Abstract
Recently synthesized proteins are sorted at the trans-Golgi network into specialized routes for exocytosis. Surprisingly little is known about the underlying molecular machinery. Here, we present a visual screen to search for proteins involved in cargo sorting and vesicle formation. We expressed a GFP-tagged plasma membrane protein in the yeast deletion library and identified mutants with altered marker localization. This screen revealed a requirement of several enzymes regulating the synthesis of sphingolipids and ergosterol in the correct and efficient delivery of the marker protein to the cell surface. Additionally, we identified mutants regulating the actin cytoskeleton (Rvs161p and Vrp1p), known membrane traffic regulators (Kes1p and Chs5p), and several unknown genes. This visual screening method can now be used for different cargo proteins to search in a genome-wide fashion for machinery involved in post-Golgi sorting.
Keywords: exocytosis, lipid rafts, Saccharomyces cerevisiae, sorting, Golgi
The mechanisms responsible for sorting proteins to the cell surface from the Golgi complex are poorly understood in eukaryotic cells. The trans-Golgi network (TGN) has been recognized as a major hub for sorting (1). However, there is also evidence that sorting occurs in endosomes (2). In polarized cells such as epithelial cells and neurons, biosynthetic cargo is delivered to separate membrane domains by pathways employing different sorting principles (3, 4). Recent work has demonstrated that yeast cells also have at least two separate routes to the cell surface (5–8). Little is known about the genes that are responsible for sorting and packaging surface cargo into different transport containers. Previous screens aimed at identifying this machinery relied, for example, on major growth defects and the internal accumulation of invertase, which has been later shown to be transported by the minor pathway to the plasma membrane (7, 8). These screens have mainly identified mutants that blocked endoplasmic reticulum (ER)-to-Golgi transport and delivery to the plasma membrane (9, 10). However, mutations in regulators of post-Golgi sorting and vesicle formation with few exceptions have not been detected by such screens, probably because a block in one transport route to the cell surface can be rescued by partial rerouting from the affected to the undisturbed pathway (7, 8).
Here we describe a visual screening procedure devised to circumvent this problem. We aimed at developing an assay sensitive enough to detect sorting defects within the secretory pathway and applicable to genome-wide screening. The screen takes advantage of the systematic yeast knockout array (11), which should contain the nonessential genes responsible for regulating cargo entry into specialized, partially redundant pathways. The results of this genome-wide screen demonstrate the suitability of our visual screening approach for identifying regulators of sorting and vesicle formation involved in surface delivery of biosynthetic cargo.
Materials and Methods
Supporting Information. For further details, see Supporting Text, Figs. 4–11, and Tables 2 and 3, which are published as supporting information on the PNAS web site. The database at http://tds.mpi-cbg.de/yeast contains images for mutants listed in Table 1. Images (GFP and DIC) showing large field of cells as well as images for colocalization of accumulated Fus-Mid-GFP and Sec7-DsRed or DsRed-FYVE were provided, respectively. Additionally, for each mutant, links to the literature available in PubMed or databases (YPD, SGD, GFP localization) have been created.
Table 1. List of mutants showing phenotypes I and II.
| ORF | Gene name | Function | Cargo accumulation |
|---|---|---|---|
| YCL001W-A | YCL001W-A | Uncharacterized | Golgi |
| YGL015C | YGL015C | Uncharacterized | Golgi |
| YLR296W | YLR296W | Uncharacterized | Endosome |
| YLR338W | YLR338W | Uncharacterized | Golgi/endosome |
| YOR318C | YOR318C | Uncharacterized | Endosome |
| YLR372W | SUR4 | Lipid metabolism | Vacuole |
| YDR297W | SUR2 | Lipid metabolism | Golgi/Vacuole |
| YBR183W | YPC1 | Lipid metabolism | Golgi |
| YIL124W | AYR1 | Lipid metabolism | Golgi |
| YML008C | ERG6 | Lipid metabolism | ND |
| YGL012W | ERG4 | Lipid metabolism | Golgi |
| YLR330W | CHS5 | TGN exit | Golgi |
| YPL145C | KES1 | TGN exit | Golgi |
| YCR009C | RVS161 | Actin organization | Golgi |
| YLR337C | VRP1 | Actin organization | Golgi |
| YNL153C | GIM3 | Actin organization | ND |
| YGR078C | PAC10 | Actin organization | Golgi/endosome |
| YFL023W | BUD27 | Actin organization | Golgi |
| YDR080W | VPS41 | Vacuole transport | Endosome |
| YGL124C | MON1 | Vacuole transport | Golgi/endosome |
| YBR131W | CCZ1 | Vacuole transport | Endosome |
| YML001W | YPT7 | Vacuole transport | Endosome |
| YFR019W | FAB1 | Vacuole transport | Endosome |
| YOR306C | MCH5 | Vacuole transport | Endosome |
Functional assignment for identified genes was based on description in YPD. The listed deletion strains displayed phenotypes that were verified in four independent experiments. The preferred site of internal accumulation of the Fus-Mid-GFP probe was assessed in colocalization experiments where cells were in addition transformed with either Sec7-DsRed to label the TGN or DsRed-FYVE to label endosomes. All images can be found at http://tds.mpi-cbg.de/yeast. In the erg6 and gim3 mutants, colocalization experiments were unsuccessful because of poor growth of double transformants (ND, not determined).
Yeast Strains and Plasmids. In this study, we used the pTPQ55 centromeric plasmid carrying Fus-Mid-GFP construct under the control of the inducible GALs promoter described in ref. 12. For colocalization experiments, we constructed the plasmids pTPQ128 and pTPQ127 containing Sec7-DsRed and DsRed-FYVE respectively. The cloning strategy is described in Supporting Text.
In these studies we used the yeast deletion library encompassing 4,848 single knockouts of nonessential genes (European Saccharomyces cerevisiae Archives for Functional analysis, http://web.uni-frankfurt.de/fb15/mikro/euroscarf/index.html). Deletions are in BY strains derived from S288C (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0). We also used RH690-15D [wild-type] (Mata his4, leu2, ura3, lys2, bar1), RH1965 [end4Δ] (Mata his4, leu2, ura3, lys2, bar1, end4::LEU2), and RH690-13D [lcb1-100] (Mata lcb1-100 his4 ura3 leu2 lys2 bar1) obtained from H. Riezman.
Tools Used to Transfer Yeast Cells. The yeast deletion library is organized in 96-well plates. To transfer cells we used a 96-floating pin replicator containing 23-mm-long pins (1.58 mm in diameter, V & P Scientific, San Diego, catalog no. VP 408FH). We also constructed a 96-fixed pin replicator with pins (16 mm long and 4 mm in diameter) that is able to transfer more material. Both types of pin replicators were sterilized by incubating two times for 1 min in sterile water, 1 min in bleach (10% sodium hypochlorite), and then two times in water; after this, the pin replicators were transferred to ethanol and flame sterilized three times.
For growing cells from the deletion library in liquid media standard 96-well plates were used. To grow on solid media we used Single Well OmniTrays (Nunc catalog no. 242811).
Transformation Protocol, Media, and Growth Conditions. Strains of the deletion library were thawed and transferred from 96-well plates to solid yeast extract/peptone with 2% dextrose (YPD) media using the floating pin replicator. Plates were incubated for 3 days at 24°C. The sterile 96-well plates were filled with transformation solution (100 μl per well) containing (per well) 30 μg of Sonicated Salmon Sperm DNA (Stratagene catalog no. 201190-81) denatured by incubation for 5 min at 95°C, 3 μg of the pTPQ55 plasmid in 40% PEG, 1 mM EDTA, 10 mM Tris·Cl (pH 7.5), 100 mM lithium acetate. The transformation solution was mixed well before use. The yeast cells were collected with fixed pin replicator and transferred to the 96-well plate containing the transformation solution. The pin replicator was gently agitated when pins were in the wells to transfer yeast cells from the pins to the liquid. Next, plates were incubated at room temperature overnight, and the edges of the plates were wrapped up with parafilm (American National Can, Chicago) and incubated in a water bath for 15 min at 45°C. The plates were incubated for 1 h at room temperature. The liquid was gently removed, and transformed cells were collected with the fixed pin replicator and transferred to selective plates containing solid synthetic dextrose minimal medium without uracil (SC-Ura). The plates were incubated for 4 days at 24°C, and cells were transferred with the fixed pin replicator for a second round of selection on SC-Ura (3–4 days of incubation).
Next, cells were transferred with floating pin replicator to the 96-well plates containing yeast extract/peptone with 2% raffinose (YPRaf) and incubated overnight at 24°C. For the induction of Fus-Mid-GFP expression, YPRaf was gently replaced with YPRaf containing 2% galactose. Plates were incubated at 24°C for 4 h. For taking final images, cells were grown overnight in SC-Ura or SC-Ura-Leu media containing 2% raffinose. To induce the protein expression, this media was exchanged with fresh SC-Ura or SC-Ura-Leu media containing 2% raffinose and 2% galactose, respectively.
Microscopy. For sample preparation, see Supporting Text. Microscopy was performed by using an Olympus BX61 microscope, and Olympus PlanApo ×60/1.10 oil LSM objective, RT Slider SPOT camera (Diagnostic Instruments), and metamorph software.
Colocalization Experiments. For colocalization experiments cells containing pTPQ55 were transformed with either pTPQ127 or pTPQ128 as described above. Expression of both colocalization markers (Sec7-DsRed and DsRed-FYVE) slightly affected protein transport. To avoid potential artifacts, all pictures of GFP fluorescence showing phenotype were taken when cells were containing plasmid pTPQ55 alone.
Results
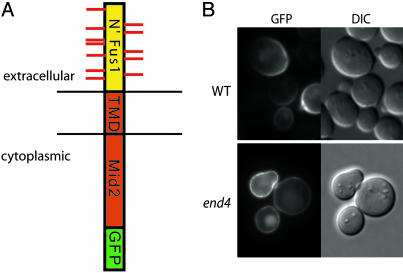
Characterization of the Marker Protein Used for Screening. The prerequisite for our screen was to design a cargo protein that could be followed by microscopy after synthesis at the ER throughout the secretory pathway to the cell surface. For this purpose, we used a GFP-tagged chimera of Fus1p and Mid2p, named Fus-Mid-GFP (Fig. 1A), that we described previously (12). This chimeric protein required O-glycosylation for sorting to the cell surface and was raft-associated by using detergent-resistance as a criterion (Fig. 4). It was efficiently transported to the plasma membrane and remained there within the time frame of observation, making it a suitable probe for our screen.
Fig. 1.
Characterization of the Fus-Mid-GFP marker. (A) Schematic representation of Fus-Mid-GFP. This construct consists of the extracellular portion of Fus1p (yellow) which is O-glycosylated (red lines) fused to the transmembrane domain (TMD) and cytoplasmic tail of Mid2p (red) followed by the GFP-tag (green). (B) Cellular localization of Fus-Mid-GFP in wild-type and end4Δ mutant cells. Note that, in wild-type cells, subtle vacuolar labeling is observed. This labeling is apparently a result of sorting of the probe directly to the vacuole, as it is also observed in the endocytosis mutant.
We used a centromeric plasmid carrying Fus-Mid-GFP that was under the control of the inducible GALs promoter (13). Inducibility was an important feature of the construct because this made it possible to “flood” the biosynthetic pathway with a wave of marker protein to be able to detect sorting delays by intracellular accumulation. This plasmid was introduced into the entire deletion library encompassing 4,848 single knockouts of nonessential genes (European Saccharomyces cerevisiae Archives for Functional analysis, http://web.uni-frankfurt.de/fb15/mikro/euroscarf/index.html). To transform the library, we developed a protocol that allowed efficient transformation directly in the 96-well plate format.
After 3–4 h of induction, robust and bright labeling of the plasma membrane was detected in wild-type cells (Fig. 1B). We also found weak labeling of the vacuole. This labeling was caused by direct biosynthetic transport of a fraction of the GFP-tagged marker to the vacuole because vacuolar staining was also observed in the endocytosis mutant end4Δ (Fig. 1B) but not in the vps1Δ mutant (Fig. 8).
Phenotype Classes and Mutant Genes Showing Internal Accumulation. The entire deletion library was first screened by individual microscopic inspection of each mutant for intracellular accumulation. Because the exocytic pathways are partially redundant, we expected that, in mutants affecting sorting, cargo would still be delivered to the cell surface, resulting in fluorescence at the plasma membrane but also showing intracellular accumulation.
We observed two different phenotypes in mutants affecting surface delivery, which we named type I and II (Fig. 2). Type I mutants show dot-like intracellular labeling in addition to plasma membrane labeling. Type II mutants exhibit exaggerated vacuolar and reduced plasma membrane fluorescence, probably because of increased missorting of Fus-Mid-GFP into a degradation rather than a surface-directed pathway. In this screen, we did not identify mutants that accumulated Fus-Mid-GFP in the ER. The reason could be that the transport of the probe out of the ER is not dependent on nonessential genes or that these genes are redundant. Alternatively, the time required for the GFP tag to become fluorescent did not allow for identification of mutants that transiently accumulated marker protein in the early phase of the secretory pathway.
Fig. 2.
Different classes of phenotypes were observed in the screen. Cells transformed with Fus-Mid-GFP were grown overnight, transferred into inducing media, incubated for 4 h, and observed by fluorescence microscopy. In wild-type cells, bright and robust plasma membrane labeling was observed. In addition, some vacuolar fluorescence was found. Type I mutants showed an internal dot-like labeling in addition to plasma membrane fluorescence. In type II mutants, the probe was mostly accumulated in the vacuole coincident with reduced plasma membrane labeling.
We found 137 mutant strains that displayed phenotype I and II (Table 2). Among these mutants there were genes that were involved in ribosome function and translation, transcription, mitochondrial function, membrane trafficking, cytoskeletal function, lipid metabolism, or unknown functions.
Here, we focused our analysis on mutants that affected genes with known function in intracellular trafficking or in lipid metabolism as well as those that were uncharacterized. This list contained 24 mutants (Table 1). The complete set of images showing phenotypes of these mutants can be found at http://tds.mpi-cgb.de/yeast.
To find out in which compartment Fus-Mid-GFP accumulated, we also performed colocalization experiments using a second fluorescent marker. For this, we coexpressed in mutant cells either Sec7-DsRed to mark the TGN or DsRed-FYVE domain, a phosphatidylinositol 3-phosphate-binding domain found on endosomal membranes, together with the Fus-Mid-GFP construct. Accumulation in the vacuole was distinguished by comparison to the differential interference contrast image (Table 1).
We measured the secretion of invertase (Fig. 11) for mutants identified in the screen (Table 1; for details, see Supporting Text). The invertase secretion assay did not reveal any transport defect in the analyzed mutants, except for a small reduction in secretion for vrp1 (79.0 ± 6.7%).
Mutants in Genes Regulating Synthesis of Lipids. We identified six mutants in genes regulating synthesis of sphingolipids (sur4/elo3, sur2/syr2, ypc1, ayr1) and ergosterol (erg6, erg4) (Fig. 5). The sur4/elo3 mutant showed the most severe phenotype with an accumulation of the marker in the vacuole and an obvious reduction in plasma membrane fluorescence intensity (Fig. 5). According to our colocalization experiments, accumulation of Fus-Mid-GFP in sur2/syr2 and ypc1 was in vacuoles and TGN, or mostly TGN, respectively (Table 1). The ergosterol synthesis mutants erg4 and erg6 showed an accumulation at the TGN (Table 1). We also identified the ayr1 mutant in which Fus-Mid-GFP accumulated in the late Golgi. The Ayr1p has 1-acyldihydroxyacetone-phosphate-reductase activity and AYR1 was also reported to interact genetically with YBR159W, the major 3-ketoreductase important for fatty acid elongation (14). Thus, defective sphingolipid and ergosterol synthesis results in inhibition of trafficking or sorting defects of Fus-Mid-GFP from the TGN.
It has been previously reported that, in a conditional mutant of the essential protein Lcb1p required for sphingolipid synthesis, raft associated Pma1p is missorted to the vacuole (15, 16). To investigate how exocytosis of our probe is affected in this mutant background, we expressed Fus-Mid-GFP in lcb1-100 at different temperatures. Similar to the sorting defect observed in sur4/elo3, Fus-Mid-GFP was no longer transported to the cell surface at the restrictive temperature but was instead missorted to the vacuole (Figs. 3 and 5). Because Fus-Mid-GFP is detergent resistant membrane (DRM)-associated like Pma1p (refs. 15 and 16 and Fig. 4), these results support a critical role of sphingolipids and ergosterol in sorting of DRM-associated cargo to the cell surface.
Fig. 3.
Cell surface delivery of Fus-Mid-GFP is blocked in a conditional-lethal sphingolipid mutant, lcb1-100. Cells were incubated at 24°C, 30°C, or 37°C and processed for fluorescence microscopy. Fus-Mid-GFP expression was induced simultaneously with the temperature shift. At a restrictive temperature, the protein was no longer transported to the cell surface but instead accumulated in the vacuole.
Mutants in Genes of Known Membrane Transport Function. Phenotype I was also observed in the chs5 and kes1 deletion mutants (Fig. 6). In agreement with a role for Chs5p and Kes1p at the late Golgi, we found the accumulation of Fus-Mid-GFP in the Sec7p-containing compartment (Table 1). Chs5p is a peripheral membrane protein of unknown molecular role important for Golgi to plasma membrane transport of Fus1p (17) and chitin synthase III (Chs3p; ref. 18), an enzyme required for synthesis of the polysaccharide chitin.
Phenotype I accumulation at the TGN was also observed in kes1 mutants (Fig. 6). Kes1p/Osh4p is a member of the oxysterol binding protein family that localizes to the TGN.
Cytoskeleton Mutants and Mutants of the Prefoldin Complex. We also found phenotype I in two mutants of genes involved in the organization of the cytoskeleton, rvs161 and vrp1 (Fig. 6). Vrp1p (verprolin), a homolog of mammalian Wiskott–Aldrich syndrome protein interacting protein (19), is an actin-binding protein.
We further observed internal accumulation in mutants of the prefoldin complex: gim3 and pac10, and of the prefoldin bud27 (Table 1). The prefoldin complex acts as a chaperone for the assembly of actin and tubulin (20, 21). We can only speculate that the trafficking defect observed in these mutants of the prefoldin complex is related to their role in organization of the cytoskeleton. Bud27/Uri (for Unconventional prefolding RBP5 Interactor) has been shown to be involved in the TOR pathway that coordinates nutrient availability with cell growth and proliferation (22). The role of this protein in membrane transport will require further investigation.
Uncharacterized Mutants. Five uncharacterized ORFs were among mutants exhibiting clearcut phenotype I (Fig. 7). Sequence analysis demonstrated that the YLR338W locus overlaps with the Vrp1p coding region, which was also isolated in our screen. Thus, two different mutants affecting production of the same protein, Vrp1p, were independently identified in our screen. YCL001W-A and YLR296W mutants show sensitivity to the anticholesterol drug Lovastatin (3-hydroxy-3-methyl-glutaryl CoA reductase inhibitor) (23).
Mutants of Genes Involved in Vacuolar Sorting. Internal accumulation of Fus-Mid-GFP at endosomes was observed in mutants involved in biosynthetic traffic to the vacuole: vps41, mon1, ccz1, mch5, ypt7, and fab1 (Table 1).
The block of protein transport in these mutants apparently in a late step of TGN-to-vacuole delivery resulted in an accumulation of cargo in an endosomal compartment.
Interestingly, we also observed another phenotype in our screen in which no vacuolar staining was seen but delivery to the cell surface was normal or enhanced (Fig. 8). These mutants represent, for the most part, genes previously linked to different aspects of vacuolar transport and function (24, 25). A number of these mutants have also been isolated in a recent genomic screen for VPS genes (26). As was the case for the screen of Bonangelino et al. (26), we also found inhibition of vacuolar transport in mutants of genes involved in N-glycosylation: och1, an α-1,6-mannosyltransferase, and genes encoding four components of a Golgi mannosyltransferase complex (mnn10, mnn11, anp1, and mnn9) (Fig. 9). Our observations imply that the addition of N-glycans is not only necessary for correct targeting of CPY, but may be required for the proper functioning of the machinery regulating the pathway because Fus-Mid-GFP itself is not N-glycosylated.
Discussion
A Visual Screening Method for Identification of Secretion Regulators. Here, we introduce a visual screening strategy to search in a genome-wide scale for genes regulating intracellular trafficking. Previously, a visual approach was used to screen the deletion library for genes involved in vacuolar homotypic fusion and centromeric cohesion (27, 28). Our screen was designed to detect phenotypes expected when cargo can be transported via two redundant exocytic routes from the Golgi to the cell surface. The probe that we used in our screen carried O-glycans, which have been shown to act as a sorting determinant for cell surface delivery of our marker protein (12) and showed properties similar to Pma1p with respect to raft-association. Previous work suggested that the secretory enzyme invertase and the bulk of exoglucanase travels via endosomes to the plasma membrane (7, 8). The other route, which is the major pathway, transports Pma1p, the plasma membrane proton ATPase probably directly from the Golgi to the cell surface. When access to the endosomes is blocked by mutants, e.g., dynamin-like vps1, invertase is shifted from a dense vesicle carrier to the light Pma1p-containing transport vesicles by which both cargo types subsequently reach their destination (7, 8). The two transport pathways employ the same docking and fusion machinery, i.e., the rab GTPase, Sec4p, the SNAREs Sso1/2p and Snc1/2p, and the exocyst-tethering complex (5, 29). However, otherwise, the mechanisms for surface delivery are poorly understood.
Lipid Metabolism. The most striking result of our efforts to obtain a global view of the genes involved in the Fus-Mid-GFP pathway from the Golgi to the cell surface was the identification of mutant strains with defects in sphingolipid and ergosterol biosynthesis. Sphingolipid synthesis up to ceramide involves 15 known enzymes (30) of which five are essential and therefore not subject of the screen. Of the remaining nonessential genes, we identified four with a sorting phenotype (Fig. 10). Here, we could show that alteration in the composition of sphingolipid molecular species had an impact on the sorting of Fus-Mid-GFP. First, elo3 mutants incapable of synthesizing C26:0 very long-chain fatty acids (VLCFAs) (31) missorted cargo to the vacuole and had weak plasma membrane staining. Thus, shortening of VL-CFAs by only two carbon atoms results in defects in protein surface delivery. Second, deletion of Sur2p abolishes hydroxylation of the sphingosine backbone and accumulated cargo in the TGN and the vacuole. Third, knockout of YPC1 encoding for an enzyme with reported ceramidase but also minor ceramide synthase activity with a substrate preference for phytoceramide (CER-B), showed defects in Golgi exit of our GFP-fusion construct (32). Intriguingly, we also identified Ayr1p in the screen. This protein has been shown to have a 1-acyldihydroxy-acetone-phosphate-reductase activity (33). However, Ayr1p also seems to be involved in fatty acid elongation because of a 3-ketoreductase activity, and could thus contribute to ceramide synthesis (14). These data suggest that the fatty acid and long chain base in the ceramide are involved in proper protein sorting. Such a model also fits with the observation that the length of the transmembrane domain determines cell surface delivery (34, 35). However, at this stage of our investigation, we cannot exclude that the phenotypes of the mentioned mutants exhibit indirect effects, for example, through accumulation of lipid metabolites that affect the regulation of membrane traffic to the cell surface.
We further identified erg4 and erg6 as phenotype I strains. These enzymes catalyze the late steps in ergosterol synthesis of which the last five involve nonessential genes. Both Erg6p and Erg4p regulate modification at position C-24 of the sterol backbone, being methyltransferases and reductases, respectively (36). Ergosterol is also required for targeting of the tryptophan permease Tat2p to the cell surface when the external tryptophan concentration is low (37).
It should be noted that ERG6 and ELO3 show strong genetic interaction and are believed to be required for formation of functional rafts (38). The maturation of GPI-anchored Gas1p is blocked in the ER in elo3Δ and more so in elo3Δerg6Δ double mutant cells. Pma1p was rapidly routed for degradation in the vacuole in elo3Δ cells and this was not drastically enhanced in elo3Δerg6Δ cells because the degradation is already rapid in the single mutant cells (38). Both Pma1p and Fus-Mid-GFP are also missorted to the vacuole when sphingolipid synthesis is blocked by the inactivation of the first enzyme in the pathway, serine-palmitoylCoA transferase (Figs. 3 and 10).
Together, these data point to an important role of sphingolipids and ergosterol in surface delivery of our DRM-associated marker protein, Fus-Mid-GFP. Previous studies identified that DRMs association in yeast begins in the ER (39). Some proteins become included into DRMs in the ER, whereas others associate later along the secretory pathway (16, 39). Although mutants identified in this screen accumulated marker protein in the Golgi, it is possible that transient accumulation of cargo occurs also in the ER but could not be detected because of the time required for the GFP tag maturation.
Actin Organization. The RVS161 gene identified in our screen has so far been implicated in endocytosis and in the generation of mating polarity in yeast (40, 41). We suggest that Rvs161p plays a direct role in exocytosis. This view is based on the fact that in our screen no internalization mutant showed TGN accumulation and phenotype I was not observed in end4Δ cells (Fig. 1B). Furthermore, rvs mutants accumulate late secretory vesicles at sites of membrane and cell wall construction (42). Rvs161p is also involved in actin regulation because the protein interacts with Myo2p, Myo1p (myosin motors), and actin (43). In yeast, the actin cytoskeleton is depolarized by NaCl stress. Rvs161p is required to repolarize the actin cytoskeleton (44). Interestingly, this requirement can be suppressed by mutations in sphingolipid biosynthesis including sur2 and sur4/elo3. These findings suggest a link between actin polarization, sphingolipids and Fus-Mid-GFP surface delivery (44). It should be noted that Rvs161p is a BAR domain protein. BAR domains are sensors of membrane curvature and could potentially be involved in increasing membrane curvature in vesicle formation (45).
The other gene involved in actin regulation that we identified was Vrp1p. This protein is part of the Arp2/3 machinery and localizes to cortical actin patches (19). In mammalian cells, a role of rafts and phosphatidylinositol 4,5-bisphosphate in formation of actin tails by the Arp2/3 complex has been suggested to drive the budding process of TGN-derived vesicles (46). Our finding that the raft-dependent cargo Fus-Mid-GFP is inefficiently transported from the TGN in vrp1 mutant cells opens the question of whether Vrp1p-dependent actin assemblies could also play a role in vesicle formation at this compartment.
Golgi Exit. Two other interesting membrane trafficking genes that we identified remain to be discussed: CHS5 and KES1. Both deletion strains lead to accumulation of Fus-Mid-GFP in the Sec7-containing compartment (Table 1). Chs5p is involved in transport of chitin synthase III (Chs3p), an enzyme required for synthesis of the polysaccharide chitin. Chs5p localizes to the TGN and is required for of the exit of Chs3p, the GPI-anchored cell wall protein Crh2p, and Fus1p from the Golgi (18, 47). We show that Fus-Mid-GFP requires functional Chs5p for undisturbed exit from the TGN en route to the cell surface. Interestingly, in a recent genome-wide screen, chs5Δ was found to be synthetic lethal with rvs161Δ (48). Whether the raft- and cytoskeleton-dependent surface transport observed for Fus-Mid-GFP coincides with the surface delivery route for Chs3p remains to be established.
Another intriguing protein implicated in Golgi function that we identified in our screen was Kes1p. Structurally, Kes1/Osh4p belongs to a family of oxysterol binding proteins and has been recently shown to bind ergosterol (49). Kes1p binds to the Golgi depending on Pik1p, a phosphatidylinositol 4-kinase required for normal Golgi structure and transport competence (50). Kes1p has also been shown to bind phosphoinositides including PI4P. Kes1p interacts genetically with Arf1p, Pik1p, and the phospholipid transfer protein Sec14p that regulate Golgi lipid composition and formation of secretory vesicles from the TGN. These data suggest a role of Kes1p in exocytosis (50) that could be mediated by the ergosterol loading capability of the protein. Involvement in formation of transport carriers to the cell surface and dependence on phosphatidylinositol 4-phosphate and the small GTPase Arf1p has also been demonstrated for the mammalian proteins FAPP1 and FAPP2 (four-phosphate adaptor proteins; ref. 51). We have recently demonstrated that FAPP2 binds to PI4P in the Golgi of epithelial MDCK cells and is involved in the machinery responsible for apical delivery of raft proteins (52). How ergosterol binding by the tunnel structure of Kes1p impinges on the sorting at the Golgi remains to be investigated.
O-Glycosylation. Previously, glycosylation has been found to be an important determinant for sorting of biosynthetic cargo to the cell surface (12). Cells lacking the protein O-mannosyltransferase Pmt4p necessary to initiate glycosylation of Fus-Mid-GFP accumulate unglycosylated Fus-Mid in late Golgi structures (12). Although O-glycosylation is required for correct sorting of Fus-Mid, our screen did not identify components regulating this modification. Unfortunately, the pmt4Δ mutant is missing from the deletion library used in the visual screen, and enzymes involved in elongation of O-glycosylation, including Ktr1p, Ktr3p, and Kre2p, have redundant functions (53).
Genome-Wide Visual Screen for Nonessential Exocytosis Regulators. The strategy presented here can now be used for systematic screening of multiple cargoes to define the pathways and the machineries responsible for sorting into different exocytic routes. The major outcome of our screen is that we identified gene products that implicate raft platforms in the sorting of proteins from the Golgi to the cell surface. As has been proposed for apical sorting in epithelial cells, raft clustering could lead to domain-induced budding and formation of the transport carrier (3, 54). What is missing are proteins, probably more than one, that regulate clustering. Of particular interest would be to define molecular requirements of the route transporting invertase because it should be governed by different principles than sorting in the route studied here. Obvious screens of this type are only a springboard for identifying these candidate genes. Further work will be required to analyze their role in the machinery involved in post-Golgi sorting events.
Supplementary Material
Acknowledgments
We are indebted to H. Riezman (University of Basel, Basel) for providing the end4Δ and lcb1-100 mutant strains and comments on the manuscript. We thank Benjamin Glick (University of Chicago, Chicago), Ruth N. Collins (Cornell University, Ithaca, NY), Claudio De Virgilio (University of Geneva, Geneva), Robert S. Fuller (University of Michigan, Ann Arbor), and Scott D. Emr (University of California at San Diego, La Jolla) for plasmids and yeast strains; Eugenio Fava, Jeremy Sanderson, and Britta Diez for advice; and Bianca Habermann for help with bioinformatics analysis. The project was supported by Transregio Grant SFB-TR13-TPA1 and European Union Grant HPRN-CT-2002-00259. P.P.H. was supported by a German–American Fulbright Grant.
Author contributions: T.J.P., R.W.K., M.B., K.S., and C.W.-S. designed research; T.J.P., R.W.K., M.G., P.P.H., Y.G., J.W., K.K., H.G., K.W., M.B., and C.W.-S. performed research; T.J.P., R.W.K., M.G., Y.G., K.W., K.S., and C.W.-S. analyzed data; and T.J.P., R.W.K., K.S., and C.W.-S. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: TGN, trans-Golgi network; ER, endoplasmic reticulum; DRM, detergent-resistant membrane.
References
- 1.Keller, P. & Simons, K. (1997) J. Cell. Sci. 110, 3001–3009. [DOI] [PubMed] [Google Scholar]
- 2.Ang, A. L., Taguchi, T., Francis, S., Folsch, H., Murrells, L. J., Pypaert, M., Warren, G. & Mellman, I. (2004) J. Cell. Biol. 167, 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuck, S. & Simons, K. (2004) J. Cell. Sci. 117, 5955–5964. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Boulan, E., Kreitze, G. & Musch, A. (2005) Nat. Rev. Mol. Cell. Biol. 6, 233–247. [DOI] [PubMed] [Google Scholar]
- 5.Harsay, E. & Bretscher, A. (1995) J. Cell Biol. 131, 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David, D., Sundarababu, S. & Gerst, J. E. (1998) J. Cell Biol. 143, 1167–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harsay, E. & Schekman, R. (2002) J. Cell Biol. 156, 271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gurunathan, S., David, D. & Gerst, J. E. (2002) EMBO J. 21, 602–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novick, P., Field, C. & Schekman, R. (1980) Cell 21, 205–215. [DOI] [PubMed] [Google Scholar]
- 10.Newman, A. P. & Ferro-Novick, S. (1987) J. Cell Biol. 105, 1587–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong, A. H., Evangelista, M., Parsons, A. B., Xu, H., Bader, G. D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C. W., Bussey, H., et al. (2001) Science 294, 2364–2368. [DOI] [PubMed] [Google Scholar]
- 12.Proszynski, T. J., Simons, K. & Bagnat, M. (2004) Mol. Biol. Cell. 15, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mumberg, D., Muller, R. & Funk, M. (1994) Nucleic Acids Res. 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han, G., Gable, K., Kohlwein, S. D., Beaudoin, F., Napier, J. A. & Dunn, T. M. (2002) J. Biol. Chem. 277, 35440–35449. [DOI] [PubMed] [Google Scholar]
- 15.Lee, M. C., Hamamoto, S. & Schekman, R. (2002) J. Biol. Chem. 277, 22395–22401. [DOI] [PubMed] [Google Scholar]
- 16.Bagnat, M., Chang, A. & Simons, K. (2001) Mol. Biol. Cell 12, 4129–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santos, B. & Snyder, M. (2003) Eukaryot. Cell 2, 821–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos, B. & Snyder, M. (1997) J. Cell Biol. 136, 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evangelista, M., Klebl, B. M., Tong, A. H., Webb, B. A., Leeuw, T., Leberer, E., Whiteway, M., Thomas, D. Y. & Boone, C. (2000) J. Cell Biol. 148, 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geissler, S., Siegers, K. & Schiebel, E. (1998) EMBO J. 17, 952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegers, K., Waldmann, T., Leroux, M. R., Grein, K., Shevchenko, A., Schiebel, E. & Hartl, F. U. (1999) EMBO J. 18, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gstaiger, M., Luke, B., Hess, D., Oakeley, E. J., Wirbelauer, C., Blondel, M., Vigneron, M., Peter, M. & Krek, W. (2003) Science 302, 1208–1212. [DOI] [PubMed] [Google Scholar]
- 23.Giaever, G., Flaherty, P., Kumm, J., Proctor, M., Nislow, C., Jaramillo, D. F., Chu, A. M., Jordan, M. I., Arkin, A. P. & Davis, R. W. (2004) Proc. Natl. Acad. Sci. USA 101, 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson, J. S., Klionsky, D. J., Banta, L. M. & Emr, S. D. (1988) Mol. Cell. Biol. 8, 4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothman, J. H. & Stevens, T. H. (1986) Cell 47, 1041–1051. [DOI] [PubMed] [Google Scholar]
- 26.Bonangelino, C. J., Chavez, E. M. & Bonifacino, J. S. (2002) Mol. Biol. Cell 13, 2486–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wickner, W. (2002) EMBO J. 21, 1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marston, A. L., Tham, W. H., Shah, H. & Amon, A. (2004) Science 303, 1367–1370. [DOI] [PubMed] [Google Scholar]
- 29.Guo, W., Roth, D., Walch-Solimena, C. & Novick, P. (1999) EMBO J. 18, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Funato, K., Vallee, B. & Riezman, H. (2002) Biochemistry 41, 15105–15114. [DOI] [PubMed] [Google Scholar]
- 31.Oh, C. S., Toke, D. A., Mandala, S. & Martin, C. E. (1997) J. Biol. Chem. 272, 17376–17384. [DOI] [PubMed] [Google Scholar]
- 32.Obeid, L. M., Okamoto, Y. & Mao, C. (2002) Biochim. Biophys. Acta 1585, 163–171. [DOI] [PubMed] [Google Scholar]
- 33.Athenstaedt, K. & Daum, G. (2000) J. Biol. Chem. 275, 235–240. [DOI] [PubMed] [Google Scholar]
- 34.Munro, S. (1995) EMBO J. 14, 4695–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rayner, J. C. & Pelham, H. R. (1997) EMBO J. 16, 1832–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daum, G., Lees, N. D., Bard, M. & Dickson, R. (1998) Yeast 14, 1471–1510. [DOI] [PubMed] [Google Scholar]
- 37.Umebayashi, K. & Nakano, A. (2003) J. Cell Biol. 161, 1117–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eisenkolb, M., Zenzmaier, C., Leitner, E. & Schneiter, R. (2002) Mol. Biol. Cell. 13, 4414–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bagnat, M., Keranen, S., Shevchenko, A. & Simons, K. (2000) Proc. Natl. Acad. Sci. USA 97, 3254–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munn, A. L., Stevenson, B. J., Geli, M. I. & Riezman, H. (1995) Mol. Biol. Cell 6, 1721–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brizzio, V., Gammie, A. E. & Rose, M. D. (1998) J. Cell Biol. 141, 567–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breton, A. M., Schaeffer, J. & Aigle, M. (2001) Yeast 18, 1053–1068. [DOI] [PubMed] [Google Scholar]
- 43.Breton, A. M. & Aigle, M. (1998) Curr. Genet. 34, 280–286. [DOI] [PubMed] [Google Scholar]
- 44.Balguerie, A., Bagnat, M., Bonneu, M., Aigle, M. & Breton, A. M. (2002) Eukaryot. Cell. 1, 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peter, B. J., Kent, H. M., Mills, I. G., Vallis, Y., Butler, P. J., Evans, P. R. & McMahon, H. T. (2004) Science 303, 495–499. [DOI] [PubMed] [Google Scholar]
- 46.Rozelle, A. L., Machesky, L. M., Yamamoto, M., Driessens, M. H., Insall, R. H., Roth, M. G., Luby-Phelps, K., Marriott, G., Hall, A. & Yin, H. L. (2000) Curr. Biol. 10, 311–320. [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez-Pena, J. M., Rodriguez, C., Alvarez, A., Nombela, C. & Arroyo, J. (2002) J. Cell Sci. 115, 2549–2558. [DOI] [PubMed] [Google Scholar]
- 48.Tong, A. H., Lesage, G., Bader, G. D., Ding, H., Xu, H., Xin, X., Young, J., Berriz, G. F., Brost, R. L., Chang, M., et al. (2004) Science 303, 808–813. [DOI] [PubMed] [Google Scholar]
- 49.Im, Y. J., Raychaudhuri, S., Prinz, W. A. & Hurley, J. H. (2005) Nature 437, 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, X., Rivas, M. P., Fang, M., Marchena, J., Mehrotra, B., Chaudhary, A., Feng, L., Prestwich, G. D. & Bankaitis, V. A. (2002) J. Cell Biol. 157, 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Godi, A., Di Campli, A., Konstantakopoulos, A., Di Tullio, G., Alessi, D. R., Kular, G. S., Daniele, T., Marra, P., Lucocq, J. M. & De Matteis, M. A. (2004) Nat. Cell Biol. 6, 393–404. [DOI] [PubMed] [Google Scholar]
- 52.Vieira, O. V., Verkade, P., Manninen, A. & Simons, K. (2005) J. Cell Biol. 170, 521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romero, P. A., Lussier, M., Veronneau, S., Sdicu, A. M., Herscovics, A. & Bussey, H. (1999) Glycobiology 9, 1045–1051. [DOI] [PubMed] [Google Scholar]
- 54.Bagnat, M. & Simons, K. (2002) Biol. Chem. 383, 1475–1480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.