so far appears to be similar to that experienced for seasonal flu: According to CDC estimates, >1 million people were infected with S-OIV between April 15 and July 24, 2009, leading to 5,011 hospitalizations and 302 deaths (http://www.cdc.gov/h1n1flu/surveillanceqa.htm). This seeming contradiction highlights the need to better understand the interaction of this pathogen with the human host.
The focus of the present study was to examine the presence of immune memory against S-OIV in the human population. Adaptive immune responses against influenza (and other pathogens) are triggered upon T-cell or B-cell receptors recognizing viral immune epitopes. Recognition of epitopes by antibodies and T cells that were induced after past influenza infections or vaccinations are key components of immune memory and protection from infection. Importantly, a virus can carry substantial sequence differences in some regions but still be recognized by the immune system if the virus retains sequence identity in regions including the immune epitopes. Therefore, we specifically examined whether there are immune epitopes in S-OIV that are likely targets of pre-existing immunity (i.e., if epitopes that were present in the H1N1 seasonal flu strains from between 1988 and 2008 are also present in the S-OIV strains).
We obtained sequence and source information on previously defined epitopes from the Immune Epitope Database (IEDB) (7, 8), which was developed for the purpose of cataloging and making available epitope information in a single repository to the scientific community. Curated epitope information from all manuscripts published to date that characterize epitopes in influenza is contained in the IEDB. Swine influenza sequence information was obtained from the National Center for Biotechnology Information (NCBI) (9) and the Global Initiative on Sharing Avian Influenza Data (GISAID) (10) influenza sequence databases. The present report analyzes how well these known influenza epitopes, and therefore the targets of immunological memory, are conserved in S-OIV isolates and experimentally validates the results.
Results
A Significant Fraction of Epitope Sequences Are Conserved in S-OIV.
The overall goal of the analysis presented in this section was to establish whether the epitopes described in the literature and presumably associated with pre-existing immunity in the general population, would be conserved in S-OIV sequences.
At the time of this writing, the epitope information available in the IEDB related to influenza A encompassed information from 594 references (journal articles and direct submissions), describing 3,724 distinct molecular structures (linear and discontinuous peptides) derived from influenza A that were tested experimentally for interaction with immune receptors. This roughly doubled the amount of information on influenza epitopes that was available in 2006 (7), emphasizing the important contribution and greatly enhanced throughput of recent influenza epitope mapping efforts, which were stepped up since the emergence of H5N1 avian flu.
We focused our analysis on influenza A epitope mapping experiments that have the most relevance for human immunity and considered only those epitopes that are present in the sequences of recently circulating strains of H1N1 (i.e., isolates from 1988 to 2008). We considered an epitope relevant if it showed recognition by antibodies or by T cells in the context of human MHC and if the epitopes were mapped in the context of whole influenza organisms or proteins. As B-cell epitopes are infrequently identified in humans (7), B-cell epitopes mapped in the context of any host organism were included. We think that this is justified, as studies have shown substantial overlap for B-cell epitopes defined in different species, presumably because the structural constraints associated with their recognition is similar across species (11).
We distinguished between two primary categories of epitopes: (i) those recognized by B cells/antibodies, and (ii) those recognized by T cells. The latter were further categorized into those recognized by CD8+ T cells in the context of major histocompatibility complex (MHC) class I molecules and those recognized by CD4+ T cells in the context of MHC class II. If multiple epitopes in the same category had sequences that were nested within each other, a representative epitope was chosen that was associated with the highest specific immune response. Full details of the specific IEDB queries are given in Materials and Methods. As shown in Table 1, a total of 26 B-cell epitopes were found in recently circulating H1N1 strains, whereas 139 CD4+ and 78 CD8+ T-cell epitopes were found.
Table 1.
Number of influenza A H1N1 epitopes in the Immune Epitope Database
| Epitope category | Recent seasonal H1N1 epitopes | Conserved in S-OIV | Conserved (%) |
|---|---|---|---|
| B-cell | 26 | 8 | 31 |
| T-cell, CD4+ | 139 | 57 | 41 |
| T-cell, CD8+ | 78 | 54 | 69 |
S-OIV, swine-origin H1N1 influenza virus.
Next, we asked how many of these experimentally defined epitopes, for which we would expect some pre-existing immunity, are totally conserved in the emerging S-OIV strains. Human S-OIV sequences were retrieved from isolates originating in Mexico, the United States, and elsewhere, as described in Materials and Methods. As shown in Table 1, a substantial number of epitopes present in recent seasonal H1N1 strains are also found to be 100% conserved in S-OIV sequences. Notably, whereas only ≈31% of the B-cell epitopes are conserved in S-OIV strains, up to 41% and 69% of the CD4+ and CD8+ T-cell epitopes, respectively, are conserved. The complete list of individual epitopes is provided in Table S1.
We also assessed whether seasonal influenza viruses of the H3N2 subtype contained additional epitopes conserved in S-OIV strains. A repeat of the analysis above showed that this was not the case, as all epitopes that are conserved between the H3N2 subtype and as S-OIV are also shared with seasonal H1N1 isolates. Overall, these data demonstrated that a sizeable fraction of epitopes derived from H1N1 seasonal flu strains and described in the literature is conserved in S-OIV sequences. This raised the possibility that some level of immunity against S-OIV sequences might exist in the general population.
Distribution of Conserved Epitopes in Different Influenza Proteins.
The proteins hemagglutinin (HA) and neuraminidase (NA), which make up the majority of the viral surface, are more variable than other influenza proteins. Therefore, it can be expected that epitopes from those proteins are less likely to be conserved across strains than others. As shown in Table 2, this is indeed the case. For T-cell epitopes, only five of 43 (12%) HA and NA epitopes are conserved, as opposed to 106 of 174 (61%) epitopes derived from other proteins. Similarly, at the level of B-cell epitopes, only a single HA/NA epitope among six is conserved (17%), whereas seven of 20 of the remaining epitopes in other proteins are conserved (35%). This shows that for both B-cell and T-cell epitopes, fewer epitopes are conserved in the virion surface antigens.
Table 2.
Distribution of epitopes among the influenza proteins
| Protein | B-cell |
T-cell, CD8+ |
T-cell, CD4+ |
Overall |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Cons. | Total | Cons. | Total | Cons. | Total | Cons. | Cons. (%) | |
| HA | 5 | 1 | 4 | 1 | 34 | 3 | 43 | 5 | 12 |
| NA | 1 | 0 | 2 | 1 | 3 | 0 | 6 | 1 | 17 |
| M1 | 4 | 1 | 17 | 13 | 28 | 14 | 49 | 28 | 57 |
| M2 | 4 | 1 | 1 | 0 | 3 | 0 | 8 | 1 | 13 |
| NS1 | 1 | 0 | 2 | 1 | 2 | 1 | 5 | 2 | 40 |
| NS2 | 0 | 0 | 1 | 1 | 1 | 0 | 2 | 1 | 50 |
| NP | 9 | 4 | 19 | 15 | 43 | 21 | 71 | 40 | 56 |
| PA | 0 | 0 | 7 | 4 | 1 | 1 | 8 | 5 | 63 |
| PB1 | 2 | 1 | 23 | 17 | 21 | 16 | 46 | 34 | 74 |
| PB1-F2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | NA |
| PB2 | 0 | 0 | 2 | 1 | 3 | 1 | 5 | 2 | 40 |
The total number of epitopes in the H1N1 seasonal flu strains from 1988–2008 (Total) as well as the number of epitopes conserved in swine-origin H1N1 influenza virus (S-OIV) (Cons.) are listed.
For B-cell responses, neutralizing epitopes are primarily located on the virion surface proteins HA and NA. That means that the majority of B-cell epitopes conserved in S-OIV are unlikely to be neutralizing. For example, the NP protein contains 9 H1N1 B-cell epitopes, four of which are conserved in S-OIV sequences. However, to be targets of directly neutralizing antibodies, epitopes need to be exposed on the virion surface. Therefore, although epitopes in the NP protein are often targeted by serological immune responses, they are not protective as they are thought to be inaccessible from the surface (12).
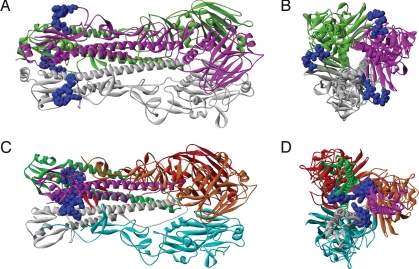
A single HA B-cell epitope is conserved in S-OIV. To determine its location in the protein, we mapped the epitope onto the three-dimensional structure of a homologous protein of the S-OIV HA trimer complex for the precursor HA (Fig. 1 A and B) and onto a homology model of the cleaved HA (Fig. 1 C and D), as the epitope is located at the N-terminal of the fusion peptide. In the precursor, the epitope is solvent accessible, with an accessible surface area (ASA) of 632 Å2, which is similar to the surface area of interfaces in known structures of antibody–peptide complexes. In the cleaved HA, the epitope is partially buried in the cavity of HA, into which the fusion peptide inserts after precursor cleavage, with an ASA of 294 Å2. The location of this epitope in the conserved HA2 glycopeptide is atypical, as most neutralizing antibodies target HA1, and it is unclear how frequent pre-existing immune responses are against this peptide. At the same time, this epitope was mapped to a monoclonal antibody that inhibited cell fusion in vitro and provided protection from challenge in vivo (13). Taken together, these data suggest that although the one B-cell epitope that is conserved might be partially exposed in some HA isoforms, the vast majority of known B-cell epitopes are not conserved in S-OIV and are unlikely to be relevant for protection.
Fig. 1.
Location of the conserved epitope in the S-OIV HA protein structure. (A and B) Quaternary structure of the HA precursor (Protein Data Bank ID: 1HA0): frontal (A) and orthogonal (B) views. Monomer chains are in white, magenta, and green. Epitopes in each monomer are identical and shown in blue. (C and D) Quaternary structure of the cleaved HA modeled for the representative swine influenza HA sequence (ACP41934.1): frontal (C) and orthogonal (D) views. HA2 chains are in white, magenta, and green; corresponding HA1 chains are in cyan, orange, and red. Epitope is located at the N-terminal of HA2 chain and is identical in each chain.
Experimental Demonstration of Memory T-Cell Response Recognizing H1N1 Influenza Sequences.
The analysis presented above suggests that a significant level of T-cell immunity might pre-exist in the general population against epitope sequences that are found totally conserved in S-OIV. To experimentally address this point, we synthesized four groups of peptide epitopes. One group corresponded to the CD4+ T-cell epitopes totally conserved in S-OIV sequences; the second group consisted of CD8+ T-cell epitopes conserved in S-OIV; and two additional groups of peptides consisted of CD4+ and CD8+ T-cell epitopes not conserved in S-OIV sequences. These four separate peptide pools were tested with peripheral blood mononuclear cells (PBMCs) from normal blood donors for induction of interferon (IFN)–γ secretion in direct ex vivo ELISPOT assays. PBMC from anonymous blood donors from the San Diego region were banked in the context of an independent study of influenza responses, and collected at least 1 year before the onset of the current S-OIV pandemic.
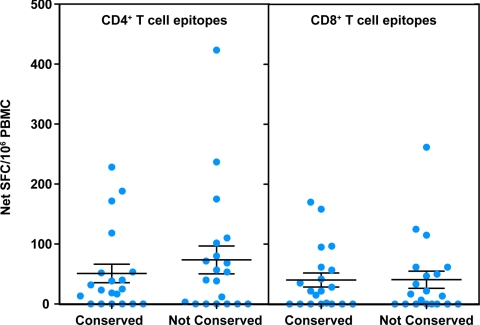
We found that, on average, responses significantly greater than zero were detected against the pool of epitopes conserved in S-OIV sequences, both in terms of CD4+ and CD8+ T-cell responses (P < 0.01 for all epitopes sets, one-tailed single sample t test) (Fig. 2). The responses to the conserved S-OIV epitope pools were similar in magnitude to the responses observed with their nonconserved counterparts, and the observed difference was not statistically significant (average of 40 spot-forming cells [SFC]/106 cells for conserved and 41 SFC for nonconserved CD8+ T-cell epitopes (P = 0.94, two-tailed paired t test), and 51 versus 74 SFC/106 for CD4+ T-cell conserved and nonconserved, respectively (P = 0.14, two-tailed paired t test). Information on age and sex was available only for half of the 20 donors (Table S2); thus investigating correlations of responses with these parameters was not possible.
Fig. 2.
Detection of pre-existing CD4+ and CD8+ T-cell immune responses to S-OIV. PBMC from normal individuals (n = 20) were stimulated with pools of either CD4+ or CD8+ T-cell epitopes from recent seasonal H1N1 influenza strains that were either absolutely conserved or not conserved in S-OIV sequences. Responses were measured through ex vivo IFN-γ ELISPOT assays. Error bars represent SEM.
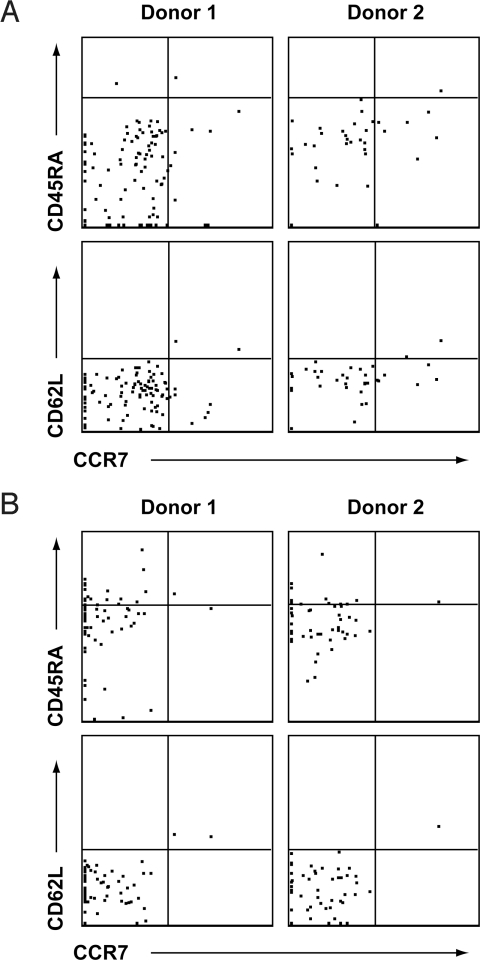
For selected donors, we sought to further document that the responding cells were of the memory phenotype. Accordingly, intracellular cytokine and cell-surface staining assays were used to characterize the IFN-γ responses against CD4+ and CD8+ T-cell epitopes conserved in S-OIV. Specifically, CD4+ or CD8+ T cells producing IFN-γ were examined for the expression of various memory markers. We found that the responding CD4+IFN-γ+ T cells, stimulated with the epitope pool conserved in S-OIV, were CD45RAmedCD62LloCCR7− (Fig. 3). Likewise, the CD8+IFN-γ+ T cells stimulated with the epitope pool conserved in S-OIV were also CD45RAmedCD62LloCCR7−. Overall, these data show that both CD4+ and CD8+ T-cell memory responses against S-OIV epitopes can be detected directly ex vivo, and that the magnitude of responses against epitopes conserved in S-OIV is at least comparable to that of other epitopes in recently circulating influenza strains.
Fig. 3.
S-OIV–specific CD4+ and CD8+ T cells appear to have an effector memory phenotype. PBMC from two representative normal donors were stimulated with either pools of (A) CD4+ or (B) CD8+ T-cell epitopes conserved in S-OIV sequences. The cells were gated for IFN-γ production and presence of the surface markers CD4 and CD8, respectively. Signal intensities of the gated cells for memory phenotypic markers CD45RA, CD62L, and CCR7 are shown.
Comparison of Literature-Defined Epitope Content for Seasonal Influenza and S-OIV at the Individual Isolate Level.
Next, we asked how the level of pre-existing immunity for an average S-OIV isolate compares to that of an average seasonal H1N1 influenza isolate. For 559 seasonal H1N1 flu isolates from 2000 to 2008, we determined the number of epitopes present in each isolate that were also present in H1N1 seasonal flu isolates of the preceding 20 years. As before, such epitopes are considered likely targets of pre-existing immune responses. Table 3 lists the median number of epitopes on a per-virus basis as 16 B cell, 93 CD4+, and 66 CD8+ T-cell epitopes. These numbers were used as a baseline against which we compared the number of epitopes conserved in S-OIV isolates, again on a per-virus basis. Interestingly, no variability in the epitope sequences within different S-OIV isolates was detected. Thus, the number of conserved epitopes in any S-OIV isolate is 8, 57, and 54 for B-cell, CD4+, and CD8+ T-cell epitopes, respectively. Thus, it would be predicted that the levels of pre-existing immunity against S-OIV, albeit reduced in comparison to the levels directed against other H1N1 strains, would still be significant, particularly in the case of T-cell epitopes, in general, and CD8+ T-cell epitopes, in particular.
Table 3.
Median number of epitopes per virus isolate found conserved in strains from preceding 20 years
| Year | Epitope category |
||
|---|---|---|---|
| B-cell | T-cell, CD8+ | T-cell, CD4+ | |
| 2000 | 16 | 67 | 84 |
| 2001 | 16 | 66 | 82 |
| 2002 | 16 | 66 | 93 |
| 2003 | 16 | 66 | 96 |
| 2004 | 18 | 65 | 92 |
| 2005 | 16 | 66 | 96 |
| 2006 | 15.5 | 66 | 93.5 |
| 2007 | 14 | 66 | 93 |
| 2008 | 16 | 68 | 87 |
| Median (2000–2008) | 16 | 66 | 93 |
| 2009, S-OIV | 8 | 54 | 57 |
Abbreviation: S-OIV, swine-origin H1N1 influenza virus.
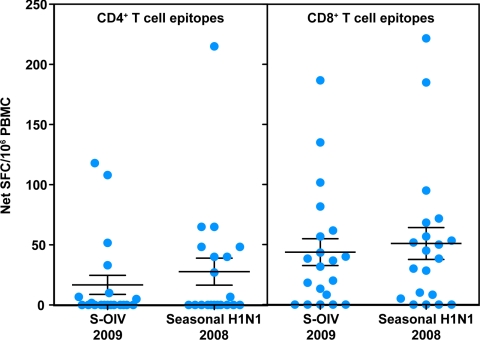
To test this hypothesis, we compared the magnitude of responses detected in the ELISPOT assay for IFN-γ release against the peptide pools corresponding to CD4+ and CD8+ T-cell epitopes conserved in S-OIV with peptide pools corresponding to CD4+ and CD8+ T-cell epitopes found in 2008 seasonal influenza. For this comparison, we used PBMC from blood donations obtained in 2007. The results obtained are shown in Fig. 4.
Fig. 4.
Comparable pre-existing CD4+ and CD8+ T-cell immunity to S-OIV and 2008 seasonal influenza. PBMC from normal individuals (n = 20) were stimulated with pools of either CD4+ or CD8+ T-cell epitopes that were conserved in S-OIV or seasonal influenza 2008 sequences. Responses were measured through ex vivo IFN-γ ELISPOT assays. Error bars represent SEM.
We found that significant CD4+ and CD8+ T-cell responses were detected to epitopes conserved in both S-OIV and 2008 seasonal influenza (P < 0.05 for all epitope sets, one-tailed single-sample t test). As predicted, the response to the epitopes in 2008 seasonal influenza was higher than the response to the S-OIV epitopes, in terms of both CD4+ and CD8+ T cells (17 versus 28 SFC/106 for CD4+ T cells, and 44 versus 51 SFC/106 for CD8+ T cells in response S-OIV and seasonal influenza epitopes, respectively); however, these differences did not reach statistical significance (P = 0.25 for CD4+ responses and P = 0.13 for CD8+ responses, according to a two-tailed paired t test). Overall, the level of pre-existing immunity was higher for CD8+ T cells compared with CD4+ T cells, as this likely reflects a higher level of conservation of the S-OIV CD8+ T-cell epitopes with recently circulating 2008 seasonal influenza.
Discussion
We provide here an analysis of immune epitopes found in the emergent S-OIV strains. We found that, overall, 49% of the epitopes reported in the literature and present in recently circulating seasonal H1N1 are also found totally conserved in S-OIV. Interestingly, the number of conserved epitopes varied greatly as a function of the class of epitopes considered. Although only 31% of the B-cell epitopes were conserved, 41% of the CD4+ and 69% of the CD8+ T-cell epitopes were conserved. It is known that crossreactive T-cell immune responses can exist even between serologically distinct influenza A strains (14, 15). Based on this observation and the data presented above, we hypothesized that it is possible that immune memory responses against S-OIV exist in the adult population, at the level of both B and T cells. However, our analysis also suggested that T cells would mediate such responses predominantly, and in particular CD8+ T cells.
In terms of B-cell responses, there is an average of only 16 known B-cell epitopes with the potential to elicit a memory immune response in a seasonal H1N1 influenza strain. As it is known that pre-existing antibodies against previous seasonal influenza strains do not provide broad protection from subsequent infections, a further drop from 16 to 8 B-cell epitopes in S-OIV is likely to mean that little memory B-cell responses against S-OIV exist in the human population. Moreover, only a single conserved epitope was found between the HA and NA surface proteins, which are the primary targets of neutralizing antibodies. This suggests that few pre-existing neutralizing antibodies will be present in the general human population, which is in agreement with the results of experimental studies that essentially found no neutralizing antibodies against S-OIV in the general human population under the age of 60 years (3, 4).
The same experimental studies indicated that an increased number of persons who contracted influenza before the 1957 flu pandemic apparently have retained antibody responses that are neutralizing against S-OIV (3, 4). The overlap between the number of epitopes found in influenza sequences in 1957 and previous years, versus S-OIV, was not significantly different from the more recently circulating strains (10 B-cell, 52 CD4+, 59 CD8+). However, this is likely due to few influenza strains from those years having been used to identify epitopes. To better address this issue, we mapped S-OIV regions homologous to identified epitopes and compared their sequence similarity in influenza strains preceding 1957 or from the past 20 years. Taking the average sequence identity, we found a significantly higher similarity between the strains preceding 1957, with 126 of all 200 epitopes and 58 of 78 epitopes from the HA and NA protein having a higher similarity hit in strains preceding 1957 (P = 3.78 E-6 according to a paired, one-tailed t test). This could explain why the presence of neutralizing antibodies in the human population depends on exposure to influenza strains preceding 1957.
In terms of T-cell responses, epitopes are more highly conserved overall. In particular, 69% of the epitopes targeted by CD8+ T-cell responses in seasonal H1N1 influenza isolates are also present in all S-OIV isolates. The comparatively lower fraction of conserved CD4+ epitopes (41%) likely reflects the relative larger size of the CD4+ T-cell epitope as compared with CD8+ T-cell epitopes, which decreases the likelihood that a CD4+ T-cell epitope is totally conserved. As predicted by the bioinformatic analysis, we were able to experimentally detect significant levels of pre-existing T-cell immunity to sequences totally conserved in S-OIV. Further analysis proved that this level of immunity is comparable to that observed in sequences conserved in the seasonal H1N1 influenza isolates. Although T-cell responses do not prevent infection, they do contribute to the clearance of infected target cells, and such pre-existing immunity may lead to a less severe course of disease (16–19).
It is tempting to speculate on the significance of our finding in terms of the disease severity and death rate of the S-OIV as compared with seasonal human H1N1 influenza virus. It was initially feared that the current S-OIV would be much more lethal than seasonal H1N1 influenza. In fact, a recent study demonstrated that a S-OIV isolate (from a hospitalized patient in the United States) caused more severe pathogenicity in infected mice, ferrets, and nonhuman primates than a currently circulating human H1N1 influenza virus (3). Not all studies in animal models unequivocally suggest increased virulence, as transmission via respitory droplets in ferrets was found to be less efficient for S-OIV compared with seasonal H1N1 by some investigators (6) but not others (5). On the other hand, S-OIV isolates were consistently found to replicate better in the lung tissue of animal models (3, 5, 6). At the same time, the potential pathogenic nature of S-OIV is not supported by the data currently available that suggests that a large number of suspected infections in the United States and a disproportionately low number of deaths are associated with S-OIV. We propose that the divergence between disease severity observed in most animal studies and that found in the human population could be due to the contribution of pre-existing T-cell–mediated immunity to lessening disease severity. This was not a component of the animal studies, as naive animals without previous exposure to seasonal H1N1 influenza were infected with the S-OIV isolate.
Clearly several mechanisms contribute to the overall infectivity and virulence of a given influenza isolate/strain. Different strains may vary in their capacity to interfere with IFN pathways based on the activity of the particular sequence of the NS1 protein, as demonstrated in the case of 1918 H1N1 (20). Also, changes in the HA receptor-binding domain can play a role in determining infectivity, as in the case of the recent H5N1 isolates (21, 22). These mechanisms are not addressed by our analysis. Our analysis does, however, demonstrate that as far as immune system recognition, the new S-OIV is not radically different from other seasonal H1N1 influenza isolates, perhaps explaining the relatively mild nature of the S-OIV strain.
A counterargument against the relevance of our findings is as follows: If conserved T-cell epitopes are indeed relevant targets of protective pre-existing immune responses, why are they not mutated in influenza in general and in S-OIV in particular, as are the neutralizing B-cell epitopes in HA and NA? There are four possible explanations. First, our analysis demonstrated that most B-cells epitope are derived from the more variable external proteins, whereas a significant fraction of T-cell epitopes are derived from the more conserved internal proteins. T-cell epitopes in the variable HA and NA proteins were just as likely to be mutated as B-cell epitopes. This suggests that mutations in internal proteins sequences may have more direct impact on viral fitness, so that the virus is less capable of avoiding immune responses against them. Second, it can be argued that influenza viruses have found alternative ways to avoid the effect of T-cell responses, notably by inhibiting IFN-related viral responses in the host (23). This would mean that the virus does not need to evade T-cell responses by mutation, as it has found other ways of escaping from the immune response. Third, antibodies provide sterilizing immunity from infection, which make it essential for the virus to escape from this response to spread. T-cell responses, on the other hand, are thought to lessen disease severity. If the virus retains the ability to infect and spread from host to host, the pressure on avoiding T-cell immune recognition may be less pronounced. Fourth, T-cell responses are much more host specific than antibody responses, as they require presentation by different types of MHC complexes. Because the S-OIV has been circulating through swine in the recent past, there has not been the selection pressure to eliminate HLA-specific peptides from the viruses.
We would like to point out several caveats that need to be kept in mind when examining the results of our study. Our analysis is based on epitope sequences that have been reported in the literature and, as such, does not include epitopes that have not yet been identified. This is a particularly serious limitation in terms of epitopes present in strains dating back to years in which epitope identification technologies were not widely available or used. A second caveat is that we consider an epitope to be conserved between two strains only if the sequence is 100% identical. Clearly, this is a very conservative assumption, as cross-reactivities between influenza epitopes with individual residue substitutions have been observed frequently (24, 25). The qualitative findings of our analysis should not be altered significantly when allowing for such cross-reactivities, however, as the relative conservation of T-cell and B-cell epitopes will likely remain unchanged.
In conclusion, we have conducted an analysis of epitopes in S-OIV based on experimentally identified epitopes cataloged in the IEDB and on viral sequences that were rapidly published in sequence databases available to scientists worldwide. An initial version of the epitope analysis in this report was published online through the IEDB to allow rapid dissemination of knowledge. Our analysis provides insights into the relative conservation of B-cell and T-cell epitopes and has allowed formulating the hypothesis experimentally tested in the present report. We hope that the analysis and datasets provided with it prove useful to experimental and computational immunologists, and that awareness is raised of the ability to track immune epitope conservation across multiple viral strains. Finally, this experience has demonstrated how “real time” exchange of information on the Internet can catalyze the scientific process in the eye of an imminent public health threat.
Materials and Methods
Querying for Epitopes with Human Relevance in the IEDB.
Epitopes derived from influenza A were retrieved from the IEDB, as described in the SI Text.
Querying for Epitopes Conserved in Circulating H1N1.
The epitopes in the human-relevant set described above were searched against H3N2 and H1N1 sequences from circulating strains between 1988 and 2008, as described in the SI Text.
Querying for Epitopes Conserved in S-OIV (H1N1 2009).
The set of circulating H1N1 epitopes were searched against all S-OIV sequences (H1N1, 2009) and the conservation of epitopes was calculated in the same manner as above.
Querying for Influenza Sequences.
Circulating H1N1 and H3N2 influenza sequences were obtained from the NCBI Influenza Virus Resource (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html). For details on the queries, see the SI Text.
At the time this analysis was performed, the NCBI database did not contain sequences from Mexican isolates. These were retrieved from the GISAID database. Table S3 lists the isolates for which sequences were obtained. All Mexican isolates were submitted by the Centers for Disease Control and Prevention (Atlanta, GA, Rebecca Garten).
Homology Modeling, Image Generation, and ASA Calculation.
The homology model for Fig. 1 was generated using the SWISS-MODEL homology-modeling server (26), as described in the SI Text.
PBMC Isolation.
PBMC were isolated from heparinized blood by gradient centrifugation with a Histopaque-1077 (Sigma-Aldrich, St. Louis, MO), suspended in fetal bovine serum (FBS) containing 10% dimethyl sulfoxide, and cryopreserved in liquid nitrogen.
Ex vivo IFN-γ ELISPOT.
IFN-γ ELISPOT assays were performed as described in ref. 27. In brief, 2 × 105 PBMC (CD8+ assays) or enriched CD4+ T cells were incubated with pools of peptides (1 μg/ml per peptide). CD4+ T cells were positively enriched by incubating PBMC with anti-CD8 microbeads (Miltenyi Biotech, Auburn, CA) for 15 min at 4 °C. After washing, PBMC were resuspended in magnetic cell sorting (MACS) buffer and passed through a magnetized LD column (Miltenyi Biotech). Enriched CD4+ T cells were collected by washing the column three times with MACS buffer. After 20 h incubation with the peptide pools at 37 °C, plates were developed, and responses calculated as described in ref. 27.
Intracellular IFN-γ Staining.
PBMC were stimulated with pools of peptides (1 μg/ml per peptide). After 2 h, brefeldin A was added, and PBMC were cultured for an additional 6 h with the peptide pools. After incubation, cells were stained for cell surface antigens CD4, CD8, CD45RA, CD62L, and CCR7. Cells were then fixed, permeabilized, and stained for intracellular IFN-γ using a Cytofix/Cytoperm kit according to manufacturer's directions (BD Biosciences). After washing, samples were resuspended in phosphate-buffered saline, and data were acquired on an LSRII flow cytometer (BD Biosciences). The frequency of CD4+ and CD8+ T cells responding to each peptide pool was quantified by determining the total number of gated CD4+ or CD8+ and IFN-γ+ T cells using FlowJo software (Tree Star, San Carlos, CA).
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health (NIH) contracts HHSN26620040006C, N01-AI30039, and N01-AI40041.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911580106/DCSupplemental.
References
- 1.Cohen J, Enserink M. Swine flu. After delays, WHO agrees: The 2009 pandemic has begun. Science. 2009;324:1496–1497. doi: 10.1126/science.324_1496. [DOI] [PubMed] [Google Scholar]
- 2.Garten RJ, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Itoh Y, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009 doi: 10.1038/nature08260. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR. 2009;58:521–524. [PubMed] [Google Scholar]
- 5.Munster VJ, et al. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science. 2009;325:481–483. doi: 10.1126/science.1177127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maines TR, et al. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science. 2009;325:484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bui HH, Peters B, Assarsson E, Mbawuike I, Sette A. Ab and T-cell epitopes of influenza A virus, knowledge and opportunities. Proc Natl Acad Sci USA. 2007;104:246–251. doi: 10.1073/pnas.0609330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters B, et al. The immune epitope database and analysis resource: From vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bao Y, et al. The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogner P, Capua I, Lipman DJ, Cox NJ, et al. A global initiative on sharing avian flu data. Nature. 2006;442:981. [Google Scholar]
- 11.Nguyen HH, et al. Heterosubtypic immunity to influenza A virus infection requires a properly diversified antibody repertoire. J Virol. 2007;81:9331–9338. doi: 10.1128/JVI.00751-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couch RB. An overview of serum antibody responses to influenza virus antigens. Dev Biol (Basel) 2003;115:25–30. [PubMed] [Google Scholar]
- 13.Prabhu N, et al. Monoclonal antibodies against the fusion peptide of hemagglutinin protect mice from lethal influenza A virus H5N1 infection. J Virol. 2009;83:2553–2562. doi: 10.1128/JVI.02165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liew FY, Russell SM, Appleyard G, Brand CM, Beale J. Cross-protection in mice infected with influenza A virus by the respiratory route is correlated with local IgA antibody rather than serum antibody or cytotoxic T-cell reactivity. Eur J Immunol. 1984;14:350–356. doi: 10.1002/eji.1830140414. [DOI] [PubMed] [Google Scholar]
- 15.Terajima M, et al. Influenza A virus matrix protein 1-specific human CD8+ T-cell response induced in trivalent inactivated vaccine recipients. J Virol. 2008;82:9283–9287. doi: 10.1128/JVI.01047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMichael AJ, Gotch FM, Noble GR, Beare PA. Cytotoxic T-cell immunity to influenza. N Engl J Med. 1983;309:13–17. doi: 10.1056/NEJM198307073090103. [DOI] [PubMed] [Google Scholar]
- 17.Webby RJ, et al. Protection and compensation in the influenza virus-specific CD8+ T-cell response. Proc Natl Acad Sci USA. 2003;100:7235–7240. doi: 10.1073/pnas.1232449100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McElhaney JE, et al. T-cell responses are better correlates of vaccine protection in the elderly. J Immunol. 2006;176:6333–6339. doi: 10.4049/jimmunol.176.10.6333. [DOI] [PubMed] [Google Scholar]
- 19.Bender BS, Croghan T, Zhang L, Small PA., Jr Transgenic mice lacking class I major histocompatibility complex-restricted T cells have delayed viral clearance and increased mortality after influenza virus challenge. J Exp Med. 1992;175:1143–1145. doi: 10.1084/jem.175.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basler CF, et al. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc Natl Acad Sci USA. 2001;98:2746–2751. doi: 10.1073/pnas.031575198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens J, et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 22.Yen HL, et al. Changes in H5N1 influenza virus hemagglutinin receptor binding domain affect systemic spread. Proc Natl Acad Sci USA. 2009;106:286–291. doi: 10.1073/pnas.0811052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Sastre A, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 24.Kreijtz JH, et al. Cross-recognition of avian H5N1 influenza virus by human cytotoxic T-lymphocyte populations directed to human influenza A virus. J Virol. 2008;82:5161–5166. doi: 10.1128/JVI.02694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee LY, et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest. 2008;118:3478–3490. doi: 10.1172/JCI32460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oseroff C, et al. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc Natl Acad Sci USA. 2005;102:13980–13985. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.