Abstract
Reggie-1 and -2 proteins (flotillin-2 and -1 respectively) form their own type of non-caveolar membrane microdomains, which are involved in important cellular processes such as T-cell activation, phagocytosis and signalling mediated by the cellular prion protein and insulin; this is consistent with the notion that reggie microdomains promote protein assemblies and signalling. While it is generally known that membrane microdomains contain large multiprotein assemblies, the exact organization of reggie microdomains remains elusive. Using chemical cross-linking approaches, we have demonstrated that reggie complexes are composed of homo- and hetero-tetramers of reggie-1 and -2. Moreover, native reggie oligomers are indeed quite stable, since non-cross-linked tetramers are resistant to 8 M urea treatment. We also show that oligomerization requires the C-terminal but not the N-terminal halves of reggie-1 and -2. Using deletion constructs, we analysed the functional relevance of the three predicted coiled-coil stretches present in the C-terminus of reggie-1. We confirmed experimentally that reggie-1 tetramerization is dependent on the presence of coiled-coil 2 and, partially, of coiled-coil 1. Furthermore, since depletion of reggie-1 by siRNA (small interfering RNA) silencing induces proteasomal degradation of reggie-2, we conclude that the protein stability of reggie-2 depends on the presence of reggie-1. Our data indicate that the basic structural units of reggie microdomains are reggie homo- and hetero-tetramers, which are dependent on the presence of reggie-1.
Keywords: cross-linking, flotillin, lipid raft, membrane microdomain, oligomerization, reggie
Abbreviations: BiFC, bimolecular fluorescence complementation; DMS, dimethyl suberimidate; DSS, disuccinimidyl suberate; DST, disuccinimidyl tartarate; EGFP, enhanced green fluorescent protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; HA, haemagglutinin; HRP, horseradish peroxidase; mAb, monoclonal antibody; MBS, m-maleimidobenzoyl-N-hydoxysuccinimide ester; N2a, neuro 2a; pAb, polyclonal antibody; NCAM, neural cell adhesion molecule; RT, reverse transcription; siRNA, small interfering RNA; SPFH, stomatin/prohibitin/flotillin/HflK/C
INTRODUCTION
Reggie-1 and -2 (flotillin-2 and -1 respectively) are two highly conserved lipid-raft-associated proteins which form their own type of non-caveolar microdomains [1,2], and define a novel clathrin-independent endocytic pathway in mammalian cells [3]. Reggies were first discovered as proteins up-regulated in retinal ganglion cells during axon regeneration in fish and rats [4,5]. They were identified independently as proteins contained in the ‘floating’ lipid raft fraction from mouse lung tissue, and called flotillin-1 and -2 [6]. To date, reggie proteins have been identified in all metazoans, and reggie-like proteins have been found in bacteria, fungi and higher plants [7,8]. Reggies and reggie-like proteins belong to the so-called SPFH (stomatin/prohibitin/flotillin/HflK/C) protein superfamily, whose members share a common N-terminal domain of unknown function, the SPFH domain [9]. In contrast with other SPFH proteins, the C-terminus of reggies is longer and includes a so-called ‘flotillin’ domain, which is characterized by the presence of repeats rich in glutamate and alanine, which are predicted to form three coiled-coil stretches [6,8,10].
Even though the functions of reggie proteins remain poorly understood, they have been implicated in interesting cellular processes, such as insulin signalling [11], phagocytosis [12], axon growth [4,5] and lymphocyte activation [13–17]. For instance, in Jurkat T-cells, reggies assemble into a pre-formed cap which recruits all the signalling molecules known to be involved in activation [13–16]. Using the same cells, we have shown that antibody-mediated signalling of the cellular prion protein takes place in the pre-formed reggie cap region [17]. During zebrafish development, knockdown of reggies produces severe morphological defects (E. Malaga-Trillo, E. Rivera-Milla and C. A. O. Stuermer, unpublished work). Although knockout of reggie proteins in Drosophila does not produce evident phenotypes, overexpression of reggie-1 and -2 in the eye imaginal disc leads to severe mistargeting of cell adhesion molecules, resulting in irregular ommatidial patterns [18]. Moreover, abnormal wingless signalling and wing development can be observed upon reggie overexpression [18]. Hence, various lines of evidence support the notion that reggie microdomains form scaffolds for protein assemblies, which are involved in signalling across the plasma membrane [19].
That reggie proteins form membrane microdomains is evident by their distinct punctate distribution along the plasma membrane, and is supported further by our own electron microscopy analyses that define reggies as non-caveolar clusters of a surprisingly constant average size of ≤0.1 μm [13]. However, it is not known how reggie proteins are organized within these microdomains, and the little experimental data available disagree about the exact size of reggie complexes [20,21]. In the present study, we determined the molecular size of oligomers of endogenous and exogenous reggie proteins by chemical cross-linking experiments in living cells. Our goal was to obtain direct information on the stoichiometry of reggie complexes, and to define the reggie domains required for oligomerization. We demonstrate that reggie proteins form homo- and hetero-tetramers via their C-terminal regions. Moreover, we also tested the oligomerization abilities of the three coiled-coil stretches predicted at the C-terminal half of reggie-1. In addition, siRNA (small interfering RNA) experiments reveal that reggie-2 protein undergoes proteasomal degradation upon silencing of reggie-1. Our studies show that reggie-1 and -2 are organized as homo- and hetero-tetramers at the plasma membrane, where the presence of reggie-1 is required for the stabilization of reggie-2. We have uncovered the basic building blocks of reggie microdomains, providing new insight into their formation and stabilization dynamics. Clarifying these processes is pivotal to better understand the function of reggie-1 and -2 as signalling platforms.
EXPERIMENTAL
Reagents and antibodies
Cell culture reagents were purchased from Gibco BRL. Antibodies were purchased from the following companies: α-flotillin-2/ESA (reggie-1) mAb (monoclonal antibody) and α-flotillin-1 (reggie-2) mAb from BD Biosciences, α-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mAb (6C5) from Abcam, and α-HA (haemagglutinin) mAb (3F10) and α-GFP (green fluorescent protein) mAb (7.1/13.1) from Roche. α-GFP pAb (polyclonal antibody) from Clontech, Alexa Fluor® 488-conjugated goat α-rabbit pAb, Cy-3-conjugated donkey α-rat pAb, HRP (horseradish peroxidase)-conjugated goat α-rabbit pAb and HRP-conjugated goat α-mouse pAb from Jackson ImmunoResearch, and HRP-conjugated goat α-rat pAb from Pierce Chemical. α-NCAM (neural cell adhesion molecule) pAb was kindly provided by E. Bock (The Protein Laboratory, University of Copenhagen, Copenhagen, Denmark). DSS (disuccinimidyl suberate), MBS (m-maleimidobenzoyl-N-hydoxysuccinimide ester), DMS (dimethyl suberimidate) and DST (disuccinimidyl tartarate) were purchased from Pierce Chemical. Lactacystin was kindly provided by M. Groettrup (Department of Biology/Immunology, University of Konstanz, Konstanz, Germany). All other reagents were purchased from Sigma–Aldrich.
Cell culture and transfection
N2a (neuro 2a) cells were maintained in MEM (minimal essential medium) containing 10% (v/v) fetal calf serum, L-glutamine, pyruvate and penicillin/streptomycin at 37 °C and 5% CO2, and grown on polylysine-coated coverslips (for immunofluorescence studies) or in six-well plates (for cross-linking experiments) for 24 h before transfection. N2a cells were transiently transfected with the relevant vectors using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's recommendations, and experiments were performed at 20 h after transfection.
Cloning
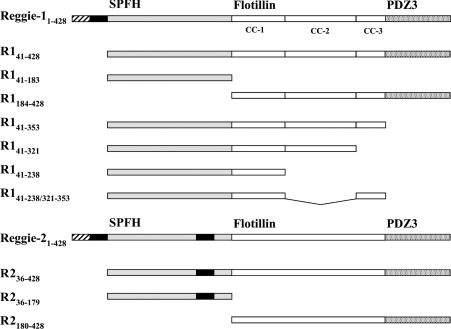
Vectors including the rat cDNA sequences of reggie-1 and -2 fused to EGFP (enhanced GFP) were generously provided by R. Tikkanen (Institute for Biochemistry II, University of Frankfurt, Frankfurt, Germany). For cloning HA- and BiFC (bimolecular fluorescence complementation)-tagged constructs (Figure 1), reggie cDNAs were amplified by PCR from the reggie–EGFP vectors described above using Pwo DNA polymerase (Peqlab Biotechnologie). N-terminally HA-tagged constructs were obtained by cloning the reggie sequences into the EcoRI and XhoI sites of pJG4-5 vector (Invitrogen). Then the HA-tagged reggie constructs were amplified by PCR and cloned into the EcoRV site of the pcDNA3 vector (Invitrogen). C-terminally HA-tagged reggie-1 and -2 were obtained by PCR with primers including the HA tag, and finally cloned into the BamHI and XhoI sites of pcDNA3 vector (Invitrogen). BiFC–reggie proteins were obtained by cloning in-frame the reggie sequences into the EcoRI and XhoI sites of pBiFC-YC155 vector (kindly provided by M. D'Silva, Department of Biology, University of Konstanz, Konstanz, Germany) [22]. These vectors coding for reggie proteins fused to the C-terminal half of YFP (yellow fluorescent protein) (amino acids 155–238).
Figure 1. Schematic representation of reggie-1 and -2 domains and deletion constructs.
The N-terminal SPFH domains are indicated in grey, the C-terminal flotillin domains are in white, and the predicted PDZ3 domains are stippled. Acylation occurs in the regions indicated by hatching, and the hydrophobic stretches are shown as black bars. The predicted coiled-coil (CC) stretches of reggie-1 are indicated.
Cross-linking experiments
Transiently transfected and non-transfected N2a cells were washed twice with PBS. Chemical cross-linkers (in DMSO) were applied to the cells at the final concentrations indicated in the Figure legends. After incubation at room temperature (22 °C) for the time indicated, cells were boiled directly in SDS/PAGE sample buffer. The resulting extracts were analysed by immunoblotting with mAbs specific for reggie-1 or -2, mAbs against the HA epitope or the GFP protein, and pAb against GFP.
Urea treatments
Non-transfected N2a cells were washed twice with PBS and lysed in PBS supplemented with the urea concentrations indicated in the corresponding Figure. After 1 h of incubation at room temperature, the resulting extracts were separated by SDS/10% PAGE and analysed by immunoblotting with a mAb specific for reggie-1. Additionally, the 6 M urea cell extract was analysed by SDS/10% PAGE supplemented with 6 M urea followed by Western blotting for detection of reggie-1.
High-speed cell-fractionation experiments
Cell extracts of transiently transfected N2a cells were prepared using non-denaturing conditions and fractionated using high-speed centrifugation. Briefly, N2a cells were washed twice with PBS and resuspended in hypo-osmotic buffer (20 mM Hepes, pH 7.0). The cell suspension was passed through a 27 gauge needle 20 times, and nuclei and unbroken cells were removed by centrifugation at 1000 g for 10 min at 4 °C. Cytosolic and membrane fractions were separated by centrifugation at 100000 g for 60 min at 4 °C. Fractions were boiled directly in SDS/PAGE sample buffer and analysed by Western blotting using mAb against the HA epitope.
Western blot analysis
For SDS/PAGE and immunoblots [23], samples containing reggie species were separated on 10 or 15% gels under reducing conditions. As a molecular marker, the Precision Plus protein standard purchased from Bio-Rad Laboratories was used. After gel electrophoresis, proteins were transferred on to Hybond-C Extra nitrocellulose membranes (Amersham Biosciences) in a tank blot apparatus. The membranes were blocked in PBS containing 5% non-fat dried milk for 1 h at room temperature and incubated with the relevant Abs, defined above and below, in blocking solution for 1 h at room temperature or overnight at 4 °C. After four washes in PBS, the blots were incubated with HRP-conjugated goat α-mouse, goat α-rat or goat α-rabbit pAbs in blocking solution for 1 h at room temperature, and, after extensive washing, developed with the use of the ECL® (enhanced chemiluminescence) substrate SuperSignal (Pierce Chemical) and Hyperfilm ECL® (Amersham Biosciences).
Immunofluorescence
Transiently transfected N2a cells were fixed using 4% (w/v) paraformaldehyde in PBS for 10 min, permeabilized in PBS containing 0.1% (v/v) Triton X-100 for 1 min at room temperature and blocked with 1% (w/v) BSA in PBS. Samples were exposed to the primary antibodies for 1 h at room temperature, washed three times with PBS, and the secondary antibodies were applied for 1 h at room temperature. After several rinses with PBS, the cells were embedded in a mixture of 5 g of Mowiol 4-88 (Hoechst), 20 ml of PBS and 10 ml of glycerol. Light-microscopical analyses were performed using a confocal laser-scanning microscope (LSM 510, Carl Zeiss) equipped with a Plan-Apochromat ×63 oil immersion objective (numerical aperture 1.4). Images were acquired with the LSM 510 software and further processed using Corel Photo-Paint 11.
RNA interference
siRNA duplexes R1.0 and R2.0 were obtained from Dharmacon and R1.1 was from Ambion. The target sequences for reggie-1 were 5′-GTTCATGGCAGACACCAAG-3′ (R1.0) and 5′-GGTGAAGATCATGACGGAG-3′ (R1.1), and 5′-CACACTGACCCTCAATGTC-3′ (R2.0) for reggie-2. A commercial siRNA against firefly luciferase (Photinus pyralis), referred to as GL2 (Dharmacon), was used as a non-specific control. N2a cells were grown in 24-well plates and transfected with duplexed siRNAs (7.5 pmol/well) using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions. Lactacystin was used at a final concentration of 10 μM 8 h post-transfection to block the 20 S subunit of the proteasome [24]. Cells treated or non-treated with lactacystin were lysed 48 h after transfection for Western blot analysis using GAPDH-, and reggie-1- and -2-specific mAbs. Total RNA extraction of the samples was obtained 48 h after transfection using the RNeasy Mini Kit (Qiagen). Then, samples were analysed by RT (reverse transcription)–PCR using the primers R1-For (5′-CAGGTGAAGATCATGACG-3′), R1-Rev (5′-ACCACAATCTCATCGAC-3′), R2-For (5′-CTTGTGGCCCAAATGAG-3′), and R2-Rev (5′-ATCTCCTGCTCCTGCAC-3′). As controls the primers GAPDH-For (5′-GGTCGGTGTGAACGGATTTGGC-3′) and GAPDH-Rev (5′-GGAGGCCATGTAGGCCATGAGG-3′), designed to amplify a fragment of the GAPDH cDNA, were used.
RESULTS
Reggie-1 and reggie-2 form homo- and hetero-tetramers
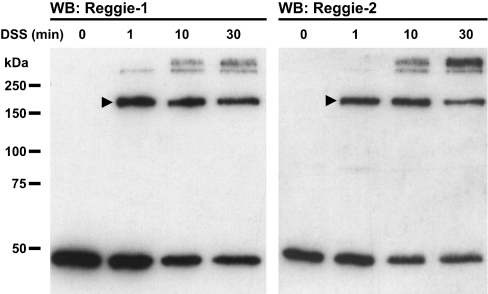
To examine the oligomerization states of endogenous reggie-1 and -2, we took advantage of DSS, a chemical compound which cross-links adjacent lysine residues within a range of 11.4 Å (1 Å=0.1 nm). DSS-treated N2a cells were analysed by Western blotting denatured cell extracts using mAbs specific for reggie-1 and -2 respectively (Figure 2). In addition to the known ∼47 kDa reggie-1 and -2 monomers [5], we observed strong bands of ∼200 kDa, corresponding to the expected size of reggie homo- and/or hetero-tetramers. Surprisingly, no bands corresponding to reggie dimers or trimers were observed, even when DSS was applied for short times (1 min; Figure 2) or at lower concentrations (50 μM; Supplementary Figure 1A at http://www.BiochemJ.org/bj/403/bj4030313add.htm). Instead, as DSS treatment time increased, additional bands of even higher molecular mass became gradually more visible at the expense of monomer and tetramer species (Figure 2). These bands might correspond to larger complexes generated by the covalent linkage of two or more reggie tetramers, although the participation of others proteins cannot be excluded. Identical results using DSS in PC12, HeLa and Jurkat T-cells (not shown) point to the widespread existence of reggie tetramers. The existence of the ∼200 kDa band was corroborated using other cross-linkers covering different distance ranges and amino acid specificities, such as DMS or MBS (Supplementary Figures 1B and 1C). The ∼200 kDa bands were not observed upon treatment with DST, a DSS analogue with a shorter distance range (6.4 Å), indicating that these bands represent the covalent binding of reggie proteins by their exposed lysine residues at a specific distance (results not shown).
Figure 2. Endogenous reggie-1 and -2 form tetramers in N2a cells after DSS-based cross-linking.
N2a cells were subjected to chemical cross-linking using 1 mM DSS for the times indicated above each lane. Reggie-1 (left-hand panel) and -2 (right-hand panel) -specific mAbs were used for detection and reveal ∼47 kDa reggie-1 and -2. Reggie tetramers migrate as ∼200 kDa bands (arrowheads). Molecular masses are indicated in kDa. WB, Western blot.
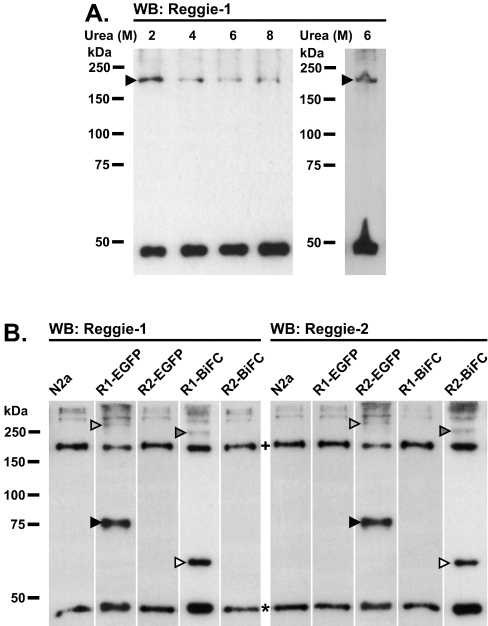
To rule out that the complexes observed are induced by experimental cross-linking, we tested for the formation of stable reggie oligomers in the absence of chemical cross-linkers and under increasing urea concentrations. Therefore urea-treated N2a cell extracts were analysed by Western blotting using a specific α-reggie-1 mAb (Figure 3A). Notably, oligomers of ∼200 kDa containing reggie-1 were observed even at 8 M urea, indicating that putative reggie tetramers are very stable. Furthermore, co-migration of the cross-linked and urea-resistant reggie complexes at ∼200 kDa strongly suggests that these complexes are identical. The oligomers detected here represent reggie tetramers and not aberrantly migrating dimers or trimers, since 6 M urea cell extracts revealed the same ∼200 kDa band when run under strong denaturing conditions (SDS/PAGE supplemented with 6 M urea; Figure 3A). Hence, endogenous reggies exist in a monomer–tetramer equilibrium.
Figure 3. Urea treatments and homotetramerization of EGFP– and BiFC–reggie-1 and -2 constructs in N2a cells.
(A) N2a cells were lysed using the concentration of urea (M) indicated above each lane. Endogenous reggie-1 tetramers migrate as ∼200 kDa bands (arrowheads) in normal SDS/PAGE (left-hand panel) and in strong denaturing 6 M urea SDS/PAGE (right-hand panel). (B) Transiently transfected N2a cells expressing EGFP- or BiFC-fusion proteins indicated above each lane and non-transfected cells (N2a) were cross-linked using 1 mM DSS for 5 min after 20 h of transfection. Reggie-1 (left-hand panel) and -2 (right-hand panel) -specific mAbs used for detection reveal endogenous reggie monomers of ∼47 kDa (*), as well as the ∼60 and ∼75 kDa monomers of reggie–BiFC and reggie–EGFP fusion proteins, respectively (open and closed arrowheads respectively). Tetramers of endogenous reggie proteins of ∼200 kDa are also evident (+). Homotetramers of reggie–BiFC and reggie–EGFP migrate as ∼240 and ∼300 kDa bands respectively (dark and light grey arrowheads respectively). Molecular masses are indicated in kDa. WB, Western blot.
To gain insight into the stoichiometry and the exact composition of reggie oligomers, we performed cross-linking experiments on exogenously expressed reggie-1 and -2. We applied DSS to N2a cells transiently transfected with reggie–EGFP or –BiFC vectors, resulting in the expression of fusion proteins of easily discernable sizes. Besides the endogenous reggie monomer and tetramer bands described above, we observed additional bands of ∼75 and ∼300 kDa, corresponding to the expected sizes of the monomeric and homotetrameric reggie-1– or -2–EGFP fusion proteins (Figure 3B). Accordingly, DSS-treated cells expressing reggie-1– or -2–BiFC fusion proteins yielded additional bands of ∼60 and ∼240 kDa (Figure 3B), corroborating the existence of reggie-1 or -2 as monomers and homotetramers. To confirm that these additional bands contained reggie–EGFP and –BiFC fusion proteins, we reprobed the samples with a pAb against all fusion proteins, and we obtained the same bands (Supplementary Figure 2 at http://www.BiochemJ.org/bj/403/bj4030313add.htm). These results strengthen the notion that the ∼200 kDa bands observed upon DSS cross-linking or urea denaturation are indeed reggie tetramers.
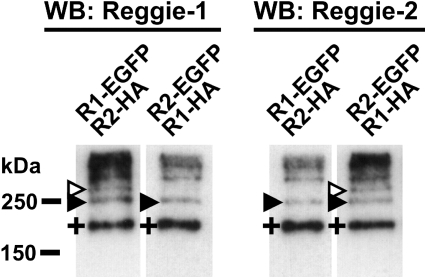
Although interactions between reggie-1 and -2 have been well studied [13,17,20], we did not observe the formation of reggie heterotetramers between endogenous and EGFP– or BiFC–reggies. The most likely explanation for this is that homotetramerization of reggie fusion proteins is favoured over hetero-tetramerization with endogenous reggies because of their faster de novo synthesis. To clarify this further, we co-transfected N2a cells with vectors encoding differentially tagged reggie-1 and -2. Interestingly, DSS treatment of cells co-expressing reggie-1–EGFP (∼75 kDa) and reggie-2–HA (∼50 kDa) or reggie-1–HA (∼50 kDa) and reggie-2–EGFP (∼75 kDa) revealed the presence of an additional band of ∼250 kDa (Figure 4), corresponding to the expected sizes of a heterotetramer formed by two molecules of each reggie. We also observed the reggie–EGFP homotetramers bands described above (∼300 kDa; Figure 3B). Reggie heterotetramers of ∼250 kDa were observed using cells transfected with different amounts of vector combinations (results not shown), and by reprobing the blots with mAbs against GFP and the HA epitope respectively (Supplementary Figure 3 at http://www.BiochemJ.org/bj/403/bj4030313add.htm). As expected, homotetramers of HA-tagged reggie proteins migrated as ∼200 kDa bands, similarly to the endogenous reggie tetramers (Supplementary Figure 3). Taken together, our results show that reggie-1 and -2 build homo- and hetero-tetramers in living cells, and that heterotetramers are formed by two molecules of reggie-1 and -2.
Figure 4. Homo- and hetero-tetramerization of HA-tagged and EGFP-fused reggie-1 and -2 in N2a cells.
N2a cells were transiently co-transfected with the constructs indicated above each lane. Cells were cross-linked at 20 h post transfection using 1 mM DSS for 5 min. Reggie-1 (left-hand panel) and -2 (right-hand panel) -specific mAbs revealed endogenous reggie tetramers of ∼200 kDa (+). Reggie–EGFP homotetramers migrate as ∼300 kDa (open arrowheads), and EGFP- and HA-containing heterotetramers migrate as ∼250 kDa bands (closed arrowheads). Molecular masses are indicated in kDa. WB, Western blot.
Mapping of reggie-oligomerization domains
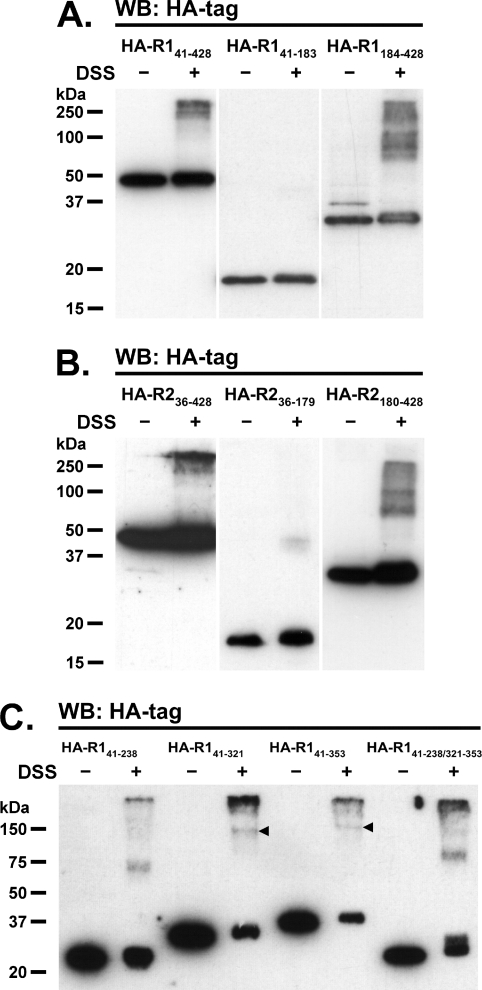
To establish whether reggie tetramerization depends on the presence of the SPFH or flotillin domains, cross-linking experiments were performed on N2a cells expressing three distinct truncated forms of HA-tagged reggies (Figure 1). These included one construct lacking the N-terminus implicated in membrane-association (HA–R141–428 and HA–R236–428), and the other two additionally lacking the flotillin (HA–R141–183 and HA–R236–179) or SPFH domains (HA–R1184–428 and HA–R2180–428). As shown in Figures 5(A) and 5(B), HA-tagged reggies lacking only the N-terminal end are seen as monomers, tetramers and higher-order oligomers, similar to full-length reggies (Figures 2 and 3). However, oligomerization of these constructs seems to be weaker than their full-length counterparts, suggesting that formation of reggie tetramers is facilitated by association to cell membranes. In addition, constructs lacking the SPFH domain are able to form comparable multimers, although tetramers are no longer the dominant forms (Figures 5A and 5B). On the other hand, HA–reggie-1 and -2 constructs lacking the flotillin domain do not form oligomers, but instead appear as monomers (Figures 5A and 5B). Thus our results indicate that the C-terminal half of reggies is necessary and sufficient for oligomerization, whereas the SPFH domain is dispensable.
Figure 5. The C-terminal halves of reggie-1 and -2 are responsible for oligomerization.
Transiently transfected N2a cells expressing HA-tagged reggie-1 (A and C) and reggie-2 (B) constructs indicated above the lanes were either cross-linked using 1 mM DSS for 5 min (+) or left untreated (−). (A and B) Samples blotted using mAb against the HA tag reveal bands indicative of oligomers of constructs including the C-terminal domain of reggie-1 (HA–R141–428 and HA–R1184–428) and reggie-2 (HA–R236–428 and HA–R2180–428). Constructs including only the SPFH domain of both reggies (HA–R141–183 and HA–R236–179) fail to form oligomers under these conditions, indicating that the C-termini are necessary and sufficient for oligomerization. (C) Arrowheads show the corresponding homotetramers of reggie-1 species including the second predicted coiled-coil stretch (HA–R141–321 and HA–R141–353). Reggie-1 constructs lacking this stretch (HA–R141–238 and HA–R141–238/321–353) fail to form homotetramers, although some smaller oligomers are also observed. Molecular masses are indicated in kDa. WB, Western blot.
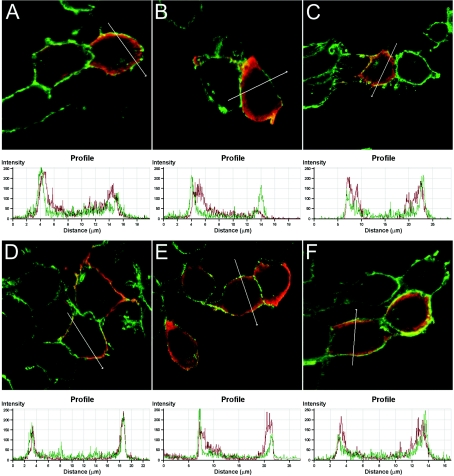
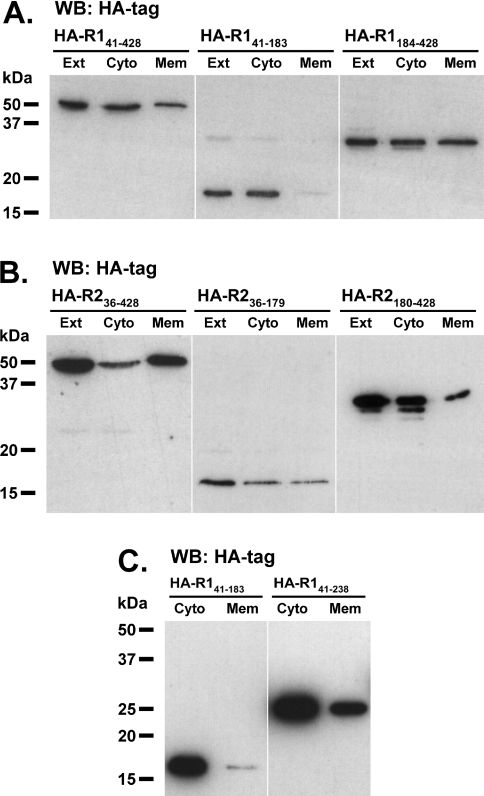
It has been established previously that acylation and hydrophobic regions determine membrane and raft association of reggie-1 and -2 [20,25]. Since our constructs containing only the C-terminal half are able to form oligomers, we tested whether they can indirectly associate to the plasma membrane via homo- and hetero-interactions with endogenous reggies. We examined the membrane association of the HA-tagged reggie constructs described above (Figure 1). Indeed, HA–reggie constructs lacking the residues for acylation and the hydrophobic domains (HA–R141–428 and HA–R236–428) can partially associate with the plasma membrane, as seen by co-localization with the cell-surface protein NCAM (Figures 6A and 6D) and their presence in membrane fractions (Figures 7A and 7B). Therefore endogenous reggies must be able to interact with these constructs and recruit them to the plasma membrane. Consequently, HA–reggie constructs including only the C-terminal half (HA–R1184–428 and HA–R2180–428) are partially associated with the plasma membrane (Figures 6C, 6F, 7A and 7B). In the case of reggie-2, it is also possible that a second hydrophobic stretch predicted in the SPFH domain (residues 134–151; Figure 1) interacts directly with the cell membrane. Accordingly, the HA–R236–179 construct containing only the reggie-2 SPFH domain co-localizes with NCAM (Figure 6E) and is present in the membrane fraction (Figure 7B); this is not the case for the corresponding reggie-1 construct (Figures 6B and 7A). Hence, our data show that the C-terminal halves mediate reggie oligomerization, and that the SPFH domains of reggie-1 and -2 differ in their ability to associate with the plasma membrane.
Figure 6. Membrane localization of HA-tagged truncated reggie constructs.
Overlay of confocal fluorescence images corresponding to staining of NCAM (green) and HA-tagged reggie constructs (red), as well as the corresponding distribution profiles. N2a cells were transiently transfected with vectors encoding HA–R141–428 (A), HA–R141–183 (B), HA–R1184–428 (C), HA–R236–428 (D), HA–R236–179 (E) and HA–R2180–428 (F) 20 h before analysis using Abs specific for NCAM and the HA tag. Note that reggie-1 constructs including the C-terminal halves (HA–R141–428 and HA–R1184–428) co-localized with NCAM, while in a construct including the SPFH domain only (HA–R141–183) failed to co-localize with NCAM at the plasma membrane as indicated in the profile graphic. All reggie-2 constructs tested (HA–R236–428, HA–R236–179 and HA–R2180–428) differentially co-localized with NCAM at the plasma membrane.
Figure 7. Cell fractionation of N2a cells expressing HA–reggie constructs.
Transiently transfected N2a cells expressing HA-tagged reggie-1 (A and C) and reggie-2 (B) constructs indicated above the lanes were fractionated by high-speed centrifugation. Aliquots of the fractions corresponding to the whole-cell extracts (Ext), cytosol (Cyto) and membrane (Mem) were analysed by Western blot (WB) using a mAb against the HA tag. (A) Reggie-1 constructs including the C-terminal halves (HA–R141–428 and HA–R1184–428) are present in both cytosolic and the membrane fractions, whereas a construct including the SPFH domain only (HA–R141–183) is not significantly present in the membrane fraction. (B) All of the reggie-2 constructs tested (HA–R236–428, HA–R236–179 and HA–R2180–428) were significantly present in both cytosolic and membrane fractions. Some small molecular mass bands were observed for HA–R1184–428 and HA–R2180–428 constructs, which might represent degradation products after the fractionation process. (C) The reggie-1 construct including the first predicted coiled-coil stretch (HA–R141–238) was significantly present in both cytosolic and membrane fractions, whereas the control construct (HA–R141–183) was largely cytosolic. Molecular masses are indicated in kDa.
Different coiled-coil regions in the C-terminal flotillin domain of reggie-1 are involved in tetramerization
Since our experiments have shown that the C-terminal half is required for reggie tetramerization, we mapped this property more precisely. As mentioned above, the C-terminal half of reggies contains a so-called ‘flotillin’ domain, which is predicted to form three adjacent coiled-coils (here termed coiled-coil 1, 2 and 3; Figure 1). To assess the specific contribution of these stretches towards tetramerization, we analysed N2a cells expressing HA-tagged reggie constructs lacking the N- and C-terminal ends and one or more of the predicted coiled-coil stretches (Figure 1). Because reggie-1 proteins are more stably expressed than their reggie-2 counterparts (see below), we focused our analyses on the coiled-coil regions of reggie-1. As expected, the reggie-1 construct containing all three coiled-coil stretches (∼37 kDa; HA–R141–353) formed tetramers upon DSS cross-linking (∼150 kDa; Figure 5C), indicating that the entire coiled-coil region is necessary and sufficient for tetramerization. Since a construct lacking coiled-coil 3 (∼33 kDa; HA–R141–321) retained the ability to form tetramers (∼130 kDa; Figure 5C), we concluded that tetramerization does not require the presence of this stretch. Notably, the construct lacking only coiled-coil 2 (∼27 kDa; HA–R141–238/321–353) did not form tetramers (Figure 5C), revealing that this stretch is essential for tetramerization. Interestingly, this construct displayed a residual ability to oligomerize, since a band corresponding in size to a trimeric species (∼80 kDa) became visible (Figure 5C). Such a phenomenon was not observed for the construct lacking only coiled-coil 3 (HA–R141–321), indicating that this additional ability to oligomerize must be encoded in coiled-coil 1. Accordingly, the construct containing only coiled-coil 1 (∼24 kDa; HA–R141–238) assembles into oligomeric species corresponding in size to trimers (∼70 kDa). To confirm that the coiled-coil 1 construct has residual oligomerization abilities, we tested whether its interaction with endogenous reggies would result in its association with cellular membranes. As shown above (Figure 7A), the control construct lacking all coiled-coil stretches (HA–R141–183) is not significantly present in the membrane fraction, whereas the construct containing only coiled-coil 1 (HA–R141–238) clearly associates with the cell membrane (Figure 7C). Taken together, these results show that the ability of reggie-1 to tetramerize is conferred by the presence of coiled-coil 2 and, partially, of coiled-coil 1.
Reggie-2 undergoes proteasomal degradation in the absence of reggie-1
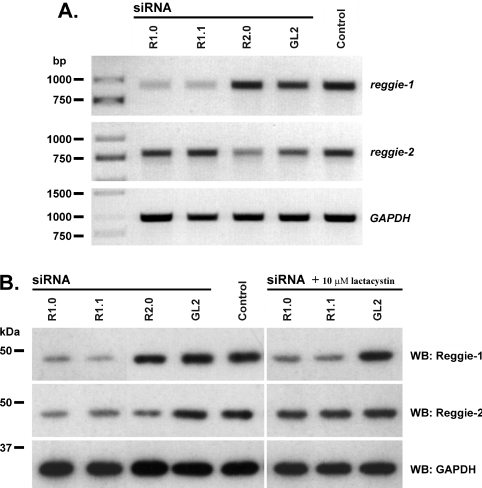
It has been reported in Drosophila mutants that the protein stability of reggie-2 requires the expression of reggie-1 [18]. Similarly, we observed that our reggie-2 constructs were not as efficiently expressed in N2a cells as our reggie-1 constructs. To clarify this phenomenon, we analysed the stability of reggie-2 upon siRNA-mediated gene silencing of reggie-1 in N2a cells. When cells were transfected with reggie-1 or reggie-2 siRNAs, the corresponding mRNA (Figure 8A) and protein levels (Figure 8B) are significantly reduced 2 days after transfection; control siRNAs produce no effect. Interestingly, transfection with reggie-1 siRNAs leads to depletion of both reggie-1 and -2 proteins (Figure 8B). RT–PCR analysis showed that reggie-1 siRNAs induce degradation of reggie-1 mRNA, whereas the reggie-2 transcripts were not affected (Figure 8A). Therefore the regulation of reggie-2 by reggie-1 occurs at the protein level. On the other hand, cells treated with reggie-2 siRNA lost the corresponding mRNA (Figure 8A) and protein (Figure 8B), but reggie-1 mRNA and protein remained unaffected. We observed identical effects using the same siRNAs in HeLa cells (results not shown).
Figure 8. siRNA-mediated knockdown of reggie-1 induces proteasomal degradation of the reggie-2 protein.
N2a cells were transfected with two different siRNAs against reggie-1 (R1.0 and R1.1), one siRNA against reggie-2 (R2.0) and a non-specific control siRNA against firefly luciferase (GL2). Additional control cells treated with the transfection reagent Lipofectamine™ 2000 alone were examined (Control). (A) RT–PCR analyses of total mRNA at 48 h post-transfection show that reggie-1 and -2 mRNAs were specifically down-regulated by the corresponding siRNAs. GAPDH was used as control for the amount of cDNA. (B) Protein analyses of siRNA-mediated knockdown by Western blotting show that both siRNAs against reggie-1 (R1.0 and R1.1) caused depletion of reggie-1 as well as reggie-2 protein, whereas siRNA against reggie-2 (R2.0) down-regulates reggie-2 protein only. Incubation with 10 μM lactacystin blocked this effect, indicating that reggie-2 underwent proteasomal degradation in the absence of reggie-1. Specific mAb against GAPDH was used as loading control. Molecular masses are indicated in kDa. WB, Western blot.
Since the proteasome system is a central component for the degradation of intracellular proteins [26], we studied its participation in the degradation of reggie-2 after reggie-1 depletion. Notably, down-regulation of reggie-2 protein by reggie-1 siRNA was no longer observed when N2a cells were incubated with 10 μM lactacystin, a 20 S proteasome inhibitor, indicating that reggie-2 degradation occurs via the proteasome system (Figure 8B). These results reveal that the presence of reggie-1 is necessary for the stability of reggie-2, which otherwise undergoes proteasomal degradation.
DISCUSSION
Altogether, we have demonstrated that the organization of reggie microdomains at the plasma membrane requires reggie protein interactions and involves the formation of reggie homo- and hetero-tetramers. We molecularly dissected the domains involved in the oligomerization process, and identified a coiled-coil stretch required for tetramerization. In addition, we showed that reggie-2 underwent proteasomal degradation in the absence of reggie-1.
Reggie proteins form homo- and hetero-tetramers in living cells
To date, the little evidence available on oligomerization of reggie proteins is vague, contradictory and relies solely on sucrose-gradient centrifugation methods. For instance, in HeLa cells, reggie-1 localizes in fractions of 70–150 kDa [20], whereas, in human erythrocytes, both reggie proteins mainly localize in fractions of 300–400 kDa [21]. On the other hand, molecular interactions between reggie-1 and reggie-2 have been examined using yeast two-hybrid assays, co-immunoprecipitation and co-localization [13,17,20]; however, no evidence for the formation of reggie hetero-oligomers has been provided so far. We assessed the oligomerization state of reggie proteins using chemical cross-linking, which is a powerful method to examine protein–protein interactions in living cells [27,28]. As reggies contain many lysine residues (34 and 30 in reggie-1 and -2 respectively), we used cell-permeable lysine-specific cross-linkers. Different homo- and hetero-bifunctional cross-linkers (DSS, MBS and DMS) with spacer arms between 12 and 9.9 Å consistently demonstrate that endogenous reggies are present as monomers and tetramers in N2a cells as well as in other cell types (PC12, HeLa and Jurkat T-cells). We did not observe reggie tetramers using DST, a lysine-specific homo-bifunctional cross-linker with a short spacer arm (6.4 Å), confirming the specificity of the cross-linking approach. We corroborated the existence of native reggie tetramers by treating N2a cells with different concentrations of urea in the absence of cross-linkers. Interestingly, urea treatments did not reveal intermediate species such as dimers and trimers of full-length reggies, indicating that reggies exist in a monomer–tetramer equilibrium, as it has been reported for other coiled-coil-containing proteins such as dynamin [29,30]. In fact, the reggie oligomers observed are formed by very stable, high-affinity protein–protein interactions, which are resistant to 8 M urea. Moreover, the identical migration patterns of reggie complexes in strong denaturing urea gels support our view that cross-linked reggie oligomers represent tetramers and not aberrantly migrating dimers or trimers, as has been shown for SDS-resistant SNARE (soluble N-ethylmaleimide-sensitive fusion protein-attachment protein receptor) complexes [31]. Since our current structural model for reggie monomers suggests that the C-terminal flotillin domain of both proteins is strongly stabilized via intramolecular interactions [8], we speculate that the only thermodynamically stable reggie-oligomerization state is the tetramer (possibly formed by a four-stranded parallel coiled-coil rod). Thus intermolecular interactions between the coiled-coil regions might drive the monomer–tetramer equilibrium, but not the formation of dimers or trimers. Intriguingly, we did not observe reggie intermediate species even at very low concentrations of homo- or hetero-bifunctional cross-linkers. Given the stochastic nature of chemical cross-linking, one might expect to find such intermediate species at low concentrations of cross-linker. However, the large number of lysine residues in the oligomerization region of reggie-1 and -2 might require even smaller titration intervals than the ones we used to obtain dimers and trimers. The confirmation that reggie-1 and -2 form homo- and hetero-tetramers was obtained by exogenous expression in N2a cells of EGFP- and BiFC-constructs as well as HA-tagged reggie proteins. We observe that cytosolic (truncated) reggie constructs oligomerize less efficiently than full-length proteins, suggesting that membrane association might facilitate reggie tetramerization. Taken together, our results indicate that reggie proteins are only present in a monomer–tetramer equilibrium, and that reggie heterotetramers are built only by two molecules of each reggie under our experimental conditions; more data will be required to better understand this restricted stoichiometry. Thus the reggie microdomains of living cells are formed by homo- and hetero-tetramers of reggie-1 and -2.
The C-terminal halves are responsible for tetramerization of reggie-1 and -2
Previous findings by Neumann-Giesen et al. [20] had suggested that the C-terminal half of reggie-1 is important for homo- and hetero-interactions with reggie-2. Using DSS-based cross-linking experiments, we have now established that the C-terminal halves of reggie-1 and -2 are necessary and sufficient for tetramerization, whereas the SPFH domains are not required for oligomerization. Moreover, we show that, within the flotillin domain of reggie-1 (residues 239–321), coiled-coil 2 is necessary for tetramerization, and coiled-coil 1 (residues 184–238) also contributes partially to oligomerization. Since expression of reggie constructs containing only C-terminal halves produces a wide continuum of multimers, it is possible that the SPFH domains restrict the oligomerization process specifically towards the formation of tetramers. In agreement with this view, expression of constructs containing the SPFH domain and either the C-terminal half or the flotillin domain indeed forms only tetramers. Because of the sequence similarity between reggie-1 and -2 at the C-terminal half [8], we expect that coiled-coil 2 of reggie-2 might likewise be involved in tetramerization, a point which requires further analysis. Since our data clearly demonstrate that the C-terminal domains of reggies mediate oligomerization, we propose the term ‘oligomerization domain’ instead of flotillin domain, which was based on sequence peculiarities and not on functional properties.
It has been reported that acylations and hydrophobic regions present in reggie proteins are required for their membrane association [20,25]. However, it is possible that protein interactions mediated by other reggie domains might strengthen its association with cellular membranes. Accordingly, we observed that reggie-1 and -2 constructs lacking the N-terminal end (acylation sites and the first hydrophobic stretch) still significantly associate with the plasma membrane. Similarly, constructs involving only the C-terminal oligomerization domains show significant association with cellular membranes. Our results suggest that the C-terminal ‘oligomerization’ domains interact with endogenous reggie proteins at the plasma membrane. In this case, overexpression of these regions might interfere with the normal function of reggie microdomains, acting as a dominant-negative. In agreement with this view, we have recently shown that overexpression of the C-terminal oligomerization domain of reggie-1 severely impairs the cytoskeleton rearrangement after T-cell activation [32]. Notably, membrane association of reggie-2 is stronger compared with reggie-1, probably due to the presence of a second hydrophobic stretch in the SPFH domain [33]. Indeed, a reggie-2 construct containing only the SPFH domain still associates with cellular membranes, whereas a similar construct of reggie-1, which does not possess any predicted second hydrophobic stretch, fails to associate. Recently, two putative CRAC (cholesterol recognition/interaction amino acid consensus) motifs have been predicted to support membrane association of reggie-1 [34]. However, these motifs might not be functionally significant, because our reggie-1 construct containing only the SPFH region does not associate with cellular membranes.
Reggie-2 needs the presence of reggie-1 to be stable at the protein level
Further evidence for the interaction between reggie-1 and reggie-2 is provided by our siRNA studies: down-regulation of reggie-1 causes the concomitant loss of reggie-2 protein, but not vice versa. This effect can be blocked using lactacystin, indicating that, in the absence of reggie-1, reggie-2 undergoes proteasomal degradation rather than non-specific blockage of translation. Since Drosophila reggie-1 is also required to maintain protein stability of reggie-2 [18], it appears that this type of protein regulation in reggie microdomains is highly conserved among metazoans. Interestingly, similar dependencies have been reported for other members of the SPFH family: prohibitin-1-null mutants in Saccharomyces cerevisiae lose prohibitin-2 protein and vice versa [35], and HflK and HflC appear to be interdependent as well [36], although data on the participation of the proteasome in these degradation processes remain elusive. It is likely that in all of these cases the stability of one of the proteins is closely related to the formation of hetero-oligomers. In the case of reggies, however, this phenomenon might be more complex, since reggie-1 and -2 can localize in different subcellular compartments in PC-3 cells during the cell cycle [37], and in cell types such as the mouse macrophage RAW 264.7 and fibroblast 3T3 (M. F. Langhorst, personal communication). Further studies are required to clarify this issue.
Our data indicate that reggie homo- and hetero-tetramers are the building blocks of larger multimeric clusters at the plasma membrane, in which reggie-2 requires reggie-1 for its stabilization. The ultimate size of reggie structures remains elusive, although from our early immunoelectron microscopy data, where the reggie microdomains were calculated roughly to be 0.05 to 0.1 μm [13], we estimate that several hundred reggie proteins contribute to form such clusters. Taken together, we have uncovered the molecular determinants for the formation and stabilization of reggie microdomains, which will be helpful for understanding the functions of reggies as scaffolding proteins involved in signalling across the plasma membrane.
Online data
Acknowledgments
We thank Matthias F. Langhorst for critical review of the manuscript pre-submission and Kay Diederichs for the constructive discussion about structural models. This work was supported by grants from the Ministerium Forschung, Wissenschaft und Kunst Baden-Württemberg (TSE programme), the Deutsche Forschungsgemeinschaft (SFB TR-11) and the Fonds der Chemischen Industrie.
References
- 1.Langhorst M. F., Reuter A., Stuermer C. A. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell. Mol. Life Sci. 2005;62:2228–2240. doi: 10.1007/s00018-005-5166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrow I. C., Parton R. G. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic. 2005;6:725–740. doi: 10.1111/j.1600-0854.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 3.Glebov O. O., Bright N. A., Nichols B. J. Flotillin-1 defines a clathrin-independent endocytic pathway in mammalian cells. Nat. Cell. Biol. 2006;8:46–54. doi: 10.1038/ncb1342. [DOI] [PubMed] [Google Scholar]
- 4.Schulte T., Paschke K. A., Laessing U., Lottspeich F., Stuermer C. A. Reggie-1 and reggie-2, two cell surface proteins expressed by retinal ganglion cells during axon regeneration. Development. 1997;124:577–587. doi: 10.1242/dev.124.2.577. [DOI] [PubMed] [Google Scholar]
- 5.Lang D. M., Lommel S., Jung M., Ankerhold R., Petrausch B., Laessing U., Wiechers M. F., Plattner H., Stuermer C. A. Identification of reggie-1 and reggie-2 as plasmamembrane-associated proteins which cocluster with activated GPI-anchored cell adhesion molecules in non-caveolar micropatches in neurons. J. Neurobiol. 1998;37:502–523. doi: 10.1002/(sici)1097-4695(199812)37:4<502::aid-neu2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 6.Bickel P. E., Scherer P. E., Schnitzer J. E., Oh P., Lisanti M. P., Lodish H. F. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J. Biol. Chem. 1997;272:13793–13802. doi: 10.1074/jbc.272.21.13793. [DOI] [PubMed] [Google Scholar]
- 7.Malaga-Trillo E., Laessing U., Lang D. M., Meyer A., Stuermer C. A. Evolution of duplicated reggie genes in zebrafish and goldfish. J. Mol. Evol. 2002;54:235–245. doi: 10.1007/s00239-001-0005-1. [DOI] [PubMed] [Google Scholar]
- 8.Rivera-Milla E., Stuermer C. A., Malaga-Trillo E. Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cell. Mol. Life Sci. 2006;63:343–357. doi: 10.1007/s00018-005-5434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tavernarakis N., Driscoll M., Kyrpides N. C. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 1999;24:425–427. doi: 10.1016/s0968-0004(99)01467-x. [DOI] [PubMed] [Google Scholar]
- 10.Schroeder W. T., Stewart-Galetka S., Mandavilli S., Parry D. A., Goldsmith L., Duvic M. Cloning and characterization of a novel epidermal cell surface antigen (ESA) J. Biol. Chem. 1994;269:19983–19991. [PubMed] [Google Scholar]
- 11.Baumann C. A., Ribon V., Kanzaki M., Thurmond D. C., Mora S., Shigematsu S., Bickel P. E., Pessin J. E., Saltiel A. R. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature. 2000;407:202–207. doi: 10.1038/35025089. [DOI] [PubMed] [Google Scholar]
- 12.Dermine J. F., Duclos S., Garin J., St-Louis F., Rea S., Parton R. G., Desjardins M. Flotillin-1-enriched lipid raft domains accumulate on maturing phagosomes. J. Biol. Chem. 2001;276:18507–18512. doi: 10.1074/jbc.M101113200. [DOI] [PubMed] [Google Scholar]
- 13.Stuermer C. A, Lang D. M., Kirsch F., Wiechers M., Deininger S. O., Plattner H. Glycosylphosphatidyl inositol-anchored proteins and fyn kinase assemble in noncaveolar plasma membrane microdomains defined by reggie-1 and -2. Mol. Biol. Cell. 2001;12:3031–3045. doi: 10.1091/mbc.12.10.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajendran L., Masilamani M., Solomon S., Tikkanen R., Stuermer C. A., Plattner H., Illges H. Asymmetric localization of flotillins/reggies in preassembled platforms confers inherent polarity to hematopoietic cells. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8241–8246. doi: 10.1073/pnas.1331629100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slaughter N., Laux I., Tu X., Whitelegge J., Zhu X., Effros R., Bickel P., Nel A. The flotillins are integral membrane proteins in lipid rafts that contain TCR-associated signaling components: implications for T-cell activation. Clin. Immunol. 2003;108:138–151. doi: 10.1016/s1521-6616(03)00097-4. [DOI] [PubMed] [Google Scholar]
- 16.Tu X., Huang A., Bae D., Slaughter N., Whitelegge J., Crother T., Bickel P. E., Nel A. Proteome analysis of lipid rafts in Jurkat cells characterizes a raft subset that is involved in NF-κB activation. J. Proteome Res. 2004;3:445–454. doi: 10.1021/pr0340779. [DOI] [PubMed] [Google Scholar]
- 17.Stuermer C. A., Langhorst M. F., Wiechers M. F., Legler D. F., Von Hanwehr S. H., Guse A. H., Plattner H. PrPc capping in T cells promotes its association with the lipid raft proteins reggie-1 and reggie-2 and leads to signal transduction. FASEB J. 2004;18:1731–1733. doi: 10.1096/fj.04-2150fje. [DOI] [PubMed] [Google Scholar]
- 18.Hoehne M., de Couet H. G., Stuermer C. A., Fischbach K. F. Loss- and gain-of-function analysis of the lipid raft proteins reggie/flotillin in Drosophila: they are posttranslationally regulated, and misexpression interferes with wing and eye development. Mol. Cell. Neurosci. 2005;30:326–338. doi: 10.1016/j.mcn.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Stuermer C. A., Plattner H. The ‘lipid raft’ microdomain proteins reggie-1 and reggie-2 (flotillins) are scaffolds for protein interaction and signalling. Biochem. Soc. Symp. 2005;72:109–118. doi: 10.1042/bss0720109. [DOI] [PubMed] [Google Scholar]
- 20.Neumann-Giesen C., Falkenbach B., Beicht P., Claasen S., Luers G., Stuermer C. A., Herzog V., Tikkanen R. Membrane and raft association of reggie-1/flotillin-2: role of myristoylation, palmitoylation and oligomerization and induction of filopodia by overexpression. Biochem. J. 2004;378:509–518. doi: 10.1042/BJ20031100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salzer U., Prohaska R. Stomatin, flotillin-1, and flotillin-2 are major integral proteins of erythrocyte lipid rafts. Blood. 2001;97:1141–1143. doi: 10.1182/blood.v97.4.1141. [DOI] [PubMed] [Google Scholar]
- 22.Hu C. D., Kerppola T. K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: a Laboratory Manual. 2nd Edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Fenteany G., Standaert R. F., Lane W. S., Choi S., Corey E. J., Schreiber S. L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 25.Morrow I. C., Rea S., Martin S., Prior I. A., Prohaska R., Hancock J. F., James D. E., Parton R. G. Flotillin-1/reggie-2 traffics to surface raft domains via a novel Golgi-independent pathway: identification of a novel membrane targeting domain and a role for palmitoylation. J. Biol. Chem. 2002;277:48834–48841. doi: 10.1074/jbc.M209082200. [DOI] [PubMed] [Google Scholar]
- 26.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin–proteasome system and onto human diseases and drug targeting. Cell Death Differ. 2005;12:1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 27.Fancy D. A. Elucidation of protein–protein interactions using chemical cross-linking or label transfer techniques. Curr. Opin. Chem. Biol. 2000;4:28–33. doi: 10.1016/s1367-5931(99)00047-2. [DOI] [PubMed] [Google Scholar]
- 28.Agou F., Ye F., Veron M. In vivo protein cross-linking. Methods Mol. Biol. 2004;261:427–442. doi: 10.1385/1-59259-762-9:427. [DOI] [PubMed] [Google Scholar]
- 29.Muhlberg A. B., Warnock D. E., Schmid S. L. Domain structure and intramolecular regulation of dynamin GTPase. EMBO J. 1997;16:6676–6683. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamoto P. M., Tripet B., Litowski J., Hodges R. S., Vallee R. B. Multiple distinct coiled-coils are involved in dynamin self-assembly. J. Biol. Chem. 1999;274:10277–10286. doi: 10.1074/jbc.274.15.10277. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi T., McMahon H., Yamasaki S., Binz T., Hata Y., Sudhof T. C., Niemann H. Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins on assembly. EMBO J. 1994;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langhorst M. F., Reuter A., Luxenhofer G., Boneberg E., Legler D. F., Plattner H., Stuermer C. A. Preformed reggie/flotillin caps: stable priming platforms for macrodomain assembly in T cells. FASEB J. 2006;20:711–713. doi: 10.1096/fj.05-4760fje. [DOI] [PubMed] [Google Scholar]
- 33.Liu J., Deyoung S. M., Zhang M., Dold L. H., Saltiel A. R. The stomatin/prohibitin/flotillin/HflK/C domain of flotillin-1 contains distinct sequences that direct plasma membrane localization and protein interactions in 3T3-L1 adipocytes. J. Biol. Chem. 2005;280:16125–16134. doi: 10.1074/jbc.M500940200. [DOI] [PubMed] [Google Scholar]
- 34.Roitbak T., Surviladze Z., Tikkanen R., Wandinger-Ness A. A polycystin multiprotein complex constitutes a cholesterol-containing signalling microdomain in human kidney epithelia. Biochem. J. 2005;392:29–38. doi: 10.1042/BJ20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berger K. H., Yaffe M. P. Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:4043–4052. doi: 10.1128/mcb.18.7.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nijtmans L. G., Artal S. M., Grivell L. A., Coates P. J. The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell. Mol. Life Sci. 2002;59:143–155. doi: 10.1007/s00018-002-8411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santamaria A., Castellanos E., Gomez V., Benedit P., Renau-Piqueras J., Morote J., Reventos J., Thomson T. M., Paciucci R. PTOV1 enables the nuclear translocation and mitogenic activity of flotillin-1, a major protein of lipid rafts. Mol. Cell. Biol. 2005;25:1900–1911. doi: 10.1128/MCB.25.5.1900-1911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.