Abstract
Guiding multipotent cells into distinct lineages and controlling their expansion remain fundamental challenges in developmental and stem cell biology. Members of the Wnt pathway control many pivotal embryonic events, often promoting self-renewal or expansion of progenitor cells. In contrast, canonical Wnt ligands are thought to negatively regulate cardiomyogenesis in several species. However, the cell-autonomous role of canonical Wnt signaling within precardiac mesoderm, through its obligatory transcriptional mediator, β-catenin, is unknown. Using tissue-specific in vivo genetic manipulation, we found that β-catenin is required for development of cardiac progenitors and is a positive regulator of proliferative expansion of such progenitor cells. At discrete windows of development in embryonic stem cells, activation of canonical Wnt signaling promoted expansion of cardiac progenitors after initial commitment and was required for cardiac differentiation. Together, these data provide in vivo and in vitro evidence that canonical Wnt signaling promotes the expansion of cardiac progenitors and differentiation of cardiomyocytes.
Keywords: cardiac development, embryonic stem cells, β-catenin, second heart field
Signaling and transcriptional networks that control early cardiac commitment, expansion, and differentiation are relevant for congenital heart malformations and attempts to guide stem cells into the cardiac lineage for therapeutic purposes. Soon after gastrulation begins, vertebrate heart formation begins in the anterior mesoderm, stimulated by the interplay of inductive and repressive signals (1, 2). Responding to signals from neighboring endoderm and ectoderm, two domains of mesoderm progenitors, the first heart field and second heart field (SHF), adopt a cardiac fate, each contributing to distinct cardiac regions (3–7). Unlike the first heart field, SHF cells remain undifferentiated until they migrate into the forming heart.
Despite evolutionary conservation of most cardiac developmental pathways, conflicting roles for Wnt signaling in cardiogenesis have been reported. Canonical Wnt signaling is initiated when Wnt ligands bind to Frizzled transmembrane receptors, stabilizing protein levels of β-catenin, a transcriptional coactivator that interacts with T cell factor (TCF/LEF) transcription factors to activate Wnt target genes (8, 9). In many contexts, canonical Wnt signaling regulates proliferation or cell fate decisions during embryonic development (10–12). In flies, Wingless, the founding member of the canonical Wnt family, is required for cardiac lineage determination (13, 14). However, in frogs and chicks, overexpression of canonical Wnts in dorsal mesoderm explants inhibits cardiomyocyte commitment or differentiation, likely by affecting endoderm (15, 16). A zebrafish mutant that globally decreases transcriptional activity of the β-catenin/TCF complex caused hyperproliferation of cardiomyocytes, suggesting β-catenin negatively regulates cardiac differentiation or expansion (17). Recent evidence suggests that canonical Wnt signaling may have temporal-specific effects on pluripotent cells (18). Noncanonical Wnt signaling mediated by alterations in intracellular calcium dependent on G protein-coupled receptors may promote cardiogenesis (19), but the cell-autonomous role of either Wnt pathway within precardiac mesodermal progenitors is unknown.
Here, we demonstrate that β-catenin activity within precardiac mesoderm is required for development of the SHF and promotes proliferation of precardiac mesoderm cells in vivo, with up-regulation of one of its target genes, cyclin D2. Furthermore, after initial precardiac mesoderm differentiation in ES cells (ESC), canonical Wnt signals were required for further differentiation and proliferation of cardiac cells. These results indicate that canonical Wnt signaling can be a positive regulator of cardiogenesis during discrete windows of mammalian development.
Results
β-Catenin Regulation of SHF Progenitors.
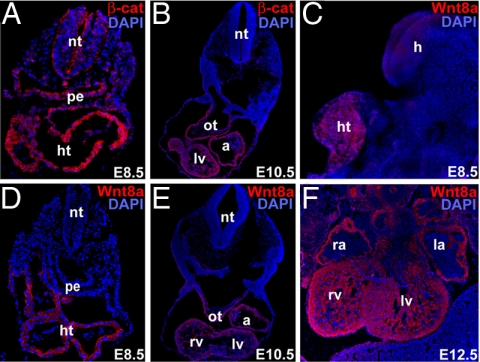
To determine whether canonical Wnt signaling might be active in the heart-forming region, we examined β-catenin protein expression in mouse embryos. At embryonic day (E)8.5, the myocardial layer was enriched in β-catenin, suggesting preferential stabilization of the protein in this region (Fig. 1A). β-Catenin protein was abundant in all chambers of the heart and in the SHF-derived outflow tract at E10.5. Of the canonical Wnt molecules expressed in the developing heart (Wnt2, Wnt7a, and Wnt8a) (20–22), Wnt8a was most specifically expressed in heart precursors at E8.5 (Fig. 1 C and D). Wnt8a was expressed initially throughout the developing heart tube. By E10.5, protein was expressed in all chambers and in the outflow tract. Wnt8a persisted in the ventricular chambers at E12.5, including the compact and trabecular layers. β-Catenin and Wnt8a expression persisted at later stages of development (Fig. 1 B, E, and F).
Fig. 1.
Wnt/β-catenin signaling is active in the developing heart. (A and B) Immunohistochemistry of mouse embryos for β-catenin protein (β-cat, red) at E8.5 (A) and E10.5 (B). (C) Lateral view of whole-mount immunostained E8.5 mouse embryo for Wnt8a (red) protein expression. (D–F) Immunohistochemistry with antibodies against Wnt8a (red) at E8.5 (D), E10.5 (E), and E12.5 (F). DAPI (blue) was used to counterstain the nuclei. All cryosections were transverse. a, atria; h, head; ht, heart tube; la, left atrium; lv, left ventricle; nt, neural tube; ot, outflow tract; pe, pharyngeal region of foregut endoderm; ra, right atrium; and rv, right ventricle.
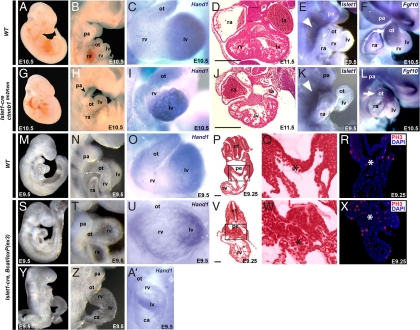
To examine the cell-autonomous role of canonical Wnt signaling in precardiac mesodermal progenitors or cardiomyocytes, we disrupted or stabilized β-catenin expression in cardiac mesodermal progenitors with ctnnb1tm2Kem (23) or β-catenin/loxP(ex3) (24) mice, respectively. In ctnnb1tm2Kem mice, Cre recombinase-mediated excision of exons 2–6 produced a β-catenin-null allele; in β-catenin/loxP(ex3) mice, cre expression activated Wnt/β-catenin signaling by generating stable β-catenin [supporting information (SI) Fig. 5A]. We deleted β-catenin in SHF progenitors before cardiac differentiation by crossing ctnnb1tm2Kem mice with Islet1-cre mice, in which Cre is expressed at E7.0–7.5 in undifferentiated mesodermal progenitors that give rise to the right ventricle (RV) and outflow tract (3). In Islet1-cre ctnnb1tm2Kem homozygous embryos, the SHF failed to form the RV, but the left ventricle (LV) was relatively normal in size (Fig. 2 A–C and G–I). In situ hybridization with a probe recognizing the LV-specific transcript Hand1 (25, 26), also expressed in the outflow tract, confirmed the identity of the residual ventricular chamber (Fig. 2 C and I). Consistent with the activity of Islet1-Cre in pharyngeal arch mesoderm, deletion of β-catenin led to hypoplasia of these structures (Fig. 2 B and H). It should be noted that this Isl1-Cre is not very active in the pharyngeal endoderm despite Isl1 mRNA expression in the endoderm. Thus, SHF mesoderm progenitors may require cell-autonomous Wnt/β-catenin signaling for specification, migration, or expansion.
Fig. 2.
β-Catenin is required for expansion of SHF cardiac progenitors. (A–F and M–R) WT embryos. (G–L) Islet1-cre, ctnnb1tm2Kem homozygous embryos. (S–A′) Islet1-cre, β-catenin/loxP(ex3) heterozygous embryos. (A and G) Lateral views of E10.5 embryos. (B and H) Lateral view focused on cardiac and pharyngeal arch (pa) regions showing hypoplasia of pharyngeal arches (pa) (arrowhead) and RV (rv). (C and I) Frontal view of E10.5 hearts hybridized with Hand1 riboprobe marking left ventricle (lv) and outflow tract (ot). (D and J) Transverse sections through the heart. (Scale bars, 250 μm.) (E, F, K, and L) Lateral views of embryos hybridized with SHF markers, Islet1 (R and K, E9.5) and Fgf10 (F and L, E10.5) riboprobes. Arrowheads indicate SHF; arrows indicate Fgf10 in ot. (M, S, and Y) Lateral views of E9.5 embryos. (N, T, and Z) Lateral view focused on cardiac and pa regions showing significantly enlarged rv region. (O, U, and A′) Frontal view of E9.0 hearts hybridized with Hand1 riboprobe showing expanded rv. (P, Q, V, and W) Transverse sections of E9.25 embryos (P and V) with enlargement of boxed areas (Q and W) showing hyperplasia of SHF cardiac progenitors (asterisk) ventral to the pharyngeal region of foregut endoderm (pe). (Scale bars, 50 μm.) (R and X) PH3 immunostaining (red) of transverse sections through the ot region. DAPI was used to counterstain the nuclei. ca, common atrium; la, left atrium; nt, neural tube; ra, right atrium.
To assess whether β-catenin was required for specification or maintenance of the initial Isl1-positive cells, we examined Isl1 expression in the Islet1-Cre/β-catenin mutant mice. Isl1 was expressed at similar levels and in a comparable domain in mutants and wild-type embryos (Fig. 2 E and K), suggesting that the initial formation and maintenance of SHF progenitors was intact. Migration also appeared intact as Fgf10, marking SHF cells that migrate into the cardiac outflow tract (3, 5, 27), was also expressed appropriately in the β-catenin mutants (Fig. 2 F and L). Finally, we observed no evidence of SHF progenitor cell death (not shown), indicating that the primary defect may lie in failure of SHF cells to differentiate and/or expand in number after specification.
To directly test this idea, we used the Islet1-Cre and β-catenin/loxP(ex3) mice to stabilize β-catenin in the Isl1-positive SHF progenitors. Gain-of-function of β-catenin in these progenitors resulted in a greatly enlarged RV segment from E8.5–9.5 compared with the LV, which was marked by Hand1 expression (Fig. 2 M–O and S–U). In some, the RV segment was so large at an early stage that protrusion of the heart tube was apparent with cardiac dysfunction resulting in developmental delay (Fig. 2 Y, Z, and A′). Most strikingly, the SHF progenitor pool dorsal to the developing heart tube was greatly expanded (Fig. 2 P, Q, V, and W) and contained an increased percentage of phosphohistone H3 (PH3)-positive cells (Fig. 2 R and X), indicating that β-catenin stabilization resulted in marked proliferation of SHF progenitors.
β-Catenin Promotes Proliferation and Differentiation of Cardiomyocytes in Vivo.
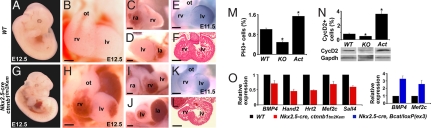
To determine whether Wnt/β-catenin signaling could also affect proliferation of cardiac cells even after they have begun to differentiate, we used a transgenic mouse line in which cre expression is under control of a ventricular-specific enhancer of the cardiac regulatory gene, Nkx2.5. Unlike the endogenous Nkx2.5 gene, which is expressed in the SHF, this Nkx2.5 enhancer is not active in undifferentiated SHF cells (28). The Cre-mediated excision event begins at E8.0–8.5 in the cells that have already arrived into the heart and have begun differentiation (28); excision of β-catenin was confirmed by western analysis as nearly complete (SI Fig. 5B).
Nkx2.5-cre, ctnnb1tm2Kem homozygous embryos died around E12.5 from cardiac dysfunction (Fig. 3 A and G), with significant reductions in ventricular size, particularly the RV (Fig. 3 B–F and H–L) and a thinner compact layer in the ventricular wall (Fig. 3 F and L). To identify the source of the myocardial alterations, we analyzed hearts by immunostaining with anti-PH3 antibody to detect alterations in cell-cycle control or by TUNEL assay to detect apoptosis. The percentage of PH3-positive cells was lower (P < 0.01) in Nkx2.5-cre, ctnnb1tm2Kem hearts and greater (P < 0.01) in Nkx2.5-cre, β-catenin/loxP(ex3) hearts than in wild-type hearts (Fig. 3M). Similarly, the percentage of cyclin D2-positive cells, a direct target of the β-catenin/TCF complex that promotes cell cycling (29), was increased in Nkx2.5-cre, β-catenin/loxP(ex3) hearts and decreased in Nkx2.5-cre, ctnnb1tm2Kem hearts (Fig. 3N Upper, P < 0.01). Total protein levels of cyclin D2 were correspondingly regulated in heart lysates (Fig. 3N Lower). Apoptosis was unaffected (not shown). Expression of numerous regulators of cardiogenesis, including BMP4, Hand2, Hrt2, Mef2c, and Sall4, was down-regulated in Nkx2.5-cre, ctnnb1tm2Kem hearts, and BMP4 and Mef2c expression was up-regulated in Nkx2.5-cre, β-catenin/loxP(ex3) hearts (Fig. 3O). Thus, Wnt/β-catenin activity within cardiac mesoderm appears to positively regulate proliferation and full differentiation of ventricular cardiomyocytes even after early commitment steps have occurred.
Fig. 3.
β-Catenin regulates proliferation of ventricular cardiomyocytes. (A–F) WT embryos. (G–L) Nkx2.5-cre, ctnnb1tm2Kem homozygous embryos. (A and G) Lateral embryonic view of E12.5 embryo. (B–D and H–J) Closeup frontal, right lateral, and left lateral views of the heart, respectively. (E and K) In situ hybridization with Hand1 riboprobe. (F and L) Transverse section of E11.5 heart. (M) Percentage of PH3-positive cells in WT, Nkx2.5-cre, ctnnb1tm2Kem homozygous (KO) and Nkx2.5-cre, β-catenin/loxP(ex3) heterozygous (Act) embryos at E11.5. (N) Percentage of Cyclin D2 (CycD2)-positive cells and Western blot of ventricles using CyclinD2 antibody in indicated mutants. GAPDH antibody was used as a control. (O) Quantitative real-time RT-PCR of indicated genes from ventricles of WT and mutant embryos at E12.5 (Left) or E13.5 (Right). ∗, P < 0.01. Error bars indicate standard deviations. la, left atrium; lv, left ventricle; ot, outflow tract; ra, right atrium; rv, right ventricle. (Scale bars, 250 μm.)
Canonical Wnt Signaling Promotes Expansion and Differentiation of Mesodermal Progenitors into Cardiomyocytes in the ESC System.
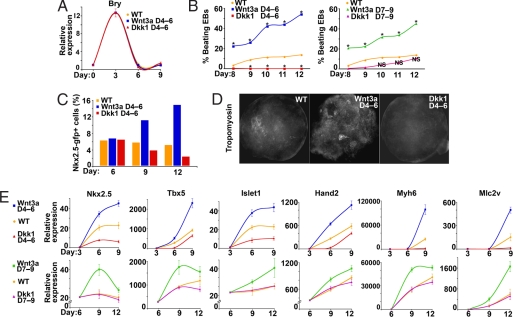
To assess the earlier function of Wnt signals in cells already committed to the mesodermal but not yet to the cardiac fate and to more precisely control the timing of Wnt stimulation or disruption, we turned to the ESC system. ESCs grown in hanging drops form clusters of cells called embryoid bodies (EBs) that randomly differentiate into derivatives of the three germ layers and can form beating cardiomyocytes, although with low frequency. The frequency is even lower when EBs are grown in suspension, which is technically much easier and would allow higher throughput generation of cardiomyocytes from ESCs.
To target canonical Wnt signaling only after mesoderm commitment, we first profiled mesoderm and cardiac gene expression during ESC differentiation in suspension, given that sensitivity to Wnt effects occurs in narrow windows of time. The early mesoderm marker Brachyury (Bry) was strongly induced on day 3 of EB differentiation (EB3) but was down-regulated afterward, suggesting mesoderm commitment by EB3 (SI Fig. 6). Other early cardiac mesoderm markers, such as the transcription factors Nkx2.5, Tbx5, Gata4, and Mesp1, were highly induced at EB4–6. Late cardiac markers, such as the sarcomeric genes Myh6, Myh7, Mlc2a, and Mlc2v, were induced by EB7–9 (SI Fig. 7).
Canonical Wnt signaling is required for general mesoderm formation in vivo (30, 31) and in EBs (32). We therefore initially targeted canonical Wnt signaling after mesoderm commitment (EB3) but before induction of early cardiac genes (EB4–5). Wnt3a, the soluble canonical Wnt ligand available, and Dickopff1 (Dkk1), an inhibitor of canonical Wnts, were used to promote or disrupt Wnt/β-catenin signaling, respectively. Bry expression was unaltered by addition of these factors at EB4–6 (Fig. 4A), suggesting that stimulating or inhibiting canonical Wnt signaling after EB3 did not alter the number of mesodermal progenitors. In contrast, stimulation of Wnt/β-catenin between EB4–6 with soluble Wnt3a increased the number of beating EBs at EB12 from 10% to >50% when cells were grown in suspension (Fig. 4B Left, P < 0.01). Inhibiting canonical Wnt signaling with Dkk-1 between EB4–6 resulted in a complete absence of beating EBs (Fig. 4B Left), suggesting that canonical Wnt signaling during this window not only promotes but also is required for cardiomyocyte formation in the ESC differentiation system.
Fig. 4.
Canonical Wnt signaling regulates cardiac expansion and differentiation in ESCs. (A) Brachyury expression profiles, determined by quantitative real-time PCR (qPCR), of undifferentiated ESCs on day 0 and EBs harvested on days 3, 6, and 9 after early Wnt3a or Dkk-1 treatment compared with control. (B) Beating EBs grown in suspension were counted from days 8 to 12. Percentage of beating EBs after early (days 4–6) or late (days 7–9) treatment with Wnt3a or Dkk-1. Asterisks indicate significant difference in beating percentage of treatment groups versus untreated EBs (P < 0.01). (C) Percentage of Nkx2.5-gfp+ cells at days 6, 9, and 12 in untreated EBs or those treated with Wnt3a or Dkk1 from days 4–6. (Bars represent percent of Nkx2.5-gfp+ cells from 4 × 104 cells analyzed). (D) Immunohistochemistry of day12 EBs with anti-Tropomyosin. (E) Cardiac gene expression after early (days 4–6, Upper) or late (days 7–9, Lower) treatment with Wnt3a or Dkk-1. EBs were harvested on days 3, 6, and 9 (early treatment) or 6, 9, and 12 (late treatment). Fold change in expression of all indicated genes (y axis) in EBs with respect to undifferentiated ESCs was assessed by qPCR. NS, not significant. ∗, P < 0.01.
The presence of beating EBs is useful for gross analyses but does not address the number of progenitors or differentiated cardiomyocytes. To more rigorously assess this aspect, we used an ESC line containing GFP under control of the endogenous Nkx2.5 regulatory elements to mark and sort early cardiac mesoderm (B.R.C. and E.C.H., unpublished results). We found that stimulation or inhibition of Wnt signaling between EB4 and EB6 did not affect the initial number of progenitors in day 6 EBs. However, flow cytometry revealed a progressive expansion of the Nkx2.5-positive pool upon Wnt3a stimulation and a decrease in the progenitor pool upon Wnt inhibition as further differentiation proceeded in EBs (Fig. 4C). The numbers of cells positive for Tropomyosin, a late cardiac sarcomeric marker, were similarly increased by the Wnt agonist by EB12 (Fig. 4D). Notably, no Tropomyosin+ cells were observed in EBs treated with the Wnt antagonist after EB4, consistent with the absence of beating EBs (Fig. 4B Left).
In agreement with a positive role for Wnt in the expansion and possibly early differentiation of the cardiac lineage, expression levels of the early cardiac genes Nkx2.5 and Tbx5 was up-regulated by exposure to Wnt3a from EB4–6 and down-regulated by similar exposure to Dkk-1 (Fig. 4E Upper). The SHF marker, Islet1, and the RV marker, Hand2 (26), showed similar responses to Wnt3a and Dkk-1, as did cardiac sarcomeric genes (Fig. 4E Upper).
To assess effects of canonical Wnt signaling after induction of early cardiac mesoderm in ESCs, we treated EBs with Wnt3a or Dkk-1 between EB 7 and EB9. Nearly 50% of EBs treated with Wnt3a and grown in suspension were beating by EB12 (Fig. 4B Right, P < 0.01), and most early and late cardiac markers were up-regulated (Fig. 4E Lower), suggesting that Wnt had a positive effect on cardiomyocyte development even at this relatively late stage. This timing of Wnt manipulation is similar to the ventricular-restricted Nkx2.5-cre transgenic line used in the in vivo studies. Approximately 10% of EBs treated with Dkk-1 were beating (vs. 13% of control EBs; Fig. 4B Right, P > 0.2), and cardiac gene expression was not changed upon Wnt inhibition (Fig. 4E Lower). Thus, after cardiac mesoderm commitment, canonical Wnt signals appear to be dispensable for full cardiac differentiation but may still recruit mesodermal precursors or expand cardiac progenitors. This result is consistent with our in vivo studies with the Nkx2.5-cre mouse described above.
Discussion
Published observations suggest that canonical Wnt signals likely play distinct roles during discrete developmental windows, first positively regulating mesoderm commitment and then possibly playing a negative role in the initial induction of cardiac progenitors (15, 16, 18, 30–32). The loss- and gain-of-function studies of canonical Wnt signaling in a spatiotemporally restricted manner described here provide compelling evidence that Wnt/β-catenin signaling is required in a cell autonomous fashion for the expansion and development of precardiac mesoderm and cardiac mesoderm in mouse. Thus, narrow developmental windows may exist during which canonical Wnt signaling sequentially inhibits then promotes cardiac development. This may be similar to the function of Bmp signaling during cardiogenesis (33).
Previous studies in other species suggested that canonical Wnt signaling inhibited early cardiac commitment in a noncell autonomous fashion, likely by acting on adjacent endoderm. Recent studies implicate activation of Sox17 as a potential mediator of this endodermal effect (34). The multiple hearts that arise with endoderm-specific deletion of β-catenin in mice is consistent with the negative effects of an endoderm-derived factor (35). In contrast, β-catenin within the mesoderm after initial cardiac commitment is clearly involved in both cardiac proliferation and differentiation, particularly in the SHF. Because Hand2 plays a critical role in SHF expansion (28, 36) and an upstream genomic region is reportedly bound by the β-catenin/TCF complex (37), it is possible that its down-regulation in part mediates the effects of β-catenin in regulating cardiac mesoderm.
The striking increase in proliferating SHF cells upon stabilization of Wnt/β-catenin suggests an instructive role in cardiac cell proliferation. This is similar to the hyperplasia of cardiac progenitors in Drosophila upon overexpression of Wg (13). This effect may be mediated through up-regulation of cyclin D2, a direct transcriptional target of β-catenin/TCF (29). These observations were supported by findings in ES-derived cells, in which Wnt signaling was necessary for cardiac differentiation from mesodermal precursors and induced expansion of cardiac precursors after their initial induction. Thus, we have shown that Wnt/β-catenin signaling is essential for development of heart progenitors in mammals and that canonical Wnt signaling can be manipulated to regulate expansion and differentiation of cardiac progenitor cells in ESCs. It will be interesting to determine whether similar Wnt signaling regulates self-renewal or expansion of cardiac progenitors in the postnatal heart.
Materials and Methods
Mouse Lines and Genetics.
The Nkx2.5-cre, ctnnb1tm2Kem or Islet1-cre, ctnnb1tm2Kem homozygous embryos were obtained by crossing Nkx2.5-cre, ctnnb1tm2Kem flox/+ or Islet1-cre, ctnnb1tm2Kem flox/+ lines with ctnnb1tm2Kem flox/flox lines, respectively. Nkx2.5-cre, β-catenin/loxP(ex3), or Islet1-cre, β-catenin/loxP(ex3) heterozygous embryos were obtained by crossing Nkx2.5-cre or Islet1-cre lines with β-catenin/loxP(ex3) flox/+ lines, respectively. Wild-type and mutant embryos were identified as described (23, 24).
Immunohistochemistry, Western Blotting, and in Situ Hybridization.
Whole or cryosectioned embryos and EBs were stained with the following antibodies: mouse anti-Islet1 (1:200) and anti-Tropomyosin (1:100; Developmental Studies Hybridoma Bank, Iowa City, IA), goat anti-Wnt8a (15 μg/ml, R&D Systems, Minneapolis, MN), goat anti-β-catenin (1:50), and rat anti-cyclin D2 (1:100) (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-phospho-histone H3 (1:100; Upstate Biotechnology, Charlottesville, VA), and horseradish peroxidase-conjugated (with TSA plus Fluorescent Systems; PerkinElmer, Waltham, MA) secondary antibodies (The Jackson Laboratory, Bar Harbor, ME). For Western blotting, lysates from E12.5 ventricles were analyzed by using antibodies against β-catenin, cyclin D2, and GAPDH (1:50, 1:150, and 1:100, respectively; Santa Cruz Biotechnology). mRNA in situ hybridization was performed with designated antisense probes as described (3, 5, 27).
ESC Culture, Flow Cytometry, and Gene Expression Analysis.
Murine ESCs were propagated undifferentiated in maintenance medium of Glasgow MEM (Sigma, St. Louis, MO; G5154) supplemented with 10% FBS (HyClone, Logan, UT; SH30071.03), 1 mM 2-mercaptoethanol (Sigma M7522), 2 mM l-glutamine (Gibco-BRL, Carlsbad, CA; 25030–081), 1 mM sodium pyruvate (GIBCO-BRL 11360-070), 0.1 mM minimum essential medium containing nonessential amino acids, and leukemia inhibitory factor (LIF)-conditioned medium (1:1,000). EBs were formed by culturing ESCs (6 × 105 per well) for 3 days in ultra-low-attachment six-well plates (Corning, Lowell, MA; 07200601) in differentiation medium (DM), which contained the same components as maintenance medium but had 20% FBS and no LIF. For early treatment, the medium was replaced with DM containing Wnt3a (150 ng/ml, R&D Systems) or Dkk1 (500 ng/ml, R&D Systems) at the start of day 4; on day 6, the medium was replaced with fresh DM. For late treatment, DM was replaced with Wnt3a- or Dkk1-containing DM at the start of day 7 and switched to fresh DM on day 9. Respective media were changed for early-, late-, and untreated EBs every 3 days. EBs were monitored for beating from day 8 to day 12 and collected between days 3 and 12 at regular intervals. At respective days of differentiation, EBs were dissociated via trypsin and passed through a nylon cell strainer. A Becton Dickinson (Franklin Lakes, NJ) FACS Diva flow cytometer and cell sorter was used for detecting Nkx2.5-GFP+ cells. For gene expression analysis, total RNA was isolated, and real-time PCR was performed with the ABI Prism system (7900HT, Applied Biosystems, Foster City, CA). TaqMan primers used in this study are listed in SI Table 1. All samples were run in triplicate. Real-time PCR data were normalized and standardized with SDS2.2 software (Applied Biosystems).
Supplementary Material
Acknowledgments
We thank E.N.O. (University of Texas Southwestern, Dallas, TX) and Sylvia Evans (University of California at San Diego, La Jolla, CA) for kindly providing Nkx2.5-cre and Islet1-cre [from Tom Jessell, Yasuto Tanabe, and Chris William (Columbia University, New York, NY)] mice, respectively. We also thank G. Howard and S. Ordway for editorial assistance, B. Taylor for manuscript preparation, J. Fish for histopathology support, and members of the Srivastava lab for helpful discussions. C.K. was supported by a fellowship from the American Heart Association; D.S. is an Established Investigator of the American Heart Association and was supported by grants from National Heart, Lung, and Blood Institute/National Institutes of Health and the March of Dimes Birth Defects Foundation. This work was also supported by National Institutes of Health/National Center for Research Resources (Grant C06 RR018928, to Gladstone Institutes).
Abbreviations
- SHF
second heart field
- En
embryonic day n
- RV
right ventricle
- ESC
ES cell
- EB
embryoid body
- EBn
day n of EB differentiation
- DM
differentiation medium.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704044104/DC1.
References
- 1.Olson EN. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srivastava D. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckingham M, Meilhac S, Zaffran S. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 5.Kelly RG, Brown NA, Buckingham ME. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 6.Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR. Dev Biol. 2001;238:97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- 7.Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Development (Cambridge, UK) 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- 8.Bejsovec A. Cell. 2005;120:11–14. doi: 10.1016/j.cell.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Logan CY, Nusse R. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 10.Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, et al. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- 11.Olson LE, Tollkuhn J, Scafoglio C, Krones A, Zhang J, Ohgi KA, Wu W, Taketo MM, Kemler R, Grosschedl R, et al. Cell. 2006;125:593–605. doi: 10.1016/j.cell.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 12.Reya T, Clevers H. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 13.Park M, Wu X, Golden K, Axelrod JD, Bodmer R. Dev Biol. 1996;177:104–116. doi: 10.1006/dbio.1996.0149. [DOI] [PubMed] [Google Scholar]
- 14.Wu X, Golden K, Bodmer R. Dev Biol. 1995;169:619–628. doi: 10.1006/dbio.1995.1174. [DOI] [PubMed] [Google Scholar]
- 15.Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. Genes Dev. 2001;15:316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider VA, Mercola M. Genes Dev. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rottbauer W, Saurin AJ, Lickert H, Shen X, Burns CG, Wo ZG, Kemler R, Kingston R, Wu C, Fishman M. Cell. 2002;111:661–672. doi: 10.1016/s0092-8674(02)01112-1. [DOI] [PubMed] [Google Scholar]
- 18.Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, Kikuchi A, Komuro I. Proc Natl Acad Sci USA. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandur P, Lasche M, Eisenberg LM, Kuhl M. Nature. 2002;418:636–641. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- 20.Bond J, Sedmera D, Jourdan J, Zhang Y, Eisenberg CA, Eisenberg LM, Gourdie RG. Dev Dyn. 2003;227:536–543. doi: 10.1002/dvdy.10333. [DOI] [PubMed] [Google Scholar]
- 21.Jaspard B, Couffinhal T, Dufourcq P, Moreau C, Duplaa C. Mech Dev. 2000;90:263–267. doi: 10.1016/s0925-4773(99)00236-1. [DOI] [PubMed] [Google Scholar]
- 22.Monkley SJ, Delaney SJ, Pennisi DJ, Christiansen JH, Wainwright BJ. Development (Cambridge, UK) 1996;122:3343–3353. doi: 10.1242/dev.122.11.3343. [DOI] [PubMed] [Google Scholar]
- 23.Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Development (Cambridge, UK) 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 24.Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srivastava D, Cserjesi P, Olson EN. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- 27.Hu T, Yamagishi H, Maeda J, McAnally J, Yamagishi C, Srivastava D. Development (Cambridge, UK) 2004;131:5491–5502. doi: 10.1242/dev.01399. [DOI] [PubMed] [Google Scholar]
- 28.McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN. Development (Cambridge, UK) 2005;132:189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- 29.Huang W, Chang HY, Fei T, Wu H, Chen YG. Oncogene. 2007;26:2471–2482. doi: 10.1038/sj.onc.1210033. [DOI] [PubMed] [Google Scholar]
- 30.Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Development (Cambridge, UK) 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 31.Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A. Nat Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 32.Lindsley RC, Gill JG, Kyba M, Murphy TL, Murphy KM. Development (Cambridge, UK) 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- 33.Yuasa S, Itabashi Y, Koshimizu U, Tanaka T, Sugimura K, Kinoshita M, Hattori F, Fukami S, Shimazaki T, Ogawa S, et al. Nat Biotechnol. 2005;23:607–611. doi: 10.1038/nbt1093. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Asakura M, Inoue H, Nakamura T, Sano M, Niu Z, Chen M, Schwartz RJ, Schneider MD. Proc Natl Acad Sci USA. 2007;104:3859–3864. doi: 10.1073/pnas.0609100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lickert H, Kutsch S, Kanzler B, Tamai Y, Taketo MM, Kemler R. Dev Cell. 2002;3:171–181. doi: 10.1016/s1534-5807(02)00206-x. [DOI] [PubMed] [Google Scholar]
- 36.Yamagishi H, Yamagishi C, Nakagawa O, Harvey RP, Olson EN, Srivastava D. Dev Biol. 2001;239:190–203. doi: 10.1006/dbio.2001.0417. [DOI] [PubMed] [Google Scholar]
- 37.Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.