Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) encodes numerous accessory proteins whose importance in the natural infection process is currently unclear. One of these accessory proteins is set apart by its function in the context of a related murine hepatitis virus (MHV) infection. SARS-CoV protein 6 increases MHV neurovirulence and accelerates MHV infection kinetics in tissue culture. Protein 6 also blocks nuclear import of macromolecules from the cytoplasm, a process known to involve its C-terminal residues interacting with cellular importins. In this study, protein 6 was expressed from plasmid DNAs and accumulated in cells prior to infection by wild-type MHV. Output of MHV progeny was significantly increased by preexisting protein 6. Protein 6 with C-terminal deletion mutations no longer interfered with nuclear import processes but still retained much of the capacity to augment MHV infections. However, some virus growth-enhancing activity could be ascribed to the C-terminal end of protein 6. To determine whether this augmentation provided by the C terminus was derived from interference with nuclear import, we evaluated the virus-modulating effects of small interfering RNAs (siRNAs) directed against importin-β mRNAs, which down-regulated classical nuclear import pathways. The siRNAs did indeed prime cells for enhanced MHV infection. Our findings indicated that protein 6-mediated nuclear import blocks augmented MHV infections but is clearly not the only way that this accessory protein operates to create a milieu conducive to robust virus growth. Thus, the SARS-CoV protein 6 accelerates MHV infections by more than one mechanism.
Severe acute respiratory syndrome coronavirus (SARS-CoV) is a zoonotic virus that is endemic in bats (33, 34). This virus can infect exotic animals marketed for consumption in southeast China (22, 58), and in doing so can acquire proximity to human populations. SARS-CoV entered human hosts in 2002 to 2003 and disseminated rapidly to more than 30 countries across five continents, killing ∼800 individuals. The SARS-CoV virion contains a ∼30-kb, plus-strand RNA genome enclosed within a pleiomorphic membrane envelope. Many of the features of this virus are known from over 25 years of research on animal coronaviruses. The 5′ ∼2/3 of the genome encodes two very large polyproteins which are rapidly proteolyzed into components functioning in viral RNA synthesis and metabolism, while the 3′ ∼1/3 includes several virion assembly subunits, notably the proteins spike (S), envelope (E), membrane (M), and nucleocapsid (N). Integrated both between and within these genes encoding virion proteins are eight so-called accessory genes (51). These accessory genes, originally designated numerically as open reading frames (ORFs) 3a through 9b, were identified in the original 2003 SARS-CoV isolate (strain Urbani) and are now known to be ubiquitous in SARS-CoVs obtained from infected bats, civit cats, raccoon dogs, and humans (33, 46, 64), suggesting evolutionary conservation of important viral functions. The majority of the accessory genes are expressed in SARS-CoV-infected cells (8, 10, 28, 37, 39, 48, 54, 70). Many are membrane proteins, and as antibodies have become available, several of the accessory proteins have been detected as copurifying with virions (26, 48, 50) and have been structurally resolved by X-ray crystallography (39).
Although these studies have provided important information about the SARS-CoV accessory proteins, their central functions remain largely unknown. Indeed, accessory protein functions may be difficult to discern, as elimination of most of the accessory genes through reverse genetics generates recombinant SARS-CoVs with tissue culture growth properties remarkably similar to native SARS viruses (1, 16, 68). Experimental rodent models for SARS-CoV infection and disease have been recently developed (35, 45, 56, 61), and under these experimental conditions, recombinant SARS-CoVs lacking a subset of accessory genes were nearly equivalent to complete SARS-CoV in virulence and pathogenicity (2, 14, 16, 68). Thus, at present there is no satisfying explanation of the roles that these evolutionarily conserved accessory proteins play in natural animal or human SARS-CoV infections.
Accessory protein 6 has received significant scrutiny, as this protein does exhibit functions that may relate to its presumed roles in SARS-CoV infections. When expressed alone from plasmid cDNA, protein 6 ablates type I interferon signaling, indicating its potential in thwarting innate immune effectors (15, 31). Notably, several different coronavirus products may have similar immune evasion activities (31), a redundancy that presumably accounts for the inapparent in vivo consequences of ORF6 elimination from SARS coronavirus. However, when evaluated in the context of heterologous murine coronavirus infections, protein 6 has clearly recognized activities. Engineered recombinant murine coronaviruses constructed to express SARS-CoV ORF6, designated rJ.2.2.6 (recombinant JHM strain 2.2-V-1 encoding SARS p6) (41), have been compared with isogenic rJ.2.2.6-KO viruses in which ORF6 is closed via knockout (KO) of the initiation codon. Relative to rJ.2.2.6-KO, the rJ.2.2.6 viruses grow more rapidly, both in tissue culture (55) and in the murine central nervous system, and have increased in vivo lethality (40, 41). These properties of protein 6 may relate to direct effects on the coronavirus replication machinery, as protein 6 colocalizes intracytoplasmically with membrane-proximal sites of viral RNA-dependent RNA synthesis and does indeed provide for earlier viral RNA synthesis in tissue culture contexts (55).
Protein 6 is a 63-amino-acid peptide that comprises a relatively hydrophobic N-terminal portion of ∼40 residues and a C-terminal hydrophilic extension. The N-terminal region may function as a signal/anchor for membrane association, as protein 6 has the biochemical characteristics of an integral membrane protein (55). This membrane-associating property of the N-terminal region is presumed to operate in assisting development of viral RNA replication sites, which are well-known to reside on the interfaces between intracellular membranes and cytosol. The C-terminal region extends from membranes into cytosolic environments (40) and displays several motifs that are vital for a separate and novel activity (15). C-terminal residues of protein 6 interact with cellular importins, mediators of protein import into the nucleus (15). Accumulating protein 6 on cytoplasmic membranes titrates importins to cytoplasmic sites and thereby thwarts cellular capacity to transport cytoplasmic cargo into the nucleus. Protein 6 has been shown to impede nuclear import of proteins such as IRF3 and STAT1, key regulators of interferon gene transcription (31). While it is reasonably assumed that this activity of the protein 6 C terminus contributes to virus growth or pathogenesis by thwarting innate immune responses, there are few supporting data. In fact, alanine substitutions engineered into the C terminus did not reduce the murine neurovirulence of rJ.2.2.6 (40), leaving it unclear whether the hydrophilic cytoplasmic tail of protein 6 makes significant contributions to the growth or virulence of this related coronavirus.
These indications that unique biological activities might be separately ascribed to the N- and C-terminal regions of protein 6 led us into investigations dissecting the relative contributions of each region to recombinant JHM virus growth. In extending a recently published report (15), we found that protein 6 blocked the nuclear accumulation of proteins relying on classical nuclear import pathways, with the C terminus being vital in blocking nuclear import. More importantly, we correlated this nuclear import blockade with enhanced murine hepatitis virus (MHV) infections, giving credence to the hypothesis that this conserved, 63-amino-acid accessory protein creates superior environments for virus growth by enriching the cytosol with proteins otherwise destined to the nucleus. We found, however, that this impediment to protein import into the nucleus made only a minor contribution to virus growth, as C-terminally truncated forms of protein 6 lacking this activity still retained the majority of virus-augmenting function. These findings indicate that this small protein 6 supports murine coronavirus infections by more than one mechanism.
MATERIALS AND METHODS
Cells.
Human epithelial kidney cells (HEK293), baby hamster kidney cells (BHK), and BHK cells stably expressing T7 polymerase gene (BSR-T7) (5) were grown and maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM Na-HEPES, 2 mM l-glutamine, and penicillin (100 U/ml)-streptomycin (100 μg/ml). Mouse fibroblast cells (17Cl-1) were grown and maintained in DMEM supplemented with 5% tryptose phosphate broth, 5% FBS, and penicillin-streptomycin.
Plasmid constructions.
Expression plasmids encoding full-length protein 6 [pCAGGS-6 and pBMN-6 (pBMN-6-IRES-eGFP)] and C-terminal deletion mutant [pBMN-6ΔC (pBMN-6ΔC-IRES-eGFP), lacking C-terminal residues 53 to 63] were constructed using standard PCR techniques. The retroviral vector pBMN-IRES-eGFP was kindly provided by Garry P. Nolan, Stanford University, Stanford, CA (30). All clones carry a hemagglutinin (HA) tag (sequence, YPYDVPDYA) at C termini for easy detection of expressed protein by Western blot and immunofluorescence assays. Renilla luciferase reporter plasmid (pRL-TK), which contains Renilla luciferase cDNA under the herpes simplex virus thymidine kinase (TK) promoter, was purchased from Promega (Madison, WI), and firefly luciferase reporter plasmid pECMVT7Luc, containing the firefly luciferase gene under control of a bacteriophage T7 promoter, has been described previously (3).
The plasmids encoding the enhanced green fluorescent protein (EGFP) trimer with or without the nuclear localization signal (NLS) were created using methods developed by Genove et al. with some modifications (18). Briefly, an EGFP cassette was PCR amplified using forward primer 5′-ATCTCGAGTGAGCAAGGGCGAGGAGC-3′ and reverse primer 5′-GAGAATTCAGCAAGGGCGAGGAGCTG-3′ and ligated into pEGFP-C1 (Clontech, Inc.) to generate a construct we designate p2xEGFP. A third EGFP cassette was PCR amplified using forward primer 5′-CTGAATTCTCCGGACTTGTACAGCAGG-3′ and reverse primer 5′-TTGTCGACGTCCGGACTTGTACAGCTCG-3′ and cloned into p2xEGFP to yield p3xEGFP. Subsequently, oligonucleotides encoding two tandem copies of the simian virus 40 (SV40) large T antigen NLS and human heterogeneous ribonucleoprotein (hnRNPA1) nuclear localization signal (M9 NLS) were subcloned at the 3′ end of the third GFP open reading frame to yield p3xEGFP-SV40 NLS and p3xEGFP-M9 NLS, respectively. The p3xEGFP-SV40 NLS encodes an ∼84-kDa fluorescent protein with a C-terminal-appended SV40 NLS (ELYKSGRRAQDPKKKRKVDPKKKRKV; the C-terminal GFP residues are shown in bold, linker residues are in italics, and the NLS is underlined). The p3xEGFP-M9 NLS encodes a fluorescent protein with C-terminal-appended M9 NLS (ELYKSGRRAQGNYNNQSSNFGPMKGGNFGGRSSGPYGGGGQYFAKPRNQGGY, with C-terminal GFP residues in bold, linker residues in italics, and the NLS underlined).
Transient transfection and infection.
293T cells were grown in 10-cm2 dishes and transiently transfected with 2.0 μg pcDNA-mCEACAM1a (MHV receptor; J. Lacny and T. M. Gallagher, unpublished data) and 1 μg of p6-encoding plasmid (pCAGGS-6/pCAGGS-6-UT [untagged]/pBMN-6/pBMN-6ΔC) or vector control, using a calcium phosphate transfection method (47). After 24 h, transfection media were removed and cells were infected with recombinant MHV strain JHM (rJ.2.2) or rJ2.2 expressing SARS-CoV protein 6 (rJ2.2.6) at a multiplicity of infection of 0.2 PFU/cell in serum-free DMEM. After a 1-h adsorption period, the inocula were removed and infections were allowed to proceed overnight in DMEM-10% FBS.
Plaque assay and immunoblotting of virion proteins.
Culture media were collected at 16 h postinfection or at other times postinfection as indicated and clarified at 2,000 × g for 20 min. In plaque assays, aliquots of clarified media were serially diluted and applied to monolayers of 17Cl-1 cells. After 1 h of adsorption, inocula were removed and cells were washed with phosphate-buffered saline (PBS) and then overlaid with DMEM containing 1% FBS and 0.5% agar (Becton Dickinson, MD). The cells were fixed at 3 days postinfection and stained with 1% crystal violet, and plaques were enumerated. For isolation and detection of virus particles, clarified medium (0.7 ml) was overlaid on 0.6-ml cushions of 20% sucrose-25 mM Na-HEPES [pH 7.4]-100 mM NaCl-0.01% bovine serum albumin. Virions were pelleted by ultracentrifugation at 200,000 × g for 30 min. Virion-containing pellets were dissolved in sodium dodecyl sulfate (SDS) solubilizer (50 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, and 0.1% bromophenol blue), fractionated using SDS-polyacrylamide gel electrophoresis (PAGE), and immunoblotted with antibodies reacting with the HA epitope (Covance, Inc., Berkeley, CA), S proteins (monoclonal antibody [MAb] 10G; provided by Fumihiro Taguchi), M proteins (MAb J.1.3), and N proteins (MAb J.3.1; provided by John O. Fleming).
Transient transfection and luciferase assay.
To evaluate the influence of protein 6 on reporter gene expression, BHK, 293T, and BSR-T7 cells were cotransfected with 1 μg of reporter plasmid (pRL-TK or pECMVT7Luc or both) and 10-fold-increasing concentrations of pCAGGS-6. The total amount of DNA was kept the same for each transfection mixture by adding empty pCAGGS-MCS vector. In 293T cells, transfections were performed with calcium phosphate transfection reagents as described elsewhere (9, 47), whereas in BHK and BSR-T7 cells, transfections were performed with Lipofectamine 2000 as recommended by the manufacturer (Invitrogen Corporation). At 24 h posttransfection (hpt), cells were rinsed twice in PBS and lysed in passive reporter lysis buffer (Promega) after resuspension at 5 × 106 cells per ml. Five-μl aliquots were assayed using luciferase assay reagent (Promega), and luminescence was recorded using a Veritas Microplate luminometer (Turner Biosystems, Sunnyvale, CA). All transfections were performed in triplicate, samples were measured in quadruplicate, and numerical uncertainties are shown by error bars.
Immunofluorescence and confocal microscopy.
To assess the nucleo-cytoplasmic distribution of NLS-containing GFP, 293T cells were grown on glass coverslips, placed into 10-cm2 dishes, and transfected with plasmids encoding reporter genes (p3xGFP-M9 NLS, p3xGFP-SV40 NLS, or p3xGFP) along with p6 expression plasmids (pCAGGS-6, pBMN-6, and pBMN-6ΔC) or vector control plasmids. Cells were fixed at 24 hpt in 4% paraformaldehyde for 10 min and permeabilized in digitonin buffer (300 mM sucrose, 100 mM KCl, 2.5 mM MgCl2, 1 mM EDTA, 10 mM HEPES [pH 6.9]) containing 5-μg/ml digitonin for 15 min at room temperature. The coverslips were incubated in blocking buffer (2% bovine serum albumin in PBS) for 15 min followed by an overnight incubation with primary antibody directed against HA tag at a dilution of 1:5,000. The coverslips were washed three times in ice-cold PBS at room temperature and incubated with fluorochrome-conjugated secondary antibodies (all from Molecular Probes, Inc., Eugene, OR) for 1 h at room temperature. The coverslips were washed three times in PBS and mounted with ProLong Gold antifade reagent (Invitrogen Corporation). Confocal images were captured using a Carl Zeiss model 510 laser-scanning confocal microscope. Digitized images were processed with Image J (National Institutes of Health).
siRNA transfection and MHV infections.
Predesigned small interfering RNAs (siRNAs; catalog number AM16704; siRNA ID 11218, 11125, and 145041) corresponding to exons 3 and 9 of human importin-β mRNA (accession number NM_002265), negative control siRNA (catalog number 4611), and scrambled control siRNA (GGCACAAUAUCAGCAGCGG; catalog number AM16104) were purchased from Ambion (Austin, TX). These siRNAs were transfected into 239T cells, in graded doses as indicated, along with pcDNA-mCEACAM1a, using Lipofectamine 2000 (Invitrogen). Typical experiments involved transfecting ∼2 × 105 293T cells with 100 nmol of siRNA and 1.5 μg of pcDNA-mCEACAM1a in 1 ml of OPTI-MEM (Life Technologies). Controls included mock transfection (transfection reagent lacking siRNA), negative control siRNA, and scrambled control siRNA. At 2 days posttransfection, cells were infected with rJ.2.2. After 16 h, supernatants were collected and infectivities were evaluated by plaque assay. Transfections were done in triplicate, and each experiment was repeated two to three times.
RESULTS
Plasmid DNAs encoding protein 6 prime cells for accelerated MHV infections.
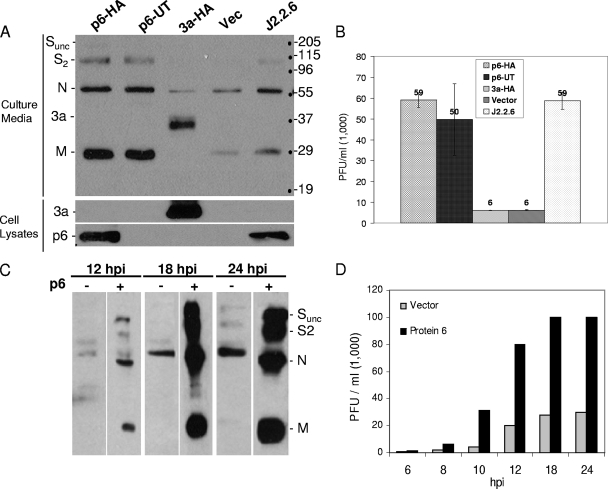
In our previous studies, we used recombinant MHVs with SARS-CoV open reading frames integrated in the genome. Using these chimeric recombinant viruses, we showed that the SARS-CoV protein 6 was a virulence factor, increasing both MHV yields in tissue culture and neurovirulence in mice (41, 55). In the present study, we employed a related but distinct approach. Here, cultured cells were transiently transfected with expression plasmids encoding wild-type and mutant protein 6 and then subsequently infected with different MHV strains. This approach was simpler than our previous approach because it did not require development of recombinant viruses and was also able to address whether protein 6 might independently modify the cell in ways that influence the subsequent infection process. 293 cells were transfected with the desired plasmids and subsequently infected with recombinant MHV (rJ2.2) or recombinant MHV expressing SARS-CoV protein 6 (rJ2.2.6). Relative to vector-transfected cells, we observed robust viral egress from p6-expressing cells, as registered by abundant Western blot signals corresponding to virion proteins S, N, and M (Fig. 1A). These secreted particles were infectious, as demonstrated by plaque assay results, revealing ∼10-fold-more infectivity in medium from p6-expressing cells relative to vector controls (Fig. 1B). Comparative analysis of HA-tagged and untagged protein 6 revealed that the HA epitope had no influence on p6 activity (Fig. 1A and B). This augmenting effect of p6 was evident as early as 8 to 10 h postinfection (Fig. 1C and D).
FIG. 1.
Effects of protein 6 on MHV infections. (A) 293 cells were cotransfected with pcDNA-mCEACAM1a and pCAGGS-6, pCAGGS-6-UT (untagged), pCAGGS-3a, or empty vector. At 24 hpt, cells were infected with rJ.2.2 or a recombinant virus expressing SARS-CoV protein 6 (rJ2.2.6) at a multiplicity of infection of ∼0.2 PFU/cell. Supernatants were collected at 16 h postinfection (hpi) and fractionated by SDS-PAGE, and virion structural proteins (marked on left) were detected by Western blotting. Positions of molecular mass standards (in kilodaltons) are listed to the right of the gels. (B) Supernatants were titrated on monolayers of 17Cl-1 cells, and plaques were enumerated on day 3 postinfection. (C) Supernatants were collected at 12, 18, and 24 h postinfection and analyzed for secreted virus particles by Western blotting. (D) Infectivities in supernatants collected at the indicated time postinfection were enumerated by plaque assay.
In a parallel experiment, 293 cells were transfected with plasmids encoding the SARS accessory proteins 3a, 3b, and 7a (24, 26, 29, 50, 69, 71) and then infected with MHV. Immunoblot analyses confirmed that the plasmids were expressed at levels comparable to protein 6; however, none had any significant impact on MHV infection kinetics (Fig. 1A and B and data not shown). These data unequivocally indicated that protein 6 creates cellular environments that benefit MHV growth; however, the underlying mechanism responsible for this accelerated infection remained unclear.
Protein 6 reduces expression from plasmids requiring transcription in the nucleus.
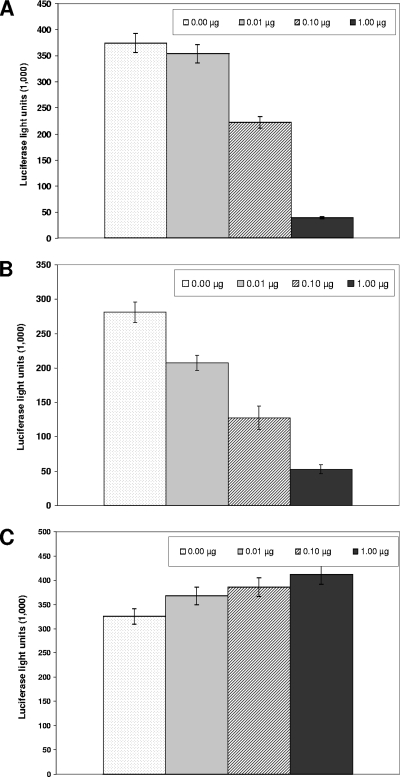
Clues to the mechanism by which protein 6 increases MHV growth came from the unexpected observation that this protein suppressed the expression of cotransfecting plasmid-based genes. An example of this phenomenon is illustrated in Fig. 2A, in which 293 cells were cotransfected with plasmids encoding Renilla luciferase and p6. p6 levels correlated with significant (∼5- to 10-fold) reductions in luciferase activities when samples were evaluated at 16 to 20 h posttransfection. Similar suppressive effects of p6 were recorded in sequential transfections in which 293 cells were first transfected with p6 expression vectors and then with luciferase reporter plasmids 18 h later (Fig. 2B). However, the reverse transfection order did not result in any p6-mediated effects on luciferase accumulations (Fig. 2C), suggesting that while p6 could inhibit expression from other plasmid DNA, it could not do so if the other plasmid had already become established in the cell nucleus.
FIG. 2.
Effects of protein 6 on reporter gene expression from cotransfected plasmids. (A) 293 cells were cotransfected with pRL-TK and with increasing amounts of pCAGGS-6. The amount of DNA for each transfection mixture was kept constant by addition of empty pCAGGS-MCS vector. Cells were lysed at 24 hpt, and Renilla luciferase luminescence was quantified. (B) 293 cells were first transfected with increasing amounts of pCAGGS-6 followed by a second transfection 18 h later to introduce pRL-TK. Luciferase luminescence was quantified at 24 hpt. (C) 293 cells were first transfected with reporter plasmid pRL-TK followed by a second transfection at 18 h with the indicated amounts of pCAGGS-6. Cells were lysed at 24 hpt, and luciferase levels were quantified.
We considered that these p6-mediated reductions in luciferase accumulation might arise from generalized suppression of transcription or translation but found no evidence to support this suggestion. Approximately 50 to 70% of 293 cells were routinely positive for p6 accumulation after plasmid transfection, as measured by immunofluorescence with anti-HA epitope antibodies, yet these cells grew normally out to 3 days posttransfection, retaining viabilities similar to control transfectants as measured by trypan blue exclusion. Pulse-labeling with [3H]uridine, in either the presence or absence of actinomycin D and with [35S]methionine, revealed that p6 effected no change in the incorporation kinetics for these radioactive RNA and protein precursors (data not shown). These findings led us to suggest that p6 specifically interfered with the cotransfected or subsequently transfected DNAs, perhaps by preventing their entrance into the cell nucleus.
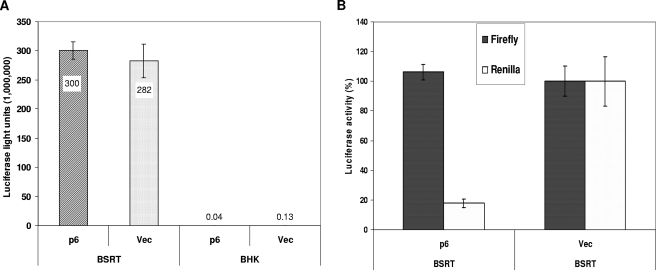
Transfected plasmid DNAs are inefficiently transported from the cell cytoplasm to the nucleus (7, 57). Numerous reports have indicated that this transport can be accomplished by DNA-binding proteins that contain nuclear localization signals. These proteins attach to plasmid DNAs and escort them through nuclear pores, using the nuclear import machinery of the cell (11-13, 59, 65). To address the suggestion that protein 6 inhibits luciferase by blocking nuclear import of its plasmid DNAs, we asked whether luciferase expressed from a cytoplasmic bacteriophage T7 promoter might be immune to p6-mediated inhibition when transfected into cells containing cytoplasmic DNA-dependent T7 RNA polymerase (6, 66). To this end, BHK cells containing a cytoplasmic T7 RNA polymerase (BSR-T7) were cotransfected with vector control or p6 plasmids along with reporter plasmids encoding firefly luciferase under T7 promoter control (3). Under these conditions, p6 was indeed unable to inhibit luciferase accumulation (Fig. 3A). Parallel transfections into parental BHK cells lacking T7 RNA polymerase only generated a background luciferase activity. These data, normalized to cotransfecting nuclear-based Renilla luciferase expression, clearly revealed that the nuclear Renilla but not cytoplasmic firefly luciferase accumulation was inhibited in BSR-T7 cells by protein 6 (Fig. 3B). Together these data suggest that the inhibitory effect of protein 6 is at the level of plasmid DNA transport from the cytoplasm to the nucleus.
FIG. 3.
Luciferase expression from nuclear and cytoplasmic promoters in protein 6-expressing cells. (A) BHK and BSR-T7 cells were cotransfected with pECMVT7Luc and pCAGGS-6 or empty vector. Cells were lysed at 24 hpt, and luciferase activities were quantified. The data represent means ± standard deviations of a representative experiment done in triplicate. (B) BSR-T7 cells were cotransfected with reporter plasmids (pRL-TK and pECMVT7Luc) and pCAGGS-6 or empty vector. At 24 hpt, cells were lysed and Renilla or firefly luciferase activities were determined. Data are presented as percent changes in luciferase activities in protein 6-expressing cells relative to empty vector-transfected cells.
Protein 6 impedes the nuclear import of NLS-containing proteins.
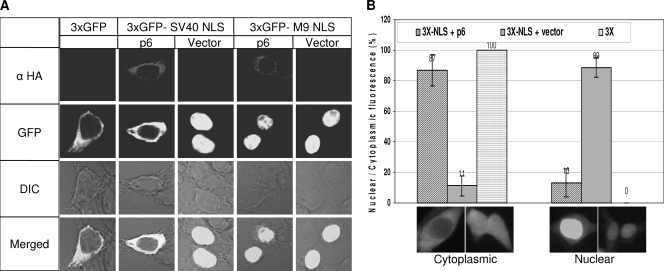
Interference with nuclear-based plasmid DNA expression represents an unconventional approach to evaluate nuclear import events. A more common technique involves the monitoring of reporter proteins with or without nuclear localization signals. Active, import factor-dependent translocation of NLS-containing proteins through nuclear pores is required for cargo larger than ∼60 kDa (19, 20). Therefore, we constructed reporter plasmids encoding three tandem copies of green fluorescent protein, with or without C-terminal-appended nuclear localization signals (SV40 NLS and M9 NLS) (27). These constructs, designated 3xGFP, 3xGFP-SV40 NLS, and 3xGFP-M9 NLS, respectively, were proteins of 84 to 90 kDa and therefore exceeded the size limit for passive diffusion into the nucleus (4). 293 cells were cotransfected with these reporter plasmids, along with vector control or p6-expressing plasmids, and then monitored 1 day later by fluorescent microscopy. As expected, the 3xGFP protein was restricted to the cytoplasm, with p6 having the expected suppressive effect on its abundance but no effect at all on its cytoplasmic distribution (Fig. 4A). In striking contrast, the 3xGFP-SV40 NLS efficiently accumulated in the nuclei of vector control-transfected cells, with p6 exerting a pronounced blockade of this nuclear accumulation (Fig. 4A). Indeed, the distribution of 3xGFP-SV40 NLS in p6-expressing cells was essentially identical to that of 3xGFP lacking an NLS, suggesting that p6 interfered with the nuclear import of classical NLS-bearing proteins. Quantitative data revealed that relative to vector-transfected cells, p6 effectively blocked the nuclear accumulation of 3xGFP-NLS in more than 85% of the cells (Fig. 4B). Notably, this p6-mediated inhibition of 3xGFP-SV40 NLS was transient. At 2 days posttransfection, significant proportions of the 3xGFP-SV40 NLS accumulated in the nuclei of p6-expressing cells (data not shown), suggesting that p6 operated to delay but not entirely eradicate the import of NLS-bearing proteins. We also found that nucleo-cytoplasmic distribution of 3xGFP-M9 NLS was not affected by p6, with clear evidence of nuclear accumulation in the presence and absence of p6 (Fig. 4A). Thus, we conclude that p6 interfered with a subset of the known nuclear import pathways (42, 63), leaving the transportin-dependent nuclear import pathway intact and perhaps also leaving slower alternate routes available for 3xGFP-SV40 NLS cargo to enter into nuclei. These observations are consistent with the absence of overt toxicity in cells accumulating protein 6.
FIG. 4.
Nucleo-cytoplasmic distribution of GFP-NLS in protein 6-expressing cells. (A) 293 cells were cotransfected with the indicated reporter plasmids and pCAGGS-6 or empty vector. Cells were fixed at 24 hpt and probed with monoclonal antibody directed against HA tag. The protein 6 expression and nucleo-cytoplasmic distribution of GFP were visualized using laser-scanning confocal microscopy, and representative images are shown. (B) The percentages of cells with nuclear and cytoplasmic GFP were calculated by counting 10 random fields from each transfection. Cells with distinct cytoplasmic fluorescence to uniform nuclear-cytoplasmic fluorescence were scored as cytoplasmic fluorescence, and cells with distinct nuclear staining were scored as nuclear fluorescence. Data represent means ± standard deviations of 10 field counts. Cells with p6 had lower GFP intensities, thus longer exposure times were employed to match the signal intensities relative to vector control transfectants.
The C terminus of protein 6 is required to retain 3xGFP-SV40 NLS in the cytoplasm but is not essential to augment MHV infections.
The SARS protein 6 primed 293 cells for efficient MHV infections and also interfered with the nuclear import of a prototype NLS-bearing protein, 3xGFP-NLS. These observations prompted us to consider whether these two activities are causally related to each other. Previous findings, notably from Frieman et al. (15), might argue for such a causal connection, as their recent research has documented that STAT1 transcription factors are cytoplasmically retained by SARS protein 6. Given that STAT1 must be in nuclei to promote expression of antiviral effector proteins, the increased MHV replication might be related to a paucity of antiviral host proteins. Fortunately, these suggestions could be readily addressed because Frieman et al. (15) had also identified the hydrophilic C terminus of protein 6 as an essential portion interfering with transport of STAT1 into nuclei.
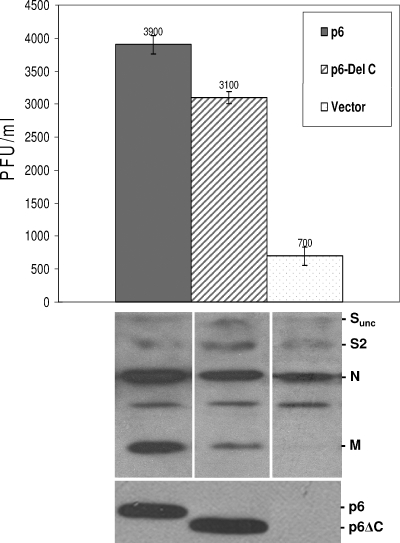
A series of protein 6 truncation mutants, generated by standard methods as described in Materials and Methods, were evaluated for their capacity to augment MHV infections and to prevent nuclear entry of 3xGFP-SV40 NLS. The most instructive p6 mutant was a C-terminal-truncated form lacking 11 amino acids, designated p6ΔC. In the MHV infection assays, we found that this mutant retained ∼80% of the complete p6 capacity to augment output of infectivity and virion particles, as measured by plaque assay and immunoblot analysis of secreted virion proteins (Fig. 5). In the transfected cells, the accumulations and subcellular localizations of these mutants were indistinguishable from the complete 6 proteins (Fig. 5 and data not shown), indicating that the incomplete augmenting activities of this mutant could not be explained by reduced abundance or position within the cells.
FIG. 5.
Effect of the C-terminal deletion of p6 on MHV infections. 293 cells were transfected with pcDNA-mCEACAM1a and plasmid encoding full-length (pBMN-6) or mutant (pBMN-6ΔC) protein 6 or empty vector. Cells were infected at 24 hpt with MHV-rJ2.2 at 0.2 PFU/cell. At 16 h postinfection, media were collected and analyzed for infectivity by plaque assay and for virion particle content by immunoblotting. The intracellular protein 6 accumulations were determined by immunoblotting (lower panel).
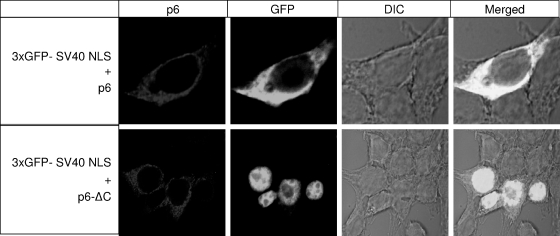
The C-terminal mutant was entirely incompetent in preventing the nuclear entry of 3xGFP-NLS (Fig. 6, lower panels). This finding confirms the data of Frieman et al. (15), and together these studies indicate that the protein 6 C terminus cytoplasmically retains both STAT1 as well as proteins with classical NLS motifs. More importantly, the collective findings suggest that a portion (∼20 to 30%) of protein 6-mediated MHV-augmenting activity is related to its ability to prevent NLS-containing proteins from entering the nucleus. The remaining portion (∼70 to 80%) of the augmenting activity presumably operates by a distinct mechanism.
FIG. 6.
Effects of the C-terminal deletion on nuclear import of 3xGFP-SV40 NLS. 293 cells were cotransfected with p3xGFP-SV40 NLS and plasmid encoding full-length (pBMN-6) or mutant (pBMN-6ΔC) protein 6. At 24 hpt, cells were fixed and probed with monoclonal antibody specific to HA tag. Images were acquired using laser-scanning confocal microscopy, and representative micrographs are shown.
siRNAs targeted to importin-β accelerate MHV infections.
It has been shown that p6 blocks nuclear import by sequestering import factors, especially importin-β, at the rough endoplasmic reticulum and Golgi membranes(15). To determine whether this mechanism at least partially contributes to the priming of cells for robust MHV growth, we attempted to reduce importin-β levels in cells without involving protein 6 at all. We reasoned that if nuclear import blockade were at least partially responsible for augmenting MHV infection, then siRNAs targeted to importin-β would exhibit MHV growth-promoting activities similar to protein 6. Thus, cells were first treated with siRNAs targeting importin-β mRNA and subsequently infected with MHV, in analogy with the experiments involving protein 6.
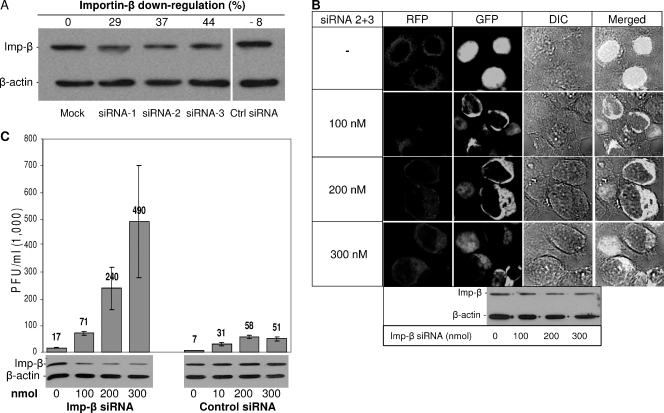
Three importin-β-specific and two different negative control siRNAs were evaluated in our experiments. Relative to the control siRNAs, 100 nM importin-β siRNAs 1, 2, and 3 reduced importin-β steady-state levels by 30 to 50% at 2 days posttransfection (Fig. 7A). These reductions did not have untoward toxic effects, as siRNA- and mock-treated cells were indistinguishable in integrity, adherence, and growth rate for up to 72 h posttransfection. Notably, these levels of importin-β reduction caused only a partial interference with the nuclear import of 3xGFP-SV40 NLS (data not shown), less stringently than that generated by protein 6. A combination of siRNAs 2 and 3, which target importin-β exons 3 and 9, was increasingly effective at preventing the nuclear localization of 3xGFP reporter protein (Fig. 7B).
FIG. 7.
Effects of importin-β down-regulation on MHV infections. (A) 293 cells were transiently transfected with siRNAs (100 nmol each) targeting exons 3 and 9 of importin-β mRNA or scrambled control siRNA. At 48 hpt, cells were lysed in 0.5% NP-40 and fractionated by SDS-PAGE followed by immunoblotting with anti-importin-β and anti-β-actin antibodies. (B) 293 cells were transiently transfected with red fluorescent protein (RFP) as a transfection control and the indicated amounts of importin-β siRNAs (siRNA 2 and 3) followed by a second transfection with p3xGFP-SV40 NLS at 48 h. After 24 h cells were fixed and nucleo-cytoplasmic distributions of 3xGFP-NLS were determined by laser-scanning confocal microscopy. Intracellular levels of importin-β and the housekeeping gene β-actin in siRNA (siRNA 2 and 3)-treated cells were determined by immunoblotting (lower panel). (C) 293 cells were transfected with pcDNA-mCEACAM1a and increasing doses of importin-β or scrambled control siRNA. At 2 day posttransfection, cells were infected with rJ.2.2 at 0.2 PFU/cell. Culture media were collected at 16 to 18 h postinfection, and secreted infectivities were determined by plaque assay. Importin-β levels (lower panel) were determined by immunoblotting.
MHV infections in these importin-β siRNA-treated cells did indeed exhibit high productivity (Fig. 7C). Interestingly, transfection with both of the control siRNAs also rendered cells more susceptible to MHV infections, but relative to these controls, the importin-β siRNAs improved MHV outputs in the range of four- to ninefold (Fig. 7C). Thus, our data revealed that reduced levels of importin-β correlated with improved MHV infections. These data suggest that protein 6 and importin-β siRNAs operate to foster MHV infections by a similar mechanism, namely, interference with protein import into the nucleus. However, protein 6 must have an additional operating mechanism to foster MHV growth, as the N-terminal p6 fragment (amino acids 1 to 52) augmented MHV growth without interfering with the NLS-GFP nuclear import.
DISCUSSION
The SARS-CoV protein 6 intercalates with intracellular membranes (17, 40), notably the endoplasmic reticulum and Golgi complex membranes, and has even been detected in association with secreted vesicles and SARS-CoV particles (25) but not with recombinant MHV particles (41). In all animal and human SARS-CoVs isolated thus far, intact ORFs for protein 6 are present, with striking conservation of amino acid sequence. Despite this abundant expression, widespread subcellular and virion distribution, maintenance in all SARS-CoVs, and knowledge of primary structure and membrane organization, the functions of protein 6 in supporting virus infection have been elusive. Minimized SARS-CoV genomes with deletions encompassing ORF6 have been constructed and evaluated, but these recombinant viruses have not yet proved informative in revealing how protein 6 influences infection (16, 68). It is not clear why currently available in vitro and in vivo SARS-CoV infection models do not reflect the natural settings where protein 6 might impact the virus. One likely possibility is that protein 6 performs in ways that are redundant with other SARS-CoV gene products (31) and therefore is frequently phenotypically masked. Interestingly, a readily discernible function was observed when ORF6 was recombined into a related mouse hepatitis coronavirus (41, 55). In this context, protein 6 increased virus virulence and growth kinetics, both in cell cultures and in mice. The mechanism for this protein 6-mediated enhancement was attributed to its putative interactions with MHV RNA replication machinery, as infection was accelerated at the level of RNA-dependent RNA synthesis and protein 6 colocalized with replicating RNAs and with viral replication factors (55). More recently, it was discovered that protein 6 binds to cellular factors that are required for protein import into the nucleus, impairing their function in trafficking cargo (15). This interesting finding clearly raises the alternative hypothesis that protein 6 modulates intracellular environments, preventing transcription factors such as IRF3 and STAT1 from entering the nucleus and thereby suppressing innate antiviral responses (15, 53). Thus, there may be two distinct operating mechanisms for protein 6 activity on coronavirus infections, and the MHV model system appears to be amenable to dissecting their relative importance.
We reasonably hypothesized that the hydrophobic, membrane-intercalating N-terminal ∼50 residues of protein 6 might be involved in the interactions with viral RNA synthetic machinery, as cytoplasmic membranes are the sites for replication of this plus-strand RNA virus (21, 52). The charge-rich C-terminal ∼15 residues of protein 6 have been clearly implicated in preventing STAT1 from entering nuclei (15), and in doing so potentially thwarting innate antiviral responses. With this knowledge, we considered whether protein 6 forms lacking their C termini might operate to accelerate rJHM infections. Thus, we constructed mutant ORF6 cDNAs and expressed them to accumulate the various truncated proteins and then infected with rJ.2.2 to determine whether cells might be primed for more rapid and efficient MHV infection. These straightforward experiments revealed convincing evidence that the complete p6 proteins could prime cells for early replication and subsequent rapid infectious progeny output (Fig. 1). In these experiments we found only modest roles for the protein 6 C termini (Fig. 5). Clearly, the primary effects of protein 6 in these experimental contexts were provided by the N-terminal hydrophobic portion (Fig. 5).
Previously, we analyzed protein 6 function in cells infected with rJ2.2.6 and reported that the C-terminal part was not required for enhancement of MHV replication in cell cultures or in mice (40). However, in the present study, using transfected cDNA, we showed that deletion of the 11 amino acids at the C terminus partially (∼20%) diminished protein 6's enhancing ability. There are several differences when protein 6 is expressed in trans compared to expression by a recombinant virus. Most importantly, protein 6 is expressed prior to MHV infection and at higher levels when expressed in trans, compared to expression in the context of recombinant virus. Thus, the role of inhibition of nuclear import may be most important in enhancing rJ2.2.6 replication at later times postinfection, when the protein is expressed at higher levels; this effect is more obvious in transfected cells when the protein is present at high levels throughout the infection. Consistent with this lack of requirement for the C-terminal part of p6 when analyzed in the context of recombinant virus, STAT1 translocation into the nucleus was only inhibited at very late times postinfection and only after syncytia had formed (H. Zhou and S. Perlman, unpublished observations).
The central activity of the protein 6 C terminus that we could identify was an effect on the cell and was largely independent of effects on the virus. In studies performed independently and concomitantly with those of Frieman et al. (15), we found that protein 6 interfered with classical NLS-containing protein import into the nucleus. Here we used proteins that could indicate the integrity of the nuclear import machinery, tandemly repeated GFPs connected to two different NLS motifs. These proteins exceeded the size limits for passive entrance into the nucleus. We found that our 3xGFP-SV40 NLS was strikingly excluded from the nucleus by overexpressed complete protein 6, but not at all by protein 6 forms lacking their C-terminal 11 amino acids (Fig. 6). Thus, it is clear that our experiments and results only modestly support the hypothesis that protein 6 supports coronavirus infection by preventing antiviral factors from accessing the nucleus and activating host antiviral gene expression. Instead, the alternative view is of protein 6 directly interacting with other viral factors, most likely RNA replicative components (55). These sorts of findings may arise because the mouse hepatitis virus we are using to evaluate the functions of SARS-CoV protein 6 encodes its own endogenous antiviral effectors. All coronaviruses encode nonstructural protein1 (nsp1) and N, which for the MHVs have recently been identified as type 1 interferon antagonists (67, 73). An additional interferon-antagonizing activity in the C terminus of SARS-CoV protein 6 might be inapparent in the context of these nsp1 and N proteins, resulting in a separate activity for protein 6 that is independent of the interferon response as the primary observed “phenotype.”
Even though the C terminus of SARS-CoV protein 6 had minimal MHV-enhancing activity, we were intrigued by its capacity to restrict admission of proteins and even plasmid DNAs (Fig. 2) into the nucleus. The cargo proteins evaluated thus far, IRF3 (32), STAT1 (49), and 3xGFP-SV40 NLS (this report), each uses the so-called “classical” nuclear import pathway. These proteins depend on importin-β and associated adaptor proteins for transport through nuclear pores and are set apart from M9 NLS-containing proteins, such as hnRNPA1 and Nup153 proteins, which use nonclassical nuclear import pathways (36, 38). Using two distinct NLS forms on our 3xGFP reporter, we found that protein 6 deterred SV40-NLS containing GFP, but not M9-NLS containing GFP, from nuclei (Fig. 4A). This demonstrates that protein 6 selectively restricts protein cargo from the nucleus, only limiting the more commonly transported proteins relying on the classical nuclear import pathway. These findings bring new appreciation for this viral protein, which may have evolved to selectively restrain those proteins conferring antiviral responses while allowing free flow of host proteins that are vital for host cell and virus growth. Notably, we have observed little cytopathology associated with abundant protein 6 expression over a 3-day period (data not shown).
Another interesting question regarding the C-terminal portion of protein 6 is whether its capacity to sequester importins (and thus block host protein cargo from entering nuclei) is truly the operating mechanism by which it modestly improves MHV growth in the 293 cells. This question was indirectly addressed by reducing importin-β activities by methods independent of protein 6 and then determining the influence, if any, on MHV growth. Therefore, in the transfections that preceded MHV infection, we replaced protein 6 with siRNAs specific for importin-β mRNAs. Our results indicated that single siRNA treatments were not sufficient, but combinations of two different siRNAs did significantly reduce the nuclear accumulation of 3xGFP-SV40 NLS (Fig. 7B). These findings could be attributed to importin-β levels, as immunoblot assays did reveal reductions of this crucial protein carrier (Fig. 7A). Here it is worth noting that the modestly reduced importin-β levels in the cultures may not accurately reflect the levels in the ∼50% of siRNA-lipofected cells; this is an important consideration because the MHV infections were targeted specifically to siRNA-positive cells via cotransfection of siRNAs with plasmids encoding the MHV receptor. In parallel experiments, cells cotransfected with MHV receptor plasmids and siRNAs did clearly enhance the production of infectious MHVs, to levels about 5-fold (ranges from 4- to 10-fold) above those generated by control siRNAs (Fig. 7C). Thus, we can suggest that a blockade of classical NLS-containing proteins from nuclear entry, either by the complete protein 6 or by siRNAs, can prime 293 cells for improved MHV production. These findings that siRNAs can functionally replace the C-terminal residues of protein 6 argue for a bifunctional accelerator of MHV growth, with the N-terminal residues making major contributions and the C-terminal residues contributing independently once the levels of protein 6 are sufficiently high to achieve titration of nuclear import factors.
Our data argue that inactivating the classical nuclear import pathway can modestly improve MHV replication. Nuclear transport machinery controls the bidirectional transport of many biologically important molecules, and these pathways must be maintained for host cells to mount effective antiviral responses. Disruption of the nuclear transport machinery is part of viral cytopathogenic effects and constitutes mechanisms by which viruses may circumvent host antiviral responses (23, 43, 44, 62). In these cases, depleting nuclei of essential gene expression factors hinders antiviral responses. At the same time, host cells impaired in generalized nuclear import pathways will develop cytosol enriched with certain proteins that may benefit infection by cytoplasmically replicating plus-strand RNA viruses. It is not yet clear whether the advantages to MHV growth derive from restricted transport of antiviral factors into host cell nuclei and/or from cytosolic enrichment of proviral proteins that normally transit into nuclei. However, the relatively profound in vitro resistances that most MHVs exhibit toward immediate-early interferons (60, 67, 72, 73) suggests that the SARS-CoV protein 6 enhances MHV infections in ways distinct from a (redundant) antagonism of interferon signaling. We are interested in determining whether the distribution of proteins in cell nuclei and cytosol might be globally altered by protein 6 in ways that create superior environments for the replication of coronaviruses.
Acknowledgments
We thank John Fleming (University of Wisconsin) and Fumihiro Taguchi (National Institute of Infectious Diseases, Tokyo, Japan) for generously providing antibodies used in this study. We also thank Heidi Olivares for technical assistance.
This work was supported in part by a grant from the NIH (PO1 A1060699).
Footnotes
Published ahead of print on 30 April 2008.
REFERENCES
- 1.Almazan, F., M. L. DeDiego, C. Galan, E. Alvarez, and L. Enjuanes. 2006. Identification of essential genes as a strategy to select a SARS candidate vaccine using a SARS-CoV infectious cDNA. Adv. Exp. Med. Biol. 581579-583. [PubMed] [Google Scholar]
- 2.Almazan, F., M. L. Dediego, C. Galan, D. Escors, E. Alvarez, J. Ortego, I. Sola, S. Zuniga, S. Alonso, J. L. Moreno, A. Nogales, C. Capiscol, and L. Enjuanes. 2006. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 8010900-10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki, Y., H. Aizaki, T. Shimoike, H. Tani, K. Ishii, I. Saito, Y. Matsuura, and T. Miyamura. 1998. A human liver cell line exhibits efficient translation of HCV RNAs produced by a recombinant adenovirus expressing T7 RNA polymerase. Virology 250140-150. [DOI] [PubMed] [Google Scholar]
- 4.Bonner, W. M. 1975. Protein migration into nuclei. I. Frog oocyte nuclei in vivo accumulate microinjected histones, allow entry to small proteins, and exclude large proteins. J. Cell Biol. 64421-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchholz, U. J., A. Bukreyev, L. Yang, E. W. Lamirande, B. R. Murphy, K. Subbarao, and P. L. Collins. 2004. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. USA 1019804-9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capecchi, M. R. 1980. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell 22479-488. [DOI] [PubMed] [Google Scholar]
- 8.Chan, W. S., C. Wu, S. C. Chow, T. Cheung, K. F. To, W. K. Leung, P. K. Chan, K. C. Lee, H. K. Ng, D. M. Au, and A. W. Lo. 2005. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS). Mod. Pathol. 181432-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 72745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Y. Y., B. Shuang, Y. X. Tan, M. J. Meng, P. Han, X. N. Mo, Q. S. Song, X. Y. Qiu, X. Luo, Q. N. Gan, X. Zhang, Y. Zheng, S. A. Liu, X. N. Wang, N. S. Zhong, and D. L. Ma. 2005. The protein X4 of severe acute respiratory syndrome-associated coronavirus is expressed on both virus-infected cells and lung tissue of severe acute respiratory syndrome patients and inhibits growth of BALB/c 3T3 cell line. Chin. Med. J. 118267-274. [PubMed] [Google Scholar]
- 11.Dean, D. A. 1997. Import of plasmid DNA into the nucleus is sequence specific. Exp. Cell Res. 230293-302. [DOI] [PubMed] [Google Scholar]
- 12.Dean, D. A., B. S. Dean, S. Muller, and L. C. Smith. 1999. Sequence requirements for plasmid nuclear import. Exp. Cell Res. 253713-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean, D. A., D. D. Strong, and W. E. Zimmer. 2005. Nuclear entry of nonviral vectors. Gene Ther. 12881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeDiego, M. L., E. Alvarez, F. Almazan, M. T. Rejas, E. Lamirande, A. Roberts, W. J. Shieh, S. R. Zaki, K. Subbarao, and L. Enjuanes. 2007. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 811701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frieman, M., B. Yount, M. Heise, S. A. Kopecky-Bromberg, P. Palese, and R. S. Baric. 2007. Severe acute respiratory coronovirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 819812-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frieman, M. B., B. Yount, A. C. Sims, D. J. Deming, T. E. Morrison, J. Sparks, M. Denison, M. Heise, and R. S. Baric. 2006. SARS coronavirus accessory ORFs encode luxury functions. Adv. Exp. Med. Biol. 581149-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng, H., Y. M. Liu, W. S. Chan, A. W. Lo, D. M. Au, M. M. Waye, and Y. Y. Ho. 2005. The putative protein 6 of the severe acute respiratory syndrome-associated coronavirus: expression and functional characterization. FEBS Lett. 5796763-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genove, G., B. S. Glick, and A. L. Barth. 2005. Brighter reporter genes from multimerized fluorescent proteins. BioTechniques 39814, 816, 818. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg, M. W., and T. D. Allen. 1995. Structural and functional organization of the nuclear envelope. Curr. Opin. Cell Biol. 7301-309. [DOI] [PubMed] [Google Scholar]
- 20.Gorlich, D., and I. W. Mattaj. 1996. Nucleocytoplasmic transport. Science 2711513-1518. [DOI] [PubMed] [Google Scholar]
- 21.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 763697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302276-278. [DOI] [PubMed] [Google Scholar]
- 23.Gustin, K. E., and P. Sarnow. 2002. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J. Virol. 768787-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, C., N. Ito, C. T. Tseng, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 807287-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, C., C. J. Peters, and S. Makino. 2007. Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells. J. Virol. 815423-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito, N., E. C. Mossel, K. Narayanan, V. L. Popov, C. Huang, T. Inoue, C. J. Peters, and S. Makino. 2005. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 793182-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalderon, D., W. D. Richardson, A. F. Markham, and A. E. Smith. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 31133-38. [DOI] [PubMed] [Google Scholar]
- 28.Keng, C. T., Y. W. Choi, M. R. Welkers, D. Z. Chan, S. Shen, S. Gee Lim, W. Hong, and Y. J. Tan. 2006. The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells. Virology 354132-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan, S., B. C. Fielding, T. H. Tan, C. F. Chou, S. Shen, S. G. Lim, W. Hong, and Y. J. Tan. 2006. Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 12220-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinoshita, S., L. Su, M. Amano, L. A. Timmerman, H. Kaneshima, and G. P. Nolan. 1997. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity 6235-244. [DOI] [PubMed] [Google Scholar]
- 31.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar, K. P., K. M. McBride, B. K. Weaver, C. Dingwall, and N. C. Reich. 2000. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 204159-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310676-679. [DOI] [PubMed] [Google Scholar]
- 34.Li, Z. F., Y. Hu, H. C. Zhan, X. X. Yun, Y. P. Du, X. M. Ke, D. X. Yu, J. D. Li, Y. C. Dai, Q. Chen, and S. Y. Yu. 2006. An epidemiological investigation of bats carrying SARS-CoV in Guangzhou and its vicinity. Nan Fang Yi Ke Da Xue Xue Bao 26949-953. (In Chinese.) [PubMed] [Google Scholar]
- 35.McCray, P. B., Jr., L. Pewe, C. Wohlford-Lenane, M. Hickey, L. Manzel, L. Shi, J. Netland, H. P. Jia, C. Halabi, C. D. Sigmund, D. K. Meyerholz, P. Kirby, D. C. Look, and S. Perlman. 2007. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michael, W. M., M. Choi, and G. Dreyfuss. 1995. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83415-422. [DOI] [PubMed] [Google Scholar]
- 37.Moshynskyy, I., S. Viswanathan, N. Vasilenko, V. Lobanov, M. Petric, L. A. Babiuk, and A. N. Zakhartchouk. 2007. Intracellular localization of the SARS coronavirus protein 9b: evidence of active export from the nucleus. Virus Res. 127116-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakielny, S., S. Shaikh, B. Burke, and G. Dreyfuss. 1999. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 181982-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson, C. A., A. Pekosz, C. A. Lee, M. S. Diamond, and D. H. Fremont. 2005. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure 1375-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Netland, J., D. Ferraro, L. Pewe, H. Olivares, T. Gallagher, and S. Perlman. 2007. Enhancement of murine coronavirus replication by SARS-CoV protein 6 requires the N terminal hydrophobic region but not C terminal sorting motifs. J. Virol. 8111520-11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pewe, L., H. Zhou, J. Netland, C. Tangudu, H. Olivares, L. Shi, D. Look, T. Gallagher, and S. Perlman. 2005. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 7911335-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philips, A. S., J. C. Kwok, and B. H. Chong. 2007. Analysis of the signals and mechanisms mediating nuclear trafficking of GATA-4: loss of DNA-binding is associated with localization in intranuclear speckles. J. Biol. Chem. 28224915-24927. [DOI] [PubMed] [Google Scholar]
- 43.Porter, F. W., Y. A. Bochkov, A. J. Albee, C. Wiese, and A. C. Palmenberg. 2006. A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 10312417-12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J. Virol. 805156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts, A., E. W. Lamirande, L. Vogel, J. P. Jackson, C. D. Paddock, J. Guarner, S. R. Zaki, T. Sheahan, R. Baric, and K. Subbarao. 2008. Animal models and vaccines for SARS-CoV infection. Virus Res. 13320-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 3001394-1399. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 3. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 48.Schaecher, S. R., J. M. Mackenzie, and A. Pekosz. 2007. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 81718-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sekimoto, T., N. Imamoto, K. Nakajima, T. Hirano, and Y. Yoneda. 1997. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 167067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen, S., P. S. Lin, Y. C. Chao, A. Zhang, X. Yang, S. G. Lim, W. Hong, and Y. J. Tan. 2005. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 330286-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 805927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spiegel, M., A. Pichlmair, L. Martinez-Sobrido, J. Cros, A. Garcia-Sastre, O. Haller, and F. Weber. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 792079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan, Y. J., E. Teng, S. Shen, T. H. Tan, P. Y. Goh, B. C. Fielding, E. E. Ooi, H. C. Tan, S. G. Lim, and W. Hong. 2004. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 786723-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tangudu, C., H. Olivares, J. Netland, S. Perlman, and T. Gallagher. 2007. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J. Virol. 811220-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tseng, C. T., C. Huang, P. Newman, N. Wang, K. Narayanan, D. M. Watts, S. Makino, M. M. Packard, S. R. Zaki, T. S. Chan, and C. J. Peters. 2007. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human angiotensin-converting enzyme 2 virus receptor. J. Virol. 811162-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tseng, W. C., F. R. Haselton, and T. D. Giorgio. 1997. Transfection by cationic liposomes using simultaneous single cell measurements of plasmid delivery and transgene expression. J. Biol. Chem. 27225641-25647. [DOI] [PubMed] [Google Scholar]
- 58.Tu, C., G. Crameri, X. Kong, J. Chen, Y. Sun, M. Yu, H. Xiang, X. Xia, S. Liu, T. Ren, Y. Yu, B. T. Eaton, H. Xuan, and L. F. Wang. 2004. Antibodies to SARS coronavirus in civets. Emerg. Infect. Dis. 102244-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vacik, J., B. S. Dean, W. E. Zimmer, and D. A. Dean. 1999. Cell-specific nuclear import of plasmid DNA. Gene Ther. 61006-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Versteeg, G. A., P. J. Bredenbeek, S. H. van den Worm, and W. J. Spaan. 2007. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology 36118-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogel, L. N., A. Roberts, C. D. Paddock, G. L. Genrich, E. W. Lamirande, S. U. Kapadia, J. K. Rose, S. R. Zaki, and K. Subbarao. 2007. Utility of the aged BALB/c mouse model to demonstrate prevention and control strategies for severe acute respiratory syndrome coronavirus (SARS-CoV). Vaccine 252173-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von Kobbe, C., J. M. van Deursen, J. P. Rodrigues, D. Sitterlin, A. Bachi, X. Wu, M. Wilm, M. Carmo-Fonseca, and E. Izaurralde. 2000. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell 61243-1252. [DOI] [PubMed] [Google Scholar]
- 63.Waldmann, I., S. Walde, and R. H. Kehlenbach. 2007. Nuclear import of c-Jun is mediated by multiple transport receptors. J. Biol. Chem. 28227685-27692. [DOI] [PubMed] [Google Scholar]
- 64.Wang, M., M. Yan, H. Xu, W. Liang, B. Kan, B. Zheng, H. Chen, H. Zheng, Y. Xu, E. Zhang, H. Wang, J. Ye, G. Li, M. Li, Z. Cui, Y. F. Liu, R. T. Guo, X. N. Liu, L. H. Zhan, D. H. Zhou, A. Zhao, R. Hai, D. Yu, Y. Guan, and J. Xu. 2005. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 111860-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson, G. L., B. S. Dean, G. Wang, and D. A. Dean. 1999. Nuclear import of plasmid DNA in digitonin-permeabilized cells requires both cytoplasmic factors and specific DNA sequences. J. Biol. Chem. 27422025-22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yap, C. C., K. Ishii, Y. Aoki, H. Aizaki, H. Tani, H. Shimizu, Y. Ueno, T. Miyamura, and Y. Matsuura. 1997. A hybrid baculovirus-T7 RNA polymerase system for recovery of an infectious virus from cDNA. Virology 231192-200. [DOI] [PubMed] [Google Scholar]
- 67.Ye, Y., K. Hauns, J. O. Langland, B. L. Jacobs, and B. G. Hogue. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 812554-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yount, B., R. S. Roberts, A. C. Sims, D. Deming, M. B. Frieman, J. Sparks, M. R. Denison, N. Davis, and R. S. Baric. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 7914909-14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu, C. J., Y. C. Chen, C. H. Hsiao, T. C. Kuo, S. C. Chang, C. Y. Lu, W. C. Wei, C. H. Lee, L. M. Huang, M. F. Chang, H. N. Ho, and F. J. Lee. 2004. Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett. 565111-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan, X., J. Li, Y. Shan, Z. Yang, Z. Zhao, B. Chen, Z. Yao, B. Dong, S. Wang, J. Chen, and Y. Cong. 2005. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 109191-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan, X., Z. Yao, Y. Shan, B. Chen, Z. Yang, J. Wu, Z. Zhao, J. Chen, and Y. Cong. 2005. Nucleolar localization of non-structural protein 3b, a protein specifically encoded by the severe acute respiratory syndrome coronavirus. Virus Res. 11470-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou, H., and S. Perlman. 2007. Mouse hepatitis virus does not induce Beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J. Virol. 81568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zust, R., L. Cervantes-Barragan, T. Kuri, G. Blakqori, F. Weber, B. Ludewig, and V. Thiel. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 3e109. [DOI] [PMC free article] [PubMed] [Google Scholar]