Abstract
To generate an extensive set of subgenomic (sg) mRNAs, nidoviruses (arteriviruses and coronaviruses) use a mechanism of discontinuous transcription. During this process, mRNAs are generated that represent the genomic 5′ sequence, the so-called leader RNA, fused at specific positions to different 3′ regions of the genome. The fusion of the leader to the mRNA bodies occurs at a short, conserved sequence element, the transcription-regulating sequence (TRS), which precedes every transcription unit in the genome and is also present at the 3′ end of the leader sequence. Here, we have used site-directed mutagenesis of the infectious cDNA clone of the arterivirus equine arteritis virus to show that sg mRNA synthesis requires a base-pairing interaction between the leader TRS and the complement of a body TRS in the viral negative strand. Mutagenesis of the body TRS of equine arteritis virus RNA7 reduced sg RNA7 transcription severely or abolished it completely. Mutations in the leader TRS dramatically influenced the synthesis of all sg mRNAs. The construction of double mutants in which a mutant leader TRS was combined with the corresponding mutant RNA7 body TRS resulted in the specific restoration of mRNA7 synthesis. The analysis of the mRNA leader–body junctions of a number of mutants with partial transcriptional activity provided support for a mechanism of discontinuous minus-strand transcription that resembles similarity-assisted, copy-choice RNA recombination.
Transcriptional regulation is one of the key processes in the controlled expression of genetic information. In general, the transcription of eukaryotic DNA genomes is regulated in the nucleus, where it is subject to pre-, co-, and posttranscriptional control mechanisms (1, 2). In contrast, the transcription of eukaryotic positive-strand RNA [(+)RNA] virus genomes occurs in the cytoplasm and relies on the process of RNA-dependent RNA synthesis. In some cases, the genome is the only viral mRNA and gene expression is regulated solely at the (post-)translational level, primarily by the synthesis and controlled processing of precursor polyproteins. Alternatively, regulation may occur at the transcriptional level, either by segmentation of the genome or by the generation of one or multiple subgenomic (sg) mRNAs. The latter strategy usually involves the recognition of internal promoter sequences by the viral RNA-dependent RNA polymerase (RdRp) complex, a process that resembles the recognition of DNA promoters by DNA-dependent RNA polymerases (3, 4).
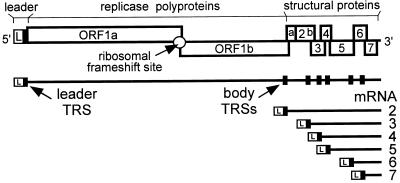
Nidoviruses (arteriviruses and coronaviruses) are mammalian (+)RNA viruses that appear to have evolved the use of sg mRNAs to regulate the expression of their polycistronic genome (5, 6). The nidovirus replicase is expressed from the genomic RNA as a polyprotein, but the structural proteins are translated from a set of six to eight sg mRNAs (Fig. 1). A key feature of these sg transcripts is the fact that their 5′ and 3′ terminal sequences are identical to those of the genome. This nested set structure results from a fusion of the sequence representing the genomic 5′ end (or “leader”) and sequences representing different 3′ regions of the genome, the so-called mRNA bodies. As a result, each of the genes in the 3′ terminal region of the genome is positioned at the 5′ end of one of the sg mRNAs.
Figure 1.
Schematic diagram of the EAV genome organization and expression. The regions of the genome specifying the leader (L) sequence, the two large replicase ORFs (ORFs 1a and 1b), and the structural genes are indicated. The nested set of seven EAV mRNAs (genome and sg mRNAs 2–6) is depicted below. The black boxes in the genomic RNA indicate the position of leader and body TRSs.
The leader sequences of arteriviruses (156–221 nt) and coronaviruses (65–98 nt) are connected to the mRNA bodies by a short, conserved sequence. In the nidovirus genome, this transcription-regulating sequence (TRS) is present both at the 3′ end of the leader sequence and at the 5′ end of the mRNA bodies. Early studies indicated that leader–body joining occurs cotranscriptionally via a unique, discontinuous transcription mechanism (7–10) in which the (+) leader TRS and the body TRS complement in the viral negative (−) strand might interact directly. The initial transcription model proposed this interaction to be part of a mechanism in which leader transcripts are used to prime the transcription of the sg mRNA bodies from a genome-length (−) strand template. However, subsequently, complementary strands of the sg mRNAs were detected in infected cells (11–13), and these were implicated in sg mRNA synthesis (14–16).
Base pairing between the (+) leader TRS and the (−) body TRSs always has been implicit in all models of nidovirus transcription (8–10, 11, 13, 16). However, the role of the TRS during discontinuous transcription could never be tested experimentally. The recent development of an infectious cDNA clone (17) for the arterivirus equine arteritis virus (EAV) allowed, for the first time, the analysis of the interaction between nidovirus leader and body TRSs by means of reverse genetics. Thus, these EAV TRSs, which consist of the conserved pentanucleotide sequence 5′-UCAAC-3′ (13, 18), were subjected to site-directed mutagenesis. Using this approach, we were able to show that EAV discontinuous mRNA synthesis is governed by a direct base-pairing interaction between the (+) leader TRS and (−) body TRSs in the viral (−) strand. Using a number of TRS mutants with reduced transcriptional activity, evidence was obtained showing that the TRS sequence at the leader–body junction of the sg mRNA is derived exclusively from the body TRS. This finding supports the idea that sg mRNAs are generated by a mechanism of discontinuous (−) strand synthesis, which is reminiscent of the process of copy-choice RNA recombination.
Methods
Mutagenesis of Full-Length cDNA Clone pEAV030.
Nucleotide substitutions in the leader TRS (nucleotides 207–211 of the EAV genome; GenBank accession no. Y07862) and the RNA7 body TRS (nucleotides 12,252–12,256) were engineered in shuttle vectors by PCR mutagenesis as described before (17). Fully sequenced restriction fragments containing the mutations were introduced into EAV full-length cDNA clone pEAV030HNB, a derivative of clone pEAV030H (17) containing engineered NcoI (nucleotides 223–228) and BspEI (nucleotides 12,228–12,233) restriction sites.
RNA Transfection.
Full-length transcripts (12.8 kb) were generated in vitro from XhoI-linearized plasmid DNA as described before (17). Baby hamster kidney cells (BHK-21; ATCC CCL10) were used for all transfection experiments by using the previously described protocol (17).
Immunofluorescence Assays (IFAs).
Transfected BHK-21 cells were seeded on coverslips and fixed with paraformaldehyde at 12 h posttransfection (19). The immunofluorescence double staining for EAV nonstructural protein 2 (nsp2) (20) and the large glycoprotein (GL) [mAb 6D10; (21)] has been described before (17). The double labeling for nsp2 and the EAV nucleocapsid protein (N) was carried out by using an anti-nsp2 rabbit serum and an anti-N mouse mAb (mAb 3E2; ref. 22). As secondary antibodies, a Cy3-conjugated donkey anti-rabbit IgG antibody and a FITC-conjugated donkey anti-mouse IgG antibody were used.
RNA Analysis.
At 12 h posttransfection, total intracellular (i.c.) RNA was isolated from BHK-21 cells transfected with transcripts from wild-type or mutant (Table 1) EAV full-length cDNA clones (17, 23). RNAs were separated in denaturing 1.5% agarose gels and hybridized (18) to a radiolabeled oligonucleotide probe complementary to the 3′ end of the genome (nucleotides 12,680–12,704), which recognizes all viral mRNAs (Fig. 1).
Table 1.
Overview of EAV full-length cDNA clone mutants with nucleotide substitutions in the leader TRS (nucleotides 207–211), the RNA7 body TRS (nucleotides 12,252–12,256), or both
| Construct | Leader TRS | RNA7 body TRS | sg mRNA7 leader–body junction |
|---|---|---|---|
| Wild type | 5′-UCAAC-3′ | 5′-UCAAC-3′ | 5′-UCAAC-3′ |
| L1 | UGAAC | UCAAC | UCAAC |
| B1 | UCAAC | UGAAC | UGAAC |
| LB1 | UGAAC | UGAAC | UGAAC |
| L2 | UCAAG | UCAAC | UCAAC |
| B2 | UCAAC | UCAAG | UCAAG |
| LB2 | UCAAG | UCAAG | UCAAG |
| L3 | UGAAG | UCAAC | ND |
| B3 | UCAAC | UGAAG | ND |
| LB3 | UGAAG | UGAAG | UGAAG |
| L4 | AGUUG | UCAAC | ND |
| B4 | UCAAC | AGUUG | ND |
| LB4 | AGUUG | AGUUG | AGUUG |
Mutations are indicated in bold typeface and are underlined. The right column shows the sequence at the mRNA7 leader–body junction, as determined by direct sequence analysis of mRNA7-specific RT-PCR products (see also Fig. 4). ND, not determined.
The sequence at the mRNA7 leader–body junction was determined by using reverse transcription–PCR (RT-PCR) and dideoxynucleotide sequencing, essentially as described previously (23). First, cDNA was generated by using a primer complementary to nucleotides 12,680–12,704 of the EAV genome and Moloney murine leukemia virus reverse transcriptase. Subsequently, a PCR was carried out by using a primer complementary to genome position 12,623–12,646 (in the mRNA7 body) and a primer corresponding to genome position 63–89 (leader). The PCR consisted of 25 cycles, each composed of 1-min denaturation at 95°C, 1-min annealing at 58°C, and 1-min extension at 72°C, after which samples were incubated at 72°C for an additional 10 min.
A similar RT-PCR was used to determine the sequences at the mRNA3 and 3.1 junction sites. In this case, the RT primer was complementary to nucleotides 10,307–10,321 and the antisense PCR primer was complementary to nucleotides 10,301–10,318. The sense PCR primer and PCR protocol were identical to those for the RNA7-specific RT-PCR, with the exception that the annealing temperature was 45°C instead of 58°C.
RNA Structure Predictions.
The structure of the 5′ terminal region of the genomes of EAV, porcine reproductive and respiratory syndrome virus, lactate-dehydrogenase elevating virus, and simian hemorrhagic fever virus (GenBank accession nos. Y07862, M96262, U15146, and L39091) were predicted by using a previously described genetic algorithm implemented in the program star (24, 25).
Results
TRS Mutagenesis Severely Affects EAV Structural Protein Expression.
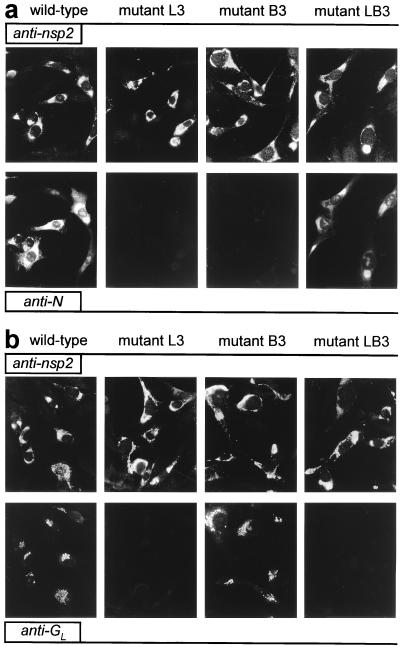
To analyze the role of the TRS in EAV sg mRNA synthesis, mutations were introduced into the full-length cDNA clone (Table 1), either in the leader TRS (L series mutants), in the RNA7 body TRS (B series mutants), or in both (LB series mutants). The RNA7 TRS was chosen because RNA7 is the most abundant sg mRNA produced by EAV. EAV replication and transcription in transfected cells initially was analyzed indirectly by using IFAs. As described before (17), a double staining with a rabbit antiserum recognizing one of the EAV replicase subunits and a mouse mAb directed against one of the viral structural proteins can be used to monitor genome replication and sg mRNA transcription, respectively. Cells were labeled for replicase cleavage product nsp2 and for either the ORF7-encoded N protein (Fig. 2a) or the ORF5-encoded glycoprotein GL (Fig. 2b), which are expressed from mRNA7 and mRNA5, respectively.
Figure 2.
Immunofluorescence analysis of BHK-21 cells transfected with wild-type EAV RNA or with mutants L3, B3, or LB3. (a) At 12 h posttransfection, cells were fixed and double-stained for nsp2 (Upper) and N (Lower) to test for genome replication and mRNA7 synthesis, respectively. (b) Double labeling for nsp2 (Upper) and GL (Lower) to monitor genome replication and mRNA5 transcription, respectively.
As an example, Fig. 2 shows the results obtained for one set of mutants (L3, B3, and LB3) in which the TRS was changed from 5′-UCAAC-3′ to 5′-UGAAG-3′. For all mutants, the nsp2 signal was identical to that of the wild-type control, indicating that they replicated their genome efficiently. However, striking effects at the level of sg mRNA transcription were revealed by the labeling for the structural proteins GL and N. Leader TRS mutant L3 appeared to be unable to generate mRNA5 and mRNA7, because GL and N synthesis could not be detected. Mutant B3, in which the mRNA7 body TRS has been changed, did not express N whereas it still expressed wild-type levels of GL, a result that suggested that mRNA7 synthesis was specifically affected in this mutant. Strikingly, mutant LB3, containing mutant leader and mRNA7 body TRSs but a wild-type mRNA5 TRS, showed efficient restoration of N expression but not of GL synthesis. Essentially similar IFA results were obtained for cells transfected with two other sets of mutants, L1/B1/LB1 and L2/B2/LB2 (data not shown).
Base Pairing Between the (+) Leader TRS and the (−) Body TRS Plays a Key Role in Arterivirus sg mRNA Transcription.
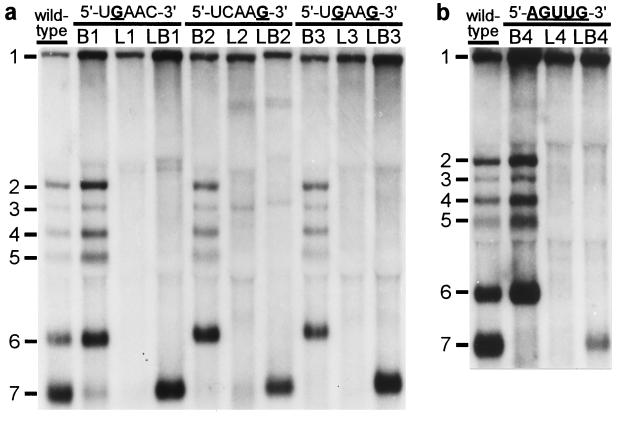
Our IFA data strongly suggested that mutations in either the genomic leader TRS or the body TRS severely affected sg mRNA synthesis. However, as illustrated by the results obtained with mutant LB3 (Fig. 2), transcription apparently could be rescued by introducing identical mutations in leader and body TRSs. Subsequently, the same set of nine mutants was subjected to a direct RNA analysis. Total i.c. RNA was isolated from transfected cells at 12 h posttransfection, and viral mRNAs were visualized by hybridization to a radiolabeled oligonucleotide probe (Fig. 3a). For the wild-type construct, the genome (mRNA1) and the six sg mRNAs (mRNAs 2–7) were readily detected. Mutants L1–L3, in which either one or both of the C residues in the leader TRS had been substituted, replicated with wild-type efficiency, which confirmed that these mutations in the 5′ end of the viral genome did not interfere with replication (Fig. 3a, lanes L1–L3). However, in agreement with our IFA data (Fig. 2), sg mRNA transcription from the 3′ terminal region of these mutant genomes was severely affected or completely abolished, which corroborated the crucial role of the leader TRS in mRNA synthesis. Remarkably, mutant L2 continued to synthesize only mRNA3 (see below) and two larger additional RNAs that were not analyzed in detail. The introduction of mutations into the RNA7 body TRS (Fig. 3a, lanes B1–B3) severely reduced or completely abolished the transcription of mRNA7, but not that of the other sg mRNAs. Finally, the simultaneous introduction of the same mutation(s) in the leader and RNA7 body TRSs (Fig. 3a, lanes LB1–LB3) specifically restored mRNA7 transcription to wild-type levels, but the synthesis of other sg mRNAs was not restored. From the results obtained with the LB double mutants, we concluded that base pairing between the (+) leader TRS and the (−) body TRS complement is an essential step in arterivirus sg mRNA transcription.
Figure 3.
Analysis of genome and sg mRNA synthesis by leader and body TRS mutants. At 12 h posttransfection, total i.c. RNA was isolated, separated in denaturing 1.5% agarose gels, and hybridized (18) to a radiolabeled oligonucleotide probe complementary to the 3′ end of the genome. (a) Analysis of constructs containing single or double C-to-G substitutions in leader TRS (L series), RNA7 body TRS (B series), or both TRSs (LB series). (b) Analysis of a similar set of mutants in which the entire TRS has been mutated (5′-UCAAC-3′ to 5′-AGUUG-3′).
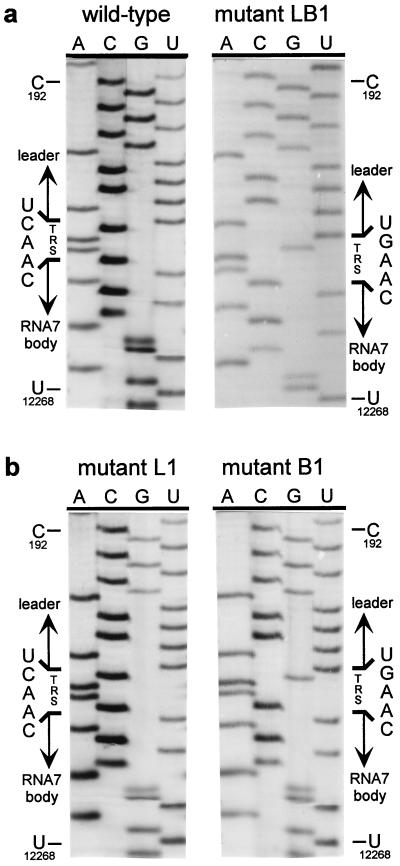
The double C → G replacement in the leader and body TRSs of mutant LB3 did not have a detectable effect on its capacity to produce sg mRNA7 (Fig. 3a). This suggested that the sequence of the TRS per se is not crucial, as long as the possibility for TRS base pairing is maintained. Therefore, we constructed a fourth set of mutants in which the entire TRS (5′-UCAAC-3′) was replaced by the sequence 5′-AGUUG-3′. As expected, mutant L4 did not synthesize any of the sg mRNAs, whereas mutant B4 produced only mRNAs 2–6 (Fig. 3b). Remarkably, the simultaneous mutagenesis of leader TRS and body TRS in construct LB4 again specifically restored RNA7 synthesis, albeit not to wild-type levels. Finally, by using RT-PCR amplification and sequence analysis, we confirmed that in all double mutants (LB1–LB4) the mRNA7 leader and body indeed had been fused at the expected position and not at an alternative site (Table 1). As an example, the results for mutant LB1 are shown in Fig. 4a.
Figure 4.
Sequence analysis of mRNA7 leader–body junction sequences. Total i.c. RNA was isolated from transfected cells and used for an mRNA7-specific RT-PCR. The leader–body junction sequences of sg mRNA7 were determined by direct sequence analysis of the RT-PCR products (see also Table 1). The position of the TRS at the mRNA7 leader–body junction is indicated, as are reference nucleotides from the leader region (genome position C-192) and the RNA7 body (U-12268). (a) Analysis of the leader–body junction sequence of the sg mRNA7 produced by the wild-type construct and by mutant LB1, containing both mutant leader and RNA7 body TRSs (5′-UGAAC-3′). (b) Sequence analysis of the leader–body junction of the mRNA7 species produced by mutants L1 and B1, containing either a mutant leader TRS (5′-UGAAC-3′) and a wild-type RNA body TRS or vice versa. In both cases, the TRS sequence at the mRNA7 leader–body junction was derived exclusively from the body TRS.
Continued RNA3 Synthesis by Mutants L2 and LB2.
The continued synthesis of mRNA3 by mutants L2 and LB2 (Fig. 3a) can be explained by our previous observation (13) that EAV produces two major mRNA3 species, one (mRNA3) derived from a consensus TRS and one (mRNA3.1) derived from a nonconsensus TRS (5′-UCAAUACCC-3′). The latter TRS lacks the C residue at position 5, but contains 5 downstream nucleotides (UACCC) that match the corresponding sequence 3′ of the leader TRS. Thus, the residue at position 5 of the mRNA3.1 TRS apparently is not involved in base pairing. Consequently, mutants L2 and LB2, which have mutations at TRS position 5, are not affected in the synthesis of mRNA3.1. An RT-PCR and sequence analysis for mutants L2 and LB2 confirmed the continued synthesis of mRNA3.1 when the leader TRS has the sequence 5′-UCAAG-3′ (data not shown).
The TRS in the sg mRNA Is Derived Exclusively from the Body TRS.
Mutant B1 and, to a lesser extent, mutants L1, L2, and B2 still produced low levels of mRNA7. The RNA7 produced by mutant B1 was detectable via hybridization (Fig. 3a, lane B1). Also, for mutants B2, L1, and L2, some RNA7 could be detected by hybridization after long exposure and, more convincingly, by RT-PCR analysis (data not shown).We took advantage of this residual mRNA7 synthesis to address another important issue. Is the TRS that forms the mRNA leader–body junction derived from the leader or from the body sequence? Because leader and body TRS normally are identical, this question cannot be answered by using wild-type sequences. However, because we mutated either the leader TRS or the RNA7 body TRS in constructs L1, L2, B1, and B2, sequence analysis of sg mRNA7 enabled us to determine the origin of the sequence at the leader–body junction site. Sequences were determined by direct analysis of mRNA7-specific RT-PCR products generated by using i.c. RNA from cells transfected with one of these four mutants (Table 1). As shown for mutants L1 and B1 in Fig. 4b, the sequences of the mRNA7 leader–body junction sites obtained in this manner were unambiguous and did not show any heterogeneity. For mutants L1 and L2, we established that point mutations in the leader TRS were not transferred to sg mRNA7, which contained the wild-type 5′-UCAAC-3′ sequence. Conversely, mRNA7 transcripts from mutants B1 and B2 contained exclusively a mutagenized TRS at their leader–body junction. Thus, in all these mutants, the mRNA7 junction site sequence was derived from the body TRS and not from the leader TRS. The implications of these findings for the mechanism of discontinuous sg RNA transcription will be discussed below.
Discussion
The results of our EAV TRS mutagenesis experiments show that the synthesis of a nested set of sg mRNAs from the nidovirus genome is governed by base pairing between sequences in the viral (+) and (−) strands, an observation that clearly rules out conventional cis-splicing as the mechanism of sg mRNA synthesis. Furthermore, we observed that sg mRNA7 synthesis can be maintained even when all nucleotides of the leader TRS and RNA7 body TRS have been substituted. This makes it highly unlikely that the primary sequence of the TRS serves as a specific recognition signal for the viral RdRp complex or for cellular proteins (26, 27) involved in discontinuous transcription.
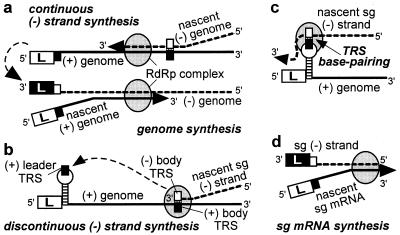
As summarized recently by Brian and Spaan (28), a polymerase jump might be at the heart of nidovirus discontinuous transcription and could be directed by TRS base pairing. This mechanism would resemble “copy-choice” RNA recombination, during which the RdRp complex and nascent transcript are released from the primary template and transcription is reinitiated on a secondary template after base pairing between this template and the 3′ of the nascent transcript. High frequency template switching is a remarkable and well documented feature of the nidovirus replicase. That the body TRS sequence is found at all sg mRNA leader-to-body junctions supports the discontinuous (−) strand transcription model of Sawicki and Sawicki (ref. 16; Fig. 5 b– d). This model proposes that the body TRS complement in the nascent (−) strand base pairs with the genomic (+) leader TRS, thereby allowing the addition of the antileader sequence. In the alternative model of discontinuous (+) strand synthesis (5, 8, 9, 28), a 3′ → 5′ exonuclease activity must be invoked (29) to explain why, after TRS base pairing, the leader TRS nucleotides are removed from the 3′ end of the nascent sg transcript to be replaced by the complement of the (−) body TRS. Such a mechanism would be without precedent among (+)RNA viruses. Instead, we consider it more plausible that nidovirus sg mRNA synthesis and genome replication are based largely on the same principles and replicase properties. Thus, (−) strand synthesis either would be continuous, yielding the full-length (−) strand template for genome replication (Fig. 5a), or discontinuous, producing a sg (−) strand template for mRNA transcription. Specific replicase subunits may direct discontinuous (−) strand synthesis as a supplement to the more basic nidovirus replicase functions, such as RdRp and helicase activities. This model also could explain the phenotype of a recently identified replicase mutant, which displays a severe sg RNA transcription defect in combination with a wild-type genome replication efficiency (17, 23).
Figure 5.
Model of nidovirus genome replication and sg mRNA transcription as proposed by Sawicki and Sawicki (16). Negative-strand synthesis is either continuous (a), yielding the genomic (−) strand, or discontinuous (b), yielding the sg (−) strand(s). The latter step involves translocation of the nascent (−) strand to the genomic leader TRS (c), where base pairing occurs and transcription is resumed to add the antileader sequence to the sg (−) strand. Subsequently, the sg (−) strand is used as the template for sg mRNA synthesis (d). The stem–loop structure in the leader TRS region is discussed in the text and Fig. 6.
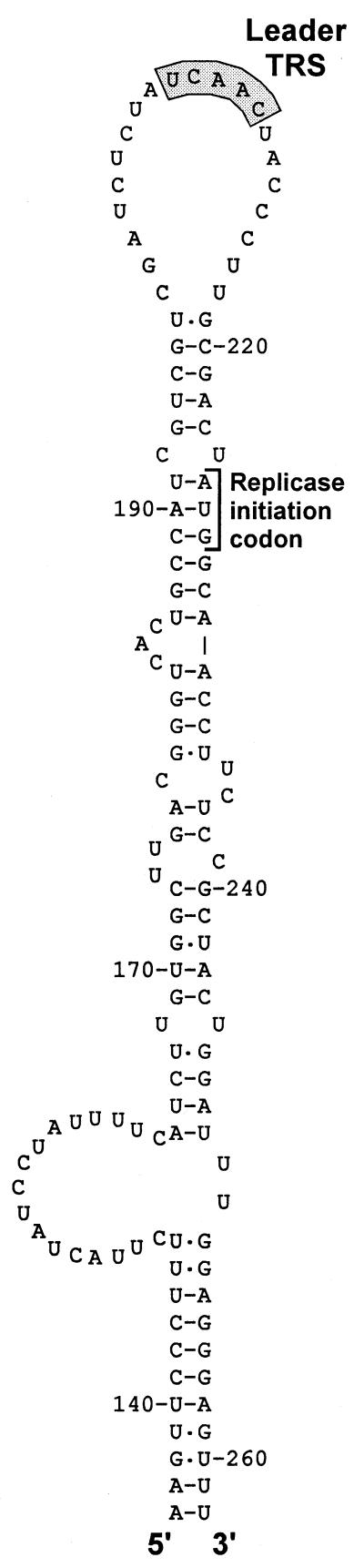
There is increasing evidence that copy-choice RNA recombination can be guided or promoted by distinctive RNA structures (30, 31). A stem–loop structure that appears to “present” the leader TRS may be involved in high-frequency leader switching between full-length and defective bovine coronavirus genomes (32). Strikingly, a similar structure can be predicted for the leader TRS region of EAV (Fig. 6) and other arteriviruses, suggesting the involvement of RNA structure in leader-to-body joining. Possibly, the (−) body TRS at the 3′ end of the nascent (−) strand can base pair with a (+) TRS only when the latter is present in the loop of a hairpin structure. In this manner, the interaction with the leader TRS could be promoted and reinitiation of transcription on, e.g., other body TRSs could be prevented. Such a mechanism would strongly resemble a number of antisense RNA-regulated control mechanisms that are based on interactions between single-stranded tails and hairpin loops (33, 34). The differential activity of the EAV body TRS elements in discontinuous transcription, which is reflected in the nonequimolar synthesis of sg mRNAs (Fig. 2), may also be determined by higher-order RNA structure. The frequency with which the RdRp complex is released from a body TRS may be influenced or determined by the local RNA structure of the template. This might also explain why the EAV genome contains about 10 5′-UCAAC-3′ elements that do not appear to function as TRS elements.
Figure 6.
RNA structure prediction for the EAV (+) leader TRS region, showing a striking hairpin that is conserved in arteriviruses and appears to “present” the leader TRS in its loop. Numbers refer to nucleotide positions in the EAV genome.
In nidoviruses, all (−) strands possess the same promoter element at their 3′ end and the rate of genomic and sg (+)RNA synthesis may be governed solely by the number of (−) strand templates. It should be noted that the leader TRS hairpin cannot be formed in the sg mRNAs, because of the absence of the sequences 3′ of the leader TRS (Fig. 6). Thus, if the leader TRS hairpin indeed directs the reinitiation of transcription (Fig. 5 b and c), the genome would be the sole template for sg (−) strand synthesis. Likewise, the initiation of (−) strand transcription may depend on cis-acting sequences or structures unique to the genomic template. In this manner, sg mRNAs could be prevented from competing with the genome for replication [e.g., by acting as replicons (11)]. This may explain why replication of transfected (synthetic) sg mRNAs in infected cells could not be achieved (refs. 35 and 36; G.v.M., W.L., and W.J.M.S., unpublished observations).
In conclusion, our data favor a model in which nidoviruses regulate their genome expression by a form of site-specific RNA recombination during (−) strand synthesis, an unprecedented mechanism that extends the known repertoire of transcriptional regulation.
Acknowledgments
We thank Leonie van Dinten, Richard Molenkamp, and Sasha Pasternak for helpful discussions and valuable technical assistance. We thank Yvonne van der Meer for the IFA-related photographic work and are grateful to Amy Glaser (Cornell University) and James MacLachlan and Udeni Balasuriya (University of California) for providing EAV GL and N-specific mAbs. We thank Stuart Siddell, Stanley Sawicki, Dorothea Sawicki, David Brian, and Kees Pleij for helpful comments on the manuscript. G.v.M. was supported by Grant 331-020 from the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (CW-NWO).
Abbreviations
- BHK
baby hamster kidney
- EAV
equine arteritis virus
- GL
large glycoprotein
- i.c.
intracellular
- IFA
immunofluorescence assay
- N
nucleocapsid protein
- nsp
nonstructural protein
- (+)RNA
positive-stranded RNA
- RdRp
RNA-dependent RNA polymerase
- RT-PCR
reverse transcription–PCR
- sg
subgenomic
- TRS
transcription-regulating sequence.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.McKnight S L, Yamamoto K R. Transcriptional Regulation. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 2.von Hippel P H. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 3.Adkins S, Stawicki S S, Faurote G, Siegel R W, Kao C C. RNA. 1998;4:455–470. [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel R W, Bellon L, Beigelman L, Kao C C. Proc Natl Acad Sci USA. 1998;95:11613–11618. doi: 10.1073/pnas.95.20.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai M M C, Cavanagh D. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snijder E J, Meulenberg J J M. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs L, Spaan W J M, Horzinek M C, van der Zeijst B A M. J Virol. 1981;39:401–406. doi: 10.1128/jvi.39.2.401-406.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baric R S, Stohlman S A, Lai M M C. J Virol. 1983;48:633–640. doi: 10.1128/jvi.48.3.633-640.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spaan W J M, Delius H, Skinner M, Armstrong J, Rottier P J M, Smeekens S, van der Zeijst B A M, Siddell S G. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.den Boon J A, Spaan W J M, Snijder E J. Virology. 1995;213:364–372. doi: 10.1006/viro.1995.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sethna P B, Hung S L, Brian D A. Proc Natl Acad Sci USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sethna P B, Hofmann M A, Brian D A. J Virol. 1991;65:320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.den Boon J A, Kleijnen M F, Spaan W J M, Snijder E J. J Virol. 1996;70:4291–4298. doi: 10.1128/jvi.70.7.4291-4298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawicki S G, Sawicki D L. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaad M C, Baric R S. J Virol. 1994;68:8169–8179. doi: 10.1128/jvi.68.12.8169-8179.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawicki S G, Sawicki D L. In: in Corona- and Related Viruses. Talbot P J, Levy G A, editors. New York: Plenum; 1995. pp. 499–505. [Google Scholar]
- 17.van Dinten L C, den Boon J A, Wassenaar A L M, Spaan W J M, Snijder E J. Proc Natl Acad Sci USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Vries A A F, Chirnside E D, Bredenbeek P J, Gravestein L A, Horzinek M C, Spaan W J M. Nucleic Acids Res. 1990;18:3241–3247. doi: 10.1093/nar/18.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Meer Y, van Tol H, Krijnse Locker J, Snijder E J. J Virol. 1998;72:6689–6698. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snijder E J, Wassenaar A L M, Spaan W J M. J Virol. 1994;68:5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaser A L, de Vries A A F, Dubovi E J. J Gen Virol. 1995;76:2223–2233. doi: 10.1099/0022-1317-76-9-2223. [DOI] [PubMed] [Google Scholar]
- 22.MacLachlan N J, Balasuriya U B, Hedges J F, Schweidler T M, McCollum W H, Timoney P J, Hullinger P J, Patton J F. J Vet Diagn Invest. 1998;10:229–236. doi: 10.1177/104063879801000302. [DOI] [PubMed] [Google Scholar]
- 23.van Marle G, van Dinten L C, Luytjes W, Spaan W J M, Snijder E J. J Virol. 1999;73:5274–5281. doi: 10.1128/jvi.73.7.5274-5281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Batenburg F H D, Gultyaev A P, Pleij C W A. J Theor Biol. 1995;174:269–280. doi: 10.1006/jtbi.1995.0098. [DOI] [PubMed] [Google Scholar]
- 25.Gultyaev A P, van Batenburg F H D, Pleij C W A. J Mol Biol. 1995;250:37–51. doi: 10.1006/jmbi.1995.0356. [DOI] [PubMed] [Google Scholar]
- 26.Li H P, Zhang X, Duncan R, Comai L, Lai M M C. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H P, Huang P, Park R S, Lai M M C. J Virol. 1999;73:772–777. doi: 10.1128/jvi.73.1.772-777.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brian D A, Spaan W J M. Semin Virol. 1997;8:101–111. doi: 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker S C, Lai M M C. EMBO J. 1990;9:4173–4179. doi: 10.1002/j.1460-2075.1990.tb07641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White K A, Morris T J. RNA. 1995;1:1029–1040. [PMC free article] [PubMed] [Google Scholar]
- 31.Nagy P D, Simon A E. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 32.Chang R Y, Krishnan R, Brian D A. J Virol. 1996;70:2720–2729. doi: 10.1128/jvi.70.5.2720-2729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner E G H, Simons R W. Annu Rev Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 34.Gerdes K, Gultyaev A P, Franch T, Pedersen K, Mikkelsen N D. Annu Rev Genet. 1997;31:1–31. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Makino S, Joo M, Makino J K. J Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang R Y, Hofmann M A, Sethna P B, Brian D A. J Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]