Abstract
Attenuation of growth factor signaling is essential for the regulation of developmental processes and tissue homeostasis in most organisms. The product of Cbl protooncogene is one such regulator, which functions as an ubiquitin ligase that ubiquitinates and promotes the degradation of a variety of cell signaling proteins. Here, we demonstrate that Grb2 bound to tyrosine-phosphorylated FRS2α forms a ternary complex with Cbl by means of its Src homology 3 domains resulting in the ubiquitination of fibroblast growth factor (FGF) receptor and FRS2α in response to FGF stimulation. These observations highlight the importance of FRS2α in the assembly of both positive (i.e., Sos, phosphatidylinositol 3-kinase) and negative (i.e., Cbl) signaling proteins to mediate a balanced FGF signal transduction. However, the partial inhibition of FGF receptor down-regulation in FRS2α−/− cells indicates that the attenuation of signaling by FGF receptor is regulated by redundant or multiple mechanisms.
The docking proteins, FRS2α and FRS2β, play a critical role in mediating the intracellular signals that are generated at the cell surface by activation of the fibroblast growth factor (FGF), nerve growth factor (NGF), or glial-derived neurotrophic factor receptors. Both FRS2α and FRS2β contain myristyl anchors and phosphotyrosine-binding domains in their N termini and multiple tyrosine phosphorylation sites in their C-terminal tails that serve as binding sites for the adaptor protein, Grb2, and for the Src homology (SH) 2 domain containing protein tyrosine phosphatase, Shp2 (1, 2). In response to FGF or NGF stimulation, Grb2 can also be recruited indirectly to FRS2α through its interaction with tyrosine-phosphorylated Shp2 molecules bound to the docking protein (2).
With mouse embryonic fibroblasts (MEFs) isolated from FRS2α−/− mouse embryos we have demonstrated that FRS2α is essential for the FGF-induced mitogen-activated protein kinase (MAPK) response, phosphatidylinositol 3-kinase (PI3-kinase) stimulation, cell proliferation, and cell migration (3). Although the recruitment of both Grb2 and Shp2 is essential for the overall effects of FGF, recruitment of Shp2 seems to play a more prominent role in stimulation of MAPK and cell proliferation (3). In addition, FRS2α−/− MEFs have also been used to demonstrate that tyrosine phosphorylation and recruitment of the docking protein Gab1 depends on tyrosine-phosphorylated FRS2α. Gab1 binds constitutively to the C-terminal SH3 (C-SH3) domain of Grb2 and its assembly in complex with Grb2/FRS2α enables tyrosine phosphorylation of Gab1, which is followed by recruitment of PI3-kinase and activation of a cell survival pathway (3, 4).
In this report we demonstrate that FGF-induced tyrosine phosphorylation of FRS2α results in complex formation with the adaptor protein Grb2 bound to Cbl by means of its SH3 domains. FGF-induced ternary complex formation among FRS2α, Grb2, and Cbl results in ubiquitination and degradation of FRS2α and FGF receptor (FGFR). Unlike the epidermal growth factor (EGF) or platelet-derived growth factor receptor, which form a direct complex with Cbl by way of its SH2-like domain, Grb2 functions as a link between Cbl and FRS2α; Grb2 is bound to FRS2α by means of its SH2 domain and to Cbl by means of its two SH3 domains. Thus, FRS2α functions as a central platform for recruitment of multiprotein complexes that are responsible for both signal activation and attenuation.
Materials and Methods
Cell Culture, Abs, and Other Reagents.
Cells were cultured in the presence of DMEM containing 10% FBS, 2 mM l-glutamine, and penicillin/streptomycin. PC12 cells were grown in DMEM supplemented with 10% FCS and 10% heat-inactivated horse serum. Generation of FRS2α−/− cells expressing wild-type or FRS2α mutants were performed as described (3). Transient transfections of HEK293 and HeLa S3 cells were performed with Lipofectamine (GIBCO) according to the manufacturer's protocols.
All retrovirus plasmids were constructed in pBABE/puro, whereas plasmids used in transient expression experiments were constructed in pRK5. FRS2α point mutants were generated with the QuickChange Site-directed Mutagenesis kit from Stratagene (1, 2). Abs against FRS2α, Grb2, phosphotyrosine (pTyr), and EGF receptor have been described (1–4). Abs against Myc, Cbl, Sos1, and horseradish peroxidase (HRP)-conjugated anti-mouse Abs were purchased from Santa Cruz Biotechnology. Anti-FLAG was purchased from Sigma. HRP anti-hemagglutinin (HA) Abs were obtained from Roche Molecular Biochemicals. HRP protein-A was obtained from Jackson ImmunoResearch.
In Vitro Binding Experiments, Immunoprecipitation, and Immunoblotting.
The expression and purification of glutathione S-transferase (GST)-Grb2, SH2, N-terminal SH3 (N-SH3), C-terminal SH3 (C-SH3), Cbl-N, and Cbl-C fusion proteins have been described (4, 5). For GST pull-down assays, ≈5 μg of recombinant protein was immobilized to glutathione-agarose beads and washed extensively before incubation with ≈2 mg of cell lysate for 2 h at 4°C. Bound proteins were washed, separated by SDS/PAGE, and transferred to nitrocellulose membranes for immunoblotting. All Abs were dissolved in 1× TBS/5% BSA, with the exception of HA-horseradish peroxidase, which was dissolved in 1× TBS/5% BSA with nonfat milk. Chemiluminescence was performed according to the manufacturer's specifications.
Receptor Endocytosis Assays.
FGF was labeled with 125I to a specific activity of ≈5 × 105 cpm/pmol based on the conventional chloramine T protocol (6). Heparin sulfate was added to labeled 125I-FGF at a final concentration of 5 μg/ml. Optimal and specific binding were determined by ligand titration and competition experiments with a 300-fold excess of unlabeled ligand (nonspecific binding accounted for less than 5% of total counts). For each experiment, cells were seeded in 24-well plates, allowed to attach, and starved overnight. Cells were incubated with 5 nM 125I-aFGF/heparin sulfate in DMEM/1% BSA for 2 h at 4°C. Unbound ligand was removed by washing with ice-cold 1× PBS. Prewarmed DMEM/1% BSA was added to the cells and incubated at 37°C for the times indicated. Samples were washed with ice-cold 1× PBS then acid-stripped with ice-cold 20 mM acetic acid/2 M NaCl for 2 min. Internalized receptors were isolated by lysis in 1 M NaOH. Cell-surface-associated and internalized radioactivities were determined by gamma counting. The results presented represent the average of quadruplicate experiments performed in duplicate.
Results
In our search for proteins that interact with Grb2 in FGF-stimulated cells, we identified several proteins that migrate on SDS/PAGE as a broad band with an apparent molecular mass of 120 kDa. We have previously demonstrated that one of these proteins is the docking protein Gab1. Gab1 is constitutively associated with the C-terminal SH3 domain of Grb2 and is recruited to the activated FGFR via Grb2 molecules bound through its SH2 domain to tyrosine-phosphorylated FRS2α (4). In the experiment presented in Fig. 1A, it is demonstrated that one of the 120-kDa proteins bound to Grb2 is the product of the Cbl protooncogene. In this experiment, lysates of unstimulated or FGF-stimulated NIH 3T3 cells were subjected to immunoprecipitation with anti-Cbl or anti-FRS2α Abs followed by immunoblotting with anti-Grb2, anti-FRS2α, or anti-pTyr Abs. This experiment shows that Cbl forms a complex with Grb2 in both unstimulated and FGF-stimulated cells, whereas only tyrosine-phosphorylated FRS2α forms a complex with Cbl and Grb2 in lysates from FGF-stimulated cells (Fig. 1A). In addition, this experiment shows that Cbl is tyrosine-phosphorylated in unstimulated cells and that FGF stimulation did not enhance the tyrosine phosphorylation of Cbl (Fig. 1A). This experiment suggests that Cbl is bound to Grb2 constitutively and that after FGF stimulation tyrosine-phosphorylated FRS2α binds to a preexisting Grb2/Cbl complex. We next explored the molecular nature of complex formation among Cbl, Grb2, and FRS2α. Lysates of unstimulated or FGF-treated NIH 3T3, MEF, or PC12 cells expressing endogenous Cbl or HEK293 cells transiently expressing Cbl were subjected to pull-down assays with GST fusion proteins of the N- or C-SH3 domains of Grb2 (Fig. 1B). Bound proteins were resolved by SDS/PAGE and immunoblotted with anti-Cbl or anti-pTyr Abs. The experiment presented in Fig. 1B shows that unlike Gab1, which binds exclusively to the C-SH3 domain of Grb2, in vitro Cbl binds efficiently to both SH3 domains of the adaptor protein. Moreover, Cbl was found to be tyrosine-phosphorylated in unstimulated cells and its tyrosine phosphorylation was not further enhanced by FGF treatment. To investigate further whether Cbl interacts constitutively with Grb2 in living cells, HEK293 cells were transiently transfected with expression vectors that direct the expression of Cbl together with HA-tagged full-length or deletion mutants of Grb2 lacking either the N- or C-terminal SH3 domain of the protein. The experiment presented in Fig. 1C shows that intact Grb2 forms a complex with Cbl. However, a deletion mutant of Grb2 lacking its C-SH3 domain binds weakly to Cbl, whereas a deletion mutant lacking the N-SH3 domain does not form a complex with Cbl. These experiments demonstrate that although the N-SH3 of Grb2 is a predominant recognition site for Cbl, both SH3 domains are required for optimal complex formation between Cbl and Grb2 in the context of living cells.
Figure 1.
Grb2 interacts with Cbl and is recruited to FRS2α in an FGF-dependent manner in vitro and in vivo. (A) Lysates from untreated or FGF-treated NIH 3T3 cells were immunoprecipitated with either anti-Cbl or anti-FRS2α Abs. Bound proteins were immunoblotted with anti-pTyr, anti-FRS2α, or anti-Grb2 Abs. (B) Lysates from untreated or FGF-stimulated NIH 3T3, MEF, or PC12 cells or lysates from HEK293 cells that were transfected with expression vector for Cbl were incubated with either GST-N-SH3 (N) or GST-C-SH3 (C) domains of Grb2. Bound proteins were detected by immunoblotting with either anti-pTyr or anti-Cbl Abs. (C) HEK293 cells were transfected with expression vectors for HA-Grb2, N-terminally truncated HA-Grb2 (HA-Grb2ΔN), or C-terminally deleted HA-Grb2 (HA-Grb2ΔC) together with expression vector of Cbl. Cell lysates were immunoprecipitated with anti-Cbl Abs and immunoblotted with either anti-HA or anti-Cbl Abs. Immunoblots of total cell lysate (TCL) are shown alongside.
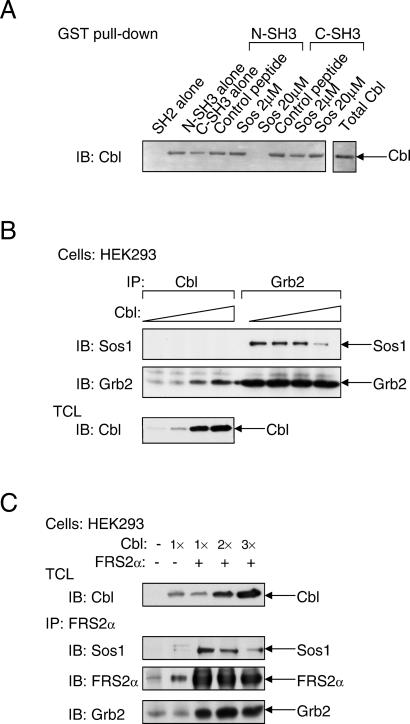
Cbl contains several proline-rich domains that could function as binding sites for the SH3 domains of Grb2. A proline-rich motif located between amino acids 494–499 (PPVPPR) has an identical sequence to a proline-rich sequence found in the guanine nucleotide exchange factor Sos1, which is known to form a complex with the N-SH3 domain of Grb2 (7, 8). To examine whether Sos1 and Cbl compete for the same binding site on Grb2, we have used a synthetic peptide from Sos1 that harbors the Grb2-N-SH3-binding motif to compete for binding of Cbl to Grb2 (Fig. 2A). As observed, 20 μM of the Sos1 peptide is sufficient to block the binding of Cbl to the N-SH3 domain of Grb2. However, this peptide did not prevent the binding of Cbl to the C-SH3 domain of Grb2. This experiment shows that Cbl and Sos1 bind to a similar region in the N-SH3 domain of Grb2. Moreover, overexpression of Cbl results in inhibition of complex formation between Sos1 and Grb2 (Fig. 2B), providing further support for the notion that Cbl and Sos1 compete for the same binding site on Grb2.
Figure 2.
Cbl and Sos compete for binding to Grb2. (A) Lysates of HEK293 cells that were transfected with expression vector for Cbl were incubated with either GST-N-SH3 or GST-C-SH3 of Grb2 in the absence or presence of varying concentrations of a Sos1 synthetic peptide derived from a region responsible for Grb2 binding or with a nonspecific control peptide. Bound Cbl was detected by immunoblotting with anti-Cbl Abs. (B) HEK293 cells were transfected with increasing amounts of expression vector for Cbl in the presence of equal amounts of expression vectors for Sos1 and Grb2. Samples were immunoprecipitated with anti-Cbl or anti-Grb2 Abs and immunoblotted with anti-Sos1 or anti-Grb2 Abs. (C) HEK293 cells were transfected with expression vectors for FGFR, Sos1, FRS2α, and increasing amounts of expression vectors for Cbl. Cell lysates were immunoprecipitated with anti-FRS2α Abs, and bound proteins were detected by immunoblotting with anti-Sos1, anti-FRS2α, or anti-Grb2 Abs.
Because Cbl associates constitutively with Grb2, we next examined whether the presence of Cbl affects the association of FGF-induced effector proteins with activated FRS2α. Lysates from FGF-stimulated HEK293 cells were subjected to immunoprecipitation with anti-FRS2α Abs followed by immunoblotting with anti-Sos1, anti-Grb2, or anti-FRS2α Abs. The experiment presented in Fig. 2C shows that in response to FGF stimulation, the recruitment of Sos1 by FRS2α in FGF-stimulated cells is decreased after Cbl overexpression, thus demonstrating that Grb2/Cbl and Grb2/Sos1 complexes compete for binding to tyrosine-phosphorylated FRS2α in vivo. On the basis of these experiments, we propose that FRS2α, through its interaction with Grb2, recruits Cbl to a multiprotein complex that is formed in response to FGF stimulation.
Cbl Forms a Complex with FRS2α Primarily Through Interactions with Grb2 Molecules Bound Directly to FRS2α.
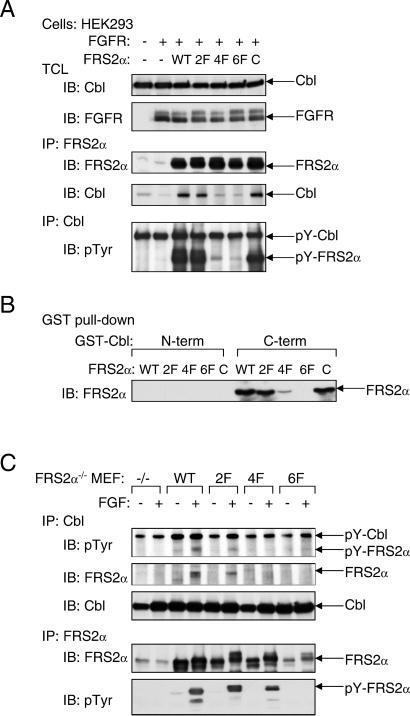
We next examined the possibility of whether Grb2-binding sites and/or Shp2-binding sites on FRS2α are responsible for recruitment of Cbl, and whether Cbl can bind directly to tyrosine-phosphorylated FRS2α by means of its SH2-like domain. We have previously demonstrated that complex formation between Grb2 and FRS2α is mediated by Y196, Y306, Y349, and Y392 of FRS2α (designated direct Grb2-binding sites; ref. 1). In addition, FRS2α recruits Grb2 indirectly by means of the protein tyrosine phosphatase Shp2 by way of residues Y436 and Y471 (designated Shp2-binding sites; ref. 2). The molecular mechanism of complex formation between Cbl and FRS2α was explored by using mutant FRS2α in which its tyrosine-phosphorylation sites were replaced by phenylalanine residues: the 2F (deficient in Shp2-binding), 4F (deficient in Grb2-binding), or the 6F (deficient in both Grb2 and Shp2 sites) mutants of FRS2α. As shown in Fig. 3A, HEK293 cells were cotransfected with an expression vector for Cbl together with expression vectors that direct the synthesis of wild-type, 2F, 4F, or the 6F mutants of FRS2α. Lysates from transfected cells were subjected to immunoprecipitation with anti-Cbl Abs followed by immunoblotting with anti-FRS2α Abs. We observed that Cbl is coimmunoprecipitated with either wild-type or the 2F mutant of FRS2α but did not coimmunoprecipitate with the 4F or the 6F FRS2α mutant. In addition, substitution of a potential binding site for the SH2-like domain of Cbl in the linker region of FRS2α at Tyr-150 with a phenylalanine residue (designated as “C”) did not affect the interaction between Cbl and FRS2α. These experiments demonstrate that the interaction between Cbl and FRS2α is indirect, with Grb2 acting as a primary link between the two proteins.
Figure 3.
FRS2α interacts with Cbl in a Grb2-dependent manner. (A) Analysis of the interaction of Cbl with wild-type FRS2α or the 2F, 4F, or 6F FRS2α mutants. Also tested was the interaction of an FRS2α mutant with a potential binding site for the SH2-like domain of Cbl (C). Interactions were analyzed by cotransfection in HEK293 cells with different expression vectors as indicated, followed by immunoblotting with anti-FRS2α, anti-Cbl, or anti-pTyr Abs. (B) The C-terminal proline-rich tail of Cbl is responsible for complex formation with FRS2α. FLAG-tagged FRS2α mutants were transiently expressed in HEK293 cells, and lysates from the transfected cells were incubated with the N- or C-terminal regions of GST-Cbl. Bound FRS2α was detected by immunoblotting with anti-FLAG Abs. (C) FRS2α-deficient fibroblasts expressing various mutants of FRS2α were generated by retroviral infection and selection. The transfected cells were matched for expression of FRS2α proteins by immunoprecipitation with anti-FRS2α Abs followed by immunoblotting with anti-FRS2α or anti-pTyr Abs. Complex formation between FRS2α and Cbl was analyzed by immunoprecipitation with anti-Cbl Abs followed by immunoblotting with anti-pTyr, anti-FRS2α or anti-Cbl Abs.
To delineate further the interaction between FRS2α and Cbl, lysates from cells expressing wild type or mutants of FRS2α were incubated with GST fusion proteins of either the N- or the C-terminal regions of Cbl. The pull-down assay presented in Fig. 3B shows that the N-terminal portion of Cbl did not interact with FRS2α, whereas the C-terminal region of Cbl (containing the proline-rich region) formed a complex with wild-type FRS2α or the 2F mutant, but not with the 4F or 6F mutant. Similar results were obtained when cell lysates from MEF derived from FRS2α−/− embryos ectopically expressing wild-type or the various mutants of FRS2α were subjected to immunoprecipitation with anti-Cbl Abs followed by immunoblotting with anti-FRS2α Abs. Specifically, wild-type FRS2α or the 2F mutant formed a complex with Cbl, whereas a complex between Cbl and the 4F or the 6F mutant could not be detected. Moreover, we were also unable to detect any direct interactions between Cbl and FGFR (data not shown). Taken together, these experiments show that Cbl is recruited to the FGFR-signaling machinery primarily by the Grb2 binding sites on FRS2α.
The Recruitment of Cbl by FRS2α Results in the Ubiquitination of FRS2α and FGFR.
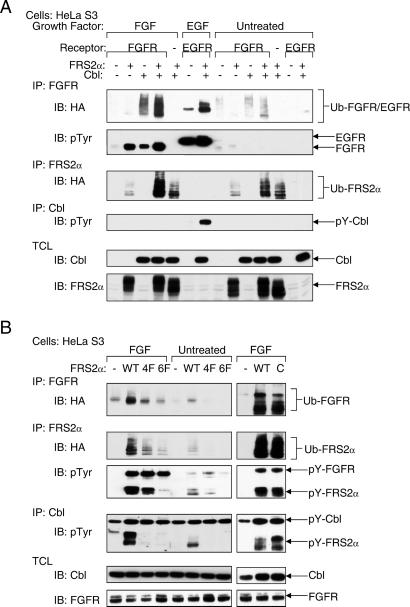
Evidence that the Cbl family of proteins functions as negative regulators of receptor tyrosine kinases came initially from genetic studies in Caenorhabditis elegans and Drosophila melanogaster (9–12). Recent biochemical and structural analyses revealed that Cbl proteins act as RING-type E3 ubiquitin ligases that direct the polyubiquitination of target proteins (reviewed in ref. 13). To examine whether the recruitment of Cbl by means of FRS2α results in ubiquitination of proteins that are in complex with FRS2α, HeLa cells were cotransfected with an expression vector for HA-tagged ubiquitin together with expression vectors for Cbl, FRS2α, or FGFR, alone or in different combinations. Lysates from unstimulated or FGF-stimulated cells were subjected to immunoprecipitation with either anti-FRS2α or anti-FGFR Abs followed by immunoblotting with anti-HA Abs to detect ubiquitinated proteins. The experiment shown in Fig. 4A demonstrates that coexpression of Cbl and FRS2α results in ubiquitination of FRS2α and FGFR. Moreover, Grb2, Shp2, and Gab1 were not ubiquitinated, indicating that only specific components of the FRS2α multiprotein complex undergo ubiquitination (data not shown). To confirm that the recruitment of ubiquitin depends on complex formation between Cbl and Grb2, the same experiment was repeated with expression vectors for the 4F, 6F, or the C FRS2α mutants. The experiment presented in Fig. 4B shows that coexpression of Cbl with either wild-type FRS2α or the FRS2α C mutant results in ubiquitination of both FRS2α and FGFR, whereas the 4F or 6F mutants of FRS2α failed to recruit the ubiquitin machinery and therefore did not promote ubiquitination of FRS2α and FGFR.
Figure 4.
Ubiquitination of FGFR depends on recruitment of Cbl by FRS2α. (A) HeLa cells were transiently transfected with plasmids encoding for the indicated proteins together with expression vector for HA-tagged ubiquitin. Ubiquitinated FGFR and FRS2α were detected by immunoprecipitation with the indicated Abs followed by immunoblotting with anti-HA, anti-pTyr, or anti-FRS2α Abs. (B) The ability of various FRS2α mutants to promote FGFR ubiquitination was analyzed by transient expression in HeLa cells of expression vectors for wild-type FRS2α and for the 4F and 6F FRS2α mutants. Protein ubiquitination was determined by immunoblotting with anti-HA Abs.
Partial Attenuation of FGFR Down-Regulation in FRS2α-Deficient Cells.
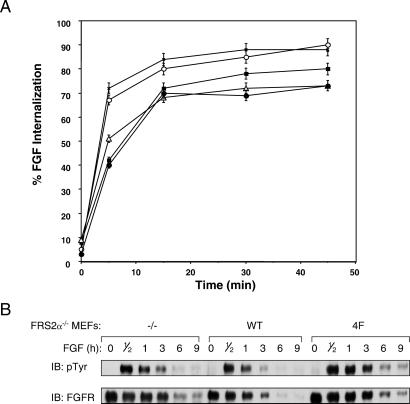
We next tested whether FRS2α-mediated ubiquitination of FGFR may play a role in the process of FGF-induced down-regulation of FGFRs. Down-regulation was determined by analyzing the effect of FRS2α deficiency on the internalization of 125I-labeled FGF into MEFs deficient in FRS2α and into MEFs expressing wild-type or the 2F, 4F, or 6F FRS2α mutants. The experiment presented in Fig. 5A shows that deficiency in FRS2α results in a small but reproducible decrease in the kinetics of internalization of 125I-FGF into mutant cells. FRS2α−/− MEFs or MEFs expressing the 4F or the 6F FRS2α mutants internalized half of 125I-FGF molecules in approximately 10 min, whereas wild-type or the 2F mutant cells internalized half of 125I-FGF molecules in ≈3 min. This pattern is consistent with the loss of Grb2 binding and recruitment of Cbl by the FRS2α mutants. There was also a reproducible decrease in the total amount of 125I-FGF molecules that were internalized into the 4F, 6F, and FRS2α−/− cells as compared with the internalization of 125I-FGF into wild-type or the 2F mutant cells. We have also examined the stability of FGFR after FGF stimulation. As shown in Fig. 5B, the half-life time of FGFR in response to FGF stimulation of MEFs expressing wild-type FRS2α is ≈2–3 h. By contrast, in MEFs deficient in FRS2α or expressing the 4F mutant, the half-life time of FGFR is extended to ≈5 h after FGF stimulation. The experiments presented in Fig. 5 show that deficiency in FRS2α results in a measurable increase in the stability of FGFR and in a decrease in the kinetics of FGF internalization into cells. However, the partial effects on FGFR stability and down-regulation indicate that redundant or other mechanisms play a role in these processes.
Figure 5.
Internalization of 125I-FGF into cells deficient in FRS2α. (A) MEFs deficient in FRS2α (■) ectopically expressing wild-type (X) FRS2α or the 2F (○), 4F (▵), or 6F (●) FRS2α mutants were assayed for 125I-FGF internalization as a function of time. The graph presented represents the average of quadruplicate experiments. (B) MEFs deficient in FRS2α ectopically expressing wild-type FRS2α or the 4F FRS2α mutant were treated with FGF and cycloheximide for the times indicated. Cell lysates were subjected to immunoprecipitation with anti-FGFR Abs followed by immunoblotting with anti-pTyr or anti-FGFR Abs.
Discussion
The docking proteins FRS2α and FRS2β play a critical role in mediating the intracellular signals initiated at the cell surface by activation of the FGF, nerve growth factor, or glial-derived neurotrophic factor receptors. We have demonstrated that the FRS2α protein functions as a site of assembly of multiprotein complexes responsible for activation of the Ras/MAPK signaling cascade, and for stimulation of PI3-kinase and the various effector proteins that are activated by the metabolites generated by this enzyme. Indeed, disruption of the frs2α gene results in embryonic lethality at embryonic day (E) 7–7.5 caused by impairment of multiple signaling pathways (3).
The experiments presented in this report illustrate that in response to FGF stimulation, Cbl is recruited by Grb2 binding to the FRS2α multiprotein complex, resulting in ubiquitination of FRS2α and FGFR. Unlike other receptor tyrosine kinases, such as EGF, platelet-derived growth factor, or CSF-1 receptors (14–16), which recruit Cbl through direct binding of its SH2-like domain to activated receptors, FGFR recruits Cbl by an alternative indirect mechanism involving the docking protein FRS2α. Specifically, Grb2 functions as a link between FRS2α and Cbl; Grb2 is bound to tyrosine-phosphorylated FRS2α by means of its SH2 domain and to a proline-rich region in the C terminus of Cbl by means of its SH3 domains. This process is highly specific, because other components of the multiprotein complex associated with FRS2α such as Grb2, Gab1, and Shp2 are not ubiquitinated. It has been demonstrated that complex formation between PI3-kinase and Cbl-b, which is mediated by binding of the SH3 domain of p85 to the proline-rich region in the C terminus of Cbl-b, also results in the ubiquitination of the regulatory p85 subunit of PI3-kinase (17). It was reported that in natural killer (NK) cells, Cbl is constitutively associated with Grb2—with a preference toward the N-SH3 domain of Grb2 (18). In addition, Cbl can form a complex with EGF receptor directly by means of its SH2-like domain and indirectly by means of Grb2 (19). A similar Grb2-mediated mechanism was recently described for the association between Cbl and hepatocyte growth factor receptor (Met), resulting in ubiquitination of Met (20). Perhaps, the general mechanism of recruitment of members of the Cbl family by adaptor and docking proteins relies on indirect interactions, whereby SH3 domains recognize the proline-rich region in the C terminus of Cbl to mediate complex formation with the target protein, resulting in its ubiquitination and subsequent degradation.
We have previously compared the capacity of the 2F, 4F, or 6F mutants of FRS2α to rescue FGF-induced responses in FRS2α−/− fibroblasts. Although a partial rescue of MAPK response and mitogenic stimulation was obtained after ectopic expression of the 4F mutant, a very weak restoration was achieved by the 2F mutant and no rescue of these responses was achieved by ectopic expression of the 6F mutant in the FRS2α−/− cells (3). This experiment shows that Shp2 binding sites on FRS2α play a critical role in MAPK stimulation and in the mitogenic responses. However, a complete rescue of these responses was achieved only by ectopic expression of wild-type FRS2α in these cells. The partial rescue achieved by ectopic expression of the 4F mutant could be due, in part, to the capacity of Grb2 molecules bound to FRS2α to recruit both positive (i.e., MAPK, PI3-kinase) and negative (i.e., Cbl) regulators of cell signaling. The total intracellular signal generated by the FRS2α multiprotein complex is determined by the balance of positive and negative regulators that are recruited by this docking protein after FGF stimulation. Elimination of a negative regulator, such as Cbl, will result in a net positive effect. At present, we do not know how the recruitment of positive and negative regulators takes place in the cell, how the exchange between positive and negative signals occurs in the context of an FRS2α multiprotein complex, and whether one or more Grb2-binding sites recruit a stoichiometrically equivalent number of Cbl molecules to overcome the signals generated by the positive regulators.
The analysis of signaling through FRS2α provides new insights as to how signals that are initiated by growth factors at the cell surface can be amplified through the recruitment of a multicomponent protein complex, to include both positive and negative regulators. The experiments presented in this report demonstrate that FRS2α plays an important role in Grb2-mediated recruitment of Cbl, resulting in ubiquitination of FGFR and FRS2α. However, the stability and down-regulation of FGFR are only partially affected by the loss of FRS2α in mutant cells. These results raise the possibility that other members of the Cbl family that are expressed in embryonic fibroblasts may compensate for the loss of Cbl by using other mechanisms for interaction with FGFR. An alternative and more likely scenario is that down-regulation of FGFR is achieved by multiple mechanisms. Indeed, it has been shown that a point mutant of FGFR deficient in binding and activation of phospholipase Cγ exhibits impaired down-regulation, suggesting that phosphoinositide metabolites may also play a role in this process (21).
Although it is widely accepted that Cbl functions as a negative regulator of receptor tyrosine kinases, the effect of the loss of Cbl on receptor tyrosine kinase ubiquitination and turnover in Cbl-deficient fibroblasts could not be demonstrated. The normal internalization of receptors in Cbl-deficient fibroblasts could reflect compensatory effects of other members of the Cbl family that are expressed in fibroblasts. It should be noted, however, that most studies describing the role of Cbl in the control of EGF receptor ubiquitination and turnover were performed with an artificial in vitro system under conditions of vast overexpression of Cbl and other proteins (19, 22). The overexpression of Cbl or Cbl mutants in these experiments may shift the equilibrium of reactions that normally occur at low stoichiometry in favor of a modified response that does not necessarily occur under normal physiological conditions. Clearly, multiple mechanisms, in addition to Cbl-induced ubiquitination (23), play a role in the control of growth factor-induced attenuation of signaling by receptor tyrosine kinases.
Acknowledgments
We thank Tony Hunter (Salk Institute) for the generous gift of the HA-ubiquitin plasmid. A.W. is a recipient of a postdoctoral fellowship from the Canadian Institutes of Health and Research (CIHR).
Abbreviations
- SH
Src homology
- FGF
fibroblast growth factor
- FGFR
FGF receptor
- MEF
mouse embryonic fibroblast
- MAPK
mitogen-activated protein kinase
- PI3-kinase
phosphatidylinositol 3-kinase
- C-SH3
C-terminal SH3
- N-SH3
N-terminal SH3
- EGF
epidermal growth factor
- pTyr
phosphotyrosine
- HA
hemagglutinin
- GST
glutathione S-transferase
References
- 1.Kouhara H, Hadari Y R, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 2.Hadari Y R, Kouhara H, Lax I, Schlessinger J. Mol Cell Biol. 1998;18:3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadari Y R, Gotoh N, Kouhara H, Lax I, Schlessinger J. Proc Natl Acad Sci USA. 2001;98:8578–8583. doi: 10.1073/pnas.161259898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ong S H, Hadari Y R, Gotoh N, Guy G R, Schlessinger J, Lax I. Proc Natl Acad Sci USA. 2001;98:6074–6079. doi: 10.1073/pnas.111114298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galisteo M L, Dikic I, Batzer A G, Langdon W Y, Schlessinger J. J Biol Chem. 1995;270:20242–20245. doi: 10.1074/jbc.270.35.20242. [DOI] [PubMed] [Google Scholar]
- 6.Yarden Y, Schlessinger J. Biochemistry. 1987;26:1443–1451. doi: 10.1021/bi00379a035. [DOI] [PubMed] [Google Scholar]
- 7.Donovan J A, Ota Y, Langdon W Y, Samelson L E. J Biol Chem. 1996;271:26369–26374. doi: 10.1074/jbc.271.42.26369. [DOI] [PubMed] [Google Scholar]
- 8.Pawson T, Schlessinger J. Curr Biol. 1993;3:434–442. doi: 10.1016/0960-9822(93)90350-w. [DOI] [PubMed] [Google Scholar]
- 9.Hime G R, Dhungat M P, Ng A, Bowtell D D L. Oncogene. 1997;14:2709–2719. doi: 10.1038/sj.onc.1201223. [DOI] [PubMed] [Google Scholar]
- 10.Meisner H, Daga A, Buxton J, Fernandez B, Chawla A, Banerjee U, Czech M P. Mol Cell Biol. 1997;17:2217–2225. doi: 10.1128/mcb.17.4.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson H, Hime G H, Lada H, Bowtell D D L. Oncogene. 2000;19:3299–3308. doi: 10.1038/sj.onc.1203624. [DOI] [PubMed] [Google Scholar]
- 12.Yoon C H, Lee J, Jongeward G D, Sternberg P W. Science. 1995;269:1102–1105. doi: 10.1126/science.7652556. [DOI] [PubMed] [Google Scholar]
- 13.Thien C B F, Langdon W Y. Nat Rev Mol Cell Biol. 2001;24:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 14.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 15.Lee P S, Wang Y, Dominguez M G, Yeung Y G, Murphy M A, Bowtell D D, Stanley E R. EMBO J. 1999;18:3616–3628. doi: 10.1093/emboj/18.13.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng N, Wang P, Jeffrey P D, Pavletich N P. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 17.Fang D, Wang H, Fang N, Altman Y, Elly C, Liu Y. J Biol Chem. 2001;276:4872–4878. doi: 10.1074/jbc.M008901200. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, Umehara H, Onoue H, Tabassam F H, Okazaki T, Kono T, Minami Y, Tanaka Y, Domae N. Mol Immunol. 2000;37:1057–1065. doi: 10.1016/s0161-5890(01)00020-7. [DOI] [PubMed] [Google Scholar]
- 19.Waterman H, Katz M, Rubin C, Shteigman K, Lavi S, Elson A, Jovin T, Yarden Y. EMBO J. 2002;21:301–313. doi: 10.1093/emboj/21.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peschard P, Fournier T M, Lamorte L, Naujokas M A, Band H, Langdon W Y, Park M. Mol Cell. 2001;8:945–1004. doi: 10.1016/s1097-2765(01)00378-1. [DOI] [PubMed] [Google Scholar]
- 21.Sorokin A, Mohammadi M, Huang J, Schlessinger J. J Biol Chem. 1994;269:17056–17061. [PubMed] [Google Scholar]
- 22.Yokouchi M, Kondo T, Houghton A, Bartikiewicz M, Horne W C, Zhang H, Yoshimura A, Baron R. J Biol Chem. 1999;247:31707–31712. doi: 10.1074/jbc.274.44.31707. [DOI] [PubMed] [Google Scholar]
- 23.Soubeyran P, Kowantez K, Szymkiewicz I, Langdon W Y, Dikic I. Nature (London) 2002;416:183–187. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]