Abstract
We have identified a novel, highly conserved protein of 14 kD copurifying with late endosomes/lysosomes on density gradients. The protein, now termed p14, is peripherally associated with the cytoplasmic face of late endosomes/lysosomes in a variety of different cell types.
In a two-hybrid screen with p14 as a bait, we identified the mitogen-activated protein kinase (MAPK) scaffolding protein MAPK/extracellular signal–regulated kinase (ERK) kinase (MEK) partner 1 (MP1) as an interacting protein. We confirmed the specificity of this interaction in vitro by glutathione S-transferase pull-down assays and by coimmunoprecipitation, cosedimentation on glycerol gradients, and colocalization. Moreover, expression of a plasma membrane–targeted p14 causes mislocalization of coexpressed MP1. In addition, we could reconstitute protein complexes containing the p14–MP1 complex associated with ERK and MEK in vitro.
The interaction between p14 and MP1 suggests a MAPK scaffolding activity localized to the cytoplasmic surface of late endosomes/lysosomes, thereby combining catalytic scaffolding and subcellular compartmentalization as means to modulate MAPK signaling within a cell.
Keywords: signal transduction scaffold, MEK, ERK, subcellular localization, endocytosis
Introduction
One of the challenges in cell biology is to identify the sequence of events following the activation of a signaling receptor. A large number of proteins involved in signal transduction have been identified, and several of these have been shown to be organized into transducing modules (for example, the mitogen-activated protein kinase [MAPK] module). However, the majority of receptors use only a limited number of modules. The key question is how the cell is able to orchestrate these modules in a way that allows specific signals to be translated into receptor-specific responses.
Membrane traffic helps to maintain the subcellular location of proteins and lipids. Within a cell, there are two separate major trafficking pathways, the secretory and the endocytic pathway, although both systems are interconnected. Intracellular trafficking plays a major role in signal transduction mainly because after ligand binding, most signaling receptors are endocytosed. For a long time, it was thought that the major impact of endocytosis on signaling is by downregulating the number of surface receptors (Di Fiore and Gill 1999). Using dominant negative dynamin in EGF- activated cells, it became clear that the EGF receptor (EGFR) activates its targets Ras, Raf, and MAPK/extracellular signal–regulated kinase (ERK) kinase (MEK) at the plasma membrane. But endocytosis has to occur to activate the MAPK ERK (Kranenburg et al. 1999). Of additional interest in this context is that members of this growth factor family that differ in their intracellular fates also differ in their signaling properties. ErbB1 for instance is routed for lysosomal degradation only when induced by EGF but recycles when binding TGF-α. In contrast, ErbB3, whose ligands are the neuregulins, is always recycled (Baulida et al. 1996). Another receptor for which membrane traffic has been shown to be important is protease-activated receptor (PAR)2, a member of the PAR family of G protein–coupled receptors. Endocytosis of activated PAR2 is necessary for the activated receptor to meet its downstream effectors Raf and ERK on the endocytic compartment. By this, activated ERK is sequestered and kept away from entering the nucleus, thereby achieving substrate selectivity (DeFea et al. 2000). These functional studies together with localization data detecting members of the MAPK module on endocytic structures (Pol et al. 1998; Kranenburg et al. 1999) emphasized the importance of endocytosis as regulator of signal transduction (Ceresa and Schmid 2000).
Within epithelial cells, an additional level of complexity is added to the organizational skills of a cell. Apical–basal polarity separates the epithelial cell into two distinct domains. Correct sorting of receptors and downstream effectors is crucial for proper flow of information (Kuwada et al. 1998; Hobert et al. 1999). Disturbance of this organization leads to pathophysiological consequences like increased autocrine stimulation of EGFR in cyst epithelia in polycystic kidney disease (Wilson 1997) and crypt epithelia of colon carcinoma (Tong et al. 1998) or ablation of an EGFR-dependent, vulva-inducing signal in Caenorhabditis elegans (Kaech et al. 1998; Whitfield et al. 1999).
Another important concept emerging during very recent years aims to explain how an activated enzyme selects the appropriate substrates by scaffolding (Pawson and Scott 1997; Whitmarsh and Davis 1998). Several proteins have been identified that influence signaling by routing certain partners of a cascade together in one complex. The cellular repertoire of such molecules spans from organizers of giant “transducisomes” like inaD to simple trimeric complex builders like MEK partner 1 (MP1). Their role in signal transduction is thought to enhance specificity and selectivity by bringing together components of a given pathway and separating them from other upstream activators as well as from downstream targets (anchoring scaffolds) or bringing the selected partners in close proximity (catalytic scaffolds) (Burack and Shaw 2000).
To better understand how a cell organizes spatiotemporal patterns of signal-transducing elements, it will be necessary to analyze protein complexes that are formed on and/or recruited to intracellular membranes upon signaling. An important question in this respect is whether scaffolding occurs on intracellular membranes. We approached this question by analyzing the proteomes of endocytic organelles by a combination of subcellular fractionation, two-dimensional gel electrophoresis (2DGE), and microsequencing.
In this report, we describe the identification of a hitherto uncharacterized protein. It localizes to late endosomes/lysosomes where it interacts with the MAPK scaffold MP1 (Schaeffer et al. 1998). Both proteins can be found in complex with elements of the MAPK module, raising the possibility that the MAPK ERK1 is recruited to and activated on late endosomes/lysosomes.
Materials and Methods
Cells and Tissue Culture
EpH4 murine mammary epithelial cells (Fialka et al. 1996), Caco-2 cells, and HeLa cells were grown in high glucose DME supplemented with 10 mM Hepes, pH 7.3, 50 IU/ml penicillin, 50 μg/ml streptomycin, and 5% FCS at 37°C, in 5% CO2 and 98% humidity. Media and reagents for tissue culture were purchased from GIBCO BRL (Life Technologies), and FCS was obtained from BioWhittaker (Boehringer).
Antibodies
Polyclonal anti-p14 antiserum was raised against a glutathione S-transferase (GST) fusion protein of p14. Anti-MP1 antibodies were raised against the peptide Kp532 (CVSDRDGVPVIKVANDSAPEHALR, amino acids 24–46, mouse MP1, sequence data available from GenBank/EMBL/DDBJ under accession no. AF082526) and affinity purified on Affi-Gel matrix (Bio-Rad Laboratories) according to the manufacturer's instructions. Polyclonal antibodies recognizing the myc epitope were obtained from Gramsch Laboratories. Antibodies specific for double phosphorylated ERK1/2 or MEK1/2 were purchased from New England BioLabs, Inc. Anti-His6 and anti-Xpress antibodies were from Invitrogen, and the anti-CD107a (LAMP-1) antibody was obtained from BD PharMingen. Polyclonal anti-GST antibodies were generated in the lab. Alexa 488™, Alexa 568™, Cy3™, and Texas red–labeled secondary antibodies were obtained from Molecular Probes, Amersham Pharmacia Biotech, and Jackson ImmunoResearch Laboratories, respectively. LysoTracker™, Red DND-99, and EGF-rhodamine were from Molecular Probes. Anti-EEA1 (Rubino et al. 2000) and anti-Rab11 (Ullrich et al. 1996) antibodies were generous gifts from Dr. Marino Zerial (European Molecular Biology Laboratory, Heidelberg, Germany).
Cell Homogenization and Membrane Preparation
EpH4 cells were homogenized and postnuclear supernatant (PNS) was prepared in homogenization buffer (250 mM sucrose, 3 mM imidazole, pH 7.4, 1 mM EDTA, containing a cocktail of protease inhibitors, 10 μg/ml aprotinin, 1 μg/ml pepstatin, 10 μg/ml leupeptin, and 1 mM Pefabloc solid compound) (Boehringer) as outlined previously (Gruenberg and Gorvel 1992; Fialka et al. 1997; Pasquali et al. 1997). Continuous sucrose gradients were used to separate different membrane compartments as described (Fialka et al. 1997). Peripheral membrane proteins were separated from integral membrane proteins by extraction with 0.1 M Na2CO3, pH 11.0 (Fujiki et al. 1982).
2DGE and Peptide Sequencing
2DGE and microsequencing of protein spots were performed as described in detail elsewhere (Fialka et al. 1997; Pasquali et al. 1997; Fialka et al. 1999).
Indirect Immunofluorescence
EpH4, HeLa, and Caco-2 cells were fixed with 4% paraformaldehyde in cytoskeleton buffer (10 mM Pipes, pH 6.8, 150 mM NaCl, 5 mM EGTA, 5 mM glucose, 5 mM MgCl2) for at least 30 min, quenched by three washes with washing buffer (cytoskeleton buffer, 50 mM NH4Cl), and permeabilized with cytoskeleton buffer supplemented with 0.3–0.5% Triton X-100. Specimens were processed for indirect immunofluorescence, mounted in Mowiol (Hoechst), and finally viewed using an Axioplan2 microscope (ZEISS). For confocal microscopy, samples were mounted in 50% glycerol, 4% n-propyl gallate (Sigma-Aldrich) in cytoskeleton buffer. Confocal images were obtained with a Leica TCS NT confocal microscope and processed using the Imaris and colocalization software packages (Bitplane AG) after deconvolution using measured point-spread functions with the Huygens software (Scientific Volume Imaging).
Proteinase K Treatment of PNS
Equal volumes of PNS from EpH4 cells (∼0.5 μg/μl protein) were treated with increasing concentrations of proteinase K (0.01–10 μg/ml; GIBCO BRL) for 20 min at room temperature. The reaction was stopped by addition of 100 mM PMSF. Undigested membrane material was pelleted at 100,000 g and proteins were analyzed by immunoblots.
Two-Hybrid Screen
A two-hybrid screen was performed using the Matchmaker Gal4 Two-Hybrid System 2 (CLONTECH Laboratories, Inc.) following the manufacturer's screening protocol. Bait constructs were generated by PCR from the original clone obtained from the United Kingdom Human Genome Mapping Project Resource Centre (I.M.A.G.E. Consortium CloneID 681056) (Lennon et al. 1996) using primers introducing an EcoRI site NH2-terminally and a PstI site COOH-terminally of the respective fragments and subsequent cloning of these fragments into pAS2-1 (CLONTECH Laboratories, Inc.). The resulting chimeric proteins consisted of the Gal4 DNA binding domain fused in frame to the full length protein, an NH2-terminal fragment of p14 (amino acids 1–48), or two different COOH-terminal fragments (C1, amino acids 43–125; and C2, amino acids 80–125). After titration of the appropriate 3-amino-1,2,4-triazole concentration to inhibit background His3 activity, the different bait constructs were introduced into yeast strain HF7c, tested for autonomous activation, and subsequently screened for interacting polypeptides using a mouse embryo Matchmaker cDNA library cloned into pACT2 (CLONTECH Laboratories, Inc.). The pAS2-1 C2 construct showed autoactivation resistant to 3-amino-1,2,4-triazole, and thus was not used for further screening.
Constructs and Transfection
Tagged versions of p14 and MP1 containing a triple myc tag at the NH2 termini of the proteins were constructed by PCR using primers introducing appropriate restriction sites (p14) or by direct cloning from one of the positive pACT2 clones (MP1) into a pBluescript SK vector containing three myc sequences preceded by a Kozak sequence (Kozak 1999) that was constructed in our laboratory. The coding sequences of the resulting chimeric proteins (myc3-p14 and myc3-MP1) were cloned into expression vectors pREP10 (Invitrogen), resulting in sense or antisense myc3-p14 constructs, and pUB6/V5-His (out of frame of the COOH-terminal V5-His tag) (Invitrogen), respectively. CAAX-tagged p14 was constructed by introducing a linker sequence encoding the last 21 amino acids of human K-ras (Choy et al. 1999) at the COOH terminus of the p14 cDNA, replacing the STOP codon. Both bona fide p14 and p14–CAAX were cloned in frame with the His6/Xpress-tag into pEF4/HisC (Invitrogen) to give rise to NH2-terminally tagged X-p14 and X-p14–CAAX expression vectors. EGFP–p14 was constructed by cloning the coding sequence of p14 into pEGFP-C1 (CLONTECH Laboratories, Inc.). Cells were transfected with the different constructs by use of Lipofectamin Plus (GIBCO BRL) following the manufacturer's suggestions and eventually selected accordingly for stable transfectants.
Recombinant proteins were constructed in pGEX6P3 (Amersham Pharmacia Biotech) or pET28 (Novagen).
Immunoprecipitation
Transfected cells were scraped in PBS and lysed by a combination of a quick freeze–thaw cycle and sonication. After centrifugation at 1,600 g, the supernatant was diluted in IP buffer (10 mM Hepes, pH 7.4, 137 mM NaCl, 4.7 mM KCl, 0.65 mM MgSO4, 1.2 mM CaCl2, 1% Triton X-100, 2 mM NaF, 20 mM β-glycerophosphate, and protease inhibitors as for cell homogenization). After preclearing with UltraLink™ immobilized protein A (Pierce Chemical Co.), the resulting supernatant was subjected to immunoprecipitation using preimmuneserum or polyclonal anti-myc antibodies and UltraLink™ immobilized protein A or protein A alone as additional negative control. After three washes with IP buffer, the samples were boiled in loading buffer and resolved on SDS-PAGE.
Glycerol Density Gradients
Total membranes from EpH4 cells were enriched by centrifugation of PNS at 100,000 g, resuspended in extraction buffer (20 mM Hepes/KOH, pH 7.0, 100 mM KCl, 1 mM DTT, 1% Triton X-100, 2 mM NaF, 20 mM β-glycerophosphate, and a cocktail of protease inhibitors as above), and extracted on ice for 30 min. The insoluble material was pelleted at 17,000 g and the resulting supernatant (containing p14 and MP1) was loaded on top of a continuous glycerol gradient (5–35% in extraction buffer). Gradients were centrifuged at 270,000 g overnight in an SW41 rotor (Beckman Coulter). Then, 600-μl fractions were collected, and proteins were precipitated and analyzed by 12.5% SDS-PAGE and immunoblots. For gradient calibration, we used a protein mix (1 μg/μl BSA, 4.5 S, 2 μg/μl aldolase, 7.3 S, 2 μg/μl catalase, 10.0 S, and 1 μg/μl thyroglobulin, 19.5 S, in homogenization buffer) the distribution of which was detected by Coomassie stain after SDS-PAGE.
In Vitro Pull-down Assay
Bacterial lysates containing recombinant proteins were prepared by sonication in PBS. GST and GST fusion proteins were bound to glutathione-Sepharose (Amersham Pharmacia Biotech), washed with PBS, and incubated with lysates containing His6-tagged recombinant proteins for 20 min at room temperature. Subsequently, the Sepharose-bound proteins were washed, resuspended in sample buffer, and analyzed by SDS-PAGE and immunoblots. Because of the reported promiscuity in the in vitro interaction of MP1 with the different isoforms of the kinases (Schaeffer et al. 1998), we used a GST–ERK2 construct that was obtained from M.J. Weber (University of Virginia, Charlottesville, VA).
Electron Microscopy
Caco-2 cells expressing EGFP–p14 were fixed in 4% paraformaldehyde/0.1% glutaraldehyde in 0.1 M phosphate buffer, pH 7.35, for 1 h at room temperature. They were then washed with 0.2 M phosphate buffer, scraped from the culture dish, and pelleted in a microfuge. The cells were then resuspended in warm gelatin (10% in phosphate buffer) and repelleted at maximum speed in the microfuge. After cooling, the gelatin-embedded cells were infiltrated with polyvinyl pyrrolidone sucrose overnight at 4°C and then processed for frozen sectioning as described (Liou et al. 1997). Ultrathin frozen sections (60–80 nm) were labeled, stained, and viewed (Jeol 1010; Centre for Microscopy and Microanalysis) according to published techniques (Parton et al. 1997).
Online Supplemental Material
Caco-2 cells stably expressing EGFP–p14 were grown on coverslips and observed using a confocal microscope. Video available at http://www.jcb.org/cgi/content/full/152/4/765/DC1 shows the enlargement of one of the EGFP–p14 labeled vesicles in Fig. 5, revealing the existence of a mobile internal vesicle.
Figure 5.
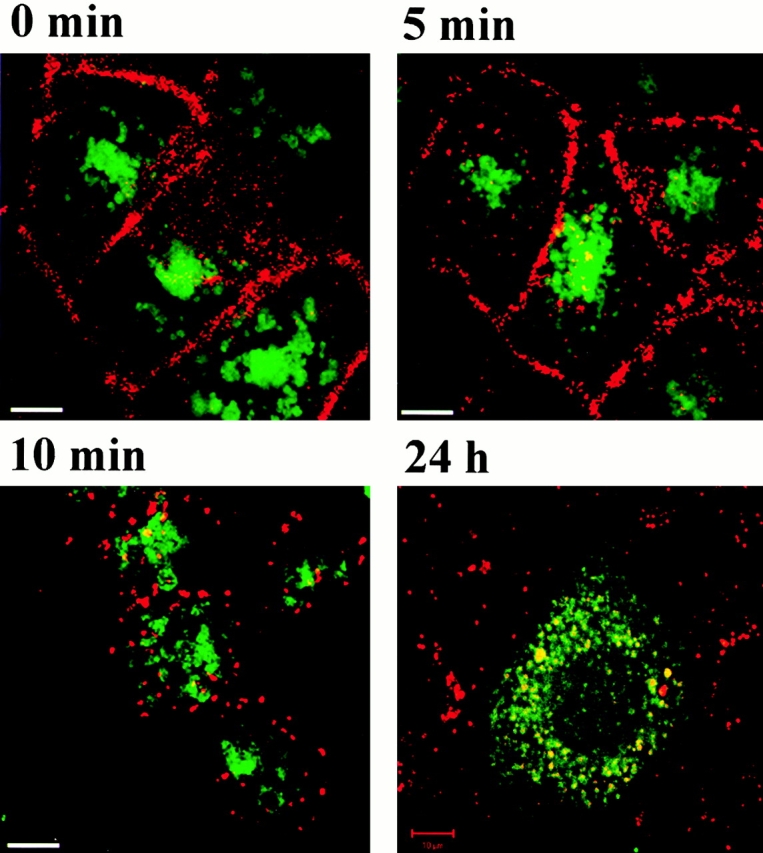
Internalized EGF-rhodamine meets p14 in a late endocytic compartment. EGF-rhodamine was internalized for 5 min, 10 min, and 24 h in EpH4 cells transiently transfected with EGFP–p14. Before internalization at 37°C, EGF-rhodamine was bound to the cell surface for 1 h at 4°C. Living cells were observed under a confocal microscope. After 24 h internalization, the EGFP-p14–containing compartment was filled with EGF-rhodamine. Video available at http://www.jcb.org/cgi/content/full/152/4/765/DC1. Bars, 10 μm.
Results
A New Highly Conserved Peripheral Membrane Protein Enriched in the Late Endosomal/Lysosomal Compartment
To investigate the protein composition of the endocytic compartment, we applied techniques already successfully used for the analysis of membrane-associated organelle proteins within the mammary epithelial cell line EpH4 (Fialka et al. 1997, Fialka et al. 1999; Pasquali et al. 1997, Pasquali et al. 1999). First, we performed sucrose density gradient centrifugation of PNSs to separate organelles derived from early and late endocytic compartments, respectively. It is important to emphasize that continuous sucrose gradients efficiently separate very light from dense fractions. In such gradients, Golgi complex membranes comigrate in fractions of medium density together with plasma membrane (both the apical and basolateral plasma membrane in epithelial cells) as well as with early endosomes. However, late endosomes which resemble a similar density as lysosomes could easily be separated and distinguished from the bulk of other membranes (Fialka et al. 1997, Fialka et al. 1999; Pasquali et al. 1999). Gradient-enriched organelle fractions were then applied to high resolution 2DGE to obtain a protein pattern allowing the identification of proteins specifically enriched in late endosomal/lysosomal fractions. Since a lot of proteins described in regulating endocytic transport (for example, Rab proteins) or scaffolding signal transducing modules (for example, Ste5p) are peripherally associated with membranes, we further characterized the fractions by carbonate extraction at high pH (Fujiki et al. 1982; Pasquali et al. 1997).
Fig. 1 A shows the analysis of the sucrose density gradient with respect to the activity of HRP internalized by fluid phase endocytosis for different time periods as well as by Western blotting for established compartment markers (Rab5 for the early endosome, Rab4, Rab11, and transferrin receptor for the recycling endosome, and Rab7 for the late endosome) (Bucci et al. 2000; Sonnichsen et al. 2000). Peak fractions for Rab5/HRP activity at 0 min chase (fraction 14–16) and Rab7/HRP activity at 20 min chase (fraction 6+7) were used for 2DGE analysis of early endosomal (Fig. 1 C) and late endosomal (Fig. 1 D) fractions, respectively. The second peak fraction of Rab5 (fraction 18–20) was omitted since it contained the bulk of plasma membrane and rough ER fractions.
Figure 1.
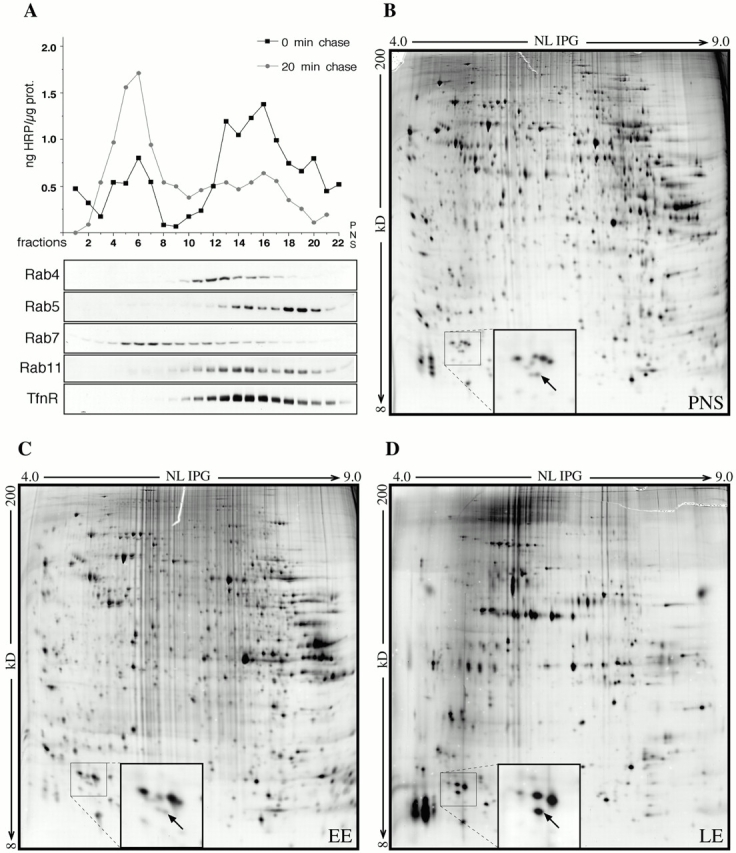
Subcellular fractionation and analysis of endosomal compartments. (A) HRP was internalized into polarized EpH4 cells for 5 min. Cells were either kept on ice (0 min chase) or incubated for another 20 min at 37°C (20 min chase). Cells were scraped, homogenized, and PNS was prepared. PNS was loaded on top of continuous sucrose gradients and separated by centrifugation as outlined in Materials and Methods. Fractions were collected, and HRP activity was measured (top) and immunoblotted for Rab4, Rab5, Rab7, Rab11, and transferrin receptor (bottom). (B–D) PNS, fractions 6+7 (LE), and fractions 18+19 (EE) were analyzed by 2DGE and compared according to enrichment of protein spots in EE and LE versus PNS, respectively. Insets show the region containing a protein (arrow) with experimental M r ∼ 14 kD and pI ∼ 4.9, specifically enriched in LE.
A large number of spots were found to be enriched in either early or late endosomal fractions (compare Fig. 1C and Fig. D). In this report, we describe the investigation of a particular protein spot with an experimental isoelectric point (pI) of ∼5.0 and a molecular mass of ∼14 kD in more detail (arrow, inset in Fig. 1, B–D). It was abundant and enriched in gradient-purified organelle fractions containing mainly late endosomes and lysosomes when compared with PNS or early endosome–containing fractions (Fig. 1, B–D). In addition, this protein was exclusively associated with membrane fractions of that density and never detected in the cytoplasmic fractions.
The spot was excised from a preparative gel (Pasquali et al. 1997), digested, and microsequenced by Edman degradation. Two peptide sequences were obtained: KETVGFGMLK and KAQALVQYLEEPLTQVA. These sequences were found to be part of open reading frames from several EST clones, originating from a variety of different species, organs, developmental stages, and cell lines. Comparison of the primary polypeptide sequences of different species revealed very high sequence conservation within multicellular organisms (Fig. 2). However, we did not detect a sequence with significant homology in the yeast genome. Comparison with databases of known proteins did not reveal homology to any other existing protein or protein domain. The calculated pI (5.3) and molecular mass (13,480 D) of the mouse protein, now termed p14, corresponded well with the experimental data obtained from our two-dimensional gels (see above; mouse sequence data for p14 are available from GenBank/EMBL/DDBJ under accession no. AJ277386). On the transcriptional level, we were able to detect a single RNA of 700 bp in EpH4 cells. We could show ubiquitous expression on a multiple tissue Northern blot from CLONTECH Laboratories, Inc. (data not shown).
Figure 2.
Sequences of p14 in multicellular organisms. Comparison of conceptual translations of EST sequences from a variety of taxons.
In a secondary analysis, we investigated the membrane topology of p14. The protein displayed features of a peripheral membrane protein since it was partially extractable by carbonate at high pH (Fig. 3 A). Furthermore, we prepared PNSs under conditions that maintain the integrity of vesicles and subjected them to proteinase K digestion. Proteinase K is not membrane permeable and thus, digests at limited concentrations only proteins accessible at the cytoplasmic face of membranes. Under these conditions, p14 was much more sensitive to digestion than known lumenal marker proteins, such as calreticulin (Krause and Michalak 1997), or even proteins on the cytoplasmic side, such as Rab5 (Somsel Rodman and Wandinger-Ness 2000) (Fig. 3 B). Calreticulin was chosen since it represented a well-characterized lumenal marker protein of the ER, the most abundant vesicle fraction in PNSs. Taken together, these results suggested a peripheral association of p14 with the cytoplasmic face of membranes.
Figure 3.
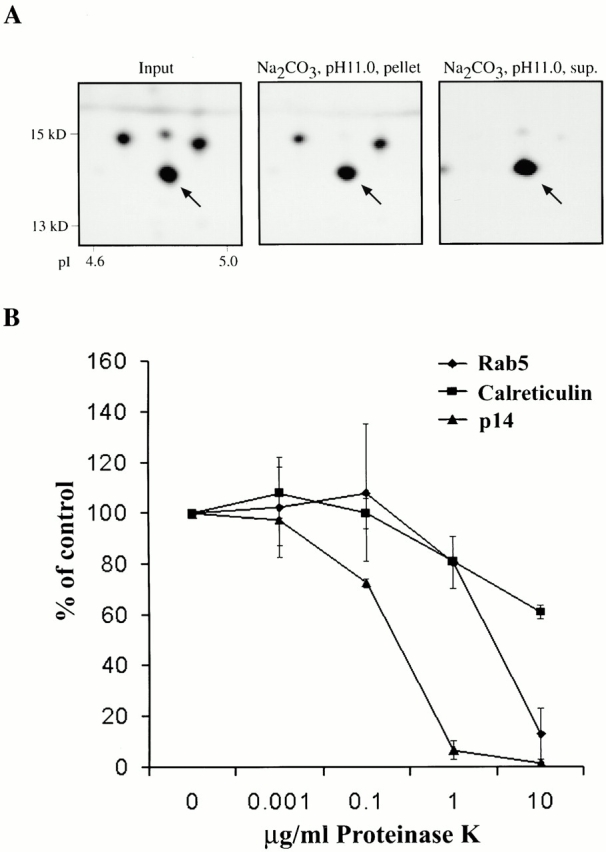
Membrane topology of p14. (A) PNS from polarized EpH4 cells was loaded on top of a continuous sucrose gradient and sedimented organelle fractions enriched in p14 were extracted with Na2CO3, pH 11.0. Input (left), carbonate insoluble (middle), and carbonate soluble (right) proteins were analyzed by 2DGE. Respective areas of high resolution two-dimensional gels are shown; arrows indicate p14. (B) PNS from polarized EpH4 cells was treated with increasing concentrations of proteinase K. Digested samples were analyzed by immunoblot, quantified, and the percentage of intact protein compared with undigested controls was plotted (numbers are an average of three independent experiments ± SD). A lumenal protein (calreticulin) and Rab5 as a protein associated with the cytoplasmic leaflet of membranes were taken as controls.
To confirm the subcellular localization of p14, we expressed an EGFP-tagged version of p14 in EpH4 cells and incubated the cells with LysoTracker™ Red DND-99, an acidotropic probe. Live microscopy revealed vesicular structures surrounded by the green fluorescence of EGFP–p14 and filled with the red fluorescence of the lysotracker (Fig. 4A, A′, and A′′), indicating a localization of p14 on acidic organelles such as late endosomes and lysosomes. To exclude mistargeting mediated by the EGFP tag or by overexpression, the analysis was extended with differently tagged versions of p14 in two other cell types (Caco-2 and HeLa) (data not shown). Another epithelial cell line, Caco-2 cells stably expressing EGFP–p14, was used for coimmunofluorescence with LAMP-1, a transmembrane glycoprotein localizing mainly to late endosomes/lysosomes. In these cells, LAMP-1 showed a complete overlap with the exogenously expressed p14 (Fig. 4B, Fig. B′, and B′′). This result was further confirmed by exploiting the well-studied intracellular trafficking of the EGFR and its ligand. After binding to the EGFR at the cell surface, EGF gets internalized together with its receptor and is transported to the early then to the late endocytic compartment and finally destined to degradation in lysosomes (Sorkin et al. 1988). EpH4 cells transiently transfected with EGFP–p14 were allowed to internalize EGF-rhodamine for various times and were then observed live under a microscope. EGF, initially separated from the green fluorescence (Fig. 5; 0, 5, and 10 min chase), was subsequently filling the compartment labeled with EGFP (Fig. 5; 24 h chase), confirming the association of p14 with late endosomes/lysosomes in living cells. In addition, HeLa cells stably transfected with EGFP–p14 were analyzed in coimmunolocalization experiments with early endocytic marker proteins. Neither EEA1 (Fig. 6A, A′, and A′′), involved in vesicular transport from the plasma membrane to the early endosomes (Rubino et al. 2000), or Rab11 (Fig. 6B, Fig. B′, and B′′), which regulates recycling through the pericentriolar recycling endosome (Ullrich et al. 1996), showed any significant coimmunolocalization with p14.
Figure 4.
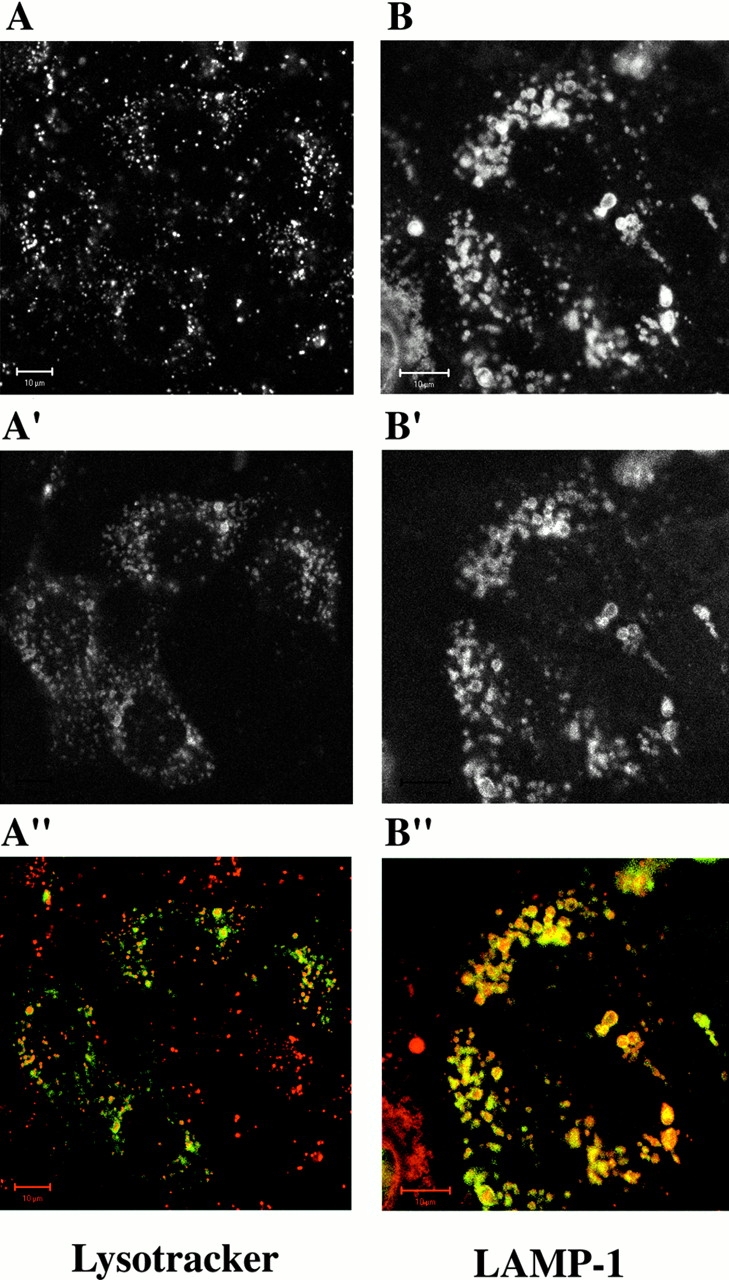
Subcellular localization of p14. EpH4 cells, transiently transfected with EGFP-p14 (A′), were treated with LysoTracker™. (A) Living cells were observed under a confocal microscope. Caco-2 cells, stably transfected with EGFP-p14 (B′), were fixed and coimmunolocalization with LAMP-1 was performed (B). Merged images in A′′ and B′′. Bars, 10 μm.
Figure 6.
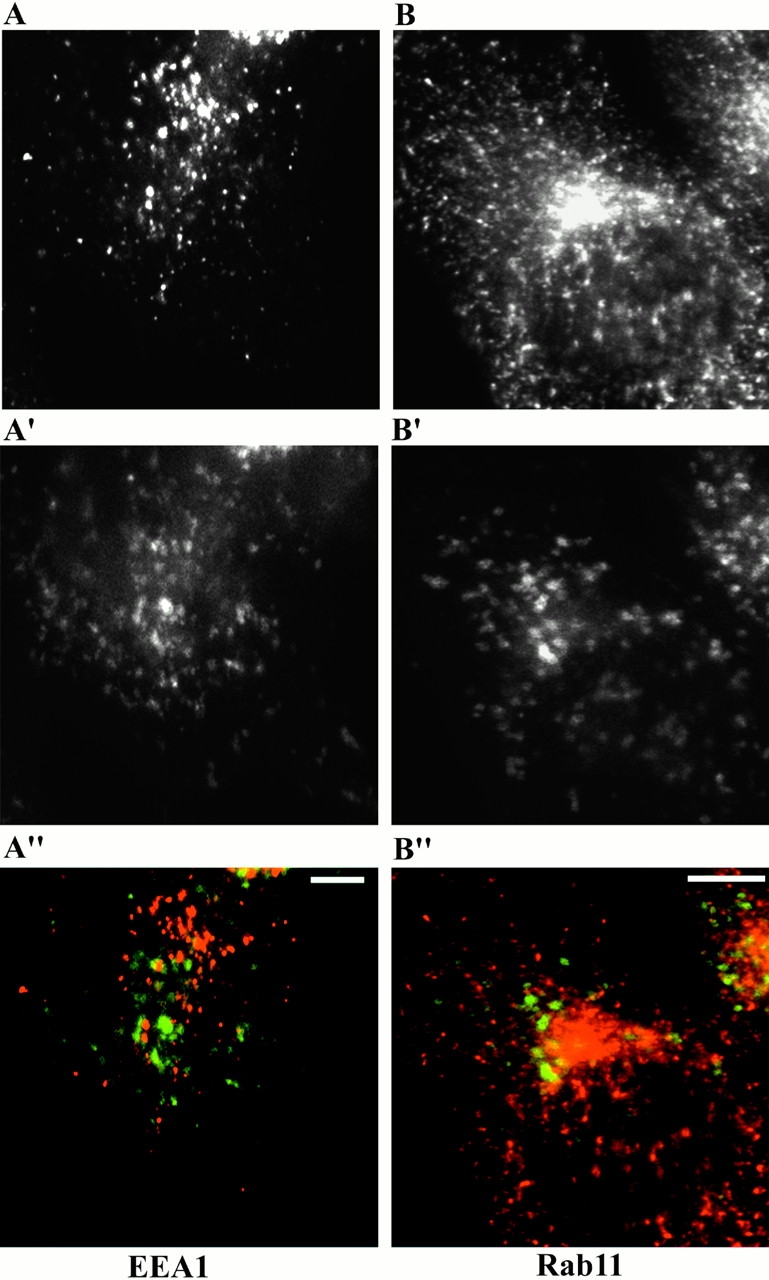
p14 does not colocalize with early endocytic markers. HeLa cells, stably transfected with EGFP–p14 (A′ and B′), were fixed and coimmunolocalization with EEA1 (A) and Rab11 (B) was performed. Merged images in A′′and B′′. Bars, 10 μm.
Immunoelectron microscopy confirmed and extended these observations. The majority of the labeling for the EGFP-tagged protein was associated with large electron-lucent structures with internal vesicular or lamellar membranes that were labeled with antibodies to the late endosomal marker, lysobisphosphatidic acid (LBPA) (Fig. 7). As shown previously (Kobayashi et al. 1998), the LBPA antibody labeled the internal membranes of the late endosomes. In contrast, EGFP–p14 was predominantly, although not exclusively (see video at http://www.jcb.org/cgi/content/full/152/4/765/DC1), associated with the cytoplasmic surface of the late endosomes. In addition, EGFP–p14 labeling was associated with putative lysosomal structures with an electron-dense core and negligible LBPA labeling (for example, Fig. 7 C). Smaller spherical LBPA-negative multivesicular endosomal structures, morphologically identifiable as endosome carrier vesicles (Clague et al. 1994), also showed low but specific labeling with the anti-GFP antibody (Fig. 7 C). Taken together, we conclude that this novel peripheral membrane protein localizes to the cytoplasmic face of late endosomal and lysosomal compartments in several different cell types and species.
Figure 7.
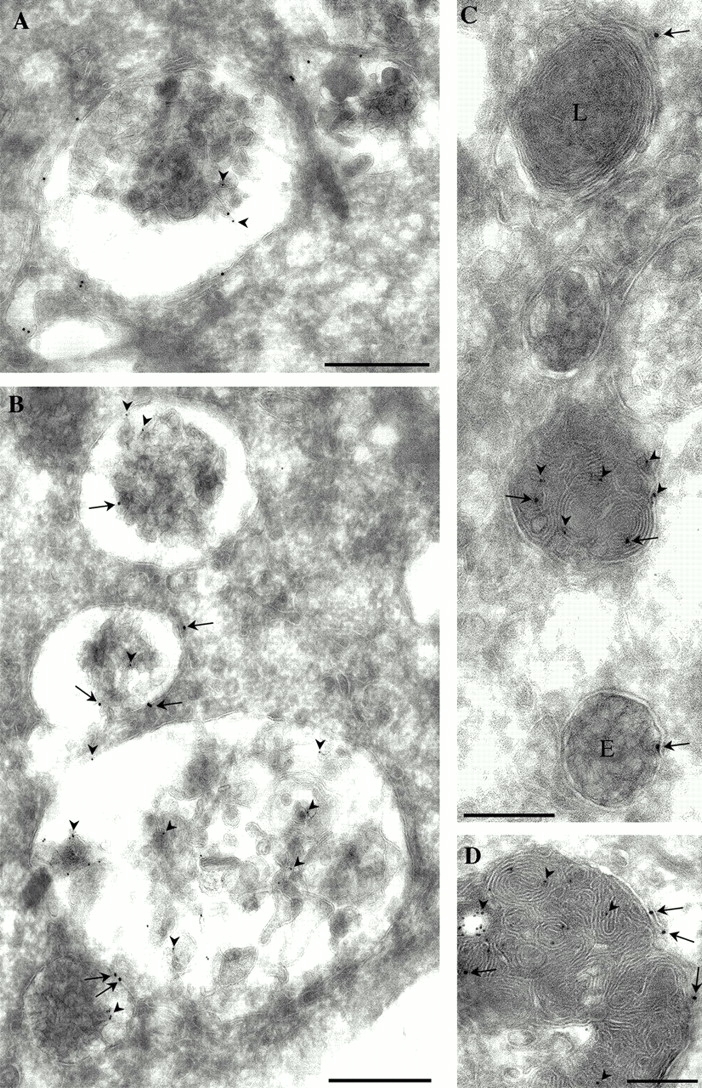
Immunoelectron microscopic localization of p14. Caco-2 cells, stably transfected with EGFP–p14, were fixed and processed for frozen sectioning. Sections were double labeled for p14 (detected with 15 nm-gold-labeled goat anti–rabbit antibodies) and LBPA (detected with 10 nm gold-labeled goat anti–mouse antibodies). Specific labeling for EGFP–p14 (large gold; arrows in B–D) is associated with large multivesicular and multilamellar late endosomes which are labeled for LBPA (arrowheads, all panels). Labeling for EGFP–p14 is predominantly around the periphery of the late endosomes with lower labeling associated with LBPA-positive internal membranes. In C, in addition to labeling of late endosomes, lower but specific labeling is associated with putative lysosomes which are electron dense and LBPA-negative (L) and with putative endosome carrier vesicles (E). Bars: (A and B) 500 nm; (C and D) 200 nm.
Interaction of p14 with MP1
To investigate the biological function of p14, we attempted to identify potential interaction partners of p14. Therefore, we performed two-hybrid screens using different parts of the p14 coding sequence (full length, an NH2-terminal, and a COOH-terminal fragment) as bait fused to the Gal4 DNA binding domain. With each construct, we screened 1.2–5.6 × 105 independent clones of a mouse embryonic library (Table ). Among positive clones, several contained the complete coding region of MP1, a protein recently identified as a scaffold protein of the MAPK pathway (Schaeffer et al. 1998). We obtained interacting MP1 clones with all three different bait constructs (Table ), suggesting that different parts of the relatively small p14 protein were involved in the interaction with MP1. Alternatively, the small overlap of the NH2- and COOH-terminal constructs (see Materials and Methods) may play a central role in the interaction with MP1.
Table 1.
Summary of Two-Hybrid Screen for p14 Interaction Partners
| Bait | Screened clones | Positive clones | Typical false positive | MP1 |
|---|---|---|---|---|
| Gal4-wt | 1.2 × 105 | 24 | 2 | 15 |
| Gal4-N | 4.8 × 105 | 5 | 0 | 1 |
| Gal4-Cl | 5.6 × 105 | 5 | 3 | 1 |
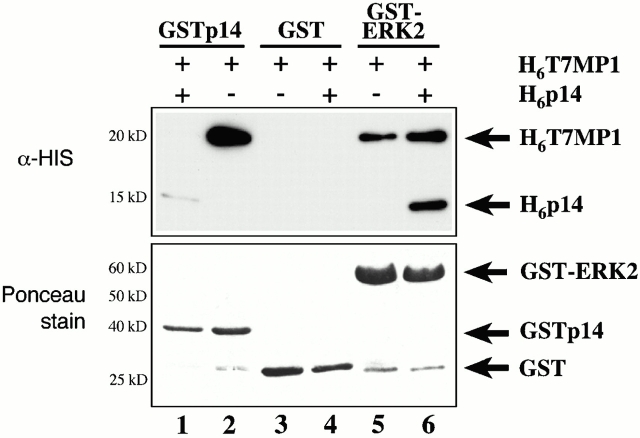
The interaction between p14 and MP1 was further confirmed with a variety of methods. First, the direct interaction between these two proteins was tested in a GST pull-down assay. GST-tagged p14 was able to directly interact with His6-T7-MP1 (Fig. 8, lane 2). The specificity of this interaction was shown by the failure of His6-T7–tagged MP1 to interact with GST alone (Fig. 8, lanes 3 and 4) and by the ability of excess His6-tagged p14 to abrogate the GST–p14/His6-T7-MP1 interaction (Fig. 8, lane 1). In addition, His6-tagged p14 weakly bound to GST–p14 (Fig. 8, lane 1), raising the possibility of a self-association.
Figure 8.
Interaction between p14 and MP1 in vitro. Recombinant GST-tagged p14 (lanes 1 and 2), GST (lanes 3 and 4), or GST–ERK2 (lanes 5 and 6) was incubated with recombinant His6-T7–tagged MP1 alone (lanes 2, 3, and 5) or with additional His6-tagged p14 (lanes 1, 4, and 6). Amounts of GST proteins are illustrated in the bottom panel by Ponceau staining.
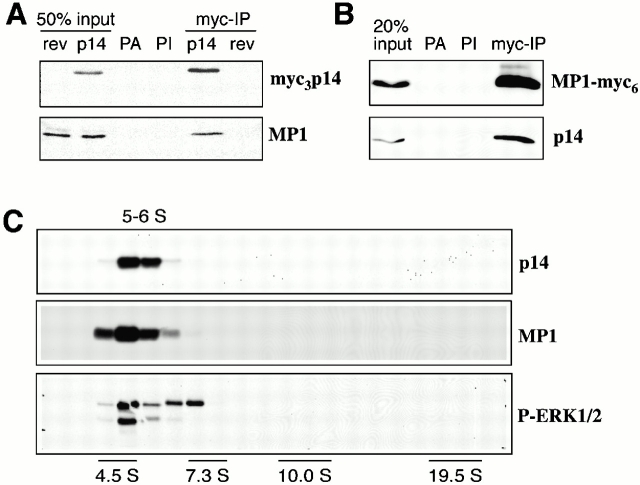
The direct interaction in the yeast two-hybrid assay and in pull-down assays in vitro demonstrated the ability of the two proteins to form a complex. Next, we tested if this complex could also form in vivo. For this purpose, we constructed HeLa cells expressing myc-tagged versions of p14 or MP1 and performed immunoprecipitations with anti-myc antibodies. In these experiments, endogenous MP1 was detectable in immunoprecipitations of myc-p14 and endogenous p14 in immunoprecipitations of myc-MP1 (Fig. 9 A), demonstrating an in vivo interaction of p14 and MP1. To further analyze a possible complex of p14–MP1 proteins, we separated proteins and protein complexes after detergent extraction of a 100,000-g PNS pellet from EpH4 cells on continuous glycerol gradients. Herein, endogenous p14 and MP1 cofractionated with a similar sedimentation coefficient of ∼5–6 S (Fig. 9 B).
Figure 9.
Interaction between p14 and MP1 in vivo I: coimmunoprecipitation and cosedimentation. (A) Extracts from HeLa cells stably transfected with myc3-p14 or a reverse myc3-p14 construct as control were subjected to immunoprecipitation using polyclonal anti-myc antibodies. 50% of the extracts from the reverse control (rev) or myc3-p14 expressing cells (p14) were analyzed as input controls. Immobilized protein A alone (PA) or with preimmune serum (PI) was used to control for unspecific binding. Coimmunoprecipitated MP1 was analyzed by immunoblotting. (B) Extracts from HeLa cells stably expressing myc3-MP1 were analyzed for coimmunoprecipitation of p14 as in A. 20% of the extract was loaded as input control. (C) Total membranes of EpH4 cells were enriched by centrifugation at 100,000 g. Extracted proteins were loaded onto a 5–35% glycerol gradient and collected fractions were analyzed by immunoblotting. S values of a mix of known proteins are indicated.
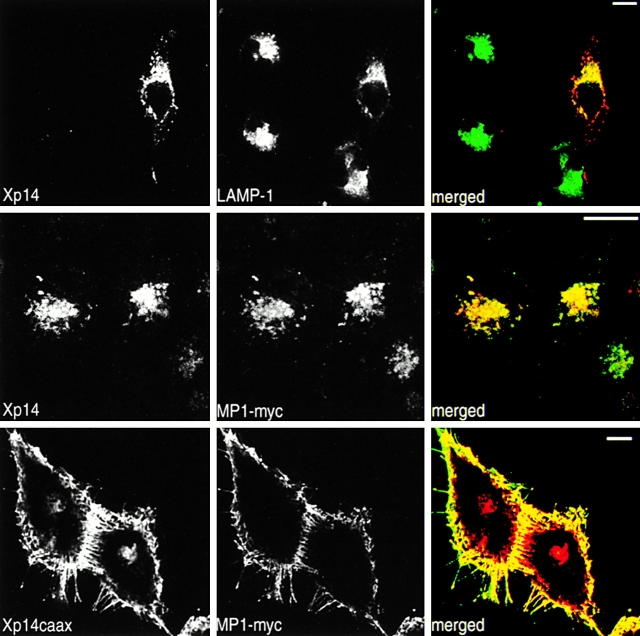
Further support for a complex formation between p14 and MP1 in vivo came from colocalization experiments. In HeLa cells overexpressing Xpress-tagged p14 and myc-tagged MP1, both proteins localized to the same perinuclear, LAMP-1–positive compartment (Fig. 10). In addition, fusing Xpress-tagged p14 to the hypervariable domain and CAAX motif of K-Ras (Choy et al. 1999), leading to plasma membrane localization of p14, resulted in efficient recruitment of coexpressed MP1 to the same sites (Fig. 10, bottom).
Figure 10.
Interaction between p14 and MP1 in vivo II: colocalization and comislocalization. HeLa cells were transiently cotransfected with X-p14 (top and middle panel) or X-p14–CAAX (bottom panel) and MP1-myc6. After 24 h incubation, the cells were fixed and processed for indirect immunofluorescence using anti–LAMP-1, anti-Xpress and anti-myc antibodies. Bars, 10 μm.
Recruitment of MAPK Cascade Components to the p14–MP1 Complex
MP1 was identified as a complex partner for MEK1 and ERK1 in vivo as well as ERK2 in vitro (Schaeffer et al. 1998). This prompted us to try to reconstitute in vitro complexes and to analyze complexes from cells for all of these components.
For the in vitro reconstitution, bacterially expressed GST-tagged ERK2 was used to pull down His6-p14, His6-T7-MP1, and His6-T7-MEK1. In this experimental setup, we failed to detect a direct interaction of MEK1 with MP1 in vitro (data not shown). The previously published MEK1–MP1 interaction (Schaeffer et al. 1998) has been obtained using proteins produced with the baculovirus expression system. Therefore, the apparent difference might be explained by the source of recombinant MP1. However, His6-T7-MP1 efficiently bound to GST–ERK2 (Fig. 8, lane 5), and this interaction was not abrogated by excess His-tagged p14. Instead, p14 bound to GST–ERK2 in addition to MP1 (Fig. 8, lane 6).
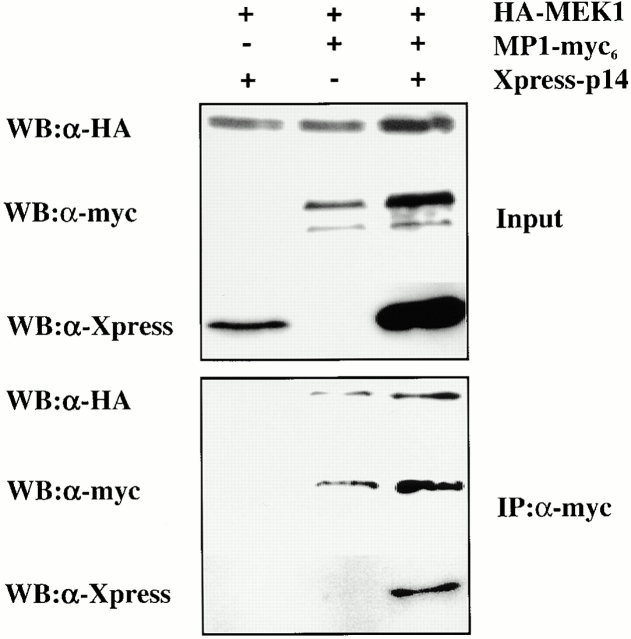
Surprisingly, our coimmunoprecipitation experiments from cell extracts had contrary results. Although we could coimmunoprecipitate HA-tagged MEK1 together with p14 and MP1 (Fig. 11), we did not succeed in recruiting Flag-tagged ERK1 to the p14–MP1–MEK1 complex (data not shown).
Figure 11.
Recruitment of MEK1 to the p14–MP1 complex. Extracts from HeLa cells transiently transfected with HA-MEK1 together with X-p14 or MP1-myc6 or both were subjected to immunoprecipitation using polyclonal anti-myc antibodies. Input and immunoprecipitates were used for immunoblot analysis using anti-HA, anti-myc, and anti-Xpress antibodies.
Interestingly, using untransfected cells we found double phosphorylated ERK1/2 cosedimenting with p14 and MP1 in glycerol gradients (Fig. 9 B) but not activated MEK1/2 (data not shown).
Taken together, the late endosomal/lysosomal peripheral membrane protein, p14, specifically interacted with MP1 and members of the MAPK cascade. However, in contrast to the constitutive interaction between p14 and MP1, MEK1 and ERK1 associate with this complex in a regulated or more dynamic fashion.
Discussion
The MAPK/ERK pathway is a ubiquitously expressed signaling module in vertebrate and invertebrate organisms which governs the proliferation, differentiation, and survival of cells. The basic setup of this pathway is a cascade of three kinases, Raf→MEK→MAPK/ERK, that sequentially activate each other and several associated proteins that modulate signal transduction. The Raf family of MAPK kinase kinases (consisting of three known members, A-Raf, B-Raf, and c-Raf) is thought to integrate upstream input signals into this biochemical signaling module. Besides other targets (Pearson et al. 2000), members of the Raf family activate the MAPK kinases MEK1 or MEK2 by dual phosphorylation. And finally, the ERK family of MAP kinases, ERK1 and ERK2 being the only known substrates of MEK, are considered the effector end with an impressive roster of >50 substrates described to date (Garrington and Johnson 1999).
To achieve an appropriate physiological response, the cell has to generate specificity within the cascade and also at its effector end. Using signaling scaffolds and intracellular membrane transport, the cell recruits several different signaling molecules of a given cascade into a multiprotein complex (Pawson and Scott 1997; Elion 1998; Zuker and Ranganathan 1999) while excluding others (Haugh et al. 1999a,Haugh et al. 1999b). Absence or mutation of scaffolds (Liao and Thorner 1980; Inouye et al. 1997) as well as irregular intracellular trafficking (Kranenburg et al. 1999; York et al. 2000; Zhang et al. 2000) results in functional incompetence of the cell to respond properly to signals.
In this report, we described our efforts in searching for and analyzing the composition of organelle fractions highly enriched in early or late endosomes/lysosomes, respectively, using a targeted proteomic approach. One of the proteins enriched on late endosomes/lysosomes was identified to be a novel, highly conserved protein. We demonstrated that this protein, which we termed p14, is peripherally associated with the cytoplasmic face of late endosomes/lysosomes.
In search of the molecular environment of this protein, we isolated the MAPK scaffold MP1 (Schaeffer et al. 1998) as interaction partner in a two-hybrid screen. We confirmed this interaction by GST pull-down assays, cofractionation, coimmunoprecipitation, colocalization, and comislocalization experiments.
The interaction between p14 and MP1 raised the possibility that MAPK kinase signaling is modulated by p14 since MP1 has been shown to specifically enhance the activation of ERK1 by MEK1 (Schaeffer et al. 1998). Therefore, we tested if MEK or ERK recruitment to MP1 is disturbed or enhanced by p14. These experiments clearly demonstrated that it is possible to reconstitute complexes that contain a member of the MAPK module together with MP1 and p14 in vitro and in vivo, indicating that p14 does not disturb the direct interaction of MP1 with the MAPK module. The difficulty of these experiments is that scaffolding activity is not constitutive but always connected to a specific biological context. Therefore, for a really meaningful interpretation of assays investigating the effect of p14 on MAPK signaling, we will first have to define the upstream signal that uses the p14–MP1 complex to achieve its proper physiological response before further exploring a possible modulating activity of p14 per se. Connected to the question of the putative involvement of p14 in regulating MAPK signaling is the question of whether the p14–MP1 interaction is regulated. Our results indicated that binding of these two proteins to each other is constitutive because the interaction occurs in vitro with bacterially expressed proteins. However, disassembly could be an induced process, but again we will have to address this issue in future studies within the connected biological context.
The localization of the p14–MP1 complex on late endosomes/lysosomes suggests a function connected to this compartment. Lysosomes have mostly been considered to be the digestive organelles of the cell. In this context, a modulation of membrane transport towards lysosomes and/or back to the late endosomal compartment could be the target of a MAPK scaffolding activity, thereby connecting regulation of membrane transport and MAPK signaling.
On the other hand, there is evidence for other functions of the lysosome because this organelle is able to fuse with the plasma membrane (Andrews 2000). Although the role of this process is not yet clear for nonhematopoetic cells, it seems clear that is has to be tightly regulated. There is evidence from hematopoetic cells that extracellular signal–regulated kinases are involved in exocytosis of a lysosome-related organelle, the secretory granule (Trotta et al. 1998; Johnson et al. 1999), suggesting that lysosomal MAPK scaffolding could be involved in regulating lysosomal secretion.
In summary, our discovery bears the potential of combining scaffolding and intracellular membrane transport as means to control the specificity in MAPK signaling in the sense of the p14–MP1 complex being a MAPK scaffold localizing to the late endosomal/lysosomal compartment. This would connect the concepts of catalytic scaffolds mainly described for the regulation of the MAPK pathway, and anchoring scaffolds, a mechanism already described for another signaling pathway, the protein kinase A (PKA) pathway. Herein, PKA is very locally activated by so-called A kinase anchoring proteins (AKAPs). These proteins are interacting with the regulatory (R) subunit of PKA at the same time as they are targeted to specific subcellular locations, thereby mediating compartmentalization of PKA activity (Faux and Scott 1996; Edwards and Scott 2000).
However, we cannot yet exclude the possibility that the p14–MP1 complex, or one of the two, could be sequestered onto the late endosomal/lysosomal compartment. This sequestration could be released upon a signal that activates effector pathways that need MP1 to achieve their specificity.
Supplemental Material
Acknowledgments
We are indebted to Robert Kurzbauer, Chas Ferguson, and Margaret Lindsay for assistance with immunoelectron microscopy. The authors also wish to thank Manuela Baccarini, Marino Zerial, and Michael J. Weber for generous supply of antibodies and constructs and for many stimulatory discussions throughout the course of this work. We wish to acknowledge Karin Paiha (I.M.P.) for assistance with the confocal analysis software. We are also grateful to Rainer Pepperkok for his help on live microscopy and Manfred Schmid and Andreas Birbach for contributing to this work. L.A. Huber and R.G. Parton dedicate this work to the memory of Thomas Kreis.
This work was supported by Boehringer Ingelheim and by grants to L.A. Huber from Johnson & Johnson Focused Giving Program and the Austrian Science Foundation (FWF, P11446-MED).
Footnotes
The online version of this article contains supplemental material.
W. Wunderlich and I. Fialka contributed equally to this work.
Abbreviations used in this paper: 2DGE, two-dimensional gel electrophoresis; EE, early endosome; EGFR, EGF receptor; ERK, extracellular signal–regulated kinase; GST, glutathione S-transferase; LBPA, lysobisphosphatidic acid; LE, late endosome; MAP, mitogen-activated protein; MAPK, MAP kinase; MEK, MAPK/ERK kinase; MP1, MEK partner 1; PAR, protease-activated receptor; pI, isoelectric point; PKA, protein kinase A; PNS, postnuclear supernatant.
References
- Andrews N.W. Regulated secretion of conventional lysosomes. Trends Cell Biol. 2000;10:316–321. doi: 10.1016/s0962-8924(00)01794-3. [DOI] [PubMed] [Google Scholar]
- Baulida J., Kraus M.H., Alimandi M., Di Fiore P.P., Carpenter G. All ErbB receptors other than the epidermal growth factor receptor are endocytosis impaired. J. Biol. Chem. 1996;271:5251–5257. doi: 10.1074/jbc.271.9.5251. [DOI] [PubMed] [Google Scholar]
- Bucci C., Thomsen P., Nicoziani P., McCarthy J., van Deurs B. Rab7a key to lysosome biogenesis. Mol. Biol. Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burack W.R., Shaw A.S. Signal transductionhanging on a scaffold. Curr. Opin. Cell Biol. 2000;12:211–216. doi: 10.1016/s0955-0674(99)00078-2. [DOI] [PubMed] [Google Scholar]
- Ceresa B.P., Schmid S.L. Regulation of signal transduction by endocytosis. Curr. Opin. Cell Biol. 2000;12:204–210. doi: 10.1016/s0955-0674(99)00077-0. [DOI] [PubMed] [Google Scholar]
- Choy E., Chiu V.K., Silletti J., Feoktistov M., Morimoto T., Michaelson D., Ivanov I.E., Philips M.R. Endomembrane trafficking of rasthe CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- Clague M.J., Urbe S., Aniento F., Gruenberg J. Vacuolar ATPase activity is required for endosomal carrier vesicle formation. J. Biol. Chem. 1994;269:21–24. [PubMed] [Google Scholar]
- DeFea K.A., Zalevsky J., Thoma M.S., Dery O., Mullins R.D., Bunnett N.W. Beta-arrestin–dependent endocytosis of proteinase-activated receptor 2 is required for intracellular targeting of activated ERK1/2. J. Cell Biol. 2000;148:1267–1281. doi: 10.1083/jcb.148.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore P.P., Gill G.N. Endocytosis and mitogenic signaling. Curr. Opin. Cell Biol. 1999;11:483–488. doi: 10.1016/s0955-0674(99)80069-6. [DOI] [PubMed] [Google Scholar]
- Edwards A.S., Scott J.D. A-kinase anchoring proteinsprotein kinase A and beyond. Curr. Opin. Cell Biol. 2000;12:217–221. doi: 10.1016/s0955-0674(99)00085-x. [DOI] [PubMed] [Google Scholar]
- Elion E.A. Routing MAP kinase cascades. Science. 1998;281:1625–1626. doi: 10.1126/science.281.5383.1625. [DOI] [PubMed] [Google Scholar]
- Faux M.C., Scott J.D. Molecular gluekinase anchoring and scaffold proteins. Cell. 1996;85:9–12. doi: 10.1016/s0092-8674(00)81075-2. [DOI] [PubMed] [Google Scholar]
- Fialka I., Schwarz H., Reichmann E., Oft M., Busslinger M., Beug H. The estrogen-dependent c-JunER protein causes a reversible loss of mammary epithelial cell polarity involving a destabilization of adherens junctions. J. Cell Biol. 1996;132:1115–1132. doi: 10.1083/jcb.132.6.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fialka I., Pasquali C., Lottspeich F., Ahorn H., Huber L.A. Subcellular fractionation of polarized epithelial cells and identification of organelle-specific proteins by two-dimensional gel electrophoresis. Electrophoresis. 1997;18:2582–2590. doi: 10.1002/elps.1150181414. [DOI] [PubMed] [Google Scholar]
- Fialka I., Pasquali C., Kurzbauer R., Lottspeich F., Huber L.A. Loss of epithelial polarity is accompanied by differential association of proteins with intracellular membranes Electrophoresis 20 1999. 331 343[published erratum at 20:1122] [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Hubbard A.L., Fowler S., Lazarow P.B. Isolation of intracellular membranes by means of sodium carbonate treatmentapplication to endoplasmic reticulum. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrington T.P., Johnson G.L. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr. Opin. Cell Biol. 1999;11:211–218. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- Gruenberg J., Gorvel J.P. In vitro reconstitution of endocytic vesicle fusion. In: Magee A.I., Wileman T., editors. Protein Targeting, A Practical Approach. Oxford University Press; Oxford: 1992. pp. 187–215. [Google Scholar]
- Haugh J.M., Huang A.C., Wiley H.S., Wells A., Lauffenburger D.A. Internalized epidermal growth factor receptors participate in the activation of p21(ras) in fibroblasts J. Biol. Chem 274 1999. 34350 34360a [DOI] [PubMed] [Google Scholar]
- Haugh J.M., Schooler K., Wells A., Wiley H.S., Lauffenburger D.A. Effect of epidermal growth factor receptor internalization on regulation of the phospholipase C-gamma1 signaling pathway J. Biol. Chem 274 1999. 8958 8965b [DOI] [PubMed] [Google Scholar]
- Hobert M.E., Friend L.A., Carlin C.R. Regulation of EGF signaling by cell polarity in MDCK kidney epithelial cells. J. Cell. Physiol. 1999;181:330–341. doi: 10.1002/(SICI)1097-4652(199911)181:2<330::AID-JCP15>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Inouye C., Dhillon N., Durfee T., Zambryski P.C., Thorner J. Mutational analysis of STE5 in the yeast Saccharomyces cerevisiaeapplication of a differential interaction trap assay for examining protein-protein interactions. Genetics. 1997;147:479–492. doi: 10.1093/genetics/147.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.L., Moore E.E., Partrick D.A., Tamura D.Y., Zallen G., Elzi D.J., Silliman C.C., Patrick D.A. Extracellular signal-related kinase 1/2 and p38 mitogen-activated protein kinase pathways serve opposite roles in neutrophil cytotoxicity Arch. Surg 134 1999. 1074 1078[published erratum at 135:989] [DOI] [PubMed] [Google Scholar]
- Kaech S.M., Whitfield C.W., Kim S.K. The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell. 1998;94:761–771. doi: 10.1016/s0092-8674(00)81735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Stang E., Fang K.S., de Moerloose P., Parton R.G., Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Kranenburg O., Verlaan I., Moolenaar W.H. Dynamin is required for the activation of mitogen-activated protein (MAP) kinase by MAP kinase kinase. J. Biol. Chem. 1999;274:35301–35304. doi: 10.1074/jbc.274.50.35301. [DOI] [PubMed] [Google Scholar]
- Krause K.H., Michalak M. Calreticulin. Cell. 1997;88:439–443. doi: 10.1016/s0092-8674(00)81884-x. [DOI] [PubMed] [Google Scholar]
- Kuwada S.K., Lund K.A., Li X.F., Cliften P., Amsler K., Opresko L.K., Wiley H.S. Differential signaling and regulation of apical vs. basolateral EGFR in polarized epithelial cells. Am. J. Physiol. 1998;275:C1419–C1428. doi: 10.1152/ajpcell.1998.275.6.C1419. [DOI] [PubMed] [Google Scholar]
- Lennon G., Auffray C., Polymeropoulos M., Soares M.B. The I.M.A.G.E. Consortiuman integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- Liao H., Thorner J. Yeast mating pheromone alpha factor inhibits adenylate cyclase. Proc. Natl. Acad. Sci. USA. 1980;77:1898–1902. doi: 10.1073/pnas.77.4.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou W., Geuze H.J., Geelen M.J., Slot J.W. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J. Cell Biol. 1997;136:61–70. doi: 10.1083/jcb.136.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R.G., Way M., Zorzi N., Stang E. Caveolin-3 associates with developing T-tubules during muscle differentiation. J. Cell Biol. 1997;136:137–154. doi: 10.1083/jcb.136.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali C., Fialka I., Huber L.A. Preparative two-dimensional gel electrophoresis of membrane proteins. Electrophoresis. 1997;18:2573–2581. doi: 10.1002/elps.1150181413. [DOI] [PubMed] [Google Scholar]
- Pasquali C., Fialka I., Huber L.A. Subcellular fractionation, electromigration analysis and mapping of organelles. J. Chromatogr. B Biomed. Sci. Appl. 1999;722:89–102. doi: 10.1016/s0378-4347(98)00314-4. [DOI] [PubMed] [Google Scholar]
- Pawson T., Scott J.D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Pearson G., Bumeister R., Henry D.O., Cobb M.H., White M.A. Uncoupling Raf1 from MEK1/2 impairs only a subset of cellular responses to Raf activation. J. Biol. Chem. 2000;275:37303–37306. doi: 10.1074/jbc.C000570200. [DOI] [PubMed] [Google Scholar]
- Pol A., Calvo M., Enrich C. Isolated endosomes from quiescent rat liver contain the signal transduction machinery. Differential distribution of activated Raf-1 and Mek in the endocytic compartment. FEBS Lett. 1998;441:34–38. doi: 10.1016/s0014-5793(98)01517-8. [DOI] [PubMed] [Google Scholar]
- Rubino M., Miaczynska M., Lippe R., Zerial M. Selective membrane recruitment of EEA1 suggests a role in directional transport of clathrin-coated vesicles to early endosomes. J. Biol. Chem. 2000;275:3745–3748. doi: 10.1074/jbc.275.6.3745. [DOI] [PubMed] [Google Scholar]
- Schaeffer H.J., Catling A.D., Eblen S.T., Collier L.S., Krauss A., Weber M.J. MP1a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- Somsel Rodman J., Wandinger-Ness A. Rab GTPases coordinate endocytosis J. Cell Sci 113Pt. 22000. 183 192 [DOI] [PubMed] [Google Scholar]
- Sonnichsen B., De Renzis S., Nielsen E., Rietdorf J., Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J. Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A.D., Teslenko L.V., Nikolsky N.N. The endocytosis of epidermal growth factor in A431 cellsa pH of microenvironment and the dynamics of receptor complex dissociation. Exp. Cell Res. 1988;175:192–205. doi: 10.1016/0014-4827(88)90266-2. [DOI] [PubMed] [Google Scholar]
- Tong W.M., Ellinger A., Sheinin Y., Cross H.S. Epidermal growth factor receptor expression in primary cultured human colorectal carcinoma cells. Br. J. Cancer. 1998;77:1792–1798. doi: 10.1038/bjc.1998.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta R., Puorro K.A., Paroli M., Azzoni L., Abebe B., Eisenlohr L.C., Perussia B. Dependence of both spontaneous and antibody-dependent, granule exocytosis-mediated NK cell cytotoxicity on extracellular signal-regulated kinases. J. Immunol. 1998;161:6648–6656. [PubMed] [Google Scholar]
- Ullrich O., Reinsch S., Urbe S., Zerial M., Parton R.G. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C.W., Benard C., Barnes T., Hekimi S., Kim S.K. Basolateral localization of the Caenorhabditis elegans epidermal growth factor receptor in epithelial cells by the PDZ protein LIN-10. Mol. Biol. Cell. 1999;10:2087–2100. doi: 10.1091/mbc.10.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh A.J., Davis R.J. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem. Sci. 1998;23:481–485. doi: 10.1016/s0968-0004(98)01309-7. [DOI] [PubMed] [Google Scholar]
- Wilson P.D. Epithelial cell polarity and disease. Am. J. Physiol. 1997;272:F434–F442. doi: 10.1152/ajprenal.1997.272.4.F434. [DOI] [PubMed] [Google Scholar]
- York R.D., Molliver D.C., Grewal S.S., Stenberg P.E., McCleskey E.W., Stork P.J. Role of phosphoinositide 3-kinase and endocytosis in nerve growth factor-induced extracellular signal-regulated kinase activation via ras and rap1. Mol. Cell. Biol. 2000;20:8069–8083. doi: 10.1128/mcb.20.21.8069-8083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Moheban D.B., Conway B.R., Bhattacharyya A., Segal R.A. Cell surface Trk receptors mediate NGF-induced survival while internalized receptors regulate NGF-induced differentiation. J. Neurosci. 2000;20:5671–5678. doi: 10.1523/JNEUROSCI.20-15-05671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker C.S., Ranganathan R. The path to specificity. Science. 1999;283:650–651. doi: 10.1126/science.283.5402.650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.